Introduction
Prostate cancer (PCa) is the most common malignant
tumor in the male genitourinary system (1). According to the Global Cancer
Observatory 2020 database, PCa is the second most frequently
diagnosed solid tumor and the fifth leading cause of cancer-related
mortality among men worldwide (2).
Prostate specific antigen (PSA), secreted by prostatic epithelial
cells, is widely used for the screening and diagnosis of PCa. The
application of PSA has greatly improved the detection rate of PCa.
However, as a prostate organ-specific but not a PCa-specific
biomarker, PSA levels can also be elevated in benign prostate
hyperplasia (BPH), prostatic inflammation or other benign diseases
(3). The low specificity of PSA
has led to unnecessary prostate biopsies and the detection of
clinically indolent tumors, which results in pain, bleeding,
infections and other complications along with over-treatment,
including surgery, radiation and additional biopsies (4). Therefore, the development of more
specific biomarkers for the early diagnosis of PCa and to guide the
decision-making for prostate biopsy is necessary.
MicroRNAs (miRNAs) are endogenous, short (18–25
nucleotides), single-stranded, non-coding RNAs widely found in both
animals and plants (5); they bind
to the 3′ untranslated region (3′ UTR) of target mRNAs leading to
their degradation or the inhibition of mRNA translation (6). miRNAs have been reported to be
associated with cancer progression, apoptosis, proliferation,
migration, metastasis and drug resistance, which indicates that
they serve a vital role in the pathogenesis of cancer and are,
therefore, good choices for the diagnosis and treatment of cancer
(7). miR-20b-5p belongs to the
tumor-related miR-106a/363 cluster, which together with the
miR-106b/25 cluster and miR-17/92 cluster forms the large miR-17
family (8). The role of miR-20b-5p
in human cancers is controversial. miR-20b-5p has been reported to
function as an oncomiR in non-small cell lung cancer (9), breast cancer (10), gastric cancer (11), esophageal cancer (12) and laryngeal squamous cell carcinoma
(13), but as a tumor-suppressor
miRNA in colon cancer (14) and
papillary thyroid carcinoma (15).
miR-20b-5p was found to promote the tumor aggression of PCa and can
predict aggressive PCa after radical prostatectomy (16,17).
In the present study, the diagnostic value and biological mechanism
of miR-20b-5p in PCa was evaluated.
Liquid biopsies, including exosomes, circulating
tumor cells and circulating nucleic acids, have been used as
minimally invasive methods to monitor patients with PCa (18). Exosomes, a type of extracellular
vesicles with a diameter of 50–150 nm, are regarded as fundamental
mediators of cell-to-cell communication, and serve a critical role
in multiple biological processes (19). In the process of cell-to-cell
communication, cell-derived exosomes transfer genetic information,
such as mRNAs and miRNAs, to neighboring cells or distant organs.
Prostatic fluid, instead of other commonly used liquid biopsies,
such as blood, urine or saliva, originates from prostatic
epithelial cells, which can directly reflect the changes in
prostate organ function, so it has a significant advantage in
screening for PCa. In the present study, exosomes were extracted
from the prostatic fluid as a source of liquid biopsy for the
detection of PCa.
Materials and methods
TCGA database
miR-20b-5p expression data from 493 human PCa
tissues and 51 adjacent normal tissues were accessed using
TCGAbiolinks package (version 2.18.0) (20) in R (version 4.0.3; http://www.r-project.org/) and were normalized and
standardized using the limma package (version 3.44.3) using the
voom method with the threshold of |log2fold change
(FC)|>1 and P<0.05 (21).
The clinical data of the 493 cases with complete clinical
information were also downloaded and were used for secondary
analysis in the present study (Table
I). The miR-20b-5p expression level data and the clinical data
of the 493 patients were combined and the clinical information was
evaluated to assess the relationship between the miR-20b-5p
expression levels and clinical variables, including age, race,
Gleason score, International Society of Urological Pathology (ISUP)
grade, Tumor-Node-Metastasis (TNM) stage, radiation, cancer subtype
and survival status. The dataset analyzed for this study can be
accessed from TCGA database (TCGA-PRAD; http://portal.gdc.cancer.gov).
 | Table I.Clinical characteristics of patients
with prostate cancer in The Cancer Genome Atlas database. |
Table I.
Clinical characteristics of patients
with prostate cancer in The Cancer Genome Atlas database.
| Clinicopathological
parameters | Patients
(n=493) | Percentage |
|---|
| Age, years |
|
|
|
<65 | 330 | 66.94 |
|
≥65 | 163 | 33.06 |
| Race |
|
|
|
White | 409 | 82.96 |
| Black
or African American | 58 | 11.76 |
|
Asian | 12 |
2.43 |
|
American Indian or Alaska
native |
1 | 0.20 |
| NA | 13 |
2.64 |
| Gleason score |
|
|
|
<7 | 45 |
9.13 |
| ≥7 | 448 | 90.87 |
| ISUP grading
group |
|
|
| 1 | 45 |
9.13 |
| 2 | 146 | 29.61 |
| 3 | 100 | 20.28 |
| 4 | 64 | 12.98 |
| 5 | 138 | 27.99 |
| T stage |
|
|
| T1 | 176 | 35.70 |
| T2 | 171 | 34.69 |
| T3 | 53 | 10.75 |
| T4 |
2 |
0.41 |
| Tx | 91 | 18.46 |
| N stage |
|
|
| N0 | 343 | 69.57 |
| N1 | 78 | 15.82 |
| Nx | 72 | 14.60 |
| M stage |
|
|
| M0 | 454 | 92.09 |
| M1 |
3 |
0.61 |
| NA | 36 |
7.30 |
| Radiation |
|
|
|
Yes | 91 | 18.46 |
| No | 361 | 73.23 |
| NA | 41 |
8.32 |
| Cancer subtype |
|
|
|
Adenocarcinoma, NOS | 481 | 97.57 |
|
Infiltrating duct carcinoma,
NOS |
9 |
1.83 |
|
Adenocarcinoma with mixed
subtypes |
3 |
0.61 |
| Survival
status |
|
|
|
Alive | 481 | 97.57 |
|
Dead | 10 |
2.03 |
| NA |
2 |
0.41 |
Cell culture and transfection
DU145 and 22Rv1 human prostate carcinoma cell lines
were purchased from the American Type Culture Collection and grown
in RPMI-1640 medium (Procell Life Science & Technology Co.,
Ltd.) supplemented with 10% fetal bovine serum (Biological
Industries Israel Beit Haemek, Ltd.), 100 U/ml penicillin and 100
µg/ml streptomycin. The RWPE-1 human prostate epithelium cell line
was purchased from Procell Life Science & Technology Co., Ltd.
and maintained in keratinocyte serum-free medium supplemented with
50 µg/ml bovine pituitary extract and 5 ng/ml epidermal growth
factor (Procell Life Science & Technology Co., Ltd.). Cells
were cultured at 37°C in a humidified atmosphere containing 5%
CO2. miR-20b-5p mimics (5′-CAAAGUGCUCAUAGUGCAGGUAG-3′),
inhibitor (5′-CUACCUGCACUAUGAGCACUUUG-3′), mimics negative control
(NC) (5′-UUCUCCGAACGUGUCACGUTT-3′) and inhibitor negative control
(INC) (5′-CAGUACUUUUGUGUAGUACAA-3′) were purchased from Applied
Biological Materials, Inc. They were transiently transfected into
DU145 cells using Lipo6000™ Transfection Reagent
(Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. Specifically, DU145 cells were seeded into
a 6-well plate and incubated at 37°C in a CO2 incubator
until cells were 70–90% confluent at the time of transfection.
miR-20b-5p mimics, inhibitor, NC and INC with a concentration of 40
nM were transfected into DU145 cells using Lipo6000™
Transfection Reagent (5 µl) at room temperature. After incubating
at 37°C for 5–8 h, the transfection solution was removed. The
subsequent experiments were performed 48 h later. Transfection
efficiency was assessed using reverse transcription-quantitative
PCR (RT-qPCR).
