Introduction
Renal cell carcinoma (RCC) is a urinary system
malignancy originating from tubular epithelial cells, and its
pathological subtypes mainly include clear, papillary and
chromophobe cell carcinoma (1,2). In
RCC epidemiology, it is estimated that RCC accounted for almost
2.2% of all cancer cases worldwide in 2020 and its incidence in
China has increased in recent years (3,4).
Most patients with RCC are asymptomatic at an early stage, while
the presence of the typical symptoms of RCC (including hematuria,
abdominal mass and flank pain) is indicative of invasion and
metastasis (5). Therefore, nearly
20% of RCC cases are diagnosed at the metastatic stage (1,6).
Furthermore, although the overall prognosis of patients with RCC is
relatively good, the heterogeneity causes different outcomes among
patients (7–9). Therefore, it is meaningful to explore
biomarkers that may assist clinicians in monitoring the development
and progression of patients with RCC.
Polo-like kinase 4 (PLK4), also referred to as
serum-inducible-kinase akin kinase, is a type of
serine/threonine-protein kinase with triple polo box architecture,
which serves as an indispensable regulator of centriole duplication
(10–12). In recent years, several studies
have demonstrated that PLK4 promotes tumorigenesis in solid tumors,
such as colorectal, prostate and non-small cell lung cancer
(13–15). For example, a previous study
indicated that PLK4 facilitated cell viability and proliferation in
colorectal cancer cells by inhibiting the Wnt/β-catenin signaling
pathway (13). An additional study
revealed that elevated PLK4 expression is associated with increased
tumor size, lymph node metastasis and inferior survival in patients
with non-small cell lung cancer (14). In terms of the biological and
mechanistic backgrounds, PLK4 upregulation drives centrosome
amplification and the cell cycle via the regulation of several
target genes, including epithelial cell transforming sequence 2
(ECT2) and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic
subunit α (PIK3CA), which further causes chromosome instability in
cancer cells (16–18). In addition, PLK4 has also been
reported to modulate epithelial-mesenchymal transition (EMT) in
various epithelial cancers, including lung squamous cell carcinoma
and epithelial ovarian cancer (19,20).
For example, one study revealed that PLK4 dysregulation was a
potential indicator of poor prognosis in lung squamous cell
carcinoma (19). Concerning the
PLK family in RCC, a previous study suggested that PLK1 is
upregulated in RCC and facilitates oncogenesis and the progression
of renal cancer (21).
Subsequently, it was hypothesized that PLK4 may have similar
potency in RCC pathogenesis, while its clinical role in patients
with RCC has not been previously identified.
The present study explored the association of PLK4
protein expression with clinicopathological characteristics and the
long-term prognosis of patients with RCC who underwent surgery, and
the results were further validated by PLK4 mRNA expression
analysis.
Materials and methods
Patients
The present study retrospectively reviewed a total
of 120 patients with RCC who underwent surgical resection at the
hospital (Jing'an District Center Hospital of Shanghai, Fudan
University, Shanghai, China) between January 2011 and June 2016.
The main screening criteria were as follows: i) Pathological
confirmation of RCC; ii) age range of 18–80 years; iii) surgical
resection for RCC; iv) retrievable carcinoma tissue specimens,
which were fixed in formalin and embedded in paraffin (FFEP) for
immunohistochemical (IHC) analysis; and v) accessible preoperative
clinical characteristics and follow-up data. Patients who had other
carcinomas or hematological malignancies (such as lung cancer,
colorectal cancer, Hodgkin's lymphoma and leukemia) were excluded.
The present study was approved by the Ethics Committee of Jing'an
District Center Hospital of Shanghai, Fudan University, Shanghai,
China [(2021) ethical approval no 13]. In addition, this was a
retrospective study and the collected data were retrieved several
years ago; therefore, the requirement for informed consent was
waived.
Data documentation and specimen
processing
The demographic information and disease
characteristics of patients with RCC were collected for analysis.
In addition, the follow-up data of patients with RCC were also
retrieved from clinical records. These data were collected for the
calculation of disease-free survival (DFS) rate and overall
survival (OS) rate. The final date of the follow-up period was June
30, 2021. The median follow-up duration was 6.9 years, and the
follow-up range was 1.2-9.9 years. The available FFEP specimens
(120 tumor tissues and 68 adjacent tissues <2 cm from the tumor
tissues) were collected to determine PLK4 protein expression and
fresh specimens frozen at −196°C in liquid nitrogen (60 tumor
tissues and 28 adjacent tissues) were collected to examine the mRNA
expression levels of PLK4. The pathology department retained core
cancer tissues of all patients, while only a small number of
adjacent tissues of some patients were retained. Therefore,
adjacent tissues were only available in some cases.
Assessment of PLK4 protein
expression
The protein expression levels of the PLK4 were
assessed by IHC as reported in a previous study (14). Briefly, the FFEP specimens, which
were fixed using 10% formalin at room temperature for 24 h, were
cut into 4-µm slices. Next, the slides were deparaffinized with
xylene. The slides were rehydrated with a descending alcohol
series. Subsequently, the slides were heated at 98°C for 10 min in
0.01 mol/l sodium citrate buffer (pH 6) for antigen retrieval. The
slides were treated with fresh 3% hydrogen peroxide to inhibit the
activity of endogenous peroxidase. After blocking with 1.5% normal
goat serum (cat. no. R37624; Invitrogen; Thermo Fisher Scientific,
Inc.) at room temperature for 20 min, the slices were incubated
with anti-PLK4 antibody (1:150; cat. no. PA5-29373; Invitrogen;
Thermo Fisher Scientific, Inc.) as the primary antibody at 4°C
overnight, and then incubated with goat anti-rabbit IgG H&L
(HRP) (1:2,000; cat. no. 31460; Invitrogen; Thermo Fisher
Scientific, Inc.) as the secondary antibody at room temperature for
60 min. Finally, 3,3′-diaminobenzidine (room temperature; 3 min;
Sangon Biotech Co., Ltd.) and hematoxylin (room temperature; 5 min;
Sangon Biotech Co., Ltd.) were used for staining. The antibodies
used in the present study were as follows: Primary antibody,
anti-PLK4 antibody (1:150; Invitrogen; Thermo Fisher Scientific,
Inc.); secondary antibody, goat anti-rabbit IgG H&L (HRP)
(1:2,000; Invitrogen; Thermo Fisher Scientific, Inc.).
