Introduction
The incidence of malignant melanoma has increased
worldwide over recent years and it is currently a significant
public health problem (1,2). Ultraviolet radiation, which directly
damages DNA, is the significant risk factor for the pathogenesis of
melanoma (1,3). The early detection of melanoma and
evaluation of melanoma tissue biomarkers are important for patient
risk stratification, personalized diagnostics and treatment
(4,5).
The current World Health Organization (WHO)
classification of skin tumors subdivides melanoma on the basis of
solar elastosis assessed by dermal elastic fibers, and measures
cumulative sun damage (CSD) (3).
According to WHO classification, there are currently 3 classes of
melanomas: Those associated with high CSD, those associated with
low CSD and nodular melanomas (3,6).
Solar elastosis is usually apparent in superficially spreading and
lentigo malignant melanoma, the so-called high CSD melanoma.
Desmoplastic melanoma is associated with increased solar elastosis.
The most common subtype of high CSD melanoma is superficially
spreading melanoma, which usually begins with early radial growth
followed by vertical growth and invasion of the dermis (3). Acral, mucosal, uveal and spitzoid
melanomas are not associated with CSD, or are characterized by low
CSD. Nodular melanomas are usually characterized as a low CSD type
with early progression to vertical growth (3).
While the advent of novel personalized treatments of
melanoma based on BRAF inhibitors and immunotherapies have reduced
mortality rate over the last decade, advanced and metastatic
melanomas still remain difficult to treat (7–10).
Therefore, early diagnostic and risk stratification for the
progression of melanoma is of particular importance. However, rare
melanoma histopathological subtypes can make diagnosis challenging
(3).
Therefore, the biomarkers of early-stage melanoma
for the prediction of melanoma clinical behavior is of particular
importance. It has been shown that such clinicopathological
characteristics such tumor size, tumor type, tumor invasiveness
(Breslow thickness, Clark level, lymphovascular invasion,
neurotropisms), ulceration and tumor mitosis activity are
significant prognostic factors for the development and progression
of melanoma (3,6,11).
In addition, it has been demonstrated that tumor-infiltrating
lymphocytes could stratify melanoma with low and high risk
progression (12–14).
The development of melanoma is closely related to
somatic and epigenetic changes. Different mutations have been
implicated in its pathogenesis and evolution. Recent genomic
classification subdivides melanoma into 4 major subtypes based on
the pattern of the most prevalent significantly mutated genes:
mutant BRAF, RAS, NF1 and triple-WT (wild type) (15). Advances in molecular pathology and
assessment of genetic biomarkers are increasingly used in clinical
practice for diagnosis, personalized treatment and the prognosis of
melanoma. Modern treatment guidelines are focused on the assessment
of genetic biomarkers of melanoma (1,16,17).
The assessment of the BRAF gene mutation is of
particular importance (3). BRAF
mutations are observed in 40–60% of all primary malignant melanoma
cases (16–20). The BRAF mutation is usually
observed in younger patients, in non CSD skin and in superficial
spreading melanoma, whereas NRAS mutational melanoma were
characterized for nodular subtype and CSD skin (1,16–20).
The BRAF gene is located on chromosome 7 and encodes a cytoplasmic
serine-threonine kinase. BRAF plays a role in MAP-kinase (MAPK)
pathway activation, contributing to cellular growth,
differentiation, survival and proliferation (21). The mutations of the BRAF gene are
generally located in codon 600 of the BRAF gene. The most common
mutation observed in up to 90% of cases, resulted of transversion
of T to A at nucleotide 1,799 position (T1799A). Less common
mutations included substitutions of V for lysine (V600K), arginine
(V600R) and leucine, V600M (22).
Previous studies showed that the BRAF V600E mutation is usually
observed in younger patients and at the body extremities, whereas
V600K mutations are associated with older patients and usually
found at the head and neck (14–19,23).
The RAS gene family includes genes that encode the G
proteins which are responsible for: cell growth, proliferation and
differentiation. RAS gene family consists of 3 main genes-NRAS,
KRAS, and HRAS (15,19). The NRAS gene is most frequently
mutated at hotspots in exon 2 (codons 12 and 13) and exon 3 (codon
61) (15,19). Recent evidence showed that in up to
20–30% of cases, NRAS mutations co-existed with BRAF mutations.
Patients with both BRAF and NRAS mutations had poorer prognoses
than those with the BRAF mutant melanoma alone (24–26).
Generally, NRAS mutations are independent of BRAF mutations, but
dual expression has been reported (25). The association of NRAS mutations
with the degree of solar elastosis suggests that NRAS is closely
related to the mutations induced by UV irradiation. Previous
studies showed that the NRAS mutation is also associated with
decreased immune responses in peritumoral melanoma tissue and a
more advanced tumor stage (26).
However, the association of NRAS mutational status with
histopathological characteristics in early-stage melanoma is still
poorly understood.
The current study's objective was to compare the
NRAS and BRAF mutation status with the clinicopathological
characteristics of patients with Stage IA-IIC melanoma.
Patients and methods
Design of the study
118 patients who underwent melanoma stage IA-IIC
surgical treatment (excision) at Riga East University Hospital,
Latvian Centre of Oncology Riga, Latvia, in 2012–2018 were
retrospectively enrolled in the study. Only patients with primary
cutaneous nodular and superficial spreading malignant invasive
melanoma were studied. Patients with nodular and superficial
spreading melanoma were defined based on gross and
histopathological examination.
Ethics
The study protocol was approved by the Central
Medical Ethics Committee of Latvia, Riga, Latvia (approval no.
01–29.1/2016-1-1 from January 2016) and the Ethical Committee of
Institute of Cardiology and Regenerative Medicine, the University
of Latvia (approval no. 12/2019; from September 2019). The study
was conducted according to The Declaration of Helsinki and Oviedo
Convention. All patients signed an informed consent to participate
in the study.
Exclusion criteria
Patients with lentigo maligna, acral lentiginous
melanomas, non-cutaneous and metastatic melanoma as well as
patients who had stage III and IV melanoma or who had undergone
neoadjuvant treatment were excluded from the study.
Clinical characteristics
The clinical characteristics of melanoma patients
such as age, gender, lesion location and size were analyzed.