RNA extraction and RT-qPCR
Total RNA was extracted from tissues, cells and
exosomes using TRIzol® reagent (Ambion; Thermo Fisher
Scientific, Inc.). The total RNA (290 ng) was reverse transcribed
using the Evo M-MLV RT Kit (Hunan Aikerui Biological Engineering
Co., Ltd.) using the following reaction conditions: 37°C for 15
min, 85°C for 5 sec and hold at 4°C. Subsequent qPCR was performed
with SYBR® Green Premix Pro Taq HS qPCR Kit (Hunan
Aikerui Biological Engineering Co., Ltd.) on a Bio-Rad CFX Connect
Real-Time PCR Detection System. The RT-qPCR conditions consisted of
initial denaturation at 95°C for 5 min, followed by 40 cycles of
95°C for 10 sec and 60°C for 1 min. miR-20b-5p expression was
normalized to U6, and retinoblastoma-associated protein (RB1)
expression was normalized to GAPDH. The primers for miR-20b-5p, U6,
RB1 and GAPDH were synthesized by Hunan Aikerui Biological
Engineering Co., Ltd., and the sequences were as follows:
miR-20b-5p specific stem-loop
RT5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT-3′,
forward, 5′-GCGCAAAGTGCTCATAGTGC-3′ and reverse,
5′-AGTGCAGGGTCCGAGGTATT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; RB1 forward,
5′-CCAGACCCAGAAGCCATTGA-3′ and reverse,
5′-TTCACAAAGTGTATTTAGCCGGAGA-3′; and GAPDH forward,
5′-CAGTATGACTCCACTCACGGC-3′ and reverse,
5′-GAGGGGCCATCCACAGTCTTC-3′. All reactions were repeated three
times, and the data were quantified using the 2−ΔΔCq
method (22).
Specimens
PCa (n=9) and BPH (n=10) tissues used in this study
were obtained from prostate biopsy specimens from patients admitted
to the Urology Department of The Second Affiliated Hospital of
Xi'an Jiaotong University (Xi'an, China) between December 2021 and
May 2022. Prostatic fluid samples were obtained at the Urology
Department of The Second Affiliated Hospital of Xi'an Jiaotong
University from patients before they underwent a prostate biopsy
between December 2021 and May 2022. A total of 10 patients with PCa
and 27 patients with negative results were enrolled. The patients
were ≥45 years old and were undergoing initial prostate biopsy for
either moderately elevated serum PSA levels (limit range, 2.0-50.0
ng/ml) and/or a suspicious digital rectal examination (DRE). Those
with a history of PCa or invasive treatment for BPH within 6 months
or taking drugs that affect serum PSA levels within the past 6
months were excluded. This study was approved by the Ethics
Committee of the Xi'an Jiaotong University Health Science Center
(approval no. 2021-1700) and was performed in accordance with the
principles of The Declaration of Helsinki. All participants
included in the study provided written informed consent. The
authors had access to information that could identify individual
participants during or after data collection. Samples were
collected by prostate massage (independently without being together
with urine or semen), which was performed by systematically
applying pressure to the prostate from the base to the apex and
from the lateral to the median line of each lobe. Samples were
collected immediately in 1.5-ml centrifuge tubes after the prostate
massage, snap frozen and kept at −80°C until further
processing.
Extraction of exosomes
The ExoQuick-TC™ Exosome Precipitation
Solution kit (System Biosciences, LLC) was used for exosome
extraction as previously described (23). Briefly, 200 µl prostatic fluid was
collected and centrifuged at 3,000 × g for 15 min at 4°C to
separate off the cells and cell debris. The supernatant was
transferred to a clean centrifuge tube, one-fifth of its volume of
ExoQuick-TC Exosome Precipitation Solution was added to it and it
was then refrigerated overnight at 4°C. The ExoQuick-TC/prostatic
fluid mixture was centrifuged at 1,500 × g for 30 min at 4°C. The
supernatant was aspirated, the residual ExoQuick-TC solution was
spun down using centrifugation at 1,500 × g for 5 min at 4°C and
all traces of fluid were removed.
Transmission electron microscopy
(TEM)
Exosomes were suspended in 100 µl of 1X PBS, then
dropped onto Formvar carbon-coated 400 mesh copper electron
microscopy grids and left to sit for 5 min at room temperature.
Samples were stained using 1% uranyl acetate for 30 sec at room
temperature, after the grids were air-dried, micrographs were
captured using a FEI TecnaiG2 spirit transmission electron
microscope at 80 kV.
Nanoparticle tracking analysis
Exosome particle size and concentration were
assessed using nanoparticle tracking analysis (NTA) using a
ZetaView PMX 110 with ZetaView 8.04.02 software (Particle Metrix
GmbH) as previously described (24). Briefly, isolated exosome samples
were diluted using 1X PBS to measure the particle size and
concentration. NTA measurement was recorded and analyzed at 11
positions. The ZetaView system was calibrated using 110 nm
polystyrene particles. The temperature was maintained at ~23°C.
Western blotting
The protein of exosomes extracted from 200 µl of
prostatic fluid and cells in 1 well of a 6-well plate was extracted
using RIPA Lysis Buffer (Beyotime Institute of Biotechnology)
according to the manufacturer's protocol. Protein concentrations
were determined using the BCA protein assay kit (Beyotime Institute
of Biotechnology). Protein samples (20 µg) in each group were
resolved on 8% gels using SDS-PAGE and transferred to
polyvinylidene fluoride membranes. After being blocked using 5%
non-fat milk for 1 h at room temperature, membranes were incubated
overnight at 4°C with the following primary antibodies: Anti-CD63
(1:2,000; WL02549; Wanleibio, Co., Ltd.), anti-tumor susceptibility
gene 101 (TSG101; 1:1,000; WL05130; Wanleibio Co., Ltd.), anti-RB1
(1:1,000; WL02216; Wanleibio, Co., Ltd.) and anti-GAPDH (1:1,000;
WL01114; Wanleibio, Co., Ltd.). Following washing of the membranes
three times with TBS-Tween-20 (0.05% Tween), the membranes were
incubated with horseradish peroxidase-conjugated goat anti-rabbit
IgG H+L secondary antibodies (1:10,000; WLA023a; Wanleibio, Co.,
Ltd.) for 2 h at room temperature, and the visualized using ECL
chemiluminescence reagent (Beyotime Institute of Biotechnology).
The protein expression levels were semi-quantified using ImageJ
software (version, 1.52a; National Institutes of Health).
Predicting the target genes of
miR-20b-5p in prostate cancer
The miRDB (25) and
miRWalk (26) databases were used
to identify the target genes of miR-20b-5p. Gene Ontology (GO)
term, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and
Disease Ontology (DO) enrichment analyses (27) were performed on the target genes
using the ClusterProfiler package (version 3.18.0) with the
threshold of |log2FC|>1 and adjusted P<0.05
(28). The Search Tool for the
Retrieval of Interacting Genes (https://string-db.org) was then used to generate a
protein-protein interaction (PPI) network of the miR-20b-5p target
genes associated with the significantly enriched pathways with the
threshold of interaction score ≥0.4. The expression levels of the
miR-20b-5p target genes in PCa and adjacent normal tissues were
determined using UALCAN (29).
Finally, the LinkedOmics Spearman's analysis tool (30) was used to assess the correlation
between the expression levels of miR-20b-5p and the potential
target genes involved in important signaling pathways with the
threshold of |rho|≥0.2 and P<0.05.
Statistical analysis
Comparisons between two groups were evaluated by
unpaired Student's t-test or the Mann-Whitney U test, and multiple
comparisons were made using a one-way ANOVA with Tukey's post hoc
test or the Kruskal-Wallis rank test with Dunn's post hoc test. A
receiver operating characteristic (ROC) curve was generated to
evaluate the diagnostic value of miR-20b-5p expression levels in
the TCGA cohort and prostatic fluid specimens, and the area under
the curve (AUC) of the ROC curve was calculated. Statistical
analysis was performed using GraphPad Prism 8 (GraphPad Software,
Inc.) or SPSS (version, 20.0; IBM Corp.). P<0.05 was considered
to indicate a statistically significant difference. The overall
experimental design flow chart is presented in Fig. 1.
