Introduction
Hepatocellular carcinoma (HCC) is one of the most
aggressive malignancies among all types of cancer and constitutes
>75% of liver cancer cases, which in turn accounts for 4.7% of
the total cancer cases in the world (1). Asia has the highest incidence of
liver cancer, with China accounting for 47% of the world's burden
(2). Over previous decades, the
incidence of HCC has increased whilst the prognosis has remained
poor, as a result of high rates of recurrence and metastasis
(3,4). At present, clinical outcomes of
patients with HCC remain unsatisfactory, although progress has been
made towards the prevention of this disease. For example, the
5-year survival of HCC was only 18% in the USA in 2014 (5). Accumulating evidence indicates that
the phenotype of each individual tumor is regulated by its tumor
microenvironment (TME) (6–8). As an immune-sensitive organ, the
liver exerts powerful metabolic functions. A number of studies have
previously found that the immune system can exert regulatory
metabolic functions in a manner that was not previously recognized,
giving rise to a novel research field called immunometabolism
(9–11). However, at present, studies that
have systematically explored this relationship between the
immunometabolism of the TME and its prognosis remain scarce.
As a transcription factor, tumor protein p53 (TP53)
inhibits cell division and survival, thereby functioning as a key
fail-safe mechanism in the cellular anticancer defense system
(12). Therefore, the TP53 gene
has been frequently found to be mutated in human malignancies,
which leads to its proposal for use as a potential predictive
marker and/or target for therapeutic intervention (13–15).
Previous studies have revealed that TP53 mutations in various
cancer types, such as breast and ovarian, are associated with
increased resistance to cancer therapies and poor prognoses
(16,17). Therefore, unraveling the
association between alterations in the TME immunometabolism
following TP53 mutation and prognosis would be of significance,
especially for HCC.
In the present study, it was hypothesized that the
overall survival (OS) time of patients with HCC harboring TP53
mutations may be influenced by immunometabolism in the TME.
Therefore, genes that are affected by TP53 mutations were
investigated, and these were used to establish an immune-metabolism
gene signature to predict the prognosis of patients with HCC in the
clinic.
Materials and methods
Collection of genome-wide mutation
data
In the present study, HCC genetic mutation profile,
transcriptomic and clinical data were all downloaded by searching
the Project ID ‘TCGA-LIHC’ using The Cancer Genome Atlas (TCGA;
http://portal.gdc.cancer.gov/). In the
section of Cases and File Counts by Experimental Strategy, 371
cases containing both clinical information and RNA sequence data
were selected and saved. Mutation data were visualized using the
‘maftools’ package (v2.12.0,
bioconductor.org/packages/release/bioc/html/maftools.html) in R
software (v4.0.3, http://mran.microsoft.com/snapshot/2020-12-04/bin/windows/base/).
MutsigCV represents mutation significance covariants.
Screening and gene enrichment analysis
of differentially expressed genes (DEGs)
The ‘Limma’ package (v3.52.4,
bioconductor.org/packages/release/bioc/html/limma.html) of R
software was used to screen for DEGs, whereas the ‘ClusterProfiler’
package (v3.0.4, http://rdocumentation.org/packages/clusterProfiler/versions/3.0.4)
was used to analyze the Kyoto Encyclopedia of Genes and Genomes
pathways and the Gene Ontology (GO) functions. P<0.05 and
−1>log(fold-change)>1 were considered to be the
thresholds.
Selection of a specific signature
Univariate and multivariate Cox regression analyses
were performed and the ‘forestplot’ package (v1.10.1, http://rdocumentation.org/packages/forestplot/versions/1.10.1)
was used to show the P-values, hazard ratios and 95% confidence
intervals of each variable. P<0.05 was considered to be the
threshold. A nomogram based on the results of the multivariate Cox
analysis, and a list of proposed risk factors found using the ‘rms’
package (v6.1-0, http://rdocumentation.org/packages/rms/versions/6.1-0),
was then constructed to predict the 1-, 3- and 5-year OS rates. If
Kaplan-Meier curves crossed over, a two-stage procedure was
performed using the ‘TSHRC’ package (v0.1-6, http://cran.r-project.org/web/packages/TSHRC/index.html)
of R software. Analyses of risk score, survival status and heatmaps
were implemented using the ‘ggrisk’ package (v1.3, http://cran.r-project.org/web/packages/ggrisk/)of R
software. The time receiver operating characteristic analysis was
used to compare the predictive accuracy of alcohol dehydrogenase 4
(ADH4). All analytical methods were performed using R software
(version 4.0.3) with the ‘ggplot2’ package (v3.3.3, http://rdocumentation.org/packages/ggplot2/versions/3.3.3).
P<0.05 was considered to indicate a statistically significant
difference.
Estimation of immune cell score and
immune infiltration
The ‘immunedeconv’ package (v2.0.3;
rdocumentation.org/packages/immunedeconv/versions/2.0.3) of R
software was used to integrate the Estimating the Proportions of
Immune and Cancer cells algorithm to estimate the immune cell score
and immune infiltration. CD274, cytotoxic T-lymphocyte-associated
protein 4 (CTLA4), hepatitis A virus cellular receptor 2 (HAVCR2),
lymphocyte activating 3 (LAG3), programmed cell death protein 1
(PDCD1), programmed cell death protein 1 ligand 2 (PDCD1LG2), T
cell immunoreceptor with Ig and immunoreceptor tyrosine-based
inhibitory motif domains, and sialic acid binding Ig-like lectin 15
were selected to be the immune checkpoint-associated relevant
transcripts, before their expression values were extracted.
Analyses were implemented using R software (version 4.0.3) with
‘ggplot2’ and ‘pheatmap’ packages (v1.0.12, http://rdocumentation.org/packages/pheatmap/versions/1.0.12).
Collection of HCC specimens
Surgically resected specimens were obtained in
October 2021 from 3 patients with HCC at Renji Hospital Affiliated
with Shanghai Jiao Tong University School of Medicine (Shanghai,
China). The inclusion criteria included: i) Pathological diagnosis
of HCC; ii) the patient had received no prior treatment; and iii)
the patient accepted surgical resection without distant disease.
The exclusion criteria included: i) Other types of primary liver
cancer; and ii) the patient refused the surgical resection. Tissue
within 2 cm from the tumor boundary is called adjacent tissue. All
specimens were conserved in liquid nitrogen. The present study was
approved by the Ethics Committee of Renji Hospital (Shanghai,
China) and written informed consent was provided by the enrolled
patients.
Immunohistochemistry
ADH4 protein expression was examined using an
immunohistochemistry assay. Fresh specimens were fixed in 4%
phosphate-buffered formalin for at least 24 h at room temperature.
All HCC tissues were paraffin-embedded and cut into 4-µm-thick
sections. The sections were then dewaxed with xylene and rehydrated
with descending graded alcohol. Activity of endogenous catalase
would be inhibited by 0.3% H2O2.
Heat-mediated antigen retrieval of tissue sections was performed at
98°C and maintained for 20 min before the sections were allowed to
cool. After blocking non-specific sites with 5% goat serum at 37°C
for 30 min, the sections were incubated with anti-ADH4 primary
antibody (cat. no. Ab137077; 1:500 dilution; Abcam) overnight at
4°C and then with HRP-conjugated goat anti-rabbit secondary
antibody (cat. no. Ab6721; 1:1,000 dilution; Abcam) for 1 h at room
temperature. Antibodies were visualized using 3,3′-diaminobenzidine
and counterstained using Meyer's Hematoxylin for 2 min at room
temperature. Sections were then dehydrated using an increasing
gradient of alcohols before being cleared in xylene. Images were
observed using a light Olympus Corporation BX51 instrument. The
difference of immunostaining results were evaluated using Image J
software (v1.52a, USA).
