Introduction
Lung cancer is one of the most common causes of
cancer-associated mortality globally (1). While tobacco smoking is the most
frequent cause of lung cancer, numerous cases are also reported in
nonsmokers that may be associated with various alternative factors,
including air pollution, environmental exposure, mutations and
single-nucleotide polymorphisms (2). Lung cancer is classified
histologically as small cell lung cancer (SCLC), which accounts for
15–20% of lung cancer cases, and non-SCLS (NSCLC) that accounts for
~80% of lung cancer cases and includes adenocarcinoma, large cell
carcinoma and squamous cell carcinoma (3,4).
Patients with both histological types have a poor prognosis, with a
5-year survival rate of only 15%. A thorough understanding of the
contributing factors, differences in histopathology and molecular
characteristics of lung cancer in smokers and nonsmokers, as well
as the role played by various carcinogenic factors may facilitate
the prevention and treatment of lung cancer (2,5).
Fumarate hydratase (FH) is a key enzyme in the
tricarboxylic acid (TCA) cycle, which catalyzes the hydration of
fumarate to form malate and is generally categorized as a tumor
suppressor. The inactivation of FH causes its substrate fumarate to
accumulate, enabling fumarate to leak out into the cytosol where it
inhibits prolyl hydroxylase enzymes and stabilizes
hypoxia-inducible factor 1, a mediator of glycolysis (6). A previous study demonstrated that the
expression of FH mRNA is downregulated in A549 lung cancer cells
compared with 16HBE non-tumorigenic bronchial epithelial cells, and
the expression of FH determined by immunohistochemistry is
significantly lower in lung cancer tissues than in normal lung
tissues and is not associated with TNM status (7). In addition, a study by Chen et
al (8) showed that the
phosphorylation of FH at serine 46 by p21-activated kinase 4
promotes tumorigenesis in lung cancer by inhibiting the
phosphorylation of FH at threonine 90.
Disabled homolog 2 (DAB2) is a tumor suppressor
protein that is downregulated in various malignancies, including
gastric (9), breast (10) and prostate cancer (11). In lung cancer, DAB2 is expressed at
low levels (12) and
hypermethylated (13,14). It is also a target of various
microRNAs (miRs), which results in DAB2 downregulation in lung
cancer (15,16).
AMP-activated protein kinase (AMPK) is an
energy-sensing protein kinase that regulates energy metabolism in
cells (17) and is activated by
various cellular processes, including oxidative stress, changes in
the AMP/ATP ratio and hypoxic conditions (18). AMPK has been reported to play a
role in the proliferation, metastasis and drug resistance of lung
cancer (19), and the activity of
AMPK has been shown to be altered in the absence of FH (20).
The present study aimed to investigate the
association of FH expression levels with the outcome of patients
with lung cancer. Furthermore, FH was knocked down in lung cancer
cells using short hairpin RNA (shRNA) or overexpressed using a
vector and the effect on migration ability was evaluated. In
addition, the potential involvement of AMPK phosphorylation and
DAB2 in the underlying mechanism was investigated. It is hoped that
the findings of the study may provide insights to aid the
development of novel strategies for lung cancer treatment.
Materials and methods
Oncomine database analysis
The expression level of FH in lung adenocarcinoma
tissue compared with normal lung tissue was analyzed using the
Oncomine online database (https://www.oncomine.org).
Patient samples
Lung cancer tissues were obtained from patients
undergoing surgical treatment at Kaohsiung Medical University
Hospital (Kaohsiung, Taiwan) and E-Da Hospital (Kaohsiung, Taiwan)
between February 2007 and February 2013. All patients with lung
cancer (adenocarcinoma and squamous cell carcinoma) who underwent a
lung resection, including 68 men and 36 women, were included in the
study. The age distribution was from 29 to 84 years old. Patients
with diseases other than lung cancer were excluded. Overall
survival (OS) was defined as the interval between the date of
diagnosis and death. IRB approval was received from the
Institutional Review Board (or Ethics Committee) of Kaohsiung
Medical University Hospital [KMUHIRB-E(I)-20180026] and the
Institutional Review Board for Human Studies of E-Da Hospital
(EMRP-098-132 and EMRP-101-040). Written informed consent was
obtained from all the patients.
Tissue microarray and
immunohistochemistry
All tissues used to create the tissue microarray
were obtained from formalin-fixed, paraffin-embedded tissue blocks.
Histopathological slides prepared from hematoxylin and
eosin-stained sections were evaluated by a pathologist who selected
representative areas of tumor or normal tissues for scoring. The
tissue microarray was constructed using Booster Arrayer & TMA
designer software (Alphelys) according to a previously described
procedure (21).
The immunohistochemical (IHC) staining of FH was
performed using a Bond-Max automated system (Leica Microsystems
GmbH) with an anti-FH antibody (GTX110128; 1:500; GeneTex, Inc.).
The relative expression of FH in the lung cancer specimens was
quantified using a TissueFAXS microscopy system and HistoQuest
software 2.0 (TissueGnostics GmbH). The IHC score of the lung
cancer tissue was calculated by multiplying the percentage (1–100%)
of positively stained cells by the intensity of staining (0, 1+, 2+
or 3+). For further statistical analysis, low and high expression
categories were established based on a receiver operating
characteristic curve analysis. Patients with lung cancer were
classified into two groups using the these scoring categories as
follows: Low FH, IHC score <45; and high FH, IHC score ≥45.