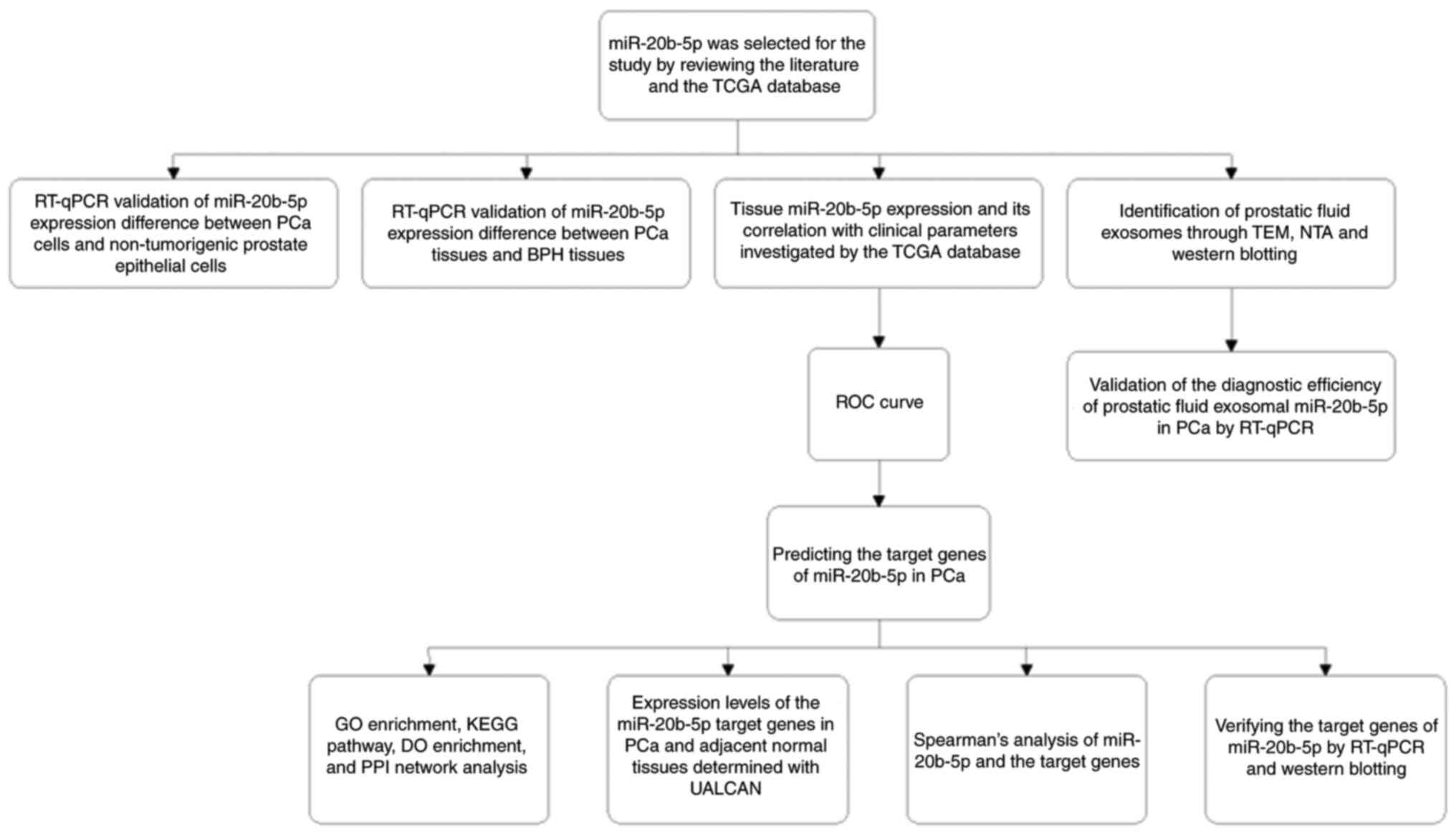 | Figure 1.Overall research design and
experimental sequence of the study. BPH, benign prostate
hyperplasia; DO, Disease Ontology; GO, Gene Ontology; KEGG, Kyoto
Encyclopedia of Genes and Genomes; miR, microRNA; NTA, nanoparticle
tracking analysis; PCa, prostate cancer; PPI, protein-protein
interaction; ROC, receiver operating characteristic; RT-qPCR,
reverse transcription-quantitative PCR; TCGA, The Cancer Genome
Atlas; TEM, transmission electron microscopy. |
Results
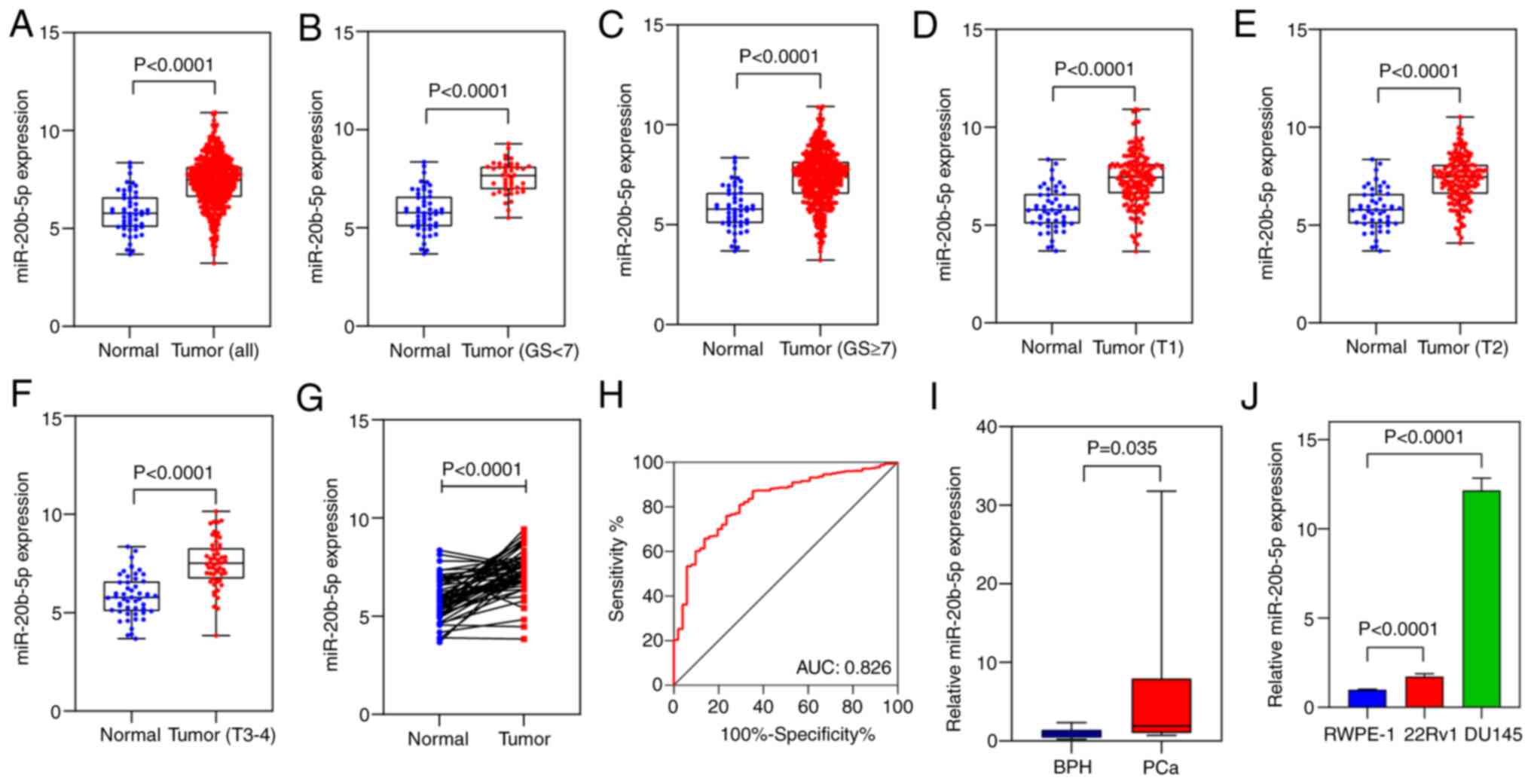
miR-20b-5p is upregulated in PCa
Data from TCGA on the miR-20b-5p expression levels
in 493 PCa and 51 normal prostate tissues were compared. The
results demonstrated significantly higher miR-20b-5p expression
levels in PCa compared with normal tissues (P<0.0001; Fig. 2A). Subgroup analysis demonstrated
significantly higher miR-20b-5p expression in all different PCa
groups compared with normal tissues (all P<0.0001; Fig. 2B-F). After matching the first
characters of the patients' identifier, 51 pairs of PCa and matched
adjacent tissues from the TCGA cohort were identified. In the
paired tissues, miR-20b-5p was significantly overexpressed in PCa
tissues compared with normal tissues (P<0.0001; Fig. 2G).