Western blotting
ADH4 protein expression of the resected tissues was
examined using western blotting. The tissues were first treated
using a lysis solution (RIPA, Radio-Immunoprecipitation Assay
buffer; Thermo Fisher Scientific, USA), which contained 1% (v/v)
protease inhibitors (cat. no. P8340; Merck KGaA) to extract protein
samples. The protein concentration was measured and normalized
using Protein Quantification kit (BCA Assay; Beyotime, China).
Western blot analysis was then performed as previously described
(18). The membranes were blocked
in Tris-Buffer Saline Tween 20 (1×TBST) containing 5% non-fat milk
at room temperature for 45 min. Then, membranes would be incubated
with anti-ADH4 primary antibody (cat. no. Ab137077; 1:1,000
dilution; Abcam) overnight at 4°C and HRP-conjugated goat
anti-rabbit secondary antibody (cat. no. Ab6721; 1:1,000 dilution;
Abcam) for 1 h at room temperature. Enhanced chemiluminescence
reagent (SuperSignal™ West Atto Ultimate Sensitivity Substrate;
Thermo Fisher Scientific, Inc.) was used for immunodetection. The
level of actin protein expression was measured as an internal
standard (anti-β actin antibody; cat. no. Ab8227; 1:1,000 dilution;
Abcam). The density of protein band was quantitatively measured
using Image J software (v1.52a, USA).
Immune checkpoint blockage (ICB)
response prediction
Potential ICB responses were predicted using the
Tumor Immune Dysfunction and Exclusion algorithm (19) and software packages ‘ggplot2’ and
‘ggpubr’ (v0.4.0, http://rdocumentation.org/packages/ggpubr/versions/0.4.0).
All analytical methods were performed using R software (version
4.0.3). P<0.05 was considered to indicate a statistically
significant difference.
Statistical analysis
All statistical analyses were performed using R
software (version 4.0.3). Unpaired Student's t-test was used to
compare any differences between the subgroups in DEGs analysis.
Data from two groups were compared in cox regression and K-M curve
analysis using the Wilcoxon rank-sum test, whereas more than three
groups were compared in immune analysis using the Kruskal-Wallis
test. Post hoc test was performed using Least Significance
Difference method. Data are presented by mean and SD. χ-square test
was used to compare significant differences between groups.
P<0.05 was considered to indicate a statistically significant
difference. Test was performed no less than three independent
repeats.
Results
TP53 is the most frequently mutated
gene in HCC
The experimental plan of the present study is
presented in Fig. 1. A total of
270 mutant genes in HCC were screened from TCGA. Mutant genes
revealed by the screen, including TP53 (28%), Titin (25%), Catenin
Beta 1 (24%), mucin 16 (16%) and piccolo presynaptic cytomatrix