Following IHC staining, IHC results were scored by
two pathologists who were blinded to the patients' information
using a light microscope using a semi-quantitative method according
to the intensity and density of stained cells. The intensity was
scored as: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). The
density was scored as: 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%)
and 4 (76–100%). The final score of the IHC assay was obtained by
multiplying the staining intensity and density scores (22). In the analysis, an IHC score of ≤3
in the tumor tissue was considered to indicate low tumor PLK4
protein expression, and an IHC score of >3 was considered to
indicate high PLK4 protein expression.
Evaluation of PLK4 mRNA
expression
The mRNA expression levels of PLK4 were evaluated by
reverse transcription-quantitative PCR (RT-qPCR). Total RNA was
obtained using a RNeasy Protect Mini Kit (Qiagen, Inc.).
Subsequently, reverse transcription was performed using a
QuantiTect Rev. Transcription Kit (Qiagen, Inc.) at 42°C for 18 min
and 95°C for 3 min. qPCR was initiated using TB Green®
Fast qPCR Mix (Takara Bio, Inc.) with the following thermocycling
conditions: 95°C for 30 sec for 1 cycle; followed by 95°C for 5 sec
and 61°C for 15 sec for 40 cycles. The relative expression levels
of PLK4 were assessed using the 2−ΔΔCq method (23), using GAPDH as the internal
reference gene. The qPCR primers were designed based on a previous
study (22). The following PCR
primers were used: PLK4 forward, 5′-CCTTATCACCTCCTCCTTC-3′ and
reverse, 5′-CCAAGTCCTTCATTTGTAACC-3′; and GAPDH forward,
5′-ACATCATCCCTGCCTCTAC-3′ and reverse, 5′-CCTGCTTCACCACCTTCT-3′.
The median of the PLK4 mRNA expression noted in the tumor tissues
was used to classify the patients into the high and low tumor PLK4
mRNA expression groups.
Analysis of PLK4, ECT2 and PIK3CA
expression using online databases
Additional data were obtained from the Gene
Expression Profiling Interactive Analysis (GEPIA) database
(http://gepia.cancer-pku.cn/index.html) to further
verify the correlation of PLK4 expression with ECT2 and PIK3CA in
patients with RCC. Furthermore, PLK4 expression data of 517
patients with RCC were obtained from GEPIA (http://gepia.cancer-pku.cn/detail.php?gene=PLK4###) to
further confirm the association between PLK4 expression and DFS
rate in patients with RCC. PLK4 expression data of 877 patients
with RCC were obtained from The Human Protein Atlas [derived from
The Cancer Genome Atlas (TCGA) analysis; available at https://www.proteinatlas.org/ENSG00000142731-PLK4/pathology/renal+cancer]
to further verify the association between PLK4 expression and OS
rate in patients with RCC.
Statistical analysis
Statistical analysis was performed using SPSS
(version 24.0; IBM Corp.), and the graphs were generated using
GraphPad Prism (version 7.01; GraphPad Software, Inc.). Normality
determination for continuous variables was performed using the
Kolmogorov-Smirnov test. Normally distributed continuous variables
are presented as the mean ± SD, continuous variables with a skewed
distribution are presented as the median and inter-quartile range
(IQR) and categorized variables are presented as the count
(percentage). The differences between the expression levels of the
tumor and adjacent tissues were compared using the Wilcoxon signed
rank test. Receiver operating characteristic (ROC) analysis was
performed to evaluate the suitability of PLK4 expression for
distinguishing tumor tissues from adjacent tissues. The association
between the PLK4 expression levels and the clinical characteristics
was analyzed using the Mann-Whitney U test for two groups and
Kruskal-Wallis test by ranks followed by Dunn's post hoc test for
three groups. The correlation of PLK4 with ECT2 and PIK3CA was
analyzed using the Pearson test. DFS and OS rates were assessed
using Kaplan-Meier curves and significant differences were
determined using the log-rank test. Prognostic factor analysis was
carried out using Cox's proportional hazard regression analysis,
and all potential factors were included in the multivariate
analysis with the forward stepwise method. P<0.050 was
considered to indicate a statistically significant difference.
Results
Patient characteristics
The data of 120 patients with RCC with a mean age of
58.1±11.7 years were retrospectively reviewed in the present study.
The participants included 34 (28.3%) female patients and 86 (71.7%)
male patients (Table I). The
primary lesions were located in the right kidney for 60 (50.0%)
patients and in the left kidney for the remaining 60 (50.0%)
patients. In addition, 99 (82.5%) patients were assessed to have an
Eastern Cooperative Oncology Group Performance Status (ECOG PS)
score of 0, whereas 21 (17.5%) patients were assessed to have an
ECOG PS score of 1 (24).
Furthermore, 56 (46.7%), 47 (39.1%) and 17 (14.2%) patients were
identified as cases with well, moderate and poor tumor
differentiation, respectively. The median tumor size of all
patients was 5.5 cm (IQR, 4.0-8.0 cm). With regard to the clinical
stage, 44 (36.7%), 43 (35.8%), 23 (19.2%), 2 (1.6%) and 8 (6.7%)
patients were assessed as T1a, T1b, T2a, T2b and T3, respectively.
Furthermore, 109 (90.8%) and 11 (9.2%) patients were diagnosed as
N0 and N1, respectively. With regard to their TNM stage, 81 (67.5%)
patients were classified as stage I, 23 (19.2%) patients as stage
II and 16 (13.3%) as stage III. The patient characteristics are
shown in Table I.
 | Table I.Characteristics of patients with
RCC. |
Table I.
Characteristics of patients with
RCC.