Various clinical factors-age, gender, length of follow-up after
surgery, recurrence or metastasis-were obtained from medical
records. Progression-free survival time was defined as local,
regional or systemic metastasis, or death from the date of surgical
excision of tumor and was estimated to be from the surgical
resection date to the first loco-regional or systemic metastasis or
death without any type of relapse. The patients were follow-up
until 1 March 2022. During follow-up, the disease progression was
estimated with at least one of these features being observed-local
recurrence, regional lymph node metastasis and distant
metastasis.
Histopathological characteristics. The
histopathological characteristics of melanoma were reviewed by 2
expert pathologists (T.Z. and S.I.) according to the current WHO
(World Health Organization) and CAP (College of American
Pathologists) guidelines (8). Such
characteristics as tumor type, ulceration, peritumoral lymphocytes,
Clark invasion level, Breslow invasion level, lymphovascular
invasion, neurotropism, regression and mitotic activity was
assessed. In addition, the excision lines and distance from the
tumor were recorded. The pTNM staging was determined on the basis
of histopathological assessment.
Evaluation and scoring of peritumoral
lymphocytes
Peritumoral lymphocytes were defined as the
lymphocytes surrounding the tumor mass. The peritumoral lymphocyte
infiltration (TIL) was scored from 0 to 3 by a previously described
method (14). The scoring was
defined as follows: 0=absence of TIL within the tumor tissue, 1=TIL
infiltrate less than 25% of the tissue, 2=TIL infiltrate 25 to 50%
of the tissue, and 3=TIL infiltrate more than 50% of the
tissue.
BRAF and NRAS mutations
evaluation
Genomic DNA was isolated from 10 µm sections, cut
from formalin-fixed paraffin-embedded tissues using
GeneRead™ DNA FFPE kit (Qiagen, Germany). The melanoma BRAF
and NRAS mutation status were assessed by digital droplet PCR
(ddPCR) using BRAF V600 (#12001037), NRAS Q61 (#12001006) and NRAS
G12/G13 (#12001627) Screening Assays (all Bio-Rad, USA) as per the
manufacturer's instructions. In addition, BRAF V600 positive
samples were tested for the presence of the BRAF V600E mutation
using the BRAFV600E Mutation Assay Kit (#1863100, Bio-Rad, USA).
Droplets were generated using the Biorad QX200 Droplet Generator
and analyzed with a QX200 Droplet Reader (Bio-Rad, USA). Absolute
quantifications of mutant and wild-type alleles were estimated by
modeling a Poisson distribution using QuantaSoftTM analysis
software version 1.7 (Bio-Rad, USA).
Statistical analysis
The results were reported as median (range).
Histopathologic and clinical characteristics were analyzed using
the χ2 or Mann-Whitney U test. Association of the
mutation status with clinical and histopathological characteristics
for categorical variables was analyzed by using Pearson
χ2 and by Mann-Whitney U test for continuous variables
to calculate statistical significance. Progression-free survival
(PFS) was estimated with the Kaplan-Meier method with the log-rank
test. Time was defined as the event of disease progression or last
follow-up visit (censored). Statistical calculations were performed
with SPSS version 21.0 (SPSS Inc., Chicago, Illinois, USA).
P-values of less than 0.05 were considered statistically
significant.
Results
General characteristics
Altogether, 118 patients were enrolled in the study.
12 patients had stage IA, 20 patients had stage IB, 18 patients had
stage IIA, 32 patients had stage IIB, and 36 patients had stage IIC
melanoma. The median age was 67 years (range 24–86). 50 patients