Furthermore, the relationship between
clinicopathological parameters and tissue miR-20b-5p expression
levels were assessed. miR-20b-5p expression levels were compared
according to age, race, Gleason score, ISUP grade, TNM stage,
radiation, cancer subtype and survival status of the patients.
However, no significant associations were found between miR-20b-5p
expression and the clinicopathological parameters (Fig. S1). An ROC curve was generated
using the tissue expression data from 493 human PCa tissues and 51
normal adjacent tissues to evaluate the diagnostic value of
miR-20b-5p. The AUC was 0.826 (95% CI, 0.771-0.881), which
indicated a marked diagnostic value (Fig. 2H). RT-qPCR data demonstrated that
miR-20b-5p expression levels were significantly higher in PCa
tissues compared with those in BPH tissues (P<0.05; Fig. 2I) and significantly higher in PCa
(DU145 and 22Rv1) cells compared with non-tumorigenic prostate
epithelial (RWPE-1) cells (P<0.0001; Fig. 2J).
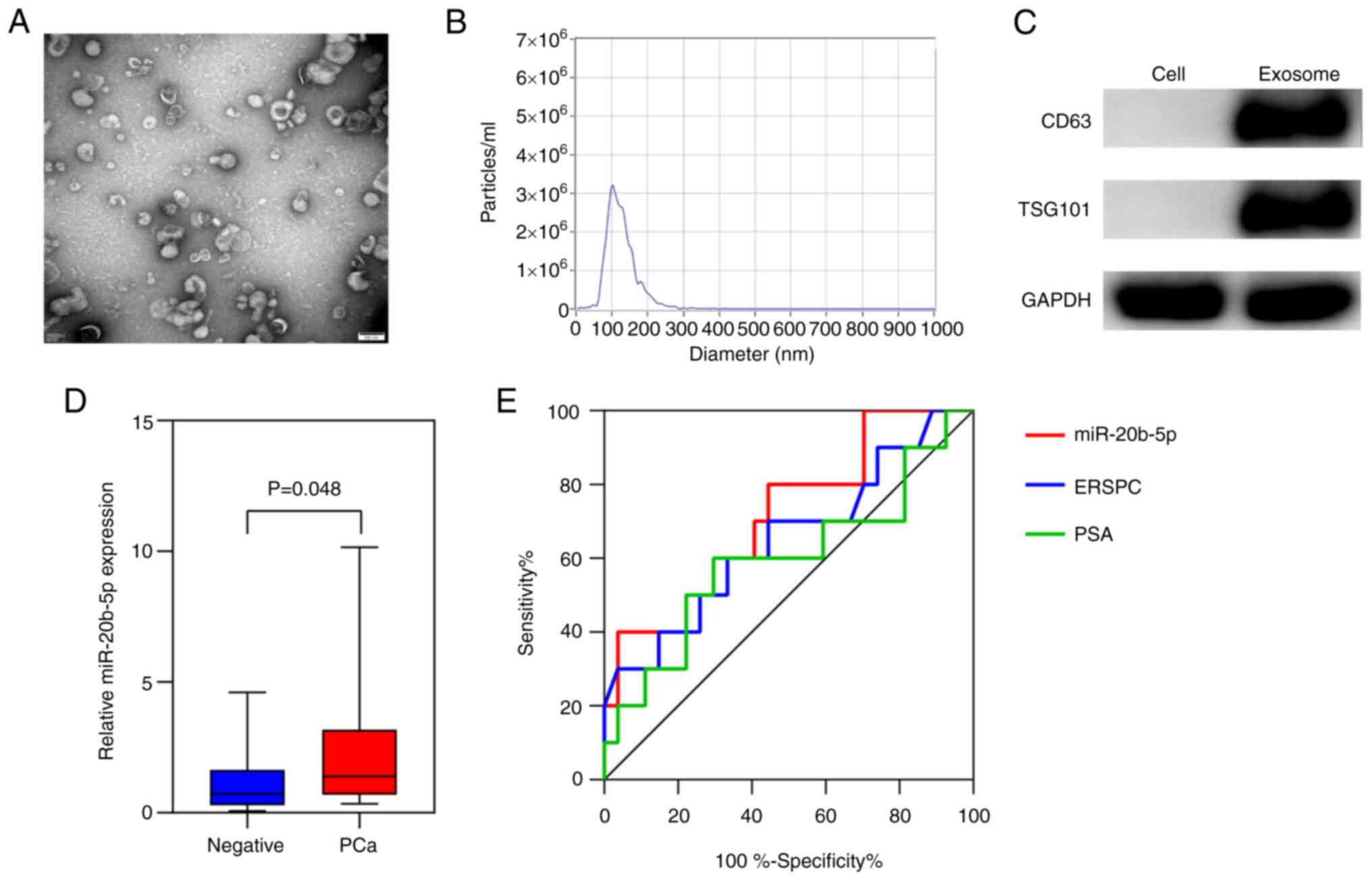
Identification of isolated
exosomes
Exosomes isolated from the prostatic fluid of the
patients were characterized using TEM, NTA and western blotting
(Fig. 3A-C). TEM demonstrated that
the exosomes were round or saucer-shaped with a 50–150 nm diameter,
which was consistent with the results from NTA. Furthermore, the
western blotting demonstrated that exosomes were positive for both
CD63 and TSG101, two typical exosome marker proteins. Therefore,
these results demonstrated that the vesicles isolated from the
prostatic fluid were exosomes.
Prostatic fluid exosomal miR-20b-5p as
a biomarker for the detection of PCa
To assess the diagnostic efficiency of prostatic
fluid exosomal miR-20b-5p in PCa, RT-qPCR was performed using
samples from 37 patients who underwent prostate biopsy with
moderately elevated serum PSA levels (limit range, 2.0-50.0 ng/ml),
including 10 patients with PCa and 27 patients that were
biopsy-negative confirmed by subsequent pathological findings. The
level of exosomal miR-20b-5p expression was significantly increased
in patients with PCa compared to patients with negative results
(P<0.05; Fig. 3D). To
evaluate the diagnostic value of miR-20b-5p for PCa, an ROC curve
was generated and the diagnostic efficiency of exosomal miR-20b-5p,
PSA and European Randomized Study of Screening for Prostate Cancer
(ERSPC) risk calculator were compared (Fig. 3E). The data show that exosomal
miR-20b-5p (AUC, 0.715; 95%CI, 0.526-0.904) was a better predictor
than both PSA (AUC, 0.596; 95%CI, 0.369-0.823) and the ERSPC risk
calculator (AUC, 0.650; 95%CI, 0.437-0.863).