Cell culture
The CL1-0, H441, H1299 and CL1-5 human lung cancer
cell lines were purchased from the Bioresource Collection and
Research Center and were maintained in Roswell Park Memorial
Institute (RPMI)-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.). A549, H250 and H460 cells were purchased from the American
Type Cell Collection and maintained in RPMI-1640 medium. All cell
lines were cultured with 5% CO2 at 37°C in a humdified
incubator. All culture media were supplemented with 10% fetal
bovine serum (FBS; Biological Industries) and 1% penicillin G,
streptomycin and amphotericin B.
Virus infection for FH knockdown or
overexpression
For the knockdown of FH in the CL1-0 and H441 lung
cancer cell lines, a pLKO.1_puro lentiviral vector (National RNAi
Core Facility Platform, Academia Sinica) expressing double-stranded
shRNA oligonucleotides targeting human FH (2 clones) was used:
Clone 1 (shFH1), IDTRCN0000052466, target sequence,
5′-GTGGTTATGTTCAACAAGTAA-3′ and oligo sequence:
5′-CCGGGTGGTTATGTTCAACAAGTAACTCGAGTTACTTGTTGAACATAACCACTTTTTG-3′;
and clone 2 (shFH2), ID TRCN0000310398, target sequence,
5′-CCCAACGATCATGTTAATAAA-3′ and oligo sequence,
5′-CGGCCCAACGATCATGTTAATAAACTCGAGTTTATTAACATGATCGTTGGGTTTTTG-3′
(National RNAi Core Facility, Academia Sinica). A pLKO.1_puro
lentiviral vector expressing shRNA targeting firefly luciferase
(shluc), which is not associated with the human genome sequence,
was used as a negative control (National RNAi Core Facility,
Academia Sinica). For the overexpression of FH in H1299 cells, a
lentivirus with a pLVX-puro backbone (#GRVL7005G) and empty control
(#GRV7006G) were purchased from Topgen Biotechnology Co., Ltd. Cell
infection was performed according to the detailed procedures
described in a previous study (22).
Transwell migration and invasion
assays
Cell migration assays were carried out using
Transwell (Costar; Corning Inc.) membrane filter inserts (diameter,
6.5 mm; pore size, 8 µm) in 24-well tissue culture plates.
Following the knockdown or overexpression of FH, CL1-0, H441 and
H1299 lung cancer cells were trypsinized, suspended in serum-free
RPMI 1640 medium and seeded (2×104 cells) on the
Transwell filter in the upper chamber, while RPMI-1640 medium
containing 10% FBS (Biological Industries) was added to the lower
chamber. The cells were then incubated for 24 h at 37°C. After
incubation, the cells were fixed with 4% formaldehyde for 10 min
and stained with crystal violet for 2 h, at room temperature.
Non-migrating cells were removed by wiping the upper side of the
filter, and the migrated cells were imaged using Olympus SZX10
stereo light microscope (Olympus Corporation) and analyzed using
ImageJ software (ij153-win-java8; National Institutes of
Health).
BioCoat Matrigel invasion chambers (Corning Inc.)
were used for the invasion assay. Prior to use, Transwell invasion
chambers were rehydrated with serum-free medium for 2 h. The
remainder of the protocol was identical to that of the migration
assay, with the exception that the lung cancer cells were not
transfected. ImageJ software was used to count the number of
invaded cells. The invasive ability was calculated based on the
percentage of invaded cells and normalized to CL1-5.
Western blotting
Western blot analysis was performed to check the
knockdown and overexpression efficiency of lentivirus infection and
to evaluate the expression of other proteins in the cells using a
previously described procedure (23). Antibodies against FH (#GTX110128;
1:2,000; GeneTex, Inc.), p-AMPK (GTX52341; 1,1,000; GeneTex, Inc.),
AMPK (#5831; 1:1,000; Cell Signaling Technology, Inc.), DAB2
(AF8064; 1:500; R&D Systems, Inc.), jagged canonical Notch
ligand 1 (JAG1; GTX31607; 1:1,000; GeneTex, Inc.),
interferon-related developmental regulator 1 (IFRD1; GTX104578;
1:1,000; GeneTex, Inc.) and actin (#A5441; 1:5,000; MilliporeSigma)
were used. The AMPK inhibitor BML-275 was purchased from Santa Cruz
Biotechnology, Inc. (sc-200689). H441 cells were treated with
BML-275 (15 µM) for 24 h at 37°C in 1% FBS-containing medium before
collection for western blot analysis. In brief, after protein
transfer, the polyvinylidene fluoride (PVDF) membrane was incubated
overnight at 4°C with primary antibodies, followed by incubation
with secondary rabbit (HRP conjugate; GTX2131101; 1:5,000; GeneTex,
Inc.) or mouse (HRP conjugate; GTX213111; 1:5,000; GeneTex, Inc.)
antibodies for 1 h at room temperature. The protein bands on the
PVDF membrane were visualized using Western LightningR
Plus-ECL enhanced chemiluminescence substrate (PerkinElmer, Inc.)
and analyzed using Image Lab software 6.0.1 (Bio-Rad Laboratories,
Inc.).
Cell proliferation assay
A cell proliferation assay was performed using
2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide
(XTT) as described in previous studies (24,25).
In brief, H441, CL1-0 and H1299 cells were seeded in 96-well plates
(3,000 cells/well). After 24–72 h, XTT (X4251; Sigma-Aldrich; Merck
KGaA) solution containing phenazine methosulfate (P9625;
Sigma-Aldrich; Merck KGaS) was added. After 30 min, the absorbance
was measured at 475 and 660 nm. The proliferation rate was
calculated as 475 nm absorbance minus non-specific reading at 660
nm absorbance.