| Variables | Patients with RCC
(N=120) |
|---|
| Age, years (mean ±
SD) | 58.1±11.7 |
| Sex, n (%) |
|
|
Female | 34 (28.3) |
|
Male | 86 (71.7) |
| Tumor location, n
(%) |
|
|
Right | 60 (50.0) |
|
Left | 60 (50.0) |
| ECOG PS score, n
(%) |
|
| 0 | 99 (82.5) |
| 1 | 21 (17.5) |
| Tumor
differentiation, n (%) |
|
|
Well | 56 (46.7) |
|
Moderate | 47 (39.1) |
|
Poor | 17 (14.2) |
| Median tumor size,
cm (IQR) | 5.5 (4.0-8.0) |
| T stage, n (%) |
|
|
T1a | 44 (36.7) |
|
T1b | 43 (35.8) |
|
T2a | 23 (19.2) |
|
T2b | 2 (1.6) |
| T3 | 8 (6.7) |
| N stage, n (%) |
|
| N0 | 109 (90.8) |
| N1 | 11 (9.2) |
| TNM stage, n
(%) |
|
| Stage
I | 81 (67.5) |
| Stage
II | 23 (19.2) |
| Stage
III | 16 (13.3) |
PLK4 protein expression
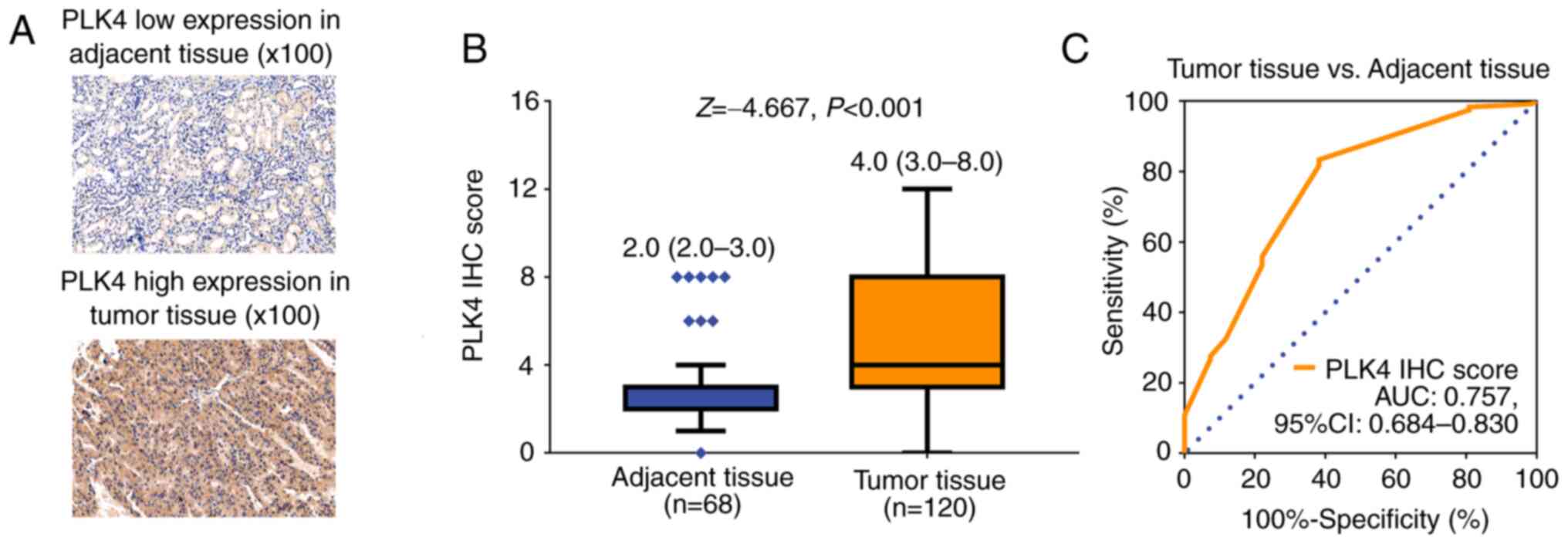
IHC staining was performed in tumor and adjacent
tissues of patients with RCC to detect PLK4 expression (Fig. 1A). Notably, PLK4 protein expression
was elevated in tumor tissues compared with in adjacent tissues
[median (IQR): 4.0 (3.0-8.0) vs. 2.0 (2.0-3.0); P<0.001;
Fig. 1B]. In addition, PLK4
protein expression could be used to differentiate tumor tissues
from adjacent tissues with an area under the curve (AUC) value of
0.757 (95% CI, 0.684-0.830; Fig.
1C).
Association of PLK4 protein expression
with clinicopathological characteristics
PLK4 protein expression was not associated with age
(P=0.224; Fig. 2A), tumor location
(P=0.141; Fig. 2B), ECOG PS score
(P>0.999; Fig. 2C) or tumor
size (P=0.151; Fig. 2D) in
patients with RCC. However, upregulation of PLK4 protein expression
was related to poor tumor differentiation (P=0.009; Fig. 2E), increased T stage (P=0.023;
Fig. 2F), N stage (P=0.014;
Fig. 2G) and TNM stage (P=0.007;
Fig. 2H).
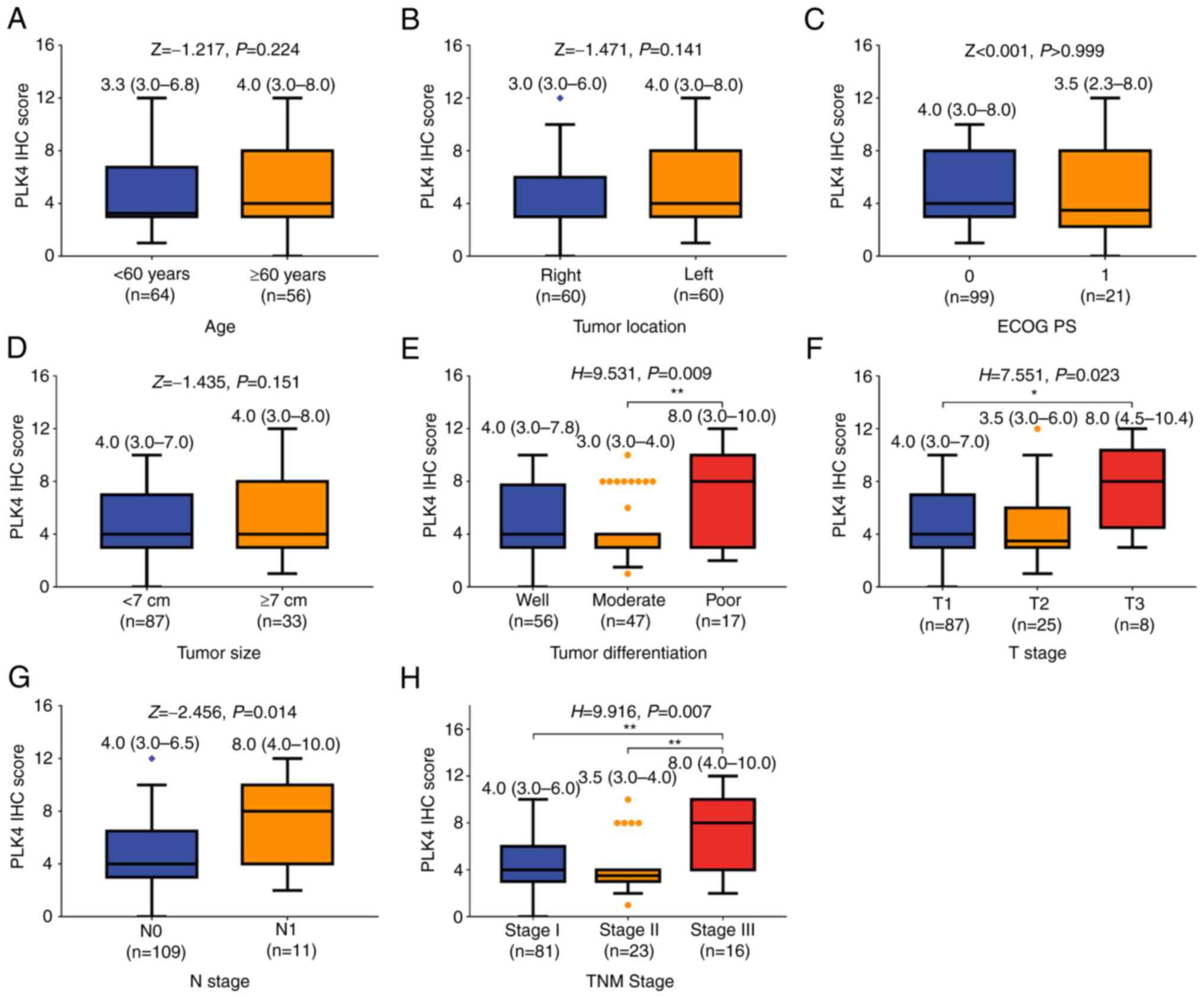 | Figure 2.Elevated PLK4 protein expression is
associated with poor tumor differentiation, and increased T, N and
TNM stages in patients with RCC. Comparison of PLK4 protein
expression in patients with RCC with different (A) age