protein (11%), had higher mutation frequencies than the rest of
screened mutant genes, which is demonstrated by the horizontal
histogram (Fig. 2A). In
particular, missense mutations appeared to be the most common type
of TP53 mutation (Fig. 2Ba).
Single-nucleotide polymorphisms showed a predominant position
compared with deletions or insertions (Fig. 2Bb). C>T was found to be the most
distinct mutation type (Fig. 2Bc).
The number of mutations per sample is shown in Fig. 2Bd. Each color in the box diagram
represents one type of mutation (Fig.
2Be). Fig. 2Bf shows the top
10 mutant genes and TP53 is ranked second. Fig. 2C presents a lollipop plot, which
shows that TP53 a highly mutated gene in HCC and missense mutations
is the most common type of TP53 mutation.
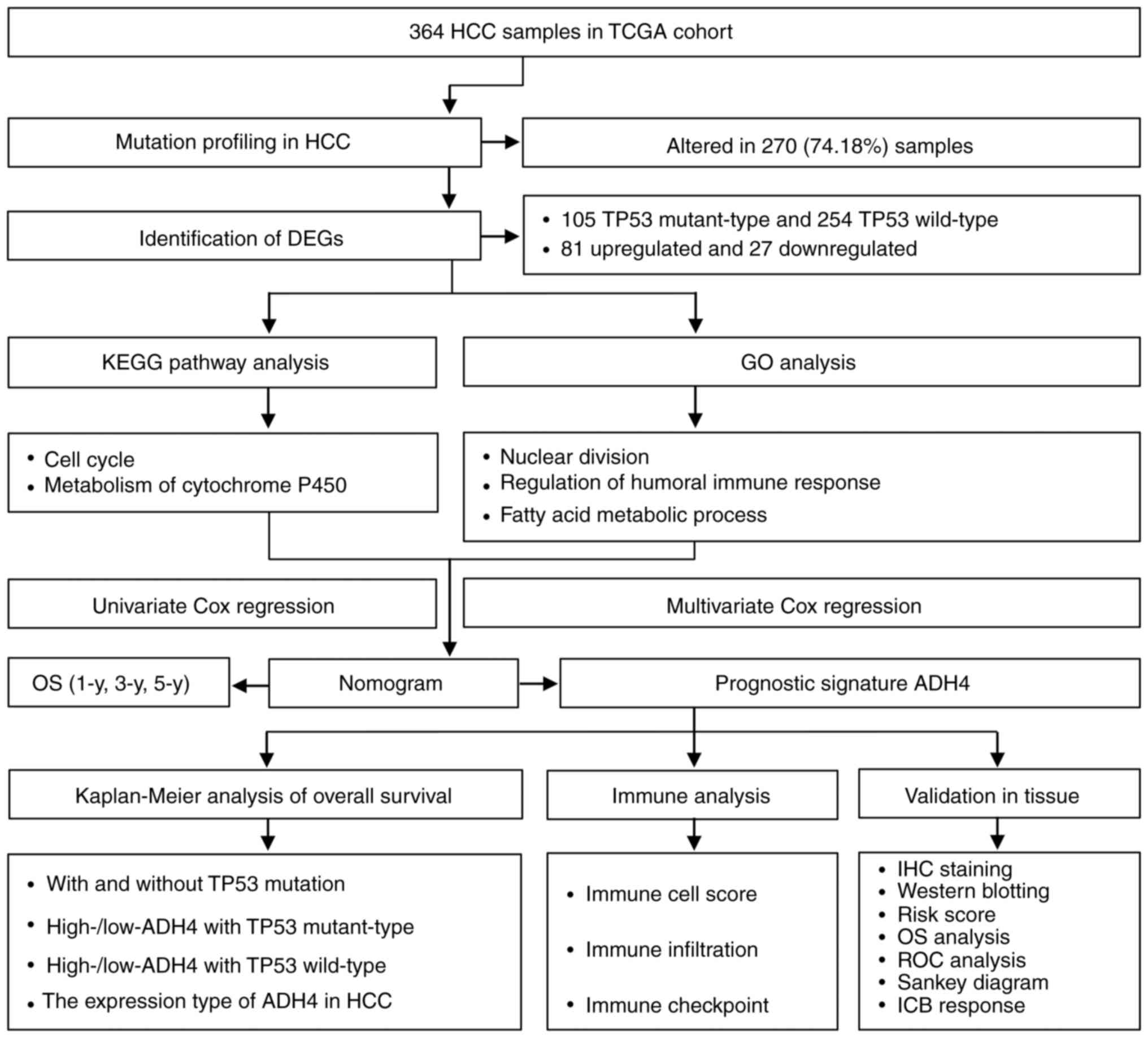 | Figure 1.Flowchart of the present study. ADH4,
alcohol dehydrogenase 4; DEG, differentially expressed gene; GO,
Gene Ontology; HCC, hepatocellular carcinoma; ICB, immune
checkpoint blockage; IHC, immunohistochemistry; KEGG, Kyoto
Encyclopedia of Genes and Genomes; OS, overall survival; ROC,
receiver operating characteristic; TCGA, The Cancer Genome Atlas;
TP53, tumor protein p53; y, years (371 cases were saved and only
364 samples were listed, because 8 cases was deleted due to
incomplete information). |
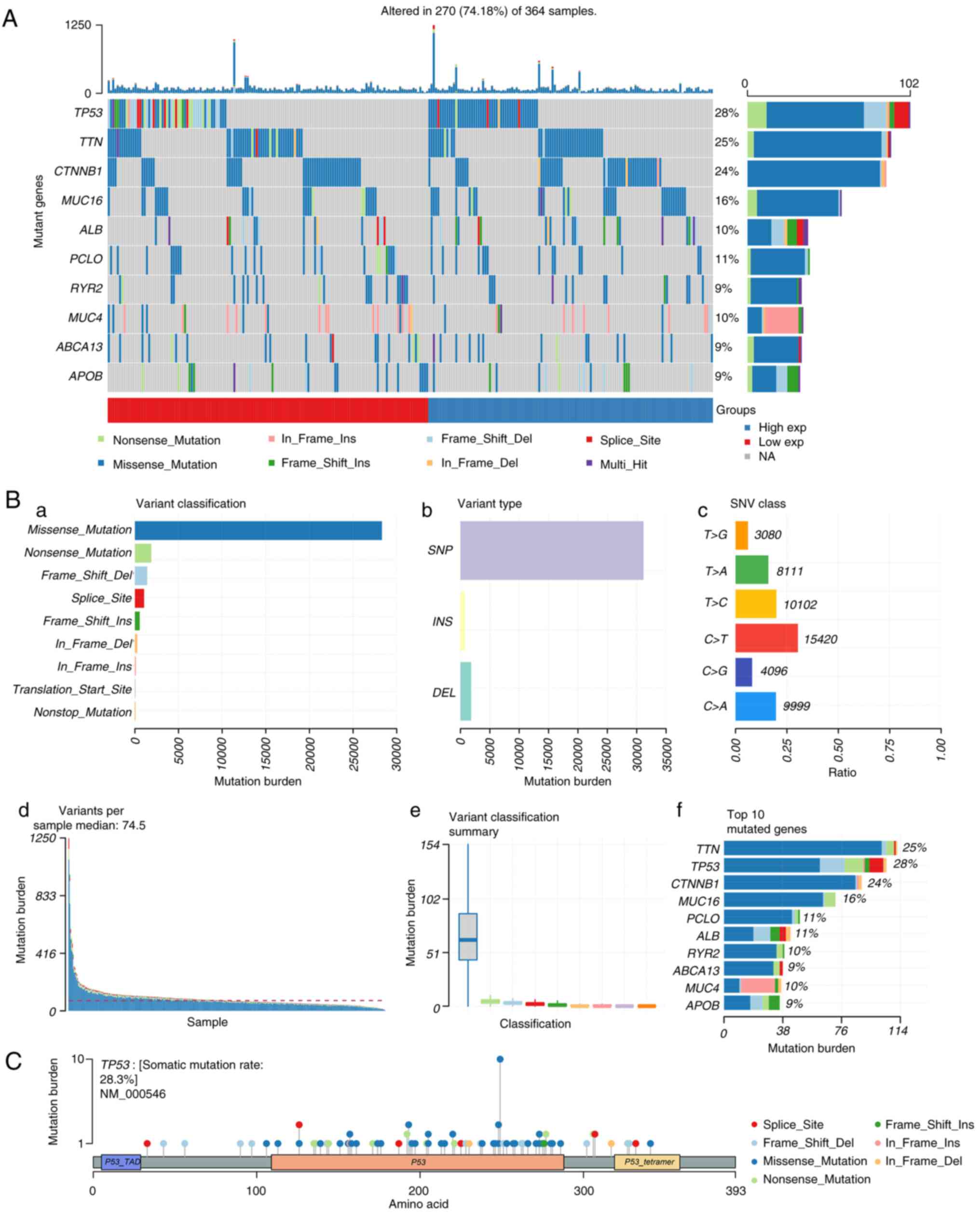 | Figure 2.Genome-wide mutation profiling in
HCC. (A) Oncoplot (left) displaying the somatic landscape of the
HCC cohort. Genes are ordered by their mutation frequency and
samples are ordered according to disease histology as indicated by
the annotation bar (bottom). The bar plot (right) shows log10
transformed Q-values estimated by MutSigCV. (B) Cohort summary plot
displaying the distribution of variants according to (a) variant
classification, (b) variant type and (c) SNV class. (d) Mutation
load of each sample and (e) variant classification type. (f) Top 10
mutant genes. (C) Lollipop plot displaying mutation distribution
and protein domains for TP53 in HCC, with labeled recurrent
hotspots. Somatic mutation rate and transcript names are indicated
by the plot title and subtitle, respectively. Del, deletion; exp,
expression; HHC, hepatocellular carcinoma; Ins, insertion; NA, not
available; SNP, single nucleotide polymorphism; SNV, single
nucleotide variant; TAD, transaction domain; TP53, tumor protein
p53. |
DEGs detected in HCC by TP53
status
DEGs between the TP53 mutant and wild-type TP53
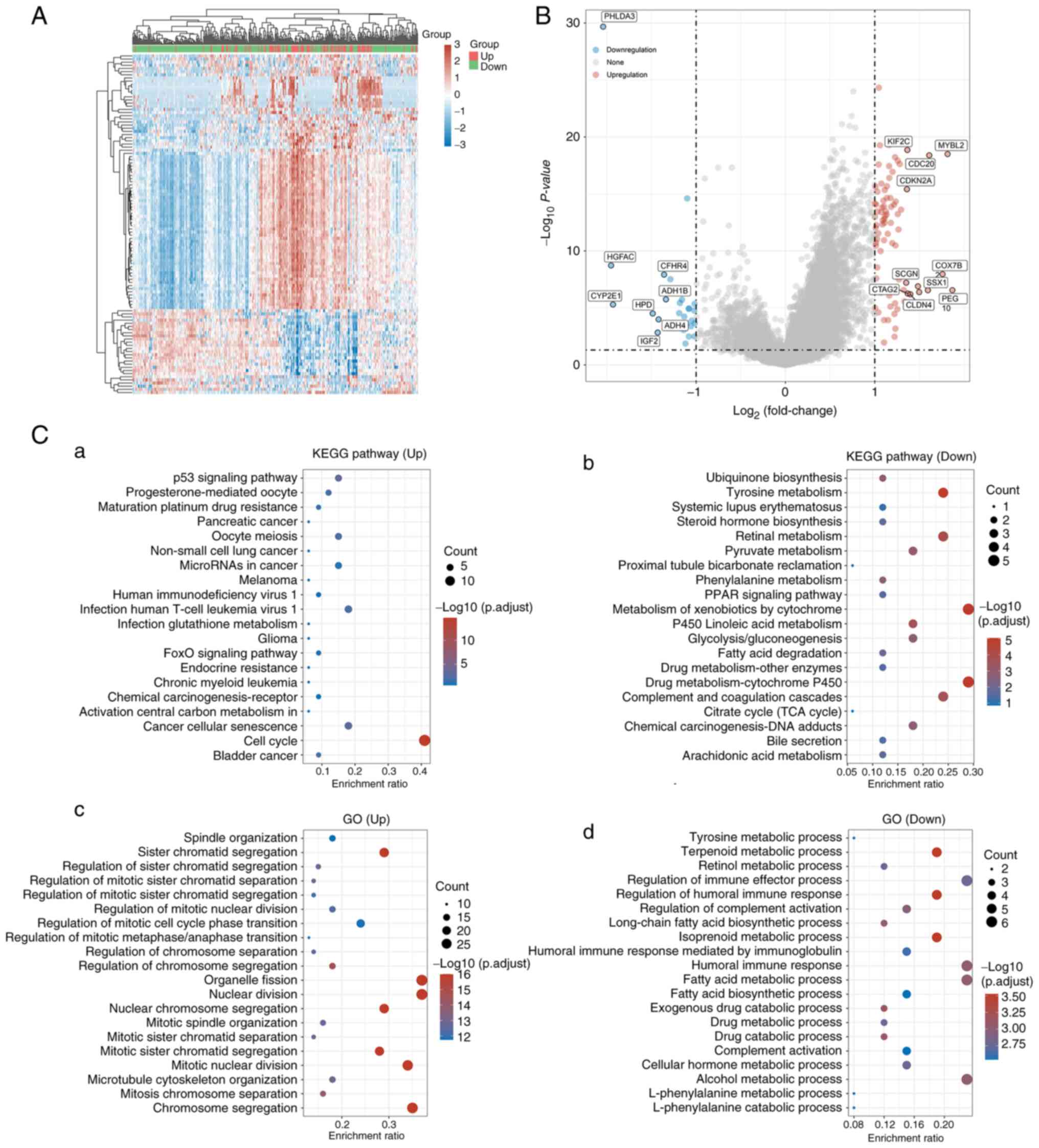
groups were next screened. In total, 81 upregulated genes and 27
downregulated genes were identified (Fig. 3A and B). Notably, ‘tyrosine
metabolism’ and ‘pyruvate metabolism’ were markedly downregulated,
in addition to ‘alcohol metabolic process’ and ‘regulation of
immune effector process’ (Fig.