were males and 68 patients were females. Primary tumor localization
was head/neck, limbs, and trunk in 18.0, 40.0, and 42.0% of
patients, respectively (Table
I).
 | Table I.Clinicopathological characteristics
of enrolled study subjects. |
Table I.
Clinicopathological characteristics
of enrolled study subjects.
| Variable | Value |
|---|
| Median age, years
(range) | 67 (24–86) |
| Sex,
male/female | 50/68 |
| Median Breslow
thickness, mm (range) | 2.4 (0.1-20) |
| Median Clark level,
n (range) | 3 (1–5) |
| Ulceration,
present/absent | 48/70 |
| LVI,
present/absent | 76/42 |
| Neurotropism,
present/absent | 6/112 |
| Solar elastosis, n
(range) | 1 (0–3) |
| Median tumor size,
cm (range) | 1.5 (0.2-20.0) |
| Median mitotic
count, /10 HPF (range) | 2 (1–18) |
| Median TIL, score
(range) | 2 (0–3) |
| BRAF mutational
status, V600 mutant/wild type | 67/51 |
| NRAS mutation
status, mutant/wild type | 35/83 |
| BRAF/NRAS co-mutant
melanoma/BRAF mutant | 26/67 |
| Stage IA, n | 12 |
| Stage IB, n | 20 |
| Stage IIA, n | 18 |
| Stage IIB, n | 32 |
| Stage IIC, n | 36 |
BRAF mutational status and its
correlation with clinicopathological characteristics
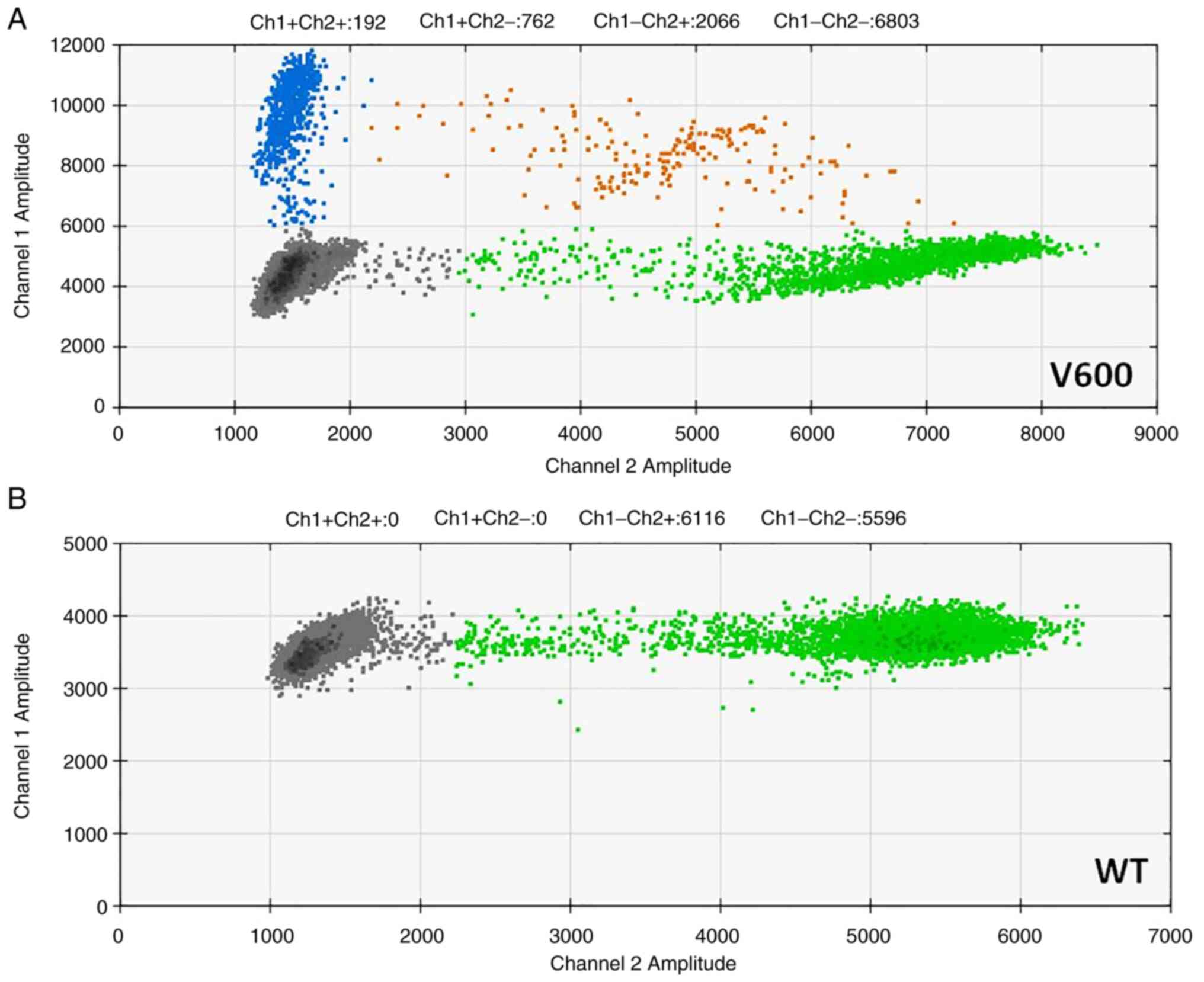
All tissues were analyzed for BRAF mutational
status, with the BRAF V600 mutation being found in 67 out of 118
patients (56.8%) (Fig. 1). From
those, 63 patients had BRAF V600E mutation and 4 patients had
another undefined V600 mutation. The associations of BRAF V600
mutational status and Breslow thickness (P=0.030), patient gender
(P=0.035; χ2=0.030), peritumoral lymphocytes
infiltration and TIL (P=0.0008) was observed (Table II). However, any association
between the disease stage, patient age, solar elastosis, mitotic
activity, Clark level of invasion and BRAF mutational status was
not demonstrated.
 | Table II.Association analysis of BRAF mutation
with clinicopathological characteristics. |
Table II.
Association analysis of BRAF mutation
with clinicopathological characteristics.
| Variable | BRAF (mutant and
wild) | BRAF (wild) | BRAF (mutant) | P-value |
|---|
| Median age, years
(range) | 67 (24–86) | 68 (44–86) | 62 (24–78) | 0.120a |
| Sex,
male/female | 50/68 | 24/27 | 26/41 | 0.035b |
| Median Breslow
thickness, mm (range) | 2.4
(0.10-20.0) | 1.90
(0.1-20.0) | 3.0 (0.2-18.0) | 0.030a |
| Median Clark level
(range) | 3.0 (1.0-5.0) | 3.0 (2.0-5.0) | 3.0 (1.0-5.0) | 0.220a |
| Ulceration,
present/absent | 48/70 | 26/25 | 22/45 | 0.120b |
| LVI,
present/absent | 76/42 | 24/27 | 52/15 | 0.280b |
| Median solar
elastosis (range) | 1.0 (0.0-3.0) | 1.0 (0.0-2.0) | 2.0 (0.0-3.0) | 0.090a |
| Median tumor size,
cm (range) | 1.5 (0.2-20.0) | 1.8 (0.7-5.0) | 1.5 (0.3-20.0) | 0.065a |
| Median mitotic
count (range) | 2.0 (1.0-18.0) | 3.0 (1.0-7.0) | 2.0 (1.0-18.0) | 0.580a |
| Median TIL, score
(range) | 2 (0.0-3.0) | 1.0 (0.0-3.0) | 2 (0.0-3.0) | 0.0008a |
| Tumor type,
nodular/superficial spreading | 68/50 | 30/21 | 38/29 | 0.460b |
The obtained results showed that the BRAF V600E
mutation is closely related to melanoma growth, since the Breslow
thickness is a major characteristic of melanoma, also incorporated
in melanoma TNM classification. In addition, a BRAF mutational
status association with peritumoral lymphocyte infiltration could
link the immune system response and tumor progression.
BRAF mutation status and PFS
All 118 patients were clinically followed up, and
there were 29 incidences of locoregional recurrence or systemic
metastasis. The PFS did not differ between wild type and BRAF
mutant melanoma (HR=1.10; 95% CI=0.40-2.50, P=0.20).