(Mann-Whitney U test), (B) tumor location (Mann-Whitney U test),
(C) ECOG PS score (Mann-Whitney U test), (D) tumor size
(Mann-Whitney U test), (E) tumor differentiation (Kruskal-Wallis
test by ranks followed by Dunn's post hoc test), (F) T stage
(Kruskal-Wallis test by ranks followed by Dunn's post hoc test),
(G) N stage (Mann-Whitney U test) and (H) TNM stage (Kruskal-Wallis
test by ranks followed by Dunn's post hoc test). *P<0.05,
**P<0.01. ECOG PS, Eastern Cooperative Oncology Group
Performance Status; IHC, immunohistochemistry; PLK4, polo-like
kinase; RCC, renal cell carcinoma. |
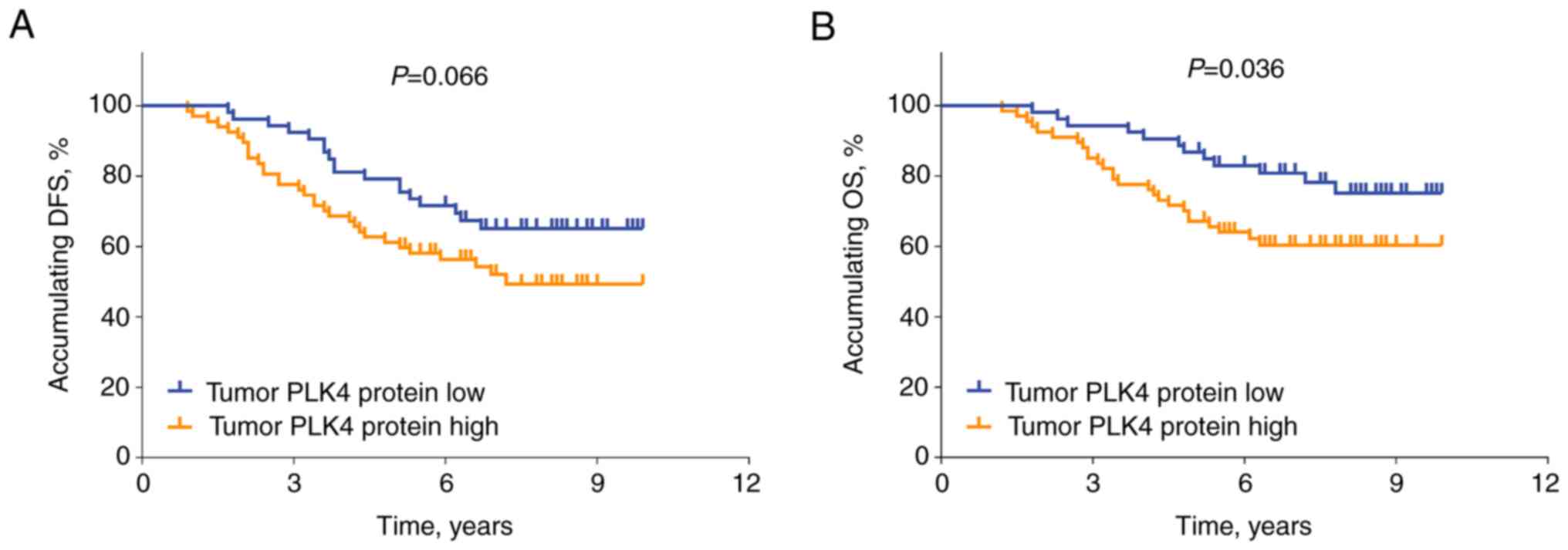
Association of PLK4 protein expression
with survival
The elevated protein expression levels of PLK4 in
tumors exhibited an associating trend (without statistical
significance) with reduced DFS rate in patients with RCC (P=0.066;
Fig. 3A). In addition, high PLK4
expression in tumors was associated with decreased OS rate in
patients with RCC (P=0.036; Fig.
3B).
Association between
clinicopathological factors and survival
Univariate Cox's regression analysis indicated that
PLK4 protein expression was not related to DFS rate in patients
with RCC [hazard ratio (HR), 1.707; P=0.070; Table II]. Additionally, tumor
differentiation (poor vs. well; P<0.001), tumor size (≥7 vs.
<7 cm; P<0.001), T stage (T2 vs. T1; P<0.001), T stage (T3
vs. T1; P<0.001), N stage (N1 vs. N0; P<0.001), TNM stage
(stage II vs. I; P<0.001) and TNM stage (stage III vs. I;
P<0.001) were linked with shortened DFS. In addition,
multivariate Cox's regression analysis demonstrated that the ECOG
PS score (1 vs. 0; HR, 2.243; P=0.025), tumor differentiation (poor
vs. well; HR, 3.185; P=0.006) and TNM stage (II vs. I, HR, 3.993,
P=0.001; III vs. I, HR, 10.488, P<0.001) were independently
associated with reduced DFS rate in patients with RCC.
 | Table II.Univariate and multivariate Cox
proportional hazards regression model analyses of factors
predicting disease-free survival. |
Table II.