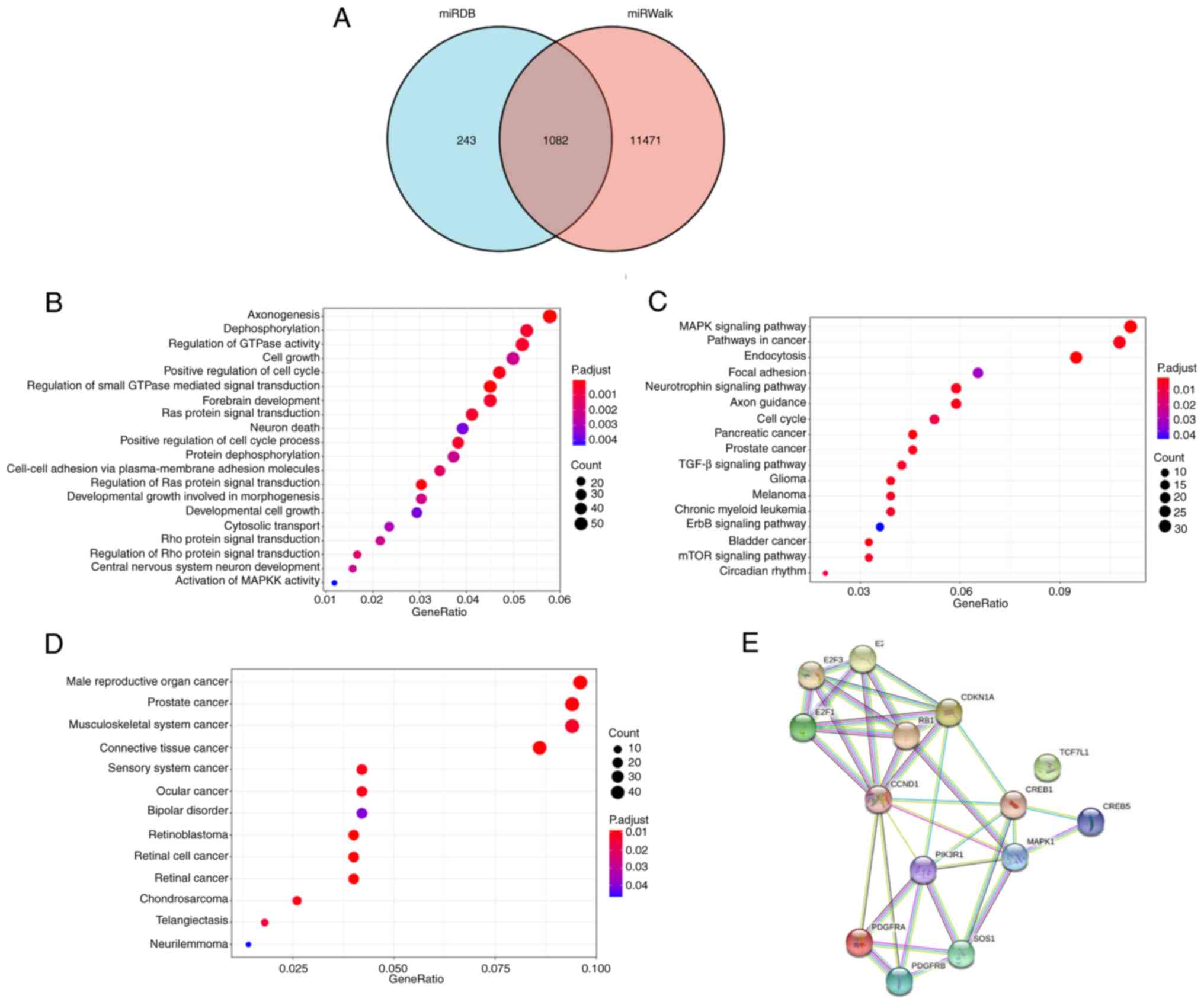
Identification of putative target
genes of miR-20b-5p in PCa
To further evaluate the potential mechanism of
miR-20b-5p in PCa, GO, KEGG and DO analyses were performed. Using
the miRDB and miRWalk databases, 1,082 overlapping target genes
were retrieved (Fig. 4A). The
biological process GO term enrichment analysis demonstrated that
the miR-20b-5p target genes were particularly enriched in
axonogenesis, dephosphorylation and regulation of GTPase activity
(Fig. 4B). KEGG and DO enrichment
analysis demonstrated that miR-20b-5p target genes participated in
multiple cancer-related pathways, including PCa (Fig. 4C and D, respectively). The
PCa-related genes included E2F1, E2F2, E2F3, RB1, SOS1, CREB1,
CREB5, CCND1, CDKN1A, MAPK1, PIK3R1, PDGFRA, PDGFRB and TCF7L1, and
a PPI network of these genes was generated (Fig. 4E). In the PPI network, 14 nodes and
36 lines illustrated strong interactions (average node degree,
5.14; enrichment P=9.55×10−15) between the potential key
target genes.
Analysis of the miR-20b-5p target
genes related to PCa
The expression levels of target genes in TCGA
dataset demonstrated that 10 of the target genes related to PCa
(RB1, SOS1, CREB1, CREB5, CCND1, CDKN1A, MAPK1, PIK3R1, PDGFRA and
TCF7L1) were significantly downregulated in PCa compared with
normal tissues and 3 of the target genes associated with PCa (E2F1,