3C). By contrast, mutant TP53 was particularly enriched for
pathways associated with the ‘cell cycle’ and cell metabolism,
‘cytochrome metabolism and drug metabolism (Fig. 3Ca and Cb). Furthermore, genes
associated with TP53 mutation tended to be more enriched for terms
associated with the metabolic process, including nuclear division
and humoral immune response (Fig. 3Cc
and Cd). These results suggested that genes associated with
TP53 mutations are likely to serve an important role in the
immunometabolism of the TME in HCC.
ADH4 is a prognostic signature for
HCC
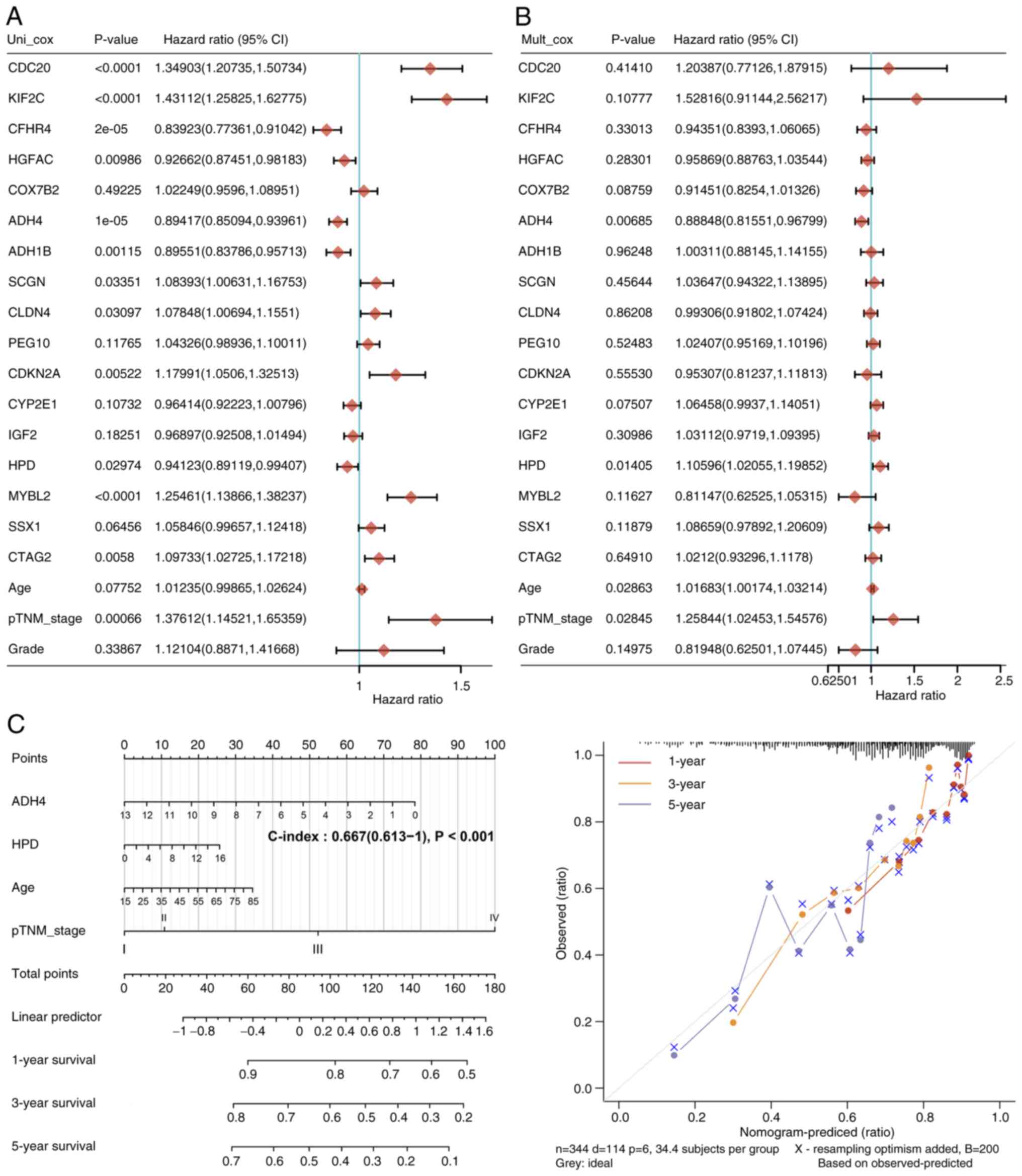
A total of 17 genes and corresponding clinical
information, including age, pTNM_stage and grade were identified
through univariate (Fig. 4A) and
multivariate (Fig. 4B) Cox
regression analyses, to build a predictive nomogram. The predictors
included ADH4, 4-hydroxyphenylpyruvate dioxygenase, patient age and
pathological Tumor-Node-Metastasis (pTNM) stage (20) (Fig.
4C), all of which met the P<0.05 criteria during risk
assessment. The P-value of ADH4 was found to be <0.001, which
suggested that ADH4 can be exploited as a distinct prognostic
signature for HCC. The calibration plots for the 1-, 3- and 5-year
OS rates were consistent compared with those predicted by the ideal
model in the entire cohort (Fig.
4C).