Univariate and multivariate Cox
proportional hazards regression model analyses of factors
predicting disease-free survival.
|
|
|
| 95% CI |
|---|
|
|
|
|
|
|---|
| Items | P-value | HR | Lower | Upper |
|---|
| Univariate Cox's
regression analysis |
|
|
|
|
| PLK4
protein (high vs. low) | 0.070 | 1.707 | 0.958 | 3.044 |
| Age
(≥60 vs. <60 years) | 0.347 | 1.306 | 0.749 | 2.278 |
| Sex
(male vs. female) | 0.626 | 1.170 | 0.622 | 2.201 |
| Tumor
location (left vs. right) | 0.401 | 1.269 | 0.728 | 2.215 |
| ECOG PS
score (1 vs. 0) | 0.094 | 1.774 | 0.908 | 3.466 |
| Tumor
differentiation |
|
|
|
|
|
Well | Ref. |
|
|
|
|
Moderate | 0.251 | 1.476 | 0.759 | 2.870 |
|
Poor | <0.001 | 7.224 | 3.520 | 14.828 |
| Tumor
size (≥7 vs. <7 cm) | <0.001 | 4.596 | 2.624 | 8.050 |
| T
stage |
|
|
|
|
|
T1 | Ref. |
|
|
|
|
T2 | <0.001 | 3.772 | 2.041 | 6.973 |
|
T3 | <0.001 | 9.714 | 4.261 | 22.145 |
| N stage
(N1 vs. N0) | <0.001 | 6.985 | 3.448 | 14.150 |
| TNM
stage |
|
|
|
|
|
Stage I | Ref. |
|
|
|
|
Stage II | <0.001 | 4.483 | 2.277 | 8.825 |
|
Stage III | <0.001 | 12.849 | 6.273 | 26.320 |
| Multivariate Cox's
regression analysis |
|
|
|
|
| ECOG PS
score (1 vs. 0) | 0.025 | 2.243 | 1.109 | 4.539 |
| Tumor
differentiation |
|
|
|
|
|
Well | Ref. |
|
|
|
|
Moderate | 0.739 | 0.874 | 0.395 | 1.933 |
|
Poor | 0.006 | 3.185 | 1.384 | 7.330 |
| TNM
stage |
|
|
|
|
|
Stage I | Ref. |
|
|
|
|
Stage II | 0.001 | 3.993 | 1.765 | 9.034 |
|
Stage III | <0.001 | 10.488 | 4.784 | 22.995 |
Furthermore, univariate Cox's regression analysis
demonstrated that PLK4 protein expression (high vs. low) was
associated with reduced OS rate in patients with RCC (HR, 2.049;
P=0.040; Table III); however, it
was not an independent prognostic factor of OS. Apart from PLK4
protein expression, it was also observed that ECOG PS score (1 vs.
0) (P=0.032), tumor differentiation (poor vs. well) (P<0.001), T
stage (T2 vs. T1) (P=0.001), T stage (T3 vs. T1) (P<0.001), TNM
stage (stage II vs. stage I) (P=0.001), and TNM stage (stage III
vs. stage I) (P<0.001) were related to shortened OS. In
addition, multivariate Cox's regression analysis demonstrated that
ECOG PS score (1 vs. 0; HR, 2.903; P=0.006), tumor differentiation
(poor vs. well; HR, 6.114; P<0.001) and TNM stage (II vs. I, HR,
2.808, P=0.030; III vs. I, HR, 8.010, P<0.001) were
independently associated with decreased OS rate in patients with
RCC.
 | Table III.Univariate and multivariate Cox
proportional hazards regression model analyses of factors
predicting overall survival. |
Table III.
Univariate and multivariate Cox
proportional hazards regression model analyses of factors
predicting overall survival.
|
|
|
| 95% CI |
|---|
|
|
|
|
|
|---|
| Items | P-value | HR | Lower | Upper |
|---|
| Univariate Cox's
regression analysis |
|
|
|
|
| PLK4
protein (high vs. low) | 0.040 | 2.049 | 1.032 | 4.065 |
| Age
(≥60 vs. <60 years) | 0.093 | 1.740 | 0.911 | 3.324 |
| Sex
(male vs. female) | 0.454 | 1.331 | 0.630 | 2.812 |
| Tumor
location (left vs. right) | 0.124 | 1.667 | 0.869 | 3.196 |
| ECOG PS
score (1 vs. 0) | 0.032 | 2.202 | 1.069 | 4.537 |
| Tumor
differentiation |
|
|
|
|
|
Well | Ref. |
|
|
|
|
Moderate | 0.487 | 1.336 | 0.590 | 3.029 |
|
Poor | <0.001 | 10.424 | 4.677 | 23.232 |
| Tumor
size (≥7 vs. <7 cm) | <0.001 | 4.315 | 2.274 | 8.188 |
| T
stage |
|
|
|
|
|
T1 | Ref. |
|
|
|
|
T2 | 0.001 | 3.296 | 1.612 | 6.740 |
|
T3 | <0.001 | 10.886 | 4.447 | 26.646 |
| N stage
(N1 vs. N0) | <0.001 | 6.245 | 2.987 | 13.058 |
| TNM
stage |
|
|
|
|
|
Stage I | Ref. |
|
|
|
|
Stage II | 0.001 | 3.737 | 1.672 | 8.355 |
|
Stage III | <0.001 | 11.158 | 5.152 | 24.167 |
| Multivariate Cox's
regression analysis |
|
|
|
|
| ECOG PS
score (1 vs. 0) | 0.006 | 2.903 | 1.354 | 6.226 |
| Tumor
differentiation |
|
|
|
|
|
Well | Ref. |
|
|
|
|
Moderate | 0.985 | 0.991 | 0.385 | 2.549 |
|
Poor | <0.001 | 6.114 | 2.402 | 15.560 |
| TNM
stage |
|
|
|
|
|
Stage I | Ref. |
|
|
|
|
Stage II | 0.030 | 2.808 | 1.104 | 7.138 |
|
Stage III | <0.001 | 8.010 | 3.385 | 18.954 |
Validation of PLK4 mRNA levels
The aforementioned observations were validated by
determining the mRNA expression levels of PLK4 in certain patients
with RCC, for which fresh specimens frozen in liquid nitrogen were
available. The mRNA expression levels of PLK4 were elevated in
tumor tissues compared with in adjacent tissues of patients with
RCC [2.275 (IQR, 1.430-2.965) vs. 1.025 (IQR, 0.675-1.568);
P<0.001; Fig. 4A]. Furthermore,
PLK4 mRNA expression possessed a good value to distinguish tumor
tissues from adjacent tissues, with an AUC value of 0.849 (95% CI,
0.770-0.927; Fig. 4B).