Metabolic profile analysis
Following the knockdown of FH, CL1-0 lung cancer
cells were seeded (1×106 cells/plate) in a 10-mm dish.
After 24 h, the medium was removed and the cells were washed with
PBS twice. The PBS was removed and 1 ml ice-cold methanol diluted
with water (80:20) was added in accordance with a previously
described method (26). The cells
were scraped from the dish and transferred to Eppendorf tubes, in
which the mixture was vortexed and put on ice for 5 min. The
samples were then centrifuged at 18,528 × g at 4°C for 10 min, and
the supernatants were collected into clean Eppendorf tubes and
subjected to speed vacuum concentration followed by lyophilization
to obtain the samples in powdered form. Metabolic data were
acquired from the samples using a Q Exactive™ Plus Orbitrap Mass
Spectrometer (Thermo Fisher Scientific, Inc.) coupled with a
Vanquish™ UPLC (Thermo Fisher Scientific, Inc.) under John Hopkins
University (Baltimore, USA) metabolomics analysis service. All
metabolomics data were normalized by the protein concentration of
each sample. Multiple reaction monitoring for fumarate and malate
was assessed at the mass transitions 115 to 100 and 133.01 to 100
m/z, respectively, with a retention time of 4 min. Other parameters
were as follows: Negative ionization mode of detection, 350°C
nitrogen gas temperature, 35 psi nebuliser pressure and an 11 l/min
sheath gas flow rate.
RT2 profiler PCR array
TRIzol® reagent (Thermo Fisher
Scientific, Inc.) was used to extract total RNA from the FH
knockdown CL1-0 cells and the respective shluc control cells in
accordance with the manufacturer's instructions. An aliquot of RNA
(2 g/sample) was processed with DNase (Merck &Co., Inc.) and
converted into cDNA using an RT2 First Strand Kit (Qiagen, Inc.).
Then, using the human-signal transduction pathway finder
RT2 profiler PCR array (PAHS-014Z; Qiagen, Inc.), 84
pathway-associated genes and 5 housekeeping genes were screened
according to the manufacturer's instructions.
Statistical analysis
All statistical analyses were performed using the
SPSS 14.0 statistical package for PC (SPSS, Inc.). Comparisons
between two groups were performed using unpaired Student's t-test
while comparisons among multiple groups were performed using
one-way ANOVA followed by Tukey's post hoc test. The relationship
between the expression of FH and invasive ability was evaluated by
linear regression. Associations of the expression of FH with age,
sex, stage, tumor size, lymph node metastasis, distant metastasis,
histologic type, tumor recurrence and smoking status were
investigated by χ2 or Fisher's exact tests. Survival
curves were generated using Kaplan-Meier estimates and the
significance of difference between curves was evaluated by log-rank
test. Furthermore, univariate and multivariate Cox regression