E2F2 and E2F3) were significantly upregulated in PCa compared with
that in normal tissues (all P<0.05; Fig. 5). Spearman correlation analysis
demonstrated that the expression of RB1, SOS1, CREB1, MAPK1,
PIK3R1, PDGFRA and PDGFRB was negatively correlated with miR-20b-5p
expression, and the expression of E2F1 was positively correlated
with miR-20b-5p expression in PCa (all P<0.05; Fig. 6).
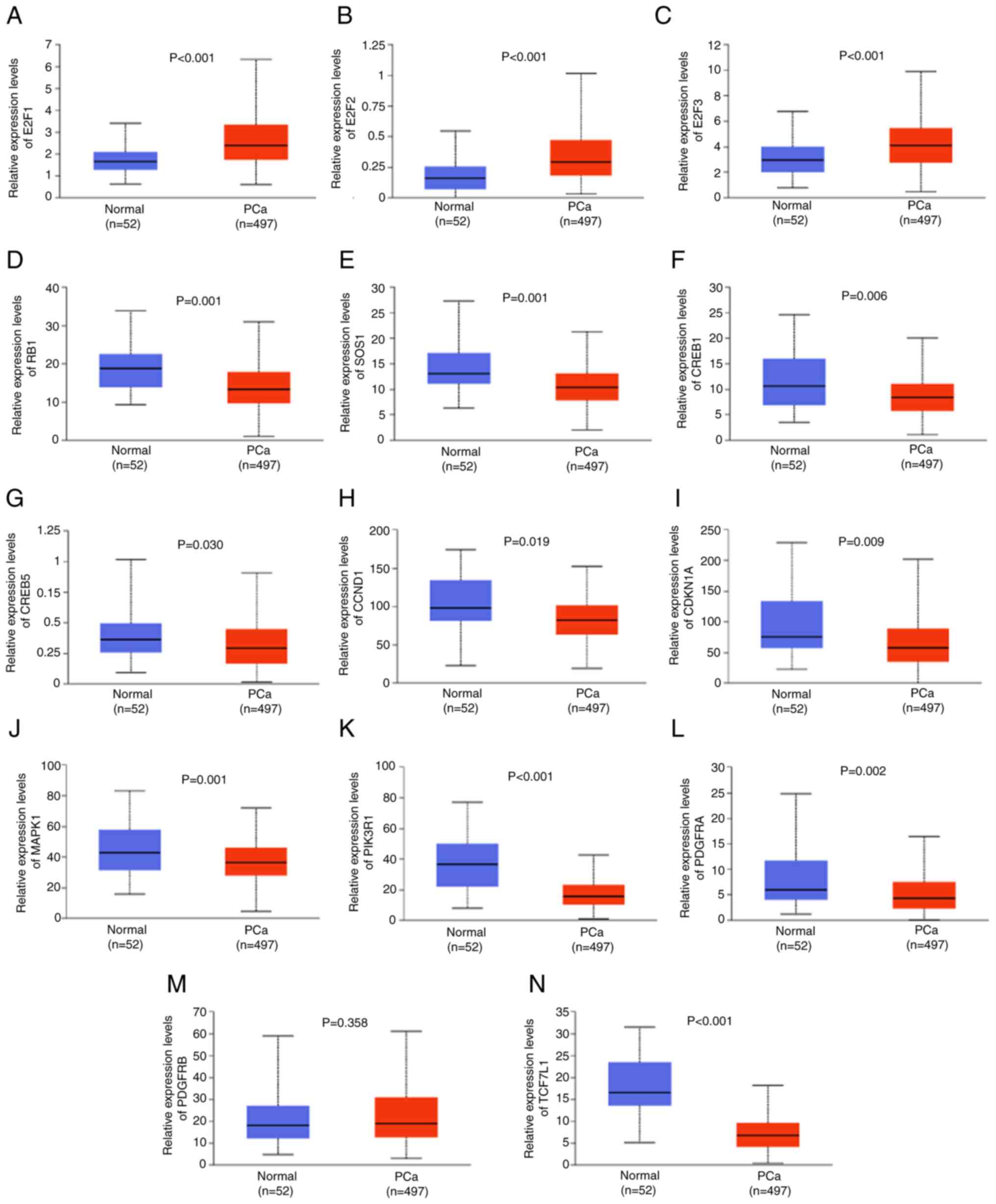 | Figure 5.Expression levels of miR-20b-5p
target genes related to PCa. Relative expression levels of (A)
E2F1, (B) E2F2, (C) E2F3, (D) RB1, (E) SOS1, (F) CREB1, (G) CREB5,
(H) CCND1, (I) CDKN1A, (J) MAPK1, (K) PIK3R1, (L) PDGFRA, (M)
PDGFRB and (N) TCF7L1 in normal and PCa tissue in The Cancer Genome
Atlas dataset. miR, microRNA; PCa, prostate cancer. |
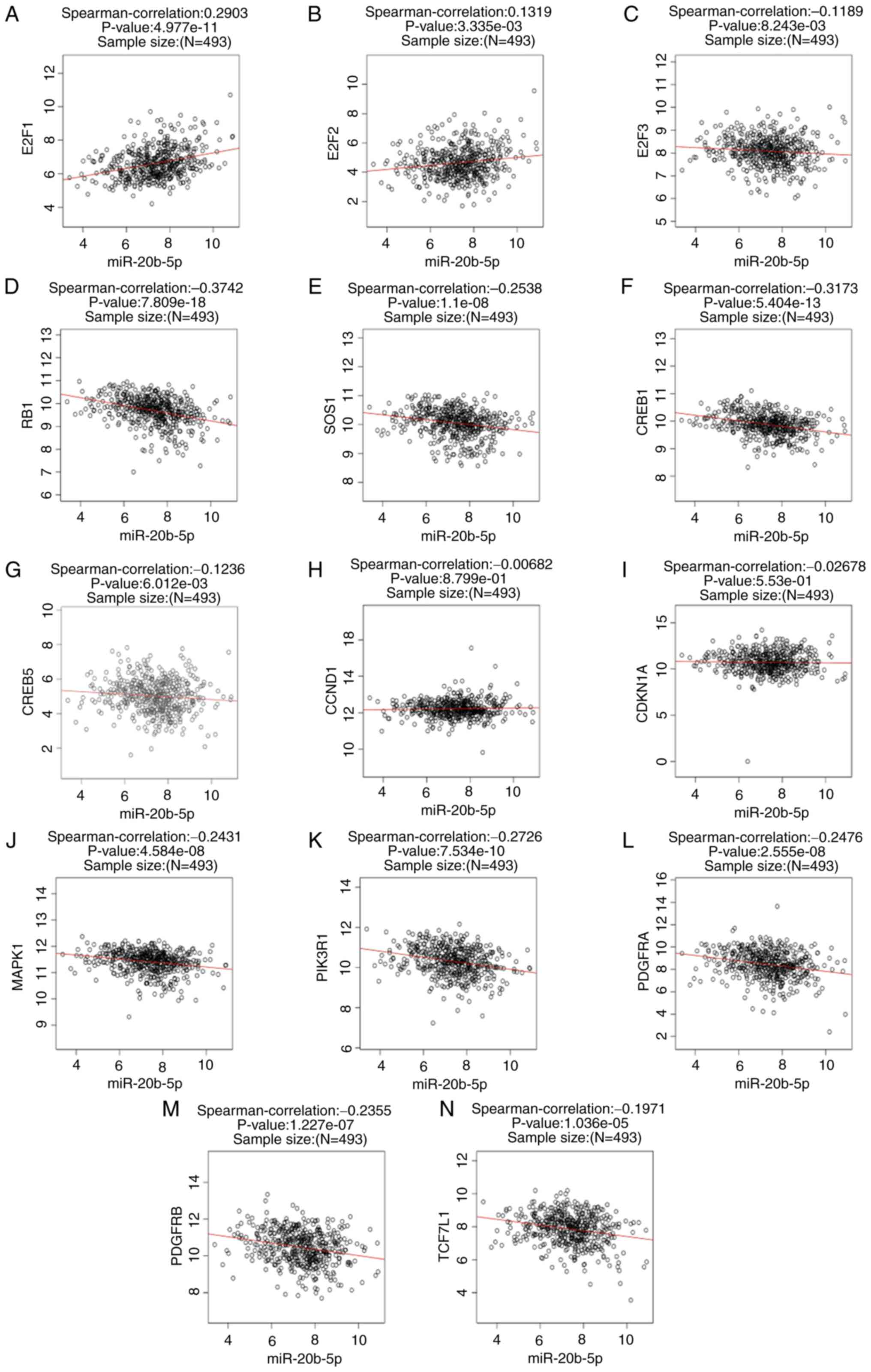 | Figure 6.Spearman's correlation analysis.
Correlation analyses between miR-20b-5p and (A) E2F1, (B) E2F2, (C)
E2F3, (D) RB1, (E) SOS1, (F) CREB1, (G) CREB5, (H) CCND1, (I)
CDKN1A, (J) MAPK1, (K) PIK3R1, (L) PDGFRA, (M) PDGFRB and (N)
TCF7L1 assessed using Spearman's correlation analysis. miR,
microRNA. |
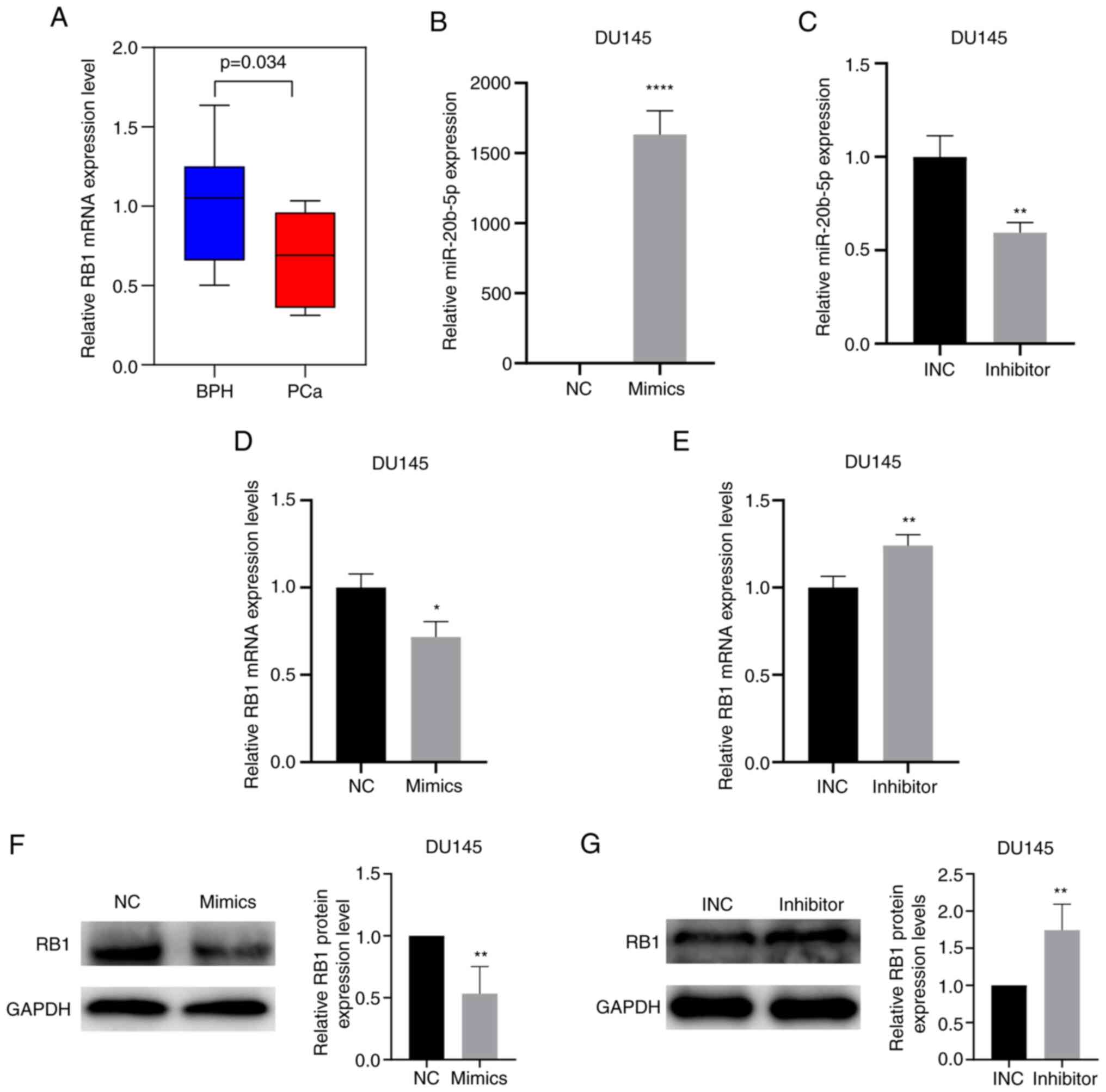
miR-20b-5p affects the mRNA and
protein expression levels of RB1
RT-qPCR data demonstrated that RB1 mRNA expression
levels were significantly lower in PCa tissues compared with those
in BPH tissues (P<0.05; Fig.