TP53 and ADH4 status are associated
with the prognosis of HCC
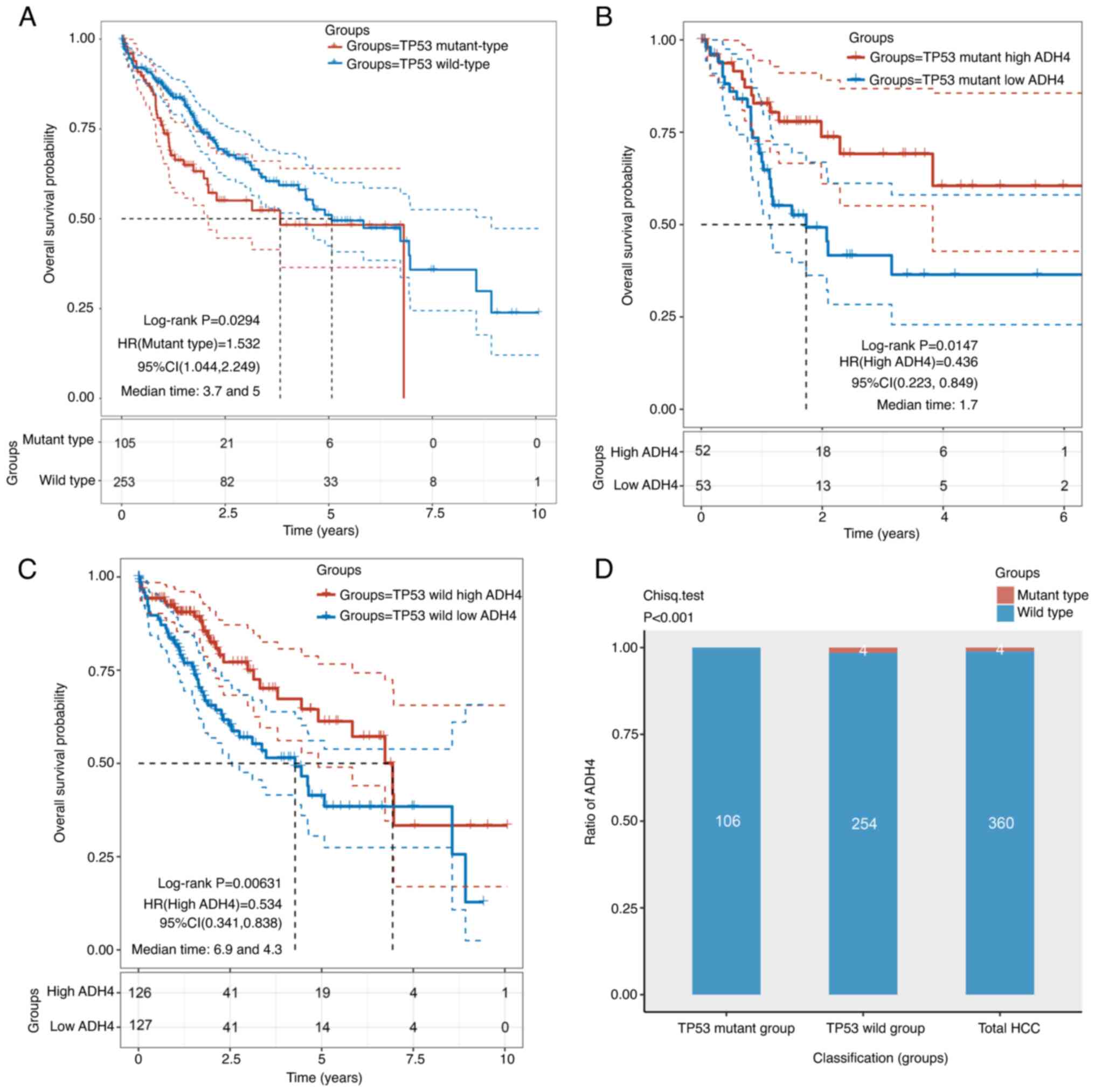
The mutant TP53 and wild-type TP53 groups included
105 and 253 patients, respectively. Patients in the mutant TP53
group displayed significantly worse OS time compared with those in
the wild-type group (Fig. 5A). To
investigate if ADH4 was independent of the TP53 mutation status,
patients with HCC were divided into high- and low-ADH4 groups based
on the TP53 mutation status. The high- and low-ADH4 groups in the
mutant TP53 group included 52 and 53 patients, respectively. By
contrast, the high- and low-ADH4 groups in the wild-type TP53 group
included 126 and 127 patients, respectively. The results revealed
that the low-ADH4 group in both the mutant and wild-type TP53
groups displayed significantly worse OS time compared with the
high-ADH4 group (Fig. 5B and C).
The crossed curves in Fig. 5 were
determined to be statistically different (Fig. 5A, P=0.0294; Fig. 5B, P=0.0147; Fig. 5C, P=0.0063;) by performing a
two-stage procedure, using the ‘TSHRC’ package of R software. In
addition, wild-type ADH4 was observed to occupy a particularly
higher proportion of patients with HCC than mutant type ADH4
(χ2 test; Fig. 5D).
Differential immune analysis
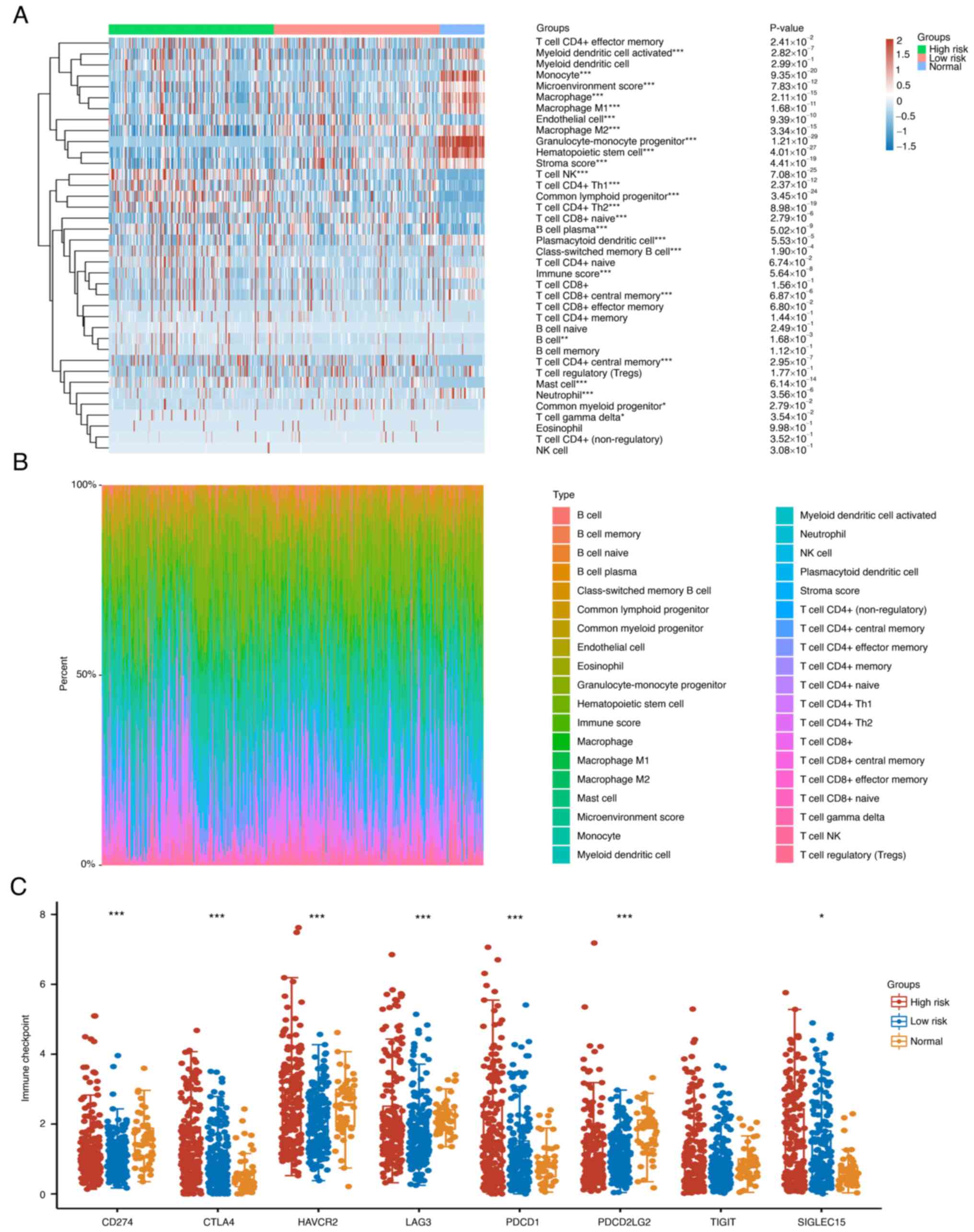
According to the results in Fig. 5, high risk group in Fig. 6 was defined as low ADH4 expression
while low risk group in Fig. 6 was
defined as high ADH4 expression. To explore the distribution of
immune scores (Fig. 6A) and immune
infiltration (Fig. 6B) in the
groups of different risks, 38 types of immune cells were compared
in total. Among them, 23 types of immune cells were found to be
statistically different from other rest 15 types of immune cells.