NRAS mutational status and its
correlation with clinicopathological characteristics
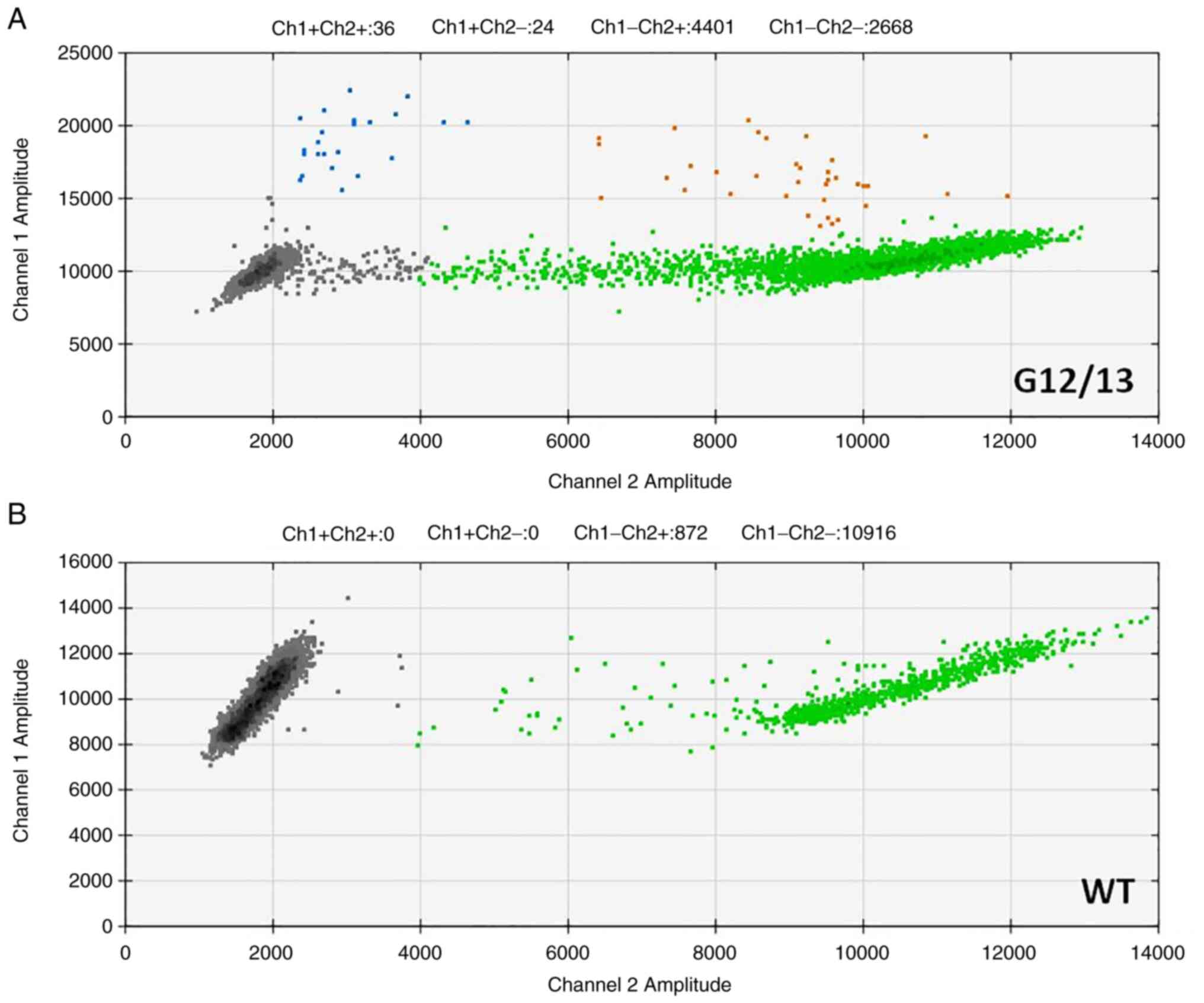
All tissues were analyzed for NRAS mutational status
(Fig. 2). NRAS mutation was found
in 35 out of 118 patients (29.6%). 26 melanoma samples (75%) were
both NRAS and BRAF co-mutant. 26 patients had NRAS Q61 mutation and
9 patients had NRAS G12, G13 mutations.
The NRAS mutational status was associated with
Breslow thickness (P=0.035), tumor type (P=0.02;
χ2=0.20), mitotic activity (P=0.025) and lymphovascular
invasion (P=0.020; χ2=0.200). However, any association
between the disease stage, Clark level of invasion, solar
elastosis, TIL, patient age, patient gender and NRAS mutational
status was not demonstrated (Table
III).
 | Table III.Association analysis of NRAS mutation
with clinicopathological characteristics. |
Table III.
Association analysis of NRAS mutation
with clinicopathological characteristics.
| Variables | NRAS (wild and
mutant) | NRAS (wild) | NRAS (mutant) | P-value |
|---|
| Median age, years
(range) | 67 (24–86) | 66 (24–83) | 68 (30–86) | 0.760a |
| Sex,
male/female | 50/68 | 35/48 | 15/20 | 0.960b |
| Median Breslow
thickness, mm (range) | 2.4
(0.10-20.0) | 1.5 (0.1-20.0) | 3.5 (0.2-20.0) | 0.035a |
| Median Clark level
(range) | 3.0 (1.0-5.0) | 3.0 (2.0-5.0) | 3.0 (1.0-5.0) | 0.220a |
| Ulceration,
present/absent | 48/70 | 27/56 | 21/14 | 0.400b |
| LVI,
present/absent | 76/42 | 45/38 | 31/4 | 0.020b |
| Median solar
elastosis (range) | 1.0 (0.0-3.0) | 1.0 (0.0-3.0) | 1.0 (0.0-3.0) | 0.720a |
| Median tumor size,
cm (range) | 1.5 (0.2-20.0) | 1.5 (0.2-7.0) | 1.6 (0.3-20.0) | 0.076a |
| Median mitotic
count (range) | 2.0 (1.0-18.0) | 2.0 (1.0-8.0) | 4.0 (1.0-18.0) | 0.025a |
| Median TIL, score
(range) | 2 (0.0-3.0) | 2.0 (0.0-3.0) | 1.0 (0.0-3.0) | 0.38a |
| Tumour type,
nodular/superficial spreading | 68/50 | 41/42 | 27/8 | 0.020b |
The obtained results showed that NRAS mutational
status was closely related to tumor growth, evaluated
histopathologically by Breslow thickness, mitotic activity and
tumor type and commonly detected clinically by gross examination.
The association of NRAS mutational status and lymphovascular
invasion could indicate that NRAS mutant melanoma has higher
metastatic potential compared to that of NRAS wild type
melanoma.