models were used to investigate the associations between
clinicopathological characteristics and OS. P<0.05 was
considered to indicate a statistically significant result.
Results
FH expression is associated with
prolonged survival in lung cancer patients
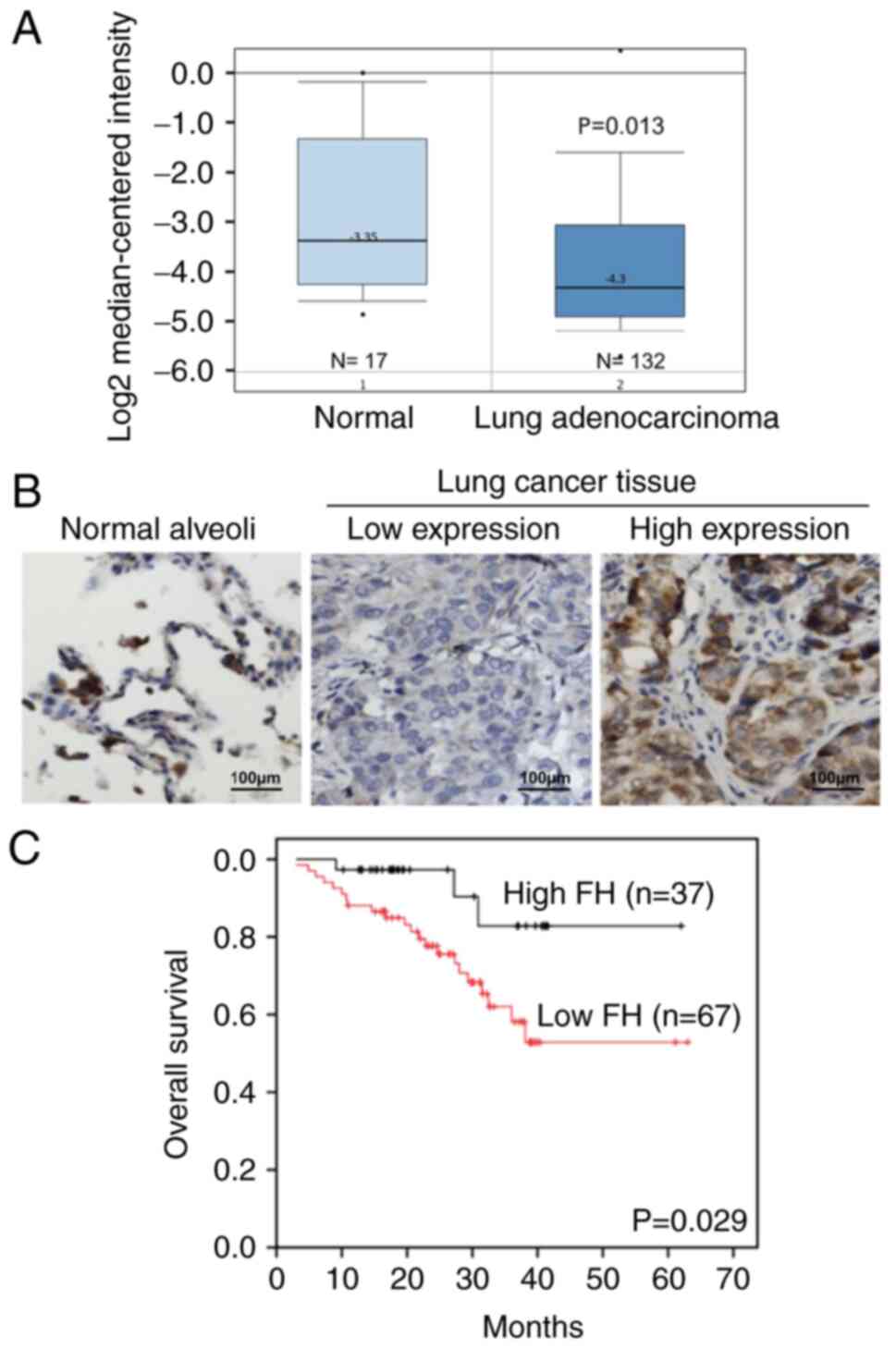
Assessment of the FH expression level in patients
with lung cancer using the Oncomine database revealed that FH mRNA
expression was significantly lower in lung adenocardinoma tissues
compared with normal lung tissues (Fig. 1A). Furthermore, the protein
expression levels of FH in lung cancer tissues collected from
patients were analyzed by IHC staining (Fig. 1B). The results showed that lung
cancer tissue had lower expression of FH compared with normal lung
alveoli. Survival analysis of the patients with lung cancer
revealed that the OS of the high FH expression group was
significantly prolonged compared with that of the low FH expression
group (P=0.029; Fig. 1C).
The associations between FH expression levels and
the clinicopathological characteristics of the patients with lung
cancer were also examined. The results revealed that low FH
expression was significantly associated with lymph node metastasis
(P=0.028) and disease recurrence (P=0.05) (Table I). Although the multivariate
analysis did not show that FH expression level was a significant
factor for OS (P=0.093), the univariate analysis indicated that FH
expression level was a significant predictor of OS for patients
with lung cancer (P=0.013; Table
II), and high FH expression was associated with lower hazard
ratio.
 | Table I.Association of FH expression with
clinicopathological characteristics in patients with lung
cancer. |
Table I.
Association of FH expression with
clinicopathological characteristics in patients with lung
cancer.
|
| FH |
|
|---|
|
|
|
|
|---|
| Variables | Low (<45),
n | High (≥45), n |
P-valuea |
|---|
| Patients | 67 | 37 |
|
| Age (years) |
|
| 0.071 |
|
≤70 | 43 | 30 |
|
|
>70 | 24 | 7 |
|
| Sex |
|
| 0.934 |
|
Female | 23 | 13 |
|
|
Male | 44 | 24 |
|
| Stage |
|
| 0.100 |
|
I/II | 42 | 29 |
|
|
III/V | 25 | 8 |
|
| T status |
|
| 0.861 |
|
T1/T2 | 57 | 31 |
|
|
T3/T4 | 10 | 6 |
|
| N status |
|
| 0.028 |
|
Negative | 36 | 28 |
|
|
Positive | 31 | 9 |
|
| M status |
|
| 0.489b |
|
Negative | 59 | 35 |
|
|
Positive | 8 | 2 |
|
| Histology |
|
| 0.023 |
|
Adenocarcinoma | 49 | 34 |
|
|
Squamous cell | 18 | 3 |
|
|
carcinoma |
|
|
|
| Recurrence |
|
| 0.05 |
| No | 30 | 24 |
|
|
Yes | 37 | 13 |
|
| Smoking status |
|
| 0.683 |
|
Never | 38 | 24 |
|
|
Former | 15 | 6 |
|
|
Current | 14 | 7 |
|
 | Table II.Univariate and multivariable analysis
of overall survival for patients with lung cancer. |
Table II.
Univariate and multivariable analysis
of overall survival for patients with lung cancer.