It was noted that there was significant immune infiltration by
cells expressing markers of B cells in the high-ADH4 group than
other immune cells in the Fig. 6B,
whilst the expression of markers of T cells was decreased,
suggesting a differential immune TME. For the expression
distribution of immune checkpoint markers in the HCC tissues,
CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2, TIGIT and SIGLEC15
were discovered to be the immune checkpoint-associated transcripts
showing a statistical difference, which was different from other
transcripts (Fig. 6C).
ADH4 expression was validated in
clinical HCC tissues
To validate the identified gene signature, ADH4
expression levels were examined using immunohistochemistry and
western blotting on pairs of HCC tissues and adjacent normal
tissues from 3 patients. The pathological diagnosis of the 3 male
patients was HCC without distant metastasis, and none had received
any prior treatment. The clinicopathological information of the 3
patients is shown in Table I. The
TP53 status of these patients was mutant type and the molecular
hallmark was a mutation at codon 249 in TP53. The results
demonstrated that ADH4 protein expression was markedly increased in
the normal tissues compared with the tumor tissues (Fig. 7A and D). The risk score of every
patient whose RNA Seq files was downloaded from TCGA was then
calculated to obtain the median cut-off point to divide them into
the high-ADH4 group (n=185) and low-ADH4 group (n=185) (Fig. 7B, top). The survival status of all
patients with HCC is shown (Fig.
7B, middle) and the prognostic ADH4 gene expression profile is
shown in the heatmap (Fig. 7B,
bottom). The Kaplan-Meier survival curves demonstrated that
patients in the low-ADH4 group had worse OS time compared with
those in the high-ADH4 group (Fig.
7C, P=0.0002). In addition, a time-dependent receiver operating
characteristic analysis suggested that ADH4 expression had accurate
predictive capabilities for 1-, 3- and 5-year OS (Fig. 7E). A Sankey diagram of ADH4
expression revealed the distribution of the same sample in
different characteristic variables including ages, pTNM_stages,
grade, ADH4 expression type and status of patients (Fig. 7F). Potential ICB responses
indicated that the distribution of the TIDE scores in the low-ADH4
group showed a worse response compared with that in the high-ADH4
group (Fig. 7G).
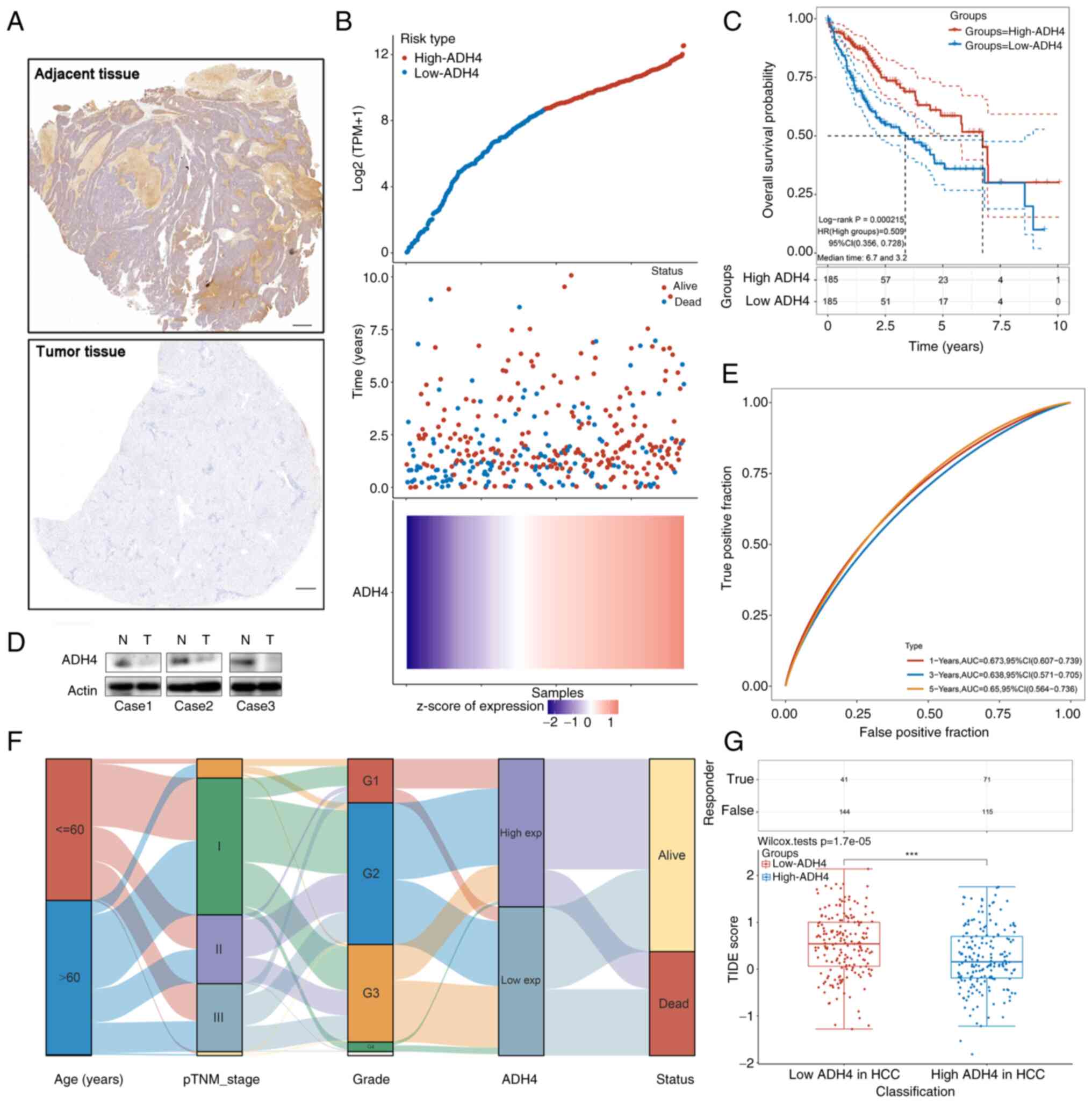 | Figure 7.Validation of gene signature in
clinical HCC tissue samples. (A) Representative images of
immunohistochemistry staining for ADH4 expression in adjacent
non-tumor tissues (top) and HCC tissues (bottom). Scale bar, 500
µm. (B) Curve of risk score (top), the survival status of the
patients (middle) and the heatmap of the ADH4 expression profile
(bottom). (C) Kaplan-Meier survival analysis of patients with HCC
by ADH4 status. Dotted lines represent the confidence interval. (D)
Western blot analysis of adjacent non-tumor and HCC tissues. (E)
Time-dependent receiver operating characteristic analysis of the
ADH4 signature. (F) Sankey diagram of ADH4 expression, which
indicates the distribution of the same sample in different
characteristic variables including ages, pTNM_stages, grade, ADH4
expression type and status of patients. (G) Potential immune
checkpoint blockage responses of HCC groups by their ADH4 status,
which indicates the distribution of immune response scores in
different groups in the prediction results (***P<0.001). The
upper table indicates the positive immune response amount in the
samples. ADH4, alcohol dehydrogenase 4; AUC, area under the curve;
exp, expression; HCC, hepatocellular carcinoma; HR, hazard ratio;
N, normal; T, tumor; pTNM, pathological Tumor-Node-Metastasis; TPM,
transcript per million; TIDE, Tumor Immune Dysfunction and
Exclusion. |
 | Table I.Clinicopathological information of 3
patients with HCC. |
Table I.