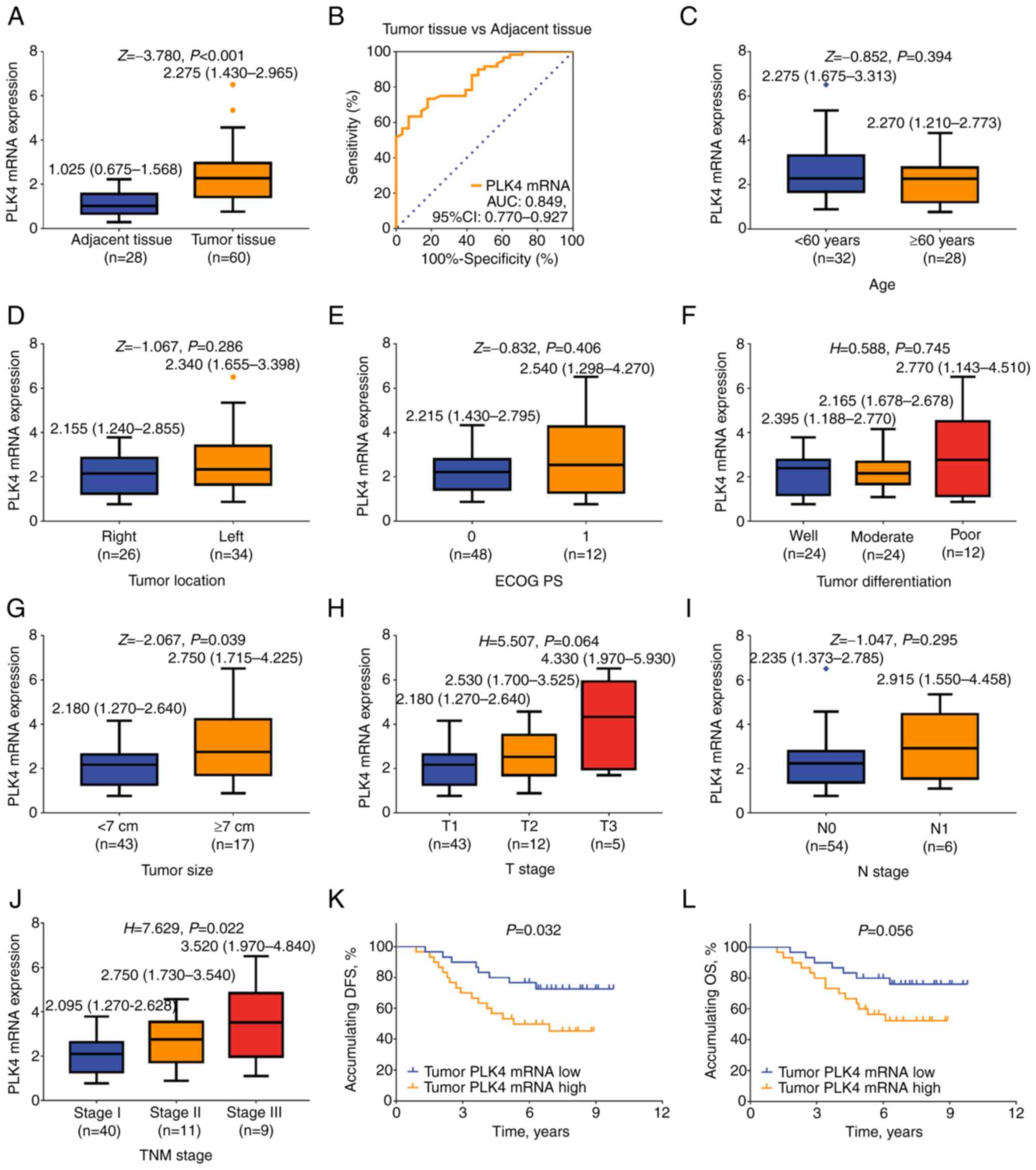 | Figure 4.Association of PLK4 mRNA expression
with clinicopathological characteristics and survival of patients
with RCC. (A) mRNA expression levels of PLK4 were assessed in tumor
and adjacent tissues of patients with RCC (paired-samples Wilcoxon
signed ranks test). (B) Diagnostic value of PLK4 in distinguishing
tumor tissues from adjacent tissues (receiver operating
characteristic analysis). Association of PLK4 mRNA expression with
(C) age (Mann-Whitney U test), (D) tumor location (Mann-Whitney U
test), (E) ECOG PS score (Mann-Whitney U test), (F) tumor
differentiation (Kruskal-Wallis test by ranks), (G) tumor size
(Mann-Whitney U test), (H) T stage (Kruskal-Wallis test by ranks),
(I) N stage (Mann-Whitney U test), (J) TNM stage (Kruskal-Wallis
test by ranks followed by Dunn's post hoc test); post hoc test
results lacked statistical significance, (K) DFS rate and (L) OS
rate in patients with RCC. DFS and OS rates were assessed using
Kaplan-Meier curves and the significant differences were determined
using the log-rank test. AUC, area under the curve; DFS,
disease-free survival; ECOG PS, Eastern Cooperative Oncology Group
Performance Status; OS, overall survival; PLK4, polo-like kinase;
RCC, renal cell carcinoma. |
The analysis of the association of the mRNA
expression levels of PLK4 with the clinicopathological
characteristics of the patients with RCC indicated that PLK4 mRNA
expression was not associated with age (P=0.394), tumor location
(P=0.286), ECOG PS score (P=0.406), tumor differentiation
(P=0.745), T stage (P=0.064) or N stage (P=0.295); however,
increased PLK4 mRNA expression levels were associated with tumor
size ≥7 cm (P=0.039) and high TNM stage (P=0.022) (Fig. 4C-J).
In addition, high PLK4 mRNA expression in tumors was
associated with reduced DFS rate (P=0.032; Fig. 4K), while it tended to be associated
with decreased OS; however, no statistically significant difference
was observed (P=0.056; Fig.
4L).
PLK4 potential target genes
Additional data were obtained from the GEPIA
database to further verify the correlation of PLK4 expression with
its potential target genes in patients with RCC. PLK4 mRNA was
positively correlated with ECT2 mRNA expression (P<0.001;
Fig. S1A) and PIK3CA mRNA
expression (P<0.001; Fig. S1B)
in patients with RCC.