|
| Univariate |
Multivariatea |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) | 1.52 | (0.68, 3.39) | 0.302 | - | - | - |
|
>70 |
|
|
|
|
|
|
|
≤70 | 1.00 |
|
| - |
|
|
| Sex |
|
|
|
|
|
|
|
Male | 1.46 | (0.46, 4.47) | 0.523 | - | - | - |
|
Female | 1.00 |
|
| - |
|
|
| T status |
|
|
|
|
|
|
|
T3/T4 | 1.11 | (0.25, 4.96) | 0.893 | - | - | - |
|
T1/T2 | 1.00 |
|
| - |
|
|
| N status |
|
|
|
|
|
|
|
Positive | 5.45 | (2.22, 13.37) | 0.010 | 4.56 | (1.84, 11.3) | 0.001 |
|
Negative | 1.00 |
|
| 1.00 |
|
|
| M status |
|
|
|
|
|
|
|
Positive | 3.37 | (1.34, 8.45) | 0.010 | 2.29 | (0.88, 5.95) | 0.089 |
|
Negative | 1.00 |
|
| 1.00 |
|
|
| Histology |
|
|
|
|
|
|
|
Squamous cell carcinoma | 1.56 | (0.62, 3.92) | 0.345 | - | - | - |
|
Adenocarcinoma | 1.00 |
|
| - |
|
|
| Smoking status |
|
|
|
|
|
|
|
Current | 2.11 | (0.79, 6.05) | 0.130 | - | - | - |
|
Former | 2.83 | (1.73, 7.14) | 0.021 | - |
|
|
|
Never | 1.00 |
|
|
|
|
|
| FH |
|
|
|
|
|
|
|
High | 0.27 | (0.08, 0.90) | 0.013 | 0.35 | (0.10, 1.19) | 0.093 |
|
Low | 1.00 |
|
| 1.00 |
|
|
FH inhibits the migration ability of
lung cancer cells
The analysis of the clinicopathological data
indicated that FH might play an important role in lung cancer
metastasis (Table I). Therefore,
the effect of FH on lung cancer cell invasion and migration was
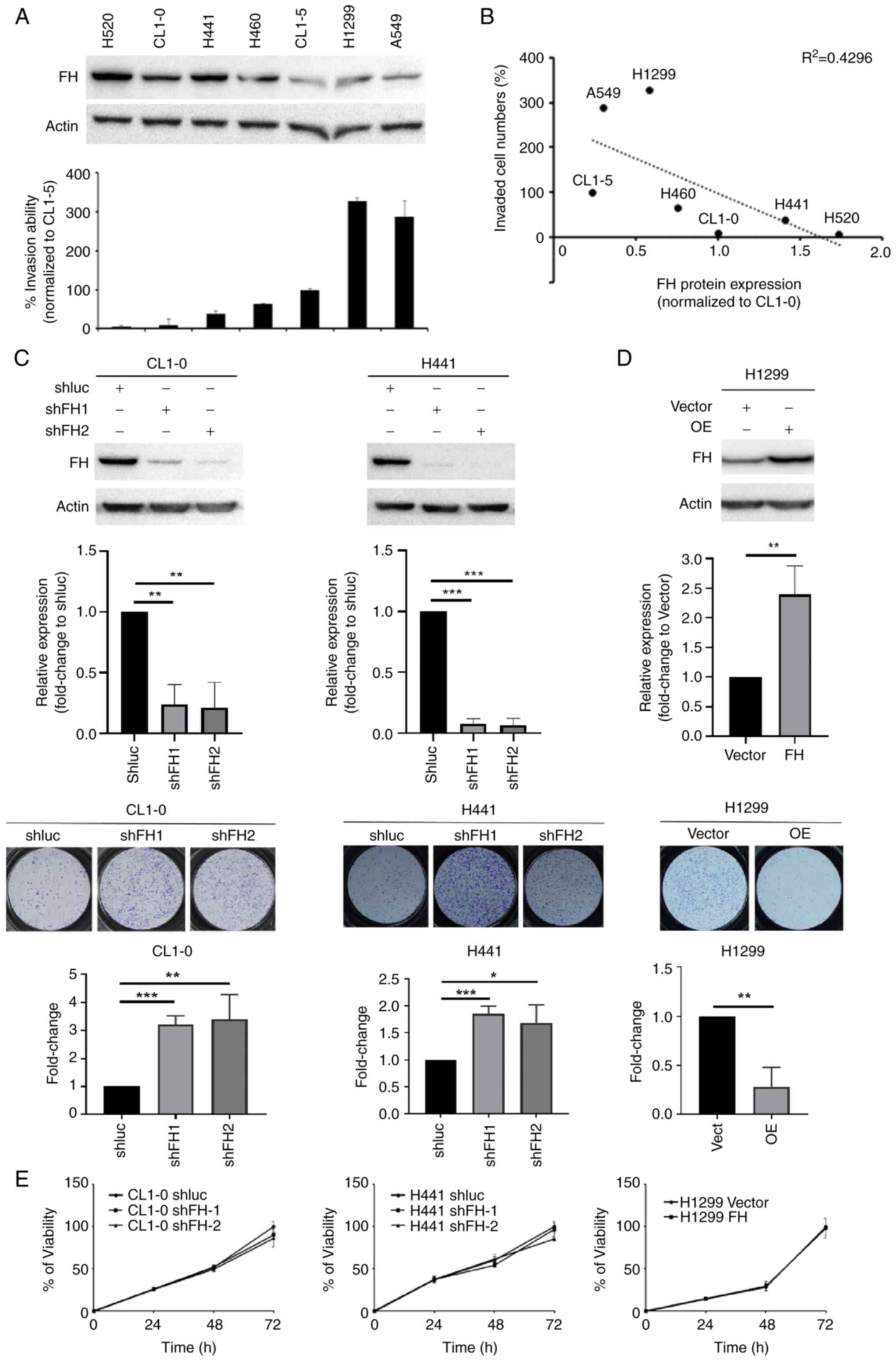
investigated in vitro. Western blotting showed that the
level of endogenous FH was downregulated in highly invasive CL1-5,
H1299 and A549 cells, and FH expression was negatively associated
with invasion ability in seven human lung cancer cell lines
(Figs. 2A, B and S1). Knockdown of FH expression in the
poorly invasive CL1-0 and H441 lung cancer cell lines significantly
increased the migration ability of the cells compared with the
respective shluc-transfected control cells (Fig. 2C). To ensure that the knockdown of
FH had an effect on the TCA cycle and reduced FH activity, the
fumarate level in FH knockdown CL1-0 cells was also measured by
metabolomics analysis and the results revealed that fumarate level
was significantly higher while the malate level was significantly
lower in the FH knockdown cells compared with the shluc control
(Fig. S2). Conversely, the
overexpression of FH in the highly invasive H1299 cell line
significantly reduced the migration ability of the cells compared
with the control cells (Fig. 2D).