7A). To assess whether miR-20b-5p affected RB1 mRNA and protein
expression levels in PCa, DU145 cells were cultured and transfected
with miR-20b-5p mimics or miR-20b-5p inhibitor and the transfection
efficiencies were assessed using RT-qPCR. Transfection of
miR-20b-5p mimics significantly increased the expression of
miR-20b-5p, while transfection of miR-20b-5p inhibitor
significantly decreased the expression of miR-20b-5p (P<0.05;
Fig. 7B and C). RB1 mRNA and
protein expression levels were assessed using RT-qPCR and western
blotting, respectively. miR-20b-5p mimics significantly reduced the
mRNA and protein expression levels of RB1 compared with the
negative control (Fig. 7D and F,
respectively); the miR-20b-5p inhibitor significantly increased the
mRNA and protein expression levels of RB1 compared with the INC
(Fig. 7E and G, respectively).
Discussion
Although the application of PSA testing has been
reported to greatly improve the detection rate of PCa, its poor
specificity and inability to identify high-grade PCa lead to
unnecessary prostate biopsies along with over-diagnosis and
over-treatment (31). Therefore,
the development of more effective biomarkers for PCa is necessary.
Several assays based on urinary exosomes have been used to detect
PCa (32); however, a recent
tracking analysis reported that urinary exosomes mainly expressed
tissue-specific genes of the bladder (33). In the present study, exosomes were
extracted from prostatic fluid, which is specifically secreted by
the prostate, as a source of liquid biopsy for detecting PCa.
In
the present study, the role of miR-20b-5p in detecting PCa was
assessed at the tissue and cellular levels, as well as using liquid
biopsy. At the tissue level, using TCGA data and tissue samples,
significantly higher expression of miR-20b-5p was demonstrated in
PCa compared with that in BPH and normal prostate tissues. No
correlation was demonstrated between miR-20b-5p expression levels
and clinicopathological parameters, which suggested that it may be
an independent predictor for PCa. ROC curve results demonstrated
the promising diagnostic value of tissue miR-20b-5p for PCa. At the
cellular level, it was demonstrated that miR-20b-5p was
significantly upregulated in PCa cells compared with
non-tumorigenic prostate epithelial cells. Furthermore, prostatic
fluid exosomal miR-20b-5p expression levels in patients with PCa
were markedly higher compared with patients with biopsy-negative
results. The use of prostatic fluid exosomal miR-20b-5p expression
levels in predicting PCa was superior to PSA and the ERSPC risk
calculator, a widely-used risk calculator which estimates the
possibility of PCa on prostate biopsy by calculating a probability
based on clinical, biochemical and image findings (34). The findings of the present study
indicated that miR-20b-5p may serve as a clinically significant
biomarker for the detection PCa and to guide the decision-making
involved in prostate biopsy.
miR-20b-5p, a member of the tumor-related
miR-106a/363 cluster, was reported to be frequently dysregulated in
numerous human malignancies (9,15,35,36).
Mechanistically, miR-20b-5p facilitated the proliferation and
inhibited the apoptosis of breast cancer stem cells through the
bidirectionally regulation of CCND1 and E2F1 (10). Conversely, miR-20b-5p was reported
to inhibit migration, invasion and the cell cycle of colon cancer
cells by regulating the CCND1/CDK4/FOXM1 axis (14). miR-20b-5p was reported to be
overexpressed in PCa tissue and in plasma samples, and was
demonstrated to promote proliferation and migration of PCa cells
(16,37). Hoey et al (17) reported that circulating miR-20b-5p
was one of the non-invasive biomarkers that predicted aggressive
PCa after radical prostatectomy, and miR-20b-5p promoted the tumor
aggression of PCa, which indicated that miR-20b-5p functioned as an
oncomiR in PCa. GO analysis in the present study included GO terms
such as axonogenesis, dephosphorylation and regulation of GTPase
activity, indicating that miR-20b-5p may have affected the
development of PCa by participation in these biological processes
and molecular functions. Among the putative target genes of
miR-20b-5p, RB1, SOS1, CREB1, MAPK1, PIK3R1 and PDGFRA were
demonstrated to be associated with miR-20b-5p, and their expression
levels were significantly lower in PCa compared with normal tissue.
Among these, RB1, a tumor suppressor that restricts the
transcription of cell cycle genes by regulating the E2F
transcription factor (38), has
been previously reported to be commonly mutated in PCa, especially
in androgen deprivation therapy-recurrent and metastatic PCa
(39). A previous study also
reported that miR-20b-5p promoted esophageal squamous cell
carcinoma cell proliferation, migration and invasion by directly
targeting RB1 (35). RT-qPCR and
western blotting results from the present study demonstrated that
miR-20b-5p overexpression significantly reduced the mRNA and
protein expression levels of RB1. These data suggested that
miR-20b-5p may serve a vital role in the promotion of the
development of PCa by reducing the expression of RB1 in PCa.
Several limitations should be noted in the present
study. First, the sample size was small when verifying the
diagnostic efficacy of prostatic fluid exosomal miR-20b-5p in
predicting PCa, and further studies using larger cohorts are needed
to validate the conclusion of the present study. Second, the role
of miR-20b-5p in PCa still needs to be evaluated using in
vivo and in vitro studies in the future.
In conclusion, the present study demonstrated that
miR-20b-5p expression was significantly upregulated in PCa at both
tissue and cellular levels. Prostatic fluid exosomal miR-20b-5p may
be used as a non-invasive and effective biomarker for the diagnosis
of PCa. RB1 is a potential target of miR-20b-5p, which may promote
the development of PCa. Further studies are needed to confirm the
findings of the present study and to explore the potential
mechanism underlying the role of miR-20b-5p in PCa.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The dataset analyzed for this study can be found in
The Cancer Genome Atlas Prostate Adenocarcinoma (TCGA-PRAD)
database (https://portal.gdc.cancer.gov). The remaining datasets
used and/or analyzed during the current study are available from
the corresponding author on reasonable request.
Authors' contributions
TYZ, TC, LDZ, ZMW and LX conceived and designed the
study, and revised the manuscript. TYZ and YBM performed the
experiments, and TYZ and MD wrote the manuscript. TYZ, MD, HW and
FL performed the statistical analysis. HW and FL collected the
clinical samples and data of the patients, and ZMW supervised the
project. TYZ and LX confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
participants. The present study was approved by the Ethics
Committee of the Xi'an Jiaotong University Health Science Center
(Xi'an, China; approval no. 2021-1700) and was conducted following
the principles of The Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gandaglia G, Leni R, Bray F, Fleshner N,
Freedland SJ, Kibel A, Stattin P, Van Poppel H and La Vecchia C:
Epidemiology and prevention of prostate cancer. Eur Urol Oncol.