Further validation of the prognostic
value of PLK4
Further survival analysis using data obtained from
GEPIA revealed that high PLK4 expression (vs. low PLK4 expression)
was associated with reduced accumulating DFS rate in patients with
RCC (P=0.029; Fig. S2A). Further
survival analysis using data from The Human Protein Atlas (derived
from TCGA analysis) revealed that high PLK4 expression (vs. low
PLK4 expression) was associated with shortened accumulating OS rate
in patients with RCC (P<0.001; Fig. S2B).
Discussion
PLK4 is located on human chromosome 4q27-28 and has
been reported to modulate centriole duplication, which affects
cancer invasion and metastasis (25). Previous studies have detected its
expression in various solid tumors, including those of the bladder
and prostate (15,18). For example, a previous study has
demonstrated elevated PLK4 expression in human prostate cancer cell
lines and tumor tissues derived from patients with prostate cancer
(15). It has also been reported
that PLK4 expression is upregulated in bladder cancer tissues
compared with in normal bladder tissues (18). The present study indicated that
both PLK4 protein and mRNA expression were upregulated in tumor
tissues compared with in adjacent tissues of patients with RCC. The
possible explanations are as follows: i) PLK4 expression was
positively associated with the number of centrioles that were
excessively amplified in cancer cells, and thus, its expression
levels were upregulated in tumor tissues compared with in adjacent
tissues of patients with RCC (26). ii) Although PLK4 could autoregulate
its stability to prevent centrosome amplification, under the
cancerous condition, it has been observed that some specific genes,
such as centrosomal protein 131, could regulate PLK4 stability and
further promote centrosome amplification (27,28).
iii) The stress-activated protein kinase pathway and P53
cooperatively suppress PLK4 activity, and the two pathways are
frequently inactive in malignant tumors (29–31).
Combining the two aforementioned aspects (ii and iii), PLK4 is
elevated in the cancerous condition of RCC.
The association of PLK4 expression with
clinicopathological features has been previously investigated
(13,32). For example, a previous study
indicated that high PLK4 expression was associated with elevated T
and TNM stages in patients with gastric cancer (32). However, the detailed association of
PLK4 with the clinicopathological characteristics of patients with
RCC is still unclear. In the present study, upregulation of PLK4
protein expression was associated with poor tumor differentiation,
and elevated T, N and TNM stages. The possible reasons for these
findings are as follows: i) PLK4 overexpression results in genomic
instability, aberrant cell cycle and tumorigenesis (9,33).
Therefore, upregulation of PLK4 expression was associated with a
higher T stage in patients with RCC. ii) As discussed previously,
PLK4 induces EMT, which accelerates epithelial cancer migration and
invasion (19,20). Consequently, elevated PLK4
expression was associated with a higher N stage in patients with
RCC. iii) The TNM stage was determined using the T and N stages in
patients with RCC. It was hypothesized that the association of PLK4
expression with TNM stage was due to the association of PLK4
expression with T and N stage.
Considering that PLK4 can regulate malignant
behavior, such as tumor migration and invasion by regulating the
actin related protein 2/3 complex, it was hypothesized that it may
be associated with the survival of patients with cancer (25). A previous study indicated that both
DFS and OS rates were reduced in patients with non-small cell lung
cancer with high PLK4 expression compared with patients with low
PLK4 expression (14). Similarly,
the present study revealed that high PLK4 expression was partially
associated with poor survival in patients with RCC, which could be
explained by PLK4 facilitating excessive centrosome amplification
(15). The latter could induce the
metastatic potential and invasion of tumor cells, which in turn may
result in poor survival of patients with RCC (34). Nevertheless, the multivariate Cox's
regression analysis demonstrated that PLK4 was not independently
associated with DFS and OS rates in patients with RCC, which could
be explained by the following: The upregulation of PLK4 protein
expression was related to increased T stage, N stage and TNM stage,
and the latter factors would weaken the association of PLK4 with
survival in the multivariate Cox's regression analysis. Therefore,
the independent prognostic value of PLK4 in patients with RCC
requires further study.
The present study demonstrated two main findings.
Firstly, a long-term follow-up duration was designed (median 6.9
years; range, 1.2-9.9 years) to increase the reliability of the
prognostic value of PLK4. Secondly, in addition to PLK4 protein
expression, the present study analyzed the mRNA expression levels
of PLK4 to further validate the clinical role of this enzyme in
patients with RCC. However, the present study has certain
limitations. Firstly, only surgically resectable patients were
enrolled, and thus, the association of PLK4 expression with the
incidence of advanced RCC requires further investigation. Secondly,
this was a retrospective study, which may cause selection bias.
Thirdly, the underlying mechanism of action of PLK4 in the
malignant behavior of RCC cells requires further exploration, which
was not included in the present study.
In conclusion, PLK4 possesses a certain clinical
utility in reflecting the clinical stage of patients with RCC,
while its prognostic value requires further validation.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WJ and JM conceived and designed the study. YZh, SZ
and YZe were involved in performing the experiments, and collected
and analyzed the data. WJ and JM confirm the authenticity of all
the raw data. All authors wrote and revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Jing'an District Center Hospital of Shanghai, Fudan
University, Shanghai, China [(2021) ethical approval no 13]. In
addition, this was a retrospective study and the collected data
were retrieved several years ago; therefore, the requirement for
informed consent was waived.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gray RE and Harris GT: Renal cell
carcinoma: Diagnosis and management. Am Fam Physician. 99:179–184.
2019.PubMed/NCBI
|
|
2
|
Padala SA and Barsouk A, Thandra KC,
Saginala K, Mohammed A, Vakiti A, Rawla P and Barsouk A:
Epidemiology of renal cell carcinoma. World J Oncol. 11:79–87.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Q, Zheng RS, Zhang SK, Zhang SW, Liu
SZ, Sun XB, Wei WW and He J: Cancer incidence and mortality of
kidney and unspecified urinary organs in China, 2015. Zhonghua
Zhong Liu Za Zhi. 42:1001–1006. 2020.(In Chinese). PubMed/NCBI
|
|
5
|
Tang T, Du X, Zhang X, Niu W, Li C and Tan
J: Computational identification and analysis of early diagnostic
biomarkers for kidney cancer. J Hum Genet. 64:1015–1022. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maestroni U, Gasparro D, Ziglioli F,
Guarino G and Campobasso D: Metastatic clear cell renal cell
carcinoma: The great pretender and the great dilemma. World J
Oncol. 12:178–182. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo Q, Zhang C, Guo X, Tao F, Xu Y, Feng
G, Han X, Ren Z, Zhang H, Zhang P, et al: Incidence of bone
metastasis and factors contributing to its development and
prognosis in newly diagnosed renal cell carcinoma: A
population-based study. Cancer Manag Res. 10:2935–2944. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lai GS, Li JR, Wang SS, Chen CS, Yang CK,
Hung SC, Cheng CL, Ou YC and Chiu KY: Tumor size significantly
affects prognosis in pathological T3a renal cell carcinoma.