This supported the patient data which showed that low FH expression
was significantly associated with lymph node metastasis in lung
cancer. The XTT assay showed that the neither the knockdown nor the
overexpression of FH in lung cancer cells affected the
proliferative potential of the cells (Fig. 2E).
FH knockdown downregulates DAB2
expression and upregulates AMPK phosphorylation
The human-signal transduction pathway finder
RT2 profiler PCR array was used to investigate the
downstream and upstream genes in FH knockdown CL1-0 cells. The
results showed that three mRNAs, namely IFRD1 (fold change 2.03),
JAG1 (fold change 2.35) and DAB2 (fold change-2.37) showed
>2-fold changes in the FH knockdown group compared with the
control group (Fig. 3A and B).
These findings were evaluated at the protein level by western
blotting and the results confirmed that DAB2 protein expression in
FH knockdown cells CL1-0 and H441 was downregulated by 0.3 and 0.4
fold, respectively, compared with the shluc control group. By
contrast, DAB2 protein level in FH overexpressed cells was
upregulated by 1-fold, compared with the vector control (Fig. 3C). However, IFRD1 and JAG1 protein
expression did not change following FH knockdown in the same manner
as the mRNA expression observed in the RT2 array
analysis. (Fig. S3). Therefore,
DAB2 was focused upon for further experiments.
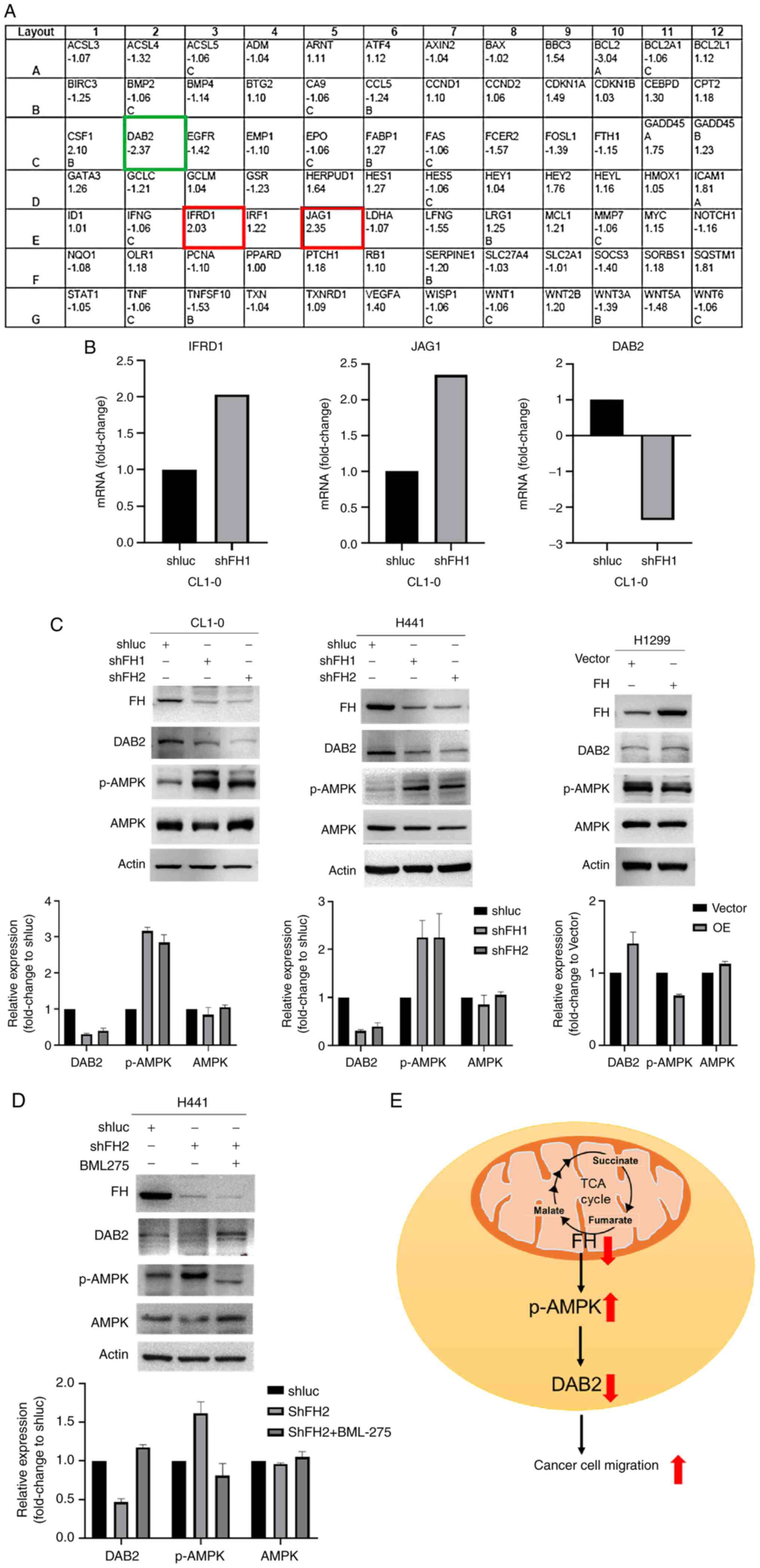 | Figure 3.FH knockdown upregulates AMPK
phosphorylation and downregulates DAB2 expression. (A) DAB2 was
downregulated in FH knockdown cells, as identified by using the
human signal transduction pathway finder RT2 profiler
PCR array. (B) Bar graphs showing the fold change in IFRD1, JAG1
and DAB2 mRNA levels compared with the shluc control group based on
the RT2 profiler PCR array results. Western blots
showing FH, DAB2, p-AMPK, and AMPK protein expression in (C) FH
knockdown CL1-0 and H441 cells and FH overexpressing H1299 cells
and (D) H441 cells treated with 15 µM BML75 (in 1% FBS containing
medium, 24 h). (E) Schematic diagram of the FH signaling pathway in
lung cancer cells. All western blots were performed twice
independently. FH, fumarate hydratase; shFH, short hairpin RNA
targeting FH; shluc, shRNA targeting firefly luciferase; DAB2,
disabled homolog 2; IFRD1, interferon-related developmental
regulator 1; JAG1, jagged canonical Notch ligand 1; p-,
phosphorylated; TCA, tricarboxylic acid cycle. |
Previous studies have shown that fumarate
accumulation affects AMPK protein expression. For example, in
kidney cancer, FH deficiency has been reported to downregulate AMPK
expression (20). Also, in renal
cancer cells, it has been reported that fumarate activates AMPK,
which protects FH-deficient cancer cells from apoptosis (27). Therefore, AMPK and p-AMPK protein
levels were evaluated by western blotting. The results demonstrated
that p-AMPK protein levels were upregulated in FH knockdown CL1-0
and H441 cells by 2- and 1.5-fold, respectively, compared with the
control shluc group (Fig. 3C)
while p-AMPK protein levels were downregulated in FH overexpressing
H1299 cells by 0.7-fold (Fig. 3C).
Furthermore, to investigate whether DAB2 was downstream or upstream
of p-AMPK, FH knockdown H441 cells were treated with the p-AMPK
inhibitor BML-275. Western blotting demonstrated that the
inhibition of p-AMPK upregulated DAB2 protein expression by