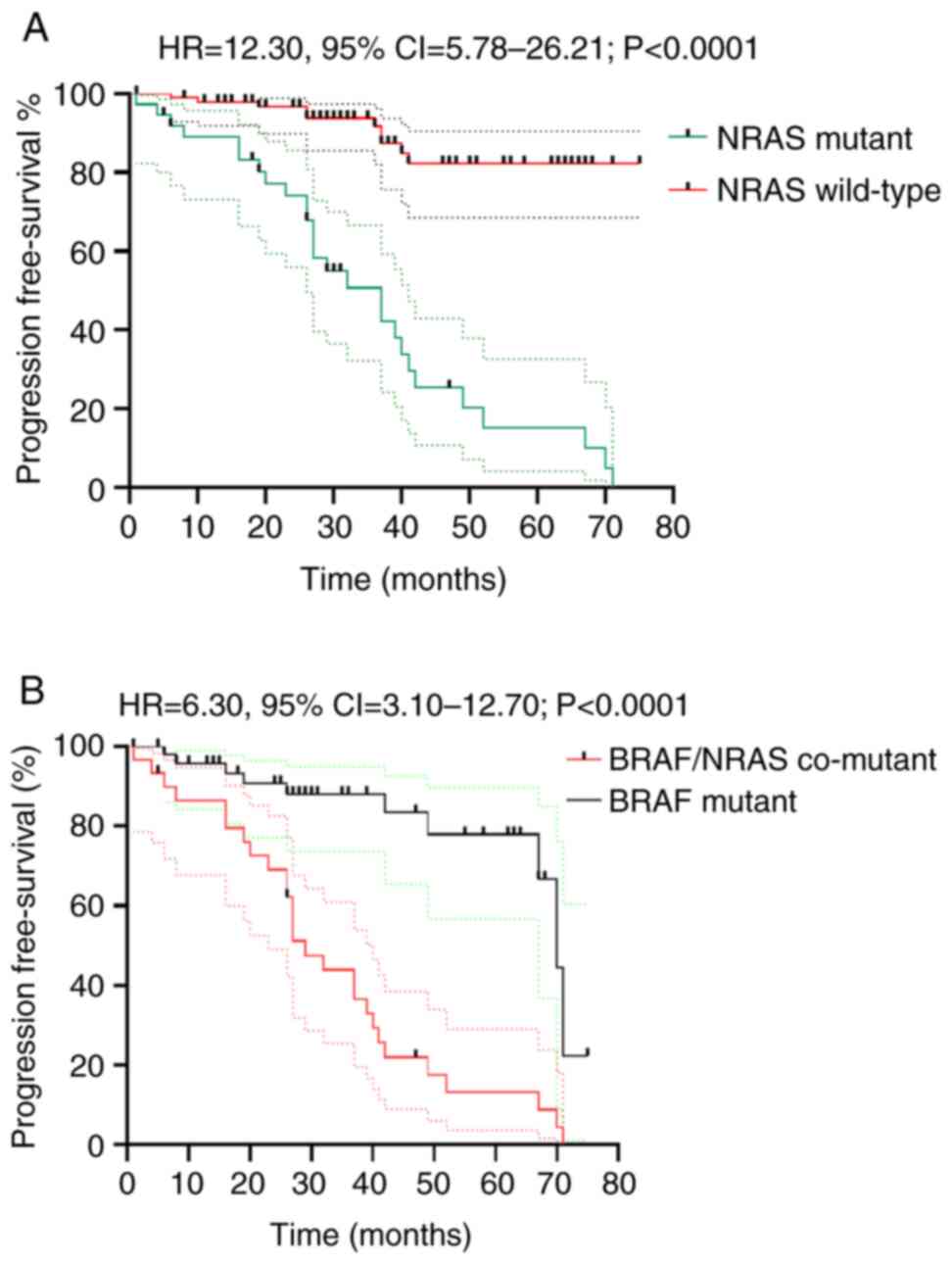
NRAS mutation status and PFS
Patients with NRAS mutant melanoma had significant
poorer PFS compared to NRAS wild melanoma (HR=12.30; 95%
CI=5.78-26.21, P<0.0001). Furthermore, the BRAF and NRAS
co-mutant melanoma had significant poorer PFS compared to the BRAF
mutant melanoma (HR=6.30; 95% CI=3.10-12.70, P<0.0001) (Fig. 3).
Discussion
In the current study, genotype-phenotype
associations were assessed in 118 patients with Stage IA-IIC
malignant invasive melanoma, according to the AJCC classification.
Histopathological examination of melanoma is currently a gold
standard for the diagnosis of melanoma. In addition, such
histopathological characteristics of invasive cutaneous melanoma as
tumor size and type, lymphovascular invasion, ulceration, Breslow
thickness, Clark invasion level, mitotic rate and disease are well
established powerful prognostic and predictive factors for melanoma
(3,6). The development of melanoma is closely
related to somatic and epigenetic changes. Activated mutations of
the oncogenes BRAF and NRAS are of particular importance in
melanoma progression (15–20). BRAF personalized treatment of
melanoma significantly improved patients' prognosis (18–20),
however, the prognostic value of BRAF and NRAS mutation and its
association with clinical and histopathological characteristics is
still controversial, especially in early-stage melanoma.
Our study showed that BRAF mutations and NRAS
mutations were identified in 56.8 and 29.6% of cases respectively.
Furthermore, while BRAF mutational status was not associated with
PFS, the NRAS mutational status did significantly correlate with
PFS. In patients with BRAS and NRAS co-mutant melanoma, the PFS was
significantly poorer compared to the BRAF mutant melanoma.
BRAF mutations in primary melanomas have been
observed at a rate of 22–72% (14–20).
In our study, the frequency of BRAF mutation falls within this
range. Over 90% of the mutations in BRAF result in substitution of
the valine at position 600, resulting in activation of the
downstream effectors of the RAS-RAF-MEK-MAPK pathway (1). The associations of BRAF V600
mutational status and Breslow thickness, patient gender, Breslow
thickness and peritumoral lymphocytes infiltration was revealed,
supporting our previous evidence (14). Previous studies showed the
different value of BRAF mutational status in association with
clinicopathological characteristics and PFS of melanoma (27–33).
Our study did not find any association between the BRAF mutational
status and PFS of melanoma.
It has been demonstrated that BRAF mutational status
correlates to younger age groups and females (34,35).
However, some studies also demonstrated associations with the male
gender (29). In our study BRAF
mutation correlated to female gender and older age. This
observation could be explained by the fact that only early-stage
melanoma patients were enrolled in our study. Some studies showed
the importance of immunological tolerance mechanisms in the
development of BRAF mutant melanoma (14,36).
This study confirmed our previous results which showed that TIL
infiltration is associated with BRAF mutational status (14,37).
It seems that the assessment of TIL is beneficial for the risk
stratification of melanoma and therefore it should be included in
the routine histopathological assessment of melanoma.
Previous studies of BRAF and NRAS com-mutant
melanoma have been discordant. NRAS gene mutation was found in
15–25% of melanoma cases (27,38).