Clinicopathological information of 3
patients with HCC.
| Case | Age, years | Sex | Ethnicity | Tumor type | Pathological
type | Edmondson-Steiner
grade (39) | TP53 status | Vital status |
|---|
| 1 | 48 | Male | Asian | Primary tumor | HCC | III | Mutation (serine
249) | Alive |
| 2 | 62 | Male | Asian | Primary tumor | HCC | III | Mutation (serine
249) | Alive |
| 3 | 55 | Male | Asian | Primary tumor | HCC | IV | Mutation (serine
249) | Alive |
Discussion
HCC is one of the main causes of cancer-associated
mortality worldwide and is associated with a poor prognosis
(21). In addition, HCC frequently
becomes aggressively malignant in a short space of time as a result
of immunosuppression and the reprogramming of metabolism (22). Accumulating evidence has indicated
that a combination of different immunotherapies and targeted
therapies can prolong the survival of patients with HCC (23,24).
In addition, metabolic reprogramming has been demonstrated to be
potentially significant for hepatocarcinogenesis and prognosis
(25,26). However, to the best of our
knowledge, the mechanism underlying this reprogramming remains
unknown (27). One study reviewed
the data from immunotherapy trials on HCC (28) and concluded that immunometabolism
in the TME exerts an influence on the prognosis of patients with
HCC. TP53 mutations result in increased mutational burden in
patients with cancer, which may in turn alter the immunometabolism
in the TME (29). Therefore, a
TP53-associated immune-metabolism signature was developed in the
present study for the prediction of HCC prognosis, which may have
an increased clinical role in the future.
A recent study has shown that TP53 mutations serve
different roles in antitumor immunity (30). In the present study, it was found
that the TP53 gene locus had a high mutation frequency in patients
with HCC, and the genes modulated downstream of the TP53 mutations
were especially enriched for GO terms associated with immune and
metabolic responses. Notably, ‘tyrosine metabolism’ and ‘pyruvate
metabolism’ were markedly downregulated, in addition to the
‘alcohol metabolic process’ and ‘regulation of immune effector
process’ (Fig. 3C), which may be
associated with alcoholic cirrhosis and tumor progression. Since
wild-type TP53 serves such fundamental roles in cancer immunity,
TP53 mutations can result in immune dysfunction, thereby promoting
tumorigenesis, cell invasion and metastasis (31). To deepen the understanding of the
changes in immunometabolism in the TME, a nomogram was constructed
using the distinctive prognostic signature of ADH4. This nomogram
was found to be an effective independent prognostic model for HCC.
In addition, it was found that other parameters, including the age
of patients and pTNM stage, also significantly affected the OS of
patients with HCC. The low-ADH4 group in both mutant and wild-type
TP53 groups displayed significantly worse OS time compared with the
high-ADH4 group. This suggested that ADH4 is a beneficial signature
for the prognosis of HCC where the type of ADH4 is suggested to be
wild-type. The ADHs belong to a large family of dehydrogenase
enzymes that are associated with good prognoses of various types of
cancer, such as colorectal and gastric cancer (32). To investigate the expression
profile of ADH4 in HCC, distributions of ADH4 mutants and wild-type
ADH4 were compared in the TP53 mutant, wild-type TP53 and total HCC
groups. The results revealed that wild-type ADH4 was present in a
markedly high proportion of patients with HCC. In conclusion, ADH4
is an immune-metabolic protective factor and low ADH4 can be
regarded as a high-risk factor for the prognosis of HCC.
Immune-metabolism relationships have been reported
in various diseases, such as cancer, metabolic syndrome and
immune-mediated diseases (33).
Changes in the TME can also alter the phenotype and function of
immune cells (34). Immune cells
have been reported to serve a variety of key roles in the
development of malignant tumors, especially in HCC (35). Tumor-infiltrating leukocytes have
been reported to impact the progression of HCC (35). In addition, another study found
that the functional interaction between tumor-infiltrating T cells
and B cells could contribute to local immune activation, improving
the prognosis of HCC (35). In the
present study, the results indicated that there was significant
immune infiltration by B cells and reduced infiltration by T cells
in the low risk group, which suggested a differential immune TME.
These results suggested that the differential prognosis is
associated with the interaction between the TME and the infiltrated
immune cells. Furthermore, CD274, CTLA4, HAVCR2, LAG3, PDCD1,
PDCD1LG2 and SIGLEC15 were revealed to be immune
checkpoint-associated transcripts, which may provide patients with
greater benefits from immunotherapy and chemotherapy (36).
The distribution of ADH4 protein expression and the
prognostic prediction value were subsequently explored in clinical
specimens in the present study. TP53 mutations in serine 249 were
revealed, which are associated with high exposure to aflatoxin and
poorly differentiated tumors (37). ADH4 protein expression was
increased in normal tissues compared with tumor tissues, and
patients in the low-ADH4 group had worse OS time compared with
those in the high-ADH4 group, which is consistent with the previous
analysis (38). This suggested
that high-risk patients with HCC are more likely to be involved in
ICB responses, which may provide a potentially novel strategy for
clinical guidance. However, it should be noted that there were
limited numbers of patients for the validation in the present
study. In addition, the lack of follow-up data on the 3 patients
sampled is also a limitation of the study. The main reason for this
limitation is that the 3 patients with HCC were diagnosed in
October 2021 and then accepted the surgery. It has been <1 year
since successful surgery and the patients are still alive at the
time of writing, according to the latest follow-up records.
Therefore, larger scale clinical trials would need to be performed
to verify the results of the present study. In addition, the
mechanism underlying the function of ADH4 and TP53 requires further
exploration using gene knockout technology applied in cell line and
animal experiments.
To conclude, the present study identified ADH4 to be
an immune-metabolism signature downstream of TP53 mutation that can
be used to independently predict the prognosis of patients with
HCC. Therefore, ADH4 may serve as an accurate biomarker for
designing novel immunotherapies.
Acknowledgements
Not applicable.
Funding
This research was supported by a grant from the National Natural
Science Foundation of China (grant no. 81874201).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ made substantial contributions to the conception
and the work of manuscript editing. HHJ made substantial
contributions to the design of the work. ZYW made substantial
contributions to the acquisition and analysis of data. BZ made
substantial contributions to conception and approved the modified
version. MBL made substantial contributions to the design of the
study. BZ and MBL confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The research received ethics approval from the
Ethics Committee of Renji Hospital (Shanghai, China) and written
informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ADH4
|
alcohol dehydrogenase 4
|
|
DEG
|
differentially expressed gene
|
|
GO
|
Gene Ontology
|
|
HCC
|
hepatocellular carcinoma
|
|
ICB
|
immune checkpoint blockage
|
|
OS
|
overall survival
|
|
TCGA
|
The Cancer Genome Atlas
|
|
TME
|
tumor microenvironment
|
|
TP53
|
tumor protein p53
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Petrick JL, Florio AA, Znaor A, Ruggieri
D, Laversanne M, Alvarez CS, Ferlay J, Valery PC, Bray F and
McGlynn KA: International trends in hepatocellular carcinoma
incidence, 1978–2012. Int J Cancer. 147:317–330. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pang Y, Liu Z, Han H, Wang B, Li W, Mao C
and Liu S: Peptide SMIM30 promotes HCC development by inducing
SRC/YES1 membrane anchoring and MAPK pathway activation. J Hepatol.