4:877–892. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saini S: PSA and beyond: Alternative
prostate cancer biomarkers. Cell Oncol (Dordr). 39:97–106. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lavallée LT, Binette A, Witiuk K, Cnossen
S, Mallick R, Fergusson DA, Momoli F, Morash C, Cagiannos I and
Breau RH: Reducing the harm of prostate cancer screening: Repeated
prostate-specific antigen testing. Mayo Clin Proc. 91:17–22. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
De Robertis M, Poeta ML, Signori E and
Fazio VM: Current understanding and clinical utility of miRNAs
regulation of colon cancer stem cells. Semin Cancer Biol.
53:232–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang H, Tang Y, Yang D and Zheng L:
MicroRNA-591 functions as a tumor suppressor in hepatocellular
carcinoma by lowering drug resistance through inhibition of
far-upstream element-binding protein 2-mediated phosphoinositide
3-kinase/Akt/mammalian target of rapamycin axis. Pharmacology.
104:173–186. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li JL, Li KZ, Xie MZ, Tang YP, Tang YL and
Hu B: Clinical significance and prognostic value of miR-28-5p in
colon cancer. Dis Markers. 2020:31598312020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li M, Zhou Y, Xia T, Zhou X, Huang Z,
Zhang H, Zhu W, Ding Q and Wang S: Circulating microRNAs from the
miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers
for breast cancer. Breast Cancer Res Treat. 170:257–270. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peng L, Li S, Li Y, Wan M, Fang X, Zhao Y,
Zuo W, Long D and Xuan Y: Regulation of BTG3 by microRNA-20b-5p in
non-small cell lung cancer. Oncol Lett. 18:137–144. 2019.PubMed/NCBI
|
|
10
|
Xia L, Li F, Qiu J, Feng Z, Xu Z, Chen Z
and Sun J: Oncogenic miR-20b-5p contributes to malignant behaviors
of breast cancer stem cells by bidirectionally regulating CCND1 and
E2F1. BMC Cancer. 20:9492020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Streleckiene G, Inciuraite R, Juzenas S,
Salteniene V, Steponaitiene R, Gyvyte U, Kiudelis G, Leja M, Ruzgys
P, Satkauskas S, et al: miR-20b and miR-451a are involved in
gastric carcinogenesis through the PI3K/AKT/mTOR signaling pathway:
Data from gastric cancer patients, Cell lines and ins-gas mouse
model. Int J Mol Sci. 21:8772020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang B, Yang J and Xiao B: MicroRNA-20b
(miR-20b) promotes the proliferation, migration, invasion, and
tumorigenicity in esophageal cancer cells via the regulation of
phosphatase and tensin homologue expression. PLoS One.
11:e01641052016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pantazis TL, Giotakis AI, Karamagkiolas S,
Giotakis I, Konstantoulakis M, Liakea A and Misiakos EP: Low
expression of miR-20b-5p indicates favorable prognosis in laryngeal
squamous cell carcinoma, especially in patients with
non-infiltrated regional lymph nodes. Am J Otolaryng.
41:1025632020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang H, Lin J, Jiang J, Ji J, Wang C and
Zhang J: miR-20b-5p functions as tumor suppressor microRNA by
targeting cyclinD1 in colon cancer. Cell Cycle. 19:2939–2954. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hong S, Yu S, Li J, Yin Y, Liu Y, Zhang Q,
Guan H, Li Y and Xiao H: MiR-20b displays tumor-suppressor
functions in papillary thyroid carcinoma by regulating the MAPK/ERK
signaling pathway. Thyroid. 26:1733–1743. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo J, Xiao Z, Yu X and Cao R: miR-20b
promotes cellular proliferation and migration by directly
regulating phosphatase and tensin homolog in prostate cancer. Oncol
Lett. 14:6895–6900. 2017.PubMed/NCBI
|
|
17
|
Hoey C, Ahmed M, Fotouhi Ghiam A, Vesprini
D, Huang X, Commisso K, Commisso A, Ray J, Fokas E, Loblaw DA, et
al: Circulating miRNAs as non-invasive biomarkers to predict
aggressive prostate cancer after radical prostatectomy. J Transl
Med. 17:1732019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu Y, Delijani K, Mecum A and Goldkorn A:
Current status of liquid biopsies for the detection and management
of prostate cancer. Cancer Manag Res. 11:5271–5291. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang H, Zhang H, Ge S, Ning T, Bai M, Li
J, Li S, Sun W, Deng T, Zhang L, et al: RETRACTED: Exosome-derived
miR-130a activates angiogenesis in gastric cancer by targeting
C-MYB in vascular endothelial cells. Mol Ther. 26:2466–2475. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res. 44:e712016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Law CW, Chen Y, Shi W and Smyth GK: voom:
Precision weights unlock linear model analysis tools for RNA-seq
read counts. Genome Biol. 15:R292014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao L, Jing Y, Wang J, Li H, Che J and
Cao B: Isolation and Identification of miRNAs in exosomes derived
from serum of colon cancer patients. J Cancer. 8:1145–1152. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bachurski D, Schuldner M, Nguyen PH, Malz
A, Reiners KS, Grenzi PC, Babatz F, Schauss AC, Hansen HP, Hallek M
and Pogge von Strandmann E: Extracellular vesicle measurements with
nanoparticle tracking analysis-an accuracy and repeatability
comparison between NanoSight NS300 and ZetaView. J Extracell
Vesicles. 8:15960162019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Y and Wang X: miRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48:D127–D131. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gene Ontology Consortium, . Gene ontology
consortium: Going forward. Nucleic Acids Res. 43:D1049–D1056. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chandrashekar DS, Bashel B, Balasubramanya
SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV and Varambally
S: UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vasaikar SV, Peter S, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tutrone R, Donovan MJ, Torkler P,
Tadigotla V, McLain T, Noerholm M, Skog J and Mckiernan J: Clinical
utility of the exosome based ExoDx prostate(IntelliScore) EPI test
in men presenting for initial biopsy with a PSA 2–10 ng/ml.
Prostate Cancer Prostatic Dis. 23:607–614. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fujita K and Nonomura N: Urinary
biomarkers of prostate cancer. Int J Urol. 25:770–779. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu Q, Cheng L, Deng C, Huang L, Li J,
Wang Y, Li M, Yang Q, Dong X, Su J, et al: The genetic source
tracking of human urinary exosomes. Proc Natl Acad Sci USA.
118:e21088761182021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kowlessur B, Phull M, Patel B, Henry M and
Lazarus J: Validating the European randomised study for screening
of prostate cancer (ERSPC) risk calculator in a contemporary South
African cohort. World J Urol. 38:1711–1718. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu J, Chen S, Niu Y, Liu M, Zhang J, Yang
Z, Gao P, Wang W, Han X and Sun G: Functional significance and
therapeutic potential of miRNA-20b-5p in esophageal squamous cell
carcinoma. Mol Ther Nucleic Acids. 21:315–331. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xue TM, Tao LD, Zhang M, Xu GC, Zhang J
and Zhang PJ: miR-20b overexpression is predictive of poor
prognosis in gastric cancer. Onco Targets Ther. 8:1871–1876. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schitcu VH, Raduly L, Nutu A, Zanoaga O,
Ciocan C, Munteanu VC, Cojocneanu R, Petrut B, Coman I, Braicu C
and Berindan-Neagoe I: MicroRNA dysregulation in prostate cancer.
Pharmgenomics Pers Med. 15:177–193. 2022.PubMed/NCBI
|
|
38
|
Dyson NJ: RB1: A prototype tumor
suppressor and an enigma. Gene Dev. 30:1492–1502. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Aparicio AM, Shen L, Tapia EL, Lu JF, Chen
HC, Zhang J, Wu G, Wang X, Troncoso P, Corn P, et al: Combined
tumor suppressor defects characterize clinically defined aggressive
variant prostate cancers. Clin Cancer Res. 22:1520–1530. 2016.
View Article : Google Scholar : PubMed/NCBI
|