In our study NRAS mutation was observed in up to 30% of melanoma
cases.
We assume therefore that the high prevalence of NRAS
mutations could be explained by the older median age of the
enrolled patients in our study, e.g., 67 years. It has been
demonstrated that patients with NRAS mutant melanoma compared with
BRAF mutant melanoma were usually older (>55 years) with a
previous history of UV exposure. NRAS mutant melanoma is commonly
found in upper extremities and characterized by increased Breslow
tumor thicknesses (1,27,38).
These results showed that NRAS mutations are
associated with Breslow thickness, nodular melanoma tumor type,
mitotic activity and lymphovascular invasion. It was assumed that
NRAS mutations in primary stage IA-IIC melanoma could have
potentially relevant predictive value. The NRAS gene is most
frequently mutated at hotspots in exon 2 (codons 12 and 13) and
exon 3 (codon 61) (38). The NRAS
mutation characteristic for nodular melanoma is localized in
sun-damaged skin (39).
Nevertheless, the value of NRAS mutation on disease
progression and prognosis is still controversial. Some studies
showed that the NRAS mutation was associated with a favorable
prognosis (40). In contrast,
other studies demonstrated that NRAS gene mutation was associated
with a poorer prognosis (38,41,42).
Other studies did not find any significant association between the
NRAS mutation and the prognosis of melanoma (37,43,44).
Similarly in stage IV melanoma, the data for the
NRAS mutation is also controversial. While one study suggested that
the NRAS-mutated tumor genotype in metastatic Stage IV melanoma was
associated with increased overall survival compared to the
BRAF-mutated and WT tumor genotypes (40). Other studies had the opposite
results and did not support this evidence (43,44).
It has been demonstrated that NRAS mutation status
was an independent predictor of shorter survival after a diagnosis
of stage IV melanoma (45). It
could be suggested that molecular mechanisms involving NRAS genetic
pathway could be different between metastatic Stage IV and
early-Stage IA-IIC melanoma. These results, in line with previous
studies, demonstrated that NRAS mutations are associated with
higher Breslow's thickness and poor disease prognosis (38,41,42).
In addition, our study showed that NRAS mutations
are associated with increased mitotic activity of the tumor and
lymphovascular invasion, which could be one of potential
explanations of the aggressive behavior of those tumors which
carried a NRAS mutation. Furthermore, it was demonstrated that NRAS
mutational status in primary Stage IA-IIC melanoma is a powerful
predictive factor, significantly associated with progression free
survival.
NRAS personalized treatment of melanoma is
challenging. Lonafarnib and tipifarnib have been studied for NRAS
mutant melanomas (1). In addition,
selective MEK inhibitors could have potential benefit in the
treatment of NRAS mutant melanoma (45). However, the potential significant
value of our study is in the finding of a significant predictive
value of NRAS mutations for Stage IA-IIC melanoma. Therefore,
routine assessment of NRAS mutations in Stage IA-IIC melanoma could
be potentially beneficial for the prediction of disease
progression. It should be stressed that our study subjects included
only those with local disease, e.g., at the time of diagnosis
patients did not have local recurrence, regional lymph node
metastasis or distant metastasis.
The value of the current study is in the
demonstration that Stage I and II NRAS mutant melanoma is
characterized by poorer progression free survival and is associated
with histopathological characteristics responsible for tumor growth
such as Breslow thickness, mitotic activity and lymphovascular
invasion. Further research for patients with advanced and
metastatic melanoma should evaluate the role of BRAF and NRAS
mutational status in disease progression.
Several limitations of our study should be
mentioned. A significantly higher number of case-cohort with equal
gender distribution would be beneficial. At the same time, the
strength of the present study was the demonstration of significant
role of NRAS mutational status in patients with early-stage IA-IIC
non-metastatic melanoma. All patients were enrolled from the single
oncology hospital, which treats up to 85% of all melanoma cases in
Latvia.
In our study BRAF and NRAS mutation status was
assessed by digital droplet PCR (ddPCR) using BRAFV600, NRAS Q61
and NRAS G12/G13 Screening Assay. The PCR testing is the gold
standard for BRAF and NRAS mutation testing according to ASCO and
CAP protocols. The immunohistochemistry is cost-effective method
compared to PCR and the value, specificity, and sensitivity of BRAF
and NRAS immunohistochemistry should be addressed in future studies
for general melanoma testing.
In conclusion, the patients with NRAS and NRAS/BRAF
co-mutant Stage IA-IIC melanoma had poorer progression free
survival when compared to the NRAS wild and BRAF mutant melanomas.
The NRAS assessment in melanoma in routine clinical practice is
beneficial for the risk stratification of disease progression. Our
results highlighted the value of NRAS personalized treatment in
patients with invasive melanoma.
Acknowledgements
The authors would like to thank Mrs. Aija Ozola and
Mr. Mohamed Omar (Latvian Biomedical Research and Study Centre,
Riga, Latvia) for their input in BRAF genetic testing.
Funding
The study was supported by the project ‘Strengthening of the
capacity of doctoral studies at the University of Latvia within the
framework of the new doctoral model’ (grant no.
8.2.2.0/20/I/006).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
All authors have contributed to and agreed on the
content of the manuscript. TZ and SI analyzed the histopathological
slides and data. DP and MK performed genetic analysis. TZ, SI and
DP wrote the manuscript. TZ and SI participated in the patient
enrollment, and data collection and analysis. SI and DP supervised
the project. All authors read and approved the final manuscript.
TZ, SI and DP confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The study protocol aimed to enroll at least 150
patients with cutaneous invasive melanoma from 2012 until 2021. The
study protocol was approved by the Central Medical Ethics Committee
of Latvia (approval no. 01-29.1/2016-1-1; January 2016) and the
Ethical Committee of the Institute of Cardiology and Regenerative
Medicine, University of Latvia (approval no. 12/2019; September
2019). The study was conducted according to The Declaration of
Helsinki and Oviedo Convention. All subjects signed informed
consent to participate in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yang K, Oak ASW, Slominski RM, Brożyna AA
and Slominski AT: Current molecular markers of melanoma and
treatment targets. Int J Mol Sci. 16:35352020. View Article : Google Scholar
|
|
2
|
Forsea AM: Melanoma epidemiology and early
detection in Europe: Diversity and disparities. Dermatol Pract
Concept. 10:e20200332020. View Article : Google Scholar
|
|
3
|
Elder DE, Massi D, Scolyer RA and Willemze
R: WHO Classification of Skin Tumours. 11. 4th edition. IARC
Publications; Geneva, CH: 2018
|
|
4
|
Shellenberger R, Nabhan M and Kakaraparthi
S: Melanoma screening: A plan for improving early detection. Ann
Med. 48:142–148. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mandalà M and Massi D: Tissue prognostic
biomarkers in primary cutaneous melanoma. Virchows Arch.