Anticancer Res. 42:2185–2191. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Demasure S, Spriet I, Debruyne PR, Laenen
A, Wynendaele W, Baldewijns M, Dumez H, Clement PM, Wildiers H,
Schöffski P, et al: Overall survival improvement in patients with
metastatic clear-cell renal cell carcinoma between 2000 and 2020: A
retrospective cohort study. Acta Oncol. 61:22–29. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao Y and Wang X: PLK4: A promising
target for cancer therapy. J Cancer Res Clin Oncol. 145:2413–2422.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mbefo MK, Paleologou KE, Boucharaba A,
Oueslati A, Schell H, Fournier M, Olschewski D, Yin G, Zweckstetter
M, Masliah E, et al: Phosphorylation of synucleins by members of
the Polo-like kinase family. J Biol Chem. 285:2807–2822. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hoffmann I: Role of polo-like kinases Plk1
and Plk4 in the initiation of centriole duplication-impact on
cancer. Cells. 11:7862022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liao Z, Zhang H, Fan P, Huang Q, Dong K,
Qi Y, Song J, Chen L, Liang H, Chen X, et al: High PLK4 expression
promotes tumor progression and induces epithelialmesenchymal
transition by regulating the Wnt/betacatenin signaling pathway in
colorectal cancer. Int J Oncol. 54:479–490. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou Q, Fan G and Dong Y: Polo-like kinase
4 correlates with greater tumor size, lymph node metastasis and
confers poor survival in non-small cell lung cancer. J Clin Lab
Anal. 34:e231522020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Singh CK, Denu RA, Nihal M, Shabbir M,
Garvey DR, Huang W, Iczkowski KA and Ahmad N: PLK4 is upregulated
in prostate cancer and its inhibition reduces centrosome
amplification and causes senescence. Prostate. 82:957–969. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Holland AJ and Cleveland DW: Polo-like
kinase 4 inhibition: A strategy for cancer therapy? Cancer Cell.
26:151–153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang X, Wei C, Liang H and Han L:
Polo-like kinase 4′s critical role in cancer development and
strategies for Plk4-targeted therapy. Front Oncol. 11:5875542021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Z, Sun H, Ma W, Wu K, Peng G, Ou T
and Wu S: Down-regulation of Polo-like kinase 4 (PLK4) induces G1
arrest via activation of the p38/p53/p21 signaling pathway in
bladder cancer. FEBS Open Bio. 11:2631–2646. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Garvey DR, Chhabra G, Ndiaye MA and Ahmad
N: Role of polo-like kinase 4 (PLK4) in epithelial cancers and
recent progress in its small molecule targeting for cancer
management. Mol Cancer Ther. 20:632–640. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dongre A and Weinberg RA: New insights
into the mechanisms of epithelial-mesenchymal transition and
implications for cancer. Nat Rev Mol Cell Biol. 20:69–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang G, Zhang Z and Liu Z: Polo-like
kinase 1 is overexpressed in renal cancer and participates in the
proliferation and invasion of renal cancer cells. Tumour Biol.
34:1887–1894. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu Z, Gu X, Zhong R and Zhong H:
Tumor-infiltrating CD45RO(+) memory cells correlate with favorable
prognosis in patients with lung adenocarcinoma. J Thorac Dis.
10:2089–2099. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oken MM, Creech RH, Tormey DC, Horton J,
Davis TE, McFadden ET and Carbone PP: Toxicity and response
criteria of the eastern cooperative oncology group. Am J Clin
Oncol. 5:649–655. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kazazian K, Go C, Wu H, Brashavitskaya O,
Xu R, Dennis JW, Gingras AC and Swallow CJ: Plk4 promotes cancer
invasion and metastasis through Arp2/3 complex regulation of the
actin cytoskeleton. Cancer Res. 77:434–447. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mittal K, Kaur J, Sharma S, Sharma N, Wei
G, Choudhary I, Imhansi-Jacob P, Maganti N, Pawar S, Rida P, et al:
Hypoxia drives centrosome amplification in cancer cells via
HIF1alpha-dependent Induction of Polo-Like Kinase 4. Mol Cancer
Res. 20:596–606. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim DH, Ahn JS, Han HJ, Kim HM, Hwang J,
Lee KH, Cha-Molstad H, Ryoo IJ, Jang JH, Ko SK, et al: Cep131
overexpression promotes centrosome amplification and colon cancer
progression by regulating Plk4 stability. Cell Death Dis.
10:5702019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Holland AJ, Lan W, Niessen S, Hoover H and
Cleveland DW: Polo-like kinase 4 kinase activity limits centrosome
overduplication by autoregulating its own stability. J Cell Biol.
188:191–198. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kurinna S, Stratton SA, Coban Z,
Schumacher JM, Grompe M, Duncan AW and Barton MC: P53 regulates a
mitotic transcription program and determines ploidy in normal mouse
liver. Hepatology. 57:2004–2013. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nakamura T, Saito H and Takekawa M: SAPK
pathways and p53 cooperatively regulate PLK4 activity and
centrosome integrity under stress. Nat Commun. 4:17752013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li J, Tan M, Li L, Pamarthy D, Lawrence TS
and Sun Y: SAK, a new polo-like kinase, is transcriptionally
repressed by p53 and induces apoptosis upon RNAi silencing.
Neoplasia. 7:312–323. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao T, Yi S, Yang X and Wu Q: Clinical
significance of polo-like kinase 4 as a marker for advanced tumor
stage and dismal prognosis in patients with surgical gastric
cancer. Technol Cancer Res Treat. 19:15330338209355312020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kahl I, Mense J, Finke C, Boller AL,
Lorber C, Győrffy B, Greve B, Götte M and Espinoza-Sánchez NA: The
cell cycle-related genes RHAMM, AURKA, TPX2, PLK1, and PLK4 are
associated with the poor prognosis of breast cancer patients. J
Cell Biochem. 123:581–600. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao JZ, Ye Q, Wang L and Lee SC:
Centrosome amplification in cancer and cancer-associated human
diseases. Biochim Biophys Acta Rev Cancer. 1876:1885662021.
View Article : Google Scholar : PubMed/NCBI
|