1.5-fold in FH knockdown cells (Fig.
3D). These results suggest that the downregulation of FH leads
to an increase in AMPK phosphorylation, which decreases DAB2
protein expression, resulting in increased cancer cell motility
(Fig. 3E).
Discussion
FH has been shown to play a role in uterine
leiomyoma, soft tissue sarcomas and type II papillary renal cell
carcinoma (28–31), and the accumulation of fumarate has
been shown to promote various signaling pathways and metabolic
changes in cancer, particularly renal cancer (32–36).
However, the role of FH in lung cancer has not been studied
extensively. Accordingly, the present study was performed to
investigate the potential function of FH in lung cancer.
Initially, the analysis of an online dataset in the
Oncomine database indicated that the expression of FH was lower in
lung cancer tissue compared with normal tissue. Therefore, the
expression of FH was evaluated in lung cancer tissue samples
collected from patients and it was observed that lung cancer tissue
had low expression levels of FH when compared with normal alveoli.
A survival analysis revealed that patients with lung cancer who had
high FH expression had an improved OS when compared with those with
low FH expression. The findings of the present study are similar
those in a previous study on lung cancer by Ming et al
(7), which suggested that the low
expression of FH could be an indicator of tumorigenesis, while the
present study found that the expression of FH was negatively
associated with lymph node metastasis and cancer recurrence. As
lung cancer is more common in Taiwanese males than females, the
proportion of males was higher than that of females in the present
study. Similar sex ratios have been reported in previous studies
(37,38).
The results of the present study showed that the
migration ability of lung cancer cells was increased while cell
proliferation was not affected when FH was knocked down, suggesting
a suppressive role of FH in metastasis. These findings are
consistent with those of studies by Sciacovelli et al
(30,39), which reported that accumulation of
fumarate without conversion to malate by FH promoted
epithelial-to-mesenchymal transition in renal cancer. Also, the low
expression level of FH in lung cancer cells implies that FH might
be a tumor suppressor (7).
Intriguingly, the phosphorylation of FH at threonine-90 has been
shown to induce the growth arrest of lung cancer cells (40). Whether this phosphorylation site of
FH contributes to the FH-mediated metastasis of lung cancer cells
remains to be investigated.
Using the human-signal transduction pathway finder
RT2 array, it was found that the DAB2 mRNA level was
decreased and JAG1 and IFRD1 mRNA levels were increased in FH
knockdown cells; however, the expression levels of JAG1 and IFRD1
proteins as determined by western blotting did not follow the same
trends as those obtained from the RT2 array. One
possible explanation for the discrepancy between JAG1 and IFRD1
mRNA and protein expression trends is post-transcriptional
modification. For example, small non-coding RNAs can perform
post-transcriptional modifications by binding to the mature RNA of
a target gene via the RNA-induced silencing complex, which causes
the destruction of the mRNA and/or the suppression of translation
(41). However, further
investigations are required to fully explain these discrepant
results.