464:265–281. 2014. View Article : Google Scholar
|
|
6
|
Elder DE, Bastian BC, Cree IA, Massi D and
Scolyer RA: The 2018 World Health Organization classification of
cutaneous, mucosal, and uveal melanoma: Detailed analysis of 9
distinct subtypes defined by their evolutionary pathway. Arch
Pathol Lab Med. 144:500–522. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fong L and Small EJ: Anti-cytotoxic
T-lymphocyte antigen-4 antibody: The first in an emerging class of
immunomodulatory antibodies for cancer treatment. J Clin Oncol.
26:5275–5283. 2008. View Article : Google Scholar
|
|
8
|
Hodi FS, O'Day SJ, McDermott DF, Weber RW,
Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel
JC, et al: Improved survival with ipilimumab in patients with
metastatic melanoma. N Engl J Med. 363:711–723. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Topalian SL, Sznol M, McDermott DF, Kluger
HM, Carvajal RD, Sharfman WH, Brahmer JR, Lawrence DP, Atkins MB,
Powderly JD, et al: Survival, durable tumor remission, and
long-term safety in patients with advanced melanoma receiving
nivolumab. J Clin Oncol. 32:1020–1030. 2014. View Article : Google Scholar
|
|
10
|
Dummer R, Ascierto PA, Gogas HJ, Arance A,
Mandala M, Liszkay G, Garbe C, Schadendorf D, Krajsova I, Gutzmer
R, et al: Encorafenib plus binimetinib versus vemurafenib or
encorafenib in patients with BRAF-mutant melanoma (COLUMBUS): A
multicentre, open-label, randomised phase 3 trial. Lancet Oncol.
19:603–615. 2018. View Article : Google Scholar
|
|
11
|
Bastian BC: The molecular pathology of
melanoma: An integrated taxonomy of melanocytic neoplasia. Annu Rev
Pathol. 9:239–271. 2014. View Article : Google Scholar
|
|
12
|
Park CK and Kim SK: Clinicopathological
significance of intratumoral and peritumoral lymphocytes and
lymphocyte score based on the histologic subtypes of cutaneous
melanoma. Oncotarget. 8:14759–14769. 2017. View Article : Google Scholar
|
|
13
|
Maibach F, Sadozai H, Seyed Jafari SM,
Hunger RE and Schenk M: Tumor-infiltrating lymphocytes and their
prognostic value in cutaneous melanoma. Front Immunol. 11:21052020.
View Article : Google Scholar
|
|
14
|
Zablocka T, Nikolajeva A, Kreismane M,
Pjanova D and Isajevs S: Addressing the importance of melanoma
tumor-infiltrating lymphocytes in disease progression and
clinicopathological characteristics. Mol Clin Oncol. 15:2552021.
View Article : Google Scholar
|
|
15
|
Cancer Genome Atlas Network, . Genomic
classification of cutaneous melanoma. Cell. 161:1681–1696. 2015.
View Article : Google Scholar
|
|
16
|
Melis C, Rogiers A, Bechter O and van den
Oord JJ: Molecular genetic and immunotherapeutic targets in
metastatic melanoma. Virchows Arch. 471:281–293. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pracht M, Mogha A, Lespagnol A, Fautrel A,
Mouchet N, Le Gall F, Paumier V, Lefeuvre-Plesse C, Rioux-Leclerc
N, Mosser J, et al: Prognostic and predictive values of oncogenic
BRAF, NRAS, c-KIT and MITF in cutaneous and mucous melanoma. J Eur
Acad Dermatol Venereol. 29:1530–1538. 2015. View Article : Google Scholar
|
|
18
|
Ny L, Hernberg M, Nyakas M, Koivunen J,
Oddershede L, Yoon M, Wang X, Guyot P and Geisler J: BRAF
mutational status as a prognostic marker for survival in malignant
melanoma: A systematic review and meta-analysis. Acta Oncol.
59:833–844. 2020. View Article : Google Scholar
|
|
19
|
Rose EE, Egyházi S, Omholt K,
Månsson-Brahme E, Platz A, Hansson J and Lundeberg J: NRAS and BRAF
mutations in melanoma tumours in relation to clinical
characteristics: A study based on mutation screening by
pyrosequencing. Melanoma Res. 6:471–478. 2006. View Article : Google Scholar
|
|
20
|
Eigentler T, Assi Z, Hassel JC,
Heinzerling L, Starz H, Berneburg M, Bauer J and Garbe C: Which
melanoma patient carries a BRAF-mutation? A comparison of
predictive models. Oncotarget. 7:36130–36137. 2016. View Article : Google Scholar
|
|
21
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Long GV, Menzies AM, Nagrial AM, Haydu LE,
Hamilton AL, Mann GJ, Hughes TM, Thompson JF, Scolyer RA and
Kefford RF: Prognostic and clinicopathologic associations of
oncogenic BRAF in metastatic melanoma. J Clin Oncol. 29:1239–1246.
2011. View Article : Google Scholar
|
|
23
|
Ito T, Tanaka Y, Murata M, Kaku-Ito Y,
Furue K and Furue M: BRAF heterogeneity in melanoma. Curr Treat
Options Oncol. 22:202021. View Article : Google Scholar
|
|
24
|
Colebatch AJ, Ferguson P, Newell F,
Kazakoff SH, Witkowski T, Dobrovic A, Johansson PA, Saw RPM,
Stretch JR, McArthur GA, et al: Molecular genomic profiling of
melanocytic nevi. J Invest Dermatol. 139:1762–1768. 2019.