73:1155–1169. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jemal A, Ward EM, Johnson CJ, Cronin KA,
Ma J, Ryerson B, Mariotto A, Lake AJ, Wilson R, Sherman RL, et al:
Annual report to the nation on the status of cancer, 1975–2014,
featuring survival. J Natl Cancer Inst. 109:djx0302017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng Y, Chen Z, Han Y, Han L, Zou X, Zhou
B, Hu R, Hao J, Bai S, Xiao H, et al: Immune suppressive landscape
in the human esophageal squamous cell carcinoma microenvironment.
Nat Commun. 11:62682020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang W and Zou W: Amino acids and their
transporters in T cell immunity and cancer therapy. Mol Cell.
80:384–395. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu J and Cai J: Dilemma and challenge of
immunotherapy for pancreatic cancer. Dig Dis Sci. 66:359–368. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Greer RL, Dong X, Moraes AC, Zielke RA,
Fernandes GR, Peremyslova E, Vasquez-Perez S, Schoenborn AA, Gomes
EP, Pereira AC, et al: Akkermansia muciniphila mediates negative
effects of IFNγ on glucose metabolism. Nat Commun. 7:133292016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu R, Chen F, Wang N, Tang D and Kang R:
ACOD1 in immunometabolism and disease. Cell Mol Immunol.
17:822–833. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mouton AJ, Li X, Hall ME and Hall JE:
Obesity, hypertension, and cardiac dysfunction: Novel roles of
immunometabolism in macrophage activation and inflammation. Circ
Res. 126:789–806. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Donehower LA, Soussi T, Korkut A, Liu Y,
Schultz A, Cardenas M, Li X, Babur O, Hsu TK, Lichtarge O, et al:
Integrated analysis of TP53 gene and pathway alterations in the
cancer genome atlas. Cell Rep. 28:1370–1384.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Olivier M, Hollstein M and Hainaut P: TP53
mutations in human cancers: Origins, consequences, and clinical
use. Cold Spring Harb Perspect Biol. 2:a0010082010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mogi A and Kuwano H: TP53 mutations in
nonsmall cell lung cancer. J Biomed Biotechnol. 2011:5839292011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Duffy MJ, Synnott NC and Crown J: Mutant
p53 as a target for cancer treatment. Eur J Cancer. 83:258–265.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Silwal-Pandit L, Langerød A and
Børresen-Dale AL: TP53 mutations in breast and ovarian cancer. Cold
Spring Harb Perspect Med. 7:a0262522017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang C, Huang X, Li Y, Chen J, Lv Y and
Dai S: Prognosis and personalized treatment prediction in
TP53-mutant hepatocellular carcinoma: An in silico strategy towards
precision oncology. Brief Bioinform. 22:bbaa1642021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meng F, Wu L, Dong L, Mitchell AV, James
Block C, Liu J, Zhang H, Lu Q, Song WM, Zhang B, et al: EGFL9
promotes breast cancer metastasis by inducing cMET activation and
metabolic reprogramming. Nat Commun. 10:50332019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Telloni SM: Tumor staging and grading: A
primer. Methods Mol Biol. 1606:1–17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pinero F, Dirchwolf M and Pessôa MG:
Biomarkers in hepatocellular carcinoma: Diagnosis, prognosis and
treatment response assessment. Cells. 9:13702020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Golonka RM and Vijay-Kumar M: Atypical
immunometabolism and metabolic reprogramming in liver cancer:
Deciphering the role of gut microbiome. Adv Cancer Res.
149:171–255. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sonbol MB, Riaz IB, Naqvi SAA, Almquist
DR, Mina S, Almasri J, Shah S, Almader-Douglas D, Uson Junior PLS,
Mahipal A, et al: Systemic therapy and sequencing options in
advanced hepatocellular carcinoma: A systematic review and network
meta-analysis. JAMA Oncol. 6:e2049302020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Llovet JM, Montal R, Sia D and Finn RS:
Molecular therapies and precision medicine for hepatocellular
carcinoma. Nat Rev Clin Oncol. 15:599–616. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Yan Q, Gong L, Xu H, Liu B, Fang
X, Yu D, Li L, Wei T, Wang Y, et al: C-terminal truncated HBx
initiates hepatocarcinogenesis by downregulating TXNIP and
reprogramming glucose metabolism. Oncogene. 40:1147–1161. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Q, Tan Y, Jiang T, Wang X, Li Q, Dong
L, Liu X and Xu G: Metabolic reprogramming and its relationship to
survival in hepatocellular carcinoma. Cells. 11:10662022.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jühling F, Hamdane N, Crouchet E, Li S, El
Saghire H, Mukherji A, Fujiwara N, Oudot MA, Thumann C, Saviano A,
et al: Targeting clinical epigenetic reprogramming for
chemoprevention of metabolic and viral hepatocellular carcinoma.
Gut. 70:157–169. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Machairas N, Tsilimigras DI and Pawlik TM:
Current landscape of immune checkpoint inhibitor therapy for
hepatocellular carcinoma. Cancers (Basel). 14:20182022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mantovani F, Collavin L and Del Sal G:
Mutant p53 as a guardian of the cancer cell. Cell Death Differ.
26:199–212. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li L, Li M and Wang X: Cancer
type-dependent correlations between TP53 mutations and antitumor
immunity. DNA Repair (Amst). 88:1027852020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Agupitan AD, Neeson P, Williams S, Howitt
J, Haupt S and Haupt Y: P53: A guardian of immunity becomes its
saboteur through mutation. Int J Mol Sci. 21:34522020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jelski W and Szmitkowski M: Alcohol
dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) in the cancer
diseases. Clin Chim Acta. 395:1–5. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lercher A, Baazim H and Bergthaler A:
Systemic immunometabolism: Challenges and opportunities. Immunity.
53:496–509. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Piñeiro Fernández J, Luddy KA, Harmon C
and O'Farrelly C: Hepatic tumor microenvironments and effects on NK
cell phenotype and function. Int J Mol Sci. 20:41312019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Garnelo M, Tan A, Her Z, Yeong J, Lim CJ,
Chen J, Lim KH, Weber A, Chow P, Chung A, et al: Interaction
between tumour-infiltrating B cells and T cells controls the
progression of hepatocellular carcinoma. Gut. 66:342–351. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Giannone G, Ghisoni E, Genta S, Scotto G,
Tuninetti V, Turinetto M and Valabrega G: Immuno-metabolism and
microenvironment in cancer: Key players for immunotherapy. Int J
Mol Sci. 21:44142020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nogueira JA, Ono-Nita SK, Nita ME, de
Souza MM, do Carmo EP, Mello ES, Scapulatempo C, Paranaguá-Vezozzo
DC, Carrilho FJ and Alves VA: 249 TP53 mutation has high prevalence
and is correlated with larger and poorly differentiated HCC in
Brazilian patients. BMC Cancer. 9:2042009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu X, Li T, Kong D, You H, Kong F and
Tang R: Prognostic implications of alcohol dehydrogenases in
hepatocellular carcinoma. BMC Cancer. 20:12042020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Edmondson HA and Steiner PE: Primary
carcinoma of the liver: A study of 100 cases among 48,900
necropsies. Cancer. 7:462–503. 1954. View Article : Google Scholar : PubMed/NCBI
|