View Article : Google Scholar
|
|
25
|
Chiappetta C, Proietti I, Soccodato V,
Puggioni C, Zaralli R, Pacini L, Porta N, Skroza N, Petrozza V,
Potenza C, et al: BRAF and NRAS mutations are heterogeneous and not
mutually exclusive in nodular melanoma. Appl Immunohistochem Mol
Morphol. 23:172–177. 2015. View Article : Google Scholar
|
|
26
|
Thomas NE, Edmiston SN, Alexander A,
Groben PA, Parrish E, Kricker A, Armstrong BK, Anton-Culver H,
Gruber SB, From L, et al: Association between NRAS and BRAF
mutational status and melanoma-specific survival among patients
with higher-risk primary melanoma. JAMA Oncol. 1:359–368. 2015.
View Article : Google Scholar
|
|
27
|
Cheng L, Lopez-Beltran A, Massari F,
MacLennan GT and Montironi R: Molecular testing for BRAF mutations
to inform melanoma treatment decisions: A move toward precision
medicine. Mod Pathol. 31:24–38. 2018. View Article : Google Scholar
|
|
28
|
Tas F and Erturk K: Clinical and
prognostic significance of BRAF V600E mutation in non-metastatic
cutaneous melanoma patients. Neoplasma. 66:631–636. 2019.
View Article : Google Scholar
|
|
29
|
Bezić J, Kuret S, Vrbičić B, Smolić J,
Borić I, Škifić I, Ledina D and Božić J: Clinicopathological
characteristics of BRAF V600E mutated melanomas in the dalmatian
region of croatia. Acta Dermatovenerol Croat. 27:225–230. 2019.
|
|
30
|
Spathis A, Katoulis AC, Damaskou V, Liakou
AI, Kottaridi C, Leventakou D, Sgouros D, Mamantopoulos A,
Rigopoulos D, Karakitsos P and Panayiotides IG: BRAF mutation
status in primary, recurrent, and metastatic malignant melanoma and
its relation to histopathological parameters. Dermatol Pract
Concept. 9:54–62. 2019.
|
|
31
|
Kim SY, Kim SN, Hahn HJ, Lee YW, Choe YB
and Ahn KJ: Metaanalysis of BRAF mutations and clinicopathologic
characteristics in primary melanoma. J Am Acad Dermatol.
72:1036–1046.e2. 2015. View Article : Google Scholar
|
|
32
|
Estrozi B, Machado J, Rodriguez R and
Bacchi CE: Clinicopathologic findings and BRAF mutation in
cutaneous melanoma in young adults. Appl Immunohistochem Mol
Morphol. 22:57–64. 2014. View Article : Google Scholar
|
|
33
|
Aksenenko MB, Kirichenko AK and Ruksha TG:
Russian study of morphological prognostic factors characterization
in BRAF-mutant cutaneous melanoma. Pathol Res Pract. 211:521–527.
2015. View Article : Google Scholar
|
|
34
|
Platz A, Egyhazi S, Ringborg U and Hansson
J: Human cutaneous melanoma; a review of NRAS and BRAF mutation
frequencies in relation to histogenetic subclass and body site. Mol
Oncol. 1:395–405. 2008. View Article : Google Scholar
|
|
35
|
Weiss SA, Han SW, Lui L, Tchack J, Shapiro
R, Berman R, Zhong J, Krogsgaard M, Osman I and Darvishian F:
Immunologic heterogeneity of tumor-infiltrating lymphocyte
composition in primary melanoma. Hum Pathol. 57:116–125. 2016.
View Article : Google Scholar
|
|
36
|
Leslie C, Bowyer SE, White A,
Grieu-Iacopetta F, Trevenen M, Iacopetta B, Amanuel B and Millward
M: FOXP3+ T regulatory lymphocytes in primary melanoma are
associated with BRAF mutation but not with response to BRAF
inhibitor. Pathology. 47:557–563. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jakob JA, Bassett RL Jr, Ng CS, Curry JL,
Joseph RW, Alvarado GC, Rohlfs ML, Richard J, Gershenwald JE, Kim
KB, et al: NRAS mutation status is an independent prognostic factor
in metastatic melanoma. Cancer. 118:4014–4023. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee JH, Choi JW and Kim YS: Frequencies of
BRAF and NRAS mutations are different in histological types and
sites of origin of cutaneous melanoma: A meta-analysis. Br J
Dermatol. 164:776–784. 2011. View Article : Google Scholar
|
|
39
|
Ugurel S, Thirumaran RK, Bloethner S, Gast
A, Sucker A, Mueller-Berghaus J, Rittgen W, Hemminki K, Becker JC,
Kumar R and Schadendorf D: B-RAF and N-RAS mutations are preserved
during short time in vitro propagation and differentially impact
prognosis. PLoS One. 2:e2362007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Devitt B, Liu W, Salemi R, Wolfe R, Kelly
J, Tzen CY, Dobrovic A and McArthur G: Clinical outcome and
pathological features associated with NRAS mutation in cutaneous
melanoma. Pigment Cell Melanoma Res. 24:666–672. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Heppt MV, Siepmann T, Engel J,
Schubert-Fritschle G, Eckel R, Mirlach L, Kirchner T, Jung A,
Gesierich A, Ruzicka T, et al: Prognostic significance of BRAF and
NRAS mutations in melanoma: A German study from routine care. BMC
Cancer. 17:5362017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ellerhorst JA, Greene VR, Ekmekcioglu S,
Warneke CL, Johnson MM, Cooke CP, Wang LE, Prieto VG, Gershenwald
JE, Wei Q and Grimm EA: Clinical correlates of NRAS and BRAF
mutations in primary human melanoma. Clin Cancer Res. 17:229–235.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Schlaak M, Bajah A, Podewski T, Kreuzberg
N, von Bartenwerffer W, Wardelmann E, Merkelbach-Bruse S, Büttner
R, Mauch C and Kurschat P: Assessment of clinical parameters
associated with mutational status in metastatic malignant melanoma:
A single-centre investigation of 141 patients. Br J Dermatol.
168:708–716. 2013. View Article : Google Scholar
|
|
44
|
Bucheit AD, Syklawer E, Jakob JA, Bassett
RL Jr, Curry JL, Gershenwald JE, Kim KB, Hwu P, Lazar AJ and Davies
MA: Clinical characteristics and outcomes with specific BRAF and
NRAS mutations in patients with metastatic melanoma. Cancer.
119:3821–3829. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Grimaldi AM, Simeone E, Festino L, Vanella
V, Strudel M and Ascierto PA: MEK inhibitors in the treatment of
metastatic melanoma and solid tumors. Am J Clin Dermatol.
18:745–754. 2017. View Article : Google Scholar
|