Previous research has shown that the downregulation
of DAB2 promotes migration in various cancers. Specifically, in
breast cancer, the knockdown of DAB2 promotes cancer cell migration
via increased Ras/MAPK signaling and the development of an
autocrine transforming growth factor signaling loop, which further
promotes epithelial-mesenchymal transition (42). In addition, the downregulation of
DAB2 in gastric cancer activates Wnt/β-catenin and Hippo/YAP
signaling pathways, which further downregulate the expression of
the epithelial marker E-cadherin and upregulate the expression of
the mesenchymal markers MMP2 and MMP9 to promote cancer cell
migration (9). Furthermore, DAB2
is suppressed by miR-106b and miR-134-5p directly binding to the 3′
untranslated region of DAB2 in hepatocellular carcinoma and lung
cancer cells, respectively, which results in the upregulation of
cell migration (16,43). These studies support the finding
that the downregulation of DAB2 in FH knockdown cells is associated
with lung cancer cell migration.
AMPK has been implicated in the progression of lung
cancer in numerous studies and has been shown to promote lung
cancer metastasis by activating various upstream mediators. In a
previous study, the inositol monophosphatase domain-containing
1-mediated activation of AMPK and its downstream effectors HEY1 and
NOTCH1 was shown to promote lung cancer metastasis (44). Other studies demonstrated that the
activation of AMPK promotes the nuclear translocation of β-catenin,
resulting in lung cancer metastasis (45,46).
The present study demonstrated that the downregulation of FH
promotes AMPK phosphorylation and contributes to lung cancer
migration. We hypothesize that in lung cancer cells, the
phosphorylation of AMPK might be increased due to mitochondrial
dysfunction caused by FH knockdown (47). miR-451 has been reported to
regulate AMPK expression in glioma cells by targeting AMPK partner
LKB1 (48). It is possible that FH
knockdown downregulates the level of miR-451 and leads to the
upregulation of AMPK activity, but further investigations are
required to verify this possibility. AMPK has been reported to
function as an epigenetic regulator and so AMPK might promote DAB2
downregulation by its hypermethylation (49), as suggested by a previous study
which showed that DAB2 is hypermethylated in non-small cell lung
cancer (14). However, the current
study focused on FH protein expression rather than the mutation
status of FH in lung cancer.
Further studies are required to determine whether
the inhibition of AMPK phosphorylation reverses the migration
ability of lung cancer cells. Furthermore, the effect of FH
knockdown on the extracellular acidification rate and oxygen
consumption rate of lung cancer cells are important aspects that
require further investigation, and the in vitro results
require confirmation by in vivo modeling. However, in
conclusion, the current study demonstrated that the low expression
of FH promotes the migration of lung cancer cells via a mechanism
involving AMPK signaling and DAB2 downregulation.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor Yi-Chen
Lee (Graduate Institute of Medicine, College of Medicine, Kaohsiung
Medical University, Kaohsiung, Taiwan) for assistance with the
statistical analysis of the clinical data. Professor Yu-Jen Cheng,
a thoracic surgeon (Department of Surgery, E-Da Hospital,
Kaohsiung, Taiwan) recruited some patients in this study; he was an
original member of the research team but was deceased before the
submission of this manuscript.
Funding
This study was funded by grants from the Ministry of Science and
Technology (grant nos. MOST110-2314-B-037-129,
MOST110-2314-B-037-084 and MOST110-2314-B-037-058) and the Center
for Intelligent Drug Systems and Smart Bio-devices (IDS2B) from the
Featured Areas Research Center Program within the framework of
Higher Education Sprout Project by the Ministry of Education,
Taiwan. This study was also supported by grants from Kaohsiung
Medical University Hospital [grant nos. KMUH110-0R43 and
KMUH-DK(A)110001] and Kaohsiung Medical University [grant nos.
KMU-DK108005, NYTU-KMU-109-IF-01, NYCU-KMU-111-I002 and
KMU-DK(A)111005].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YFY, YYW and SSFY designed the study. AV, YFY and
PYC performed the experiments and the formal analysis. YMW, SCT,
KHC, SCSH, TLC, YYW, and SSFY investigated and validated the
results. AV, YFY and PYC prepared and wrote the original draft of
the manuscript. YMW, SCT, KHC, SCSH, TLC, YYW and SSFY reviewed and
edited the manuscript. All authors read and approved the final
version of the manuscript. AV and SSFY confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
The study was conducted in accordance with the
Declaration of Helsinki and approved by the Institutional Review
Board of Kaohsiung Medical University Hospital
[KMUHIRB-E(I)-20220014] and the Institutional Review Board for
Human Studies of the E-Da Hospital (EMRP-098-132 and EMRP-101-040).
Written informed consent was obtained from all the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Akhtar N and Bansal JG: Risk factors of
lung cancer in nonsmoker. Curr Probl Cancer. 41:328–339. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ihde DC: Chemotherapy of lung cancer. N
Engl J Med. 327:1434–1441. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hoffman PC, Mauer AM and Vokes EE: Lung
cancer. Lancet. 355:479–485. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gou LY, Niu FY, Wu YL and Zhong WZ:
Differences in driver genes between smoking-related and
non-smoking-related lung cancer in the Chinese population. Cancer.
121:3069–3079. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
King A, Selak MA and Gottlieb E: Succinate
dehydrogenase and fumarate hydratase: Linking mitochondrial
dysfunction and cancer. Oncogene. 25:4675–4682. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ming Z, Jiang M, Li W, Fan N, Deng W,
Zhong Y, Zhang Y, Zhang Q and Yang S: Bioinformatics analysis and
expression study of fumarate hydratase in lung cancer. Thoracic
Cancer. 5:543–549. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen T, Wang T, Liang W, Zhao Q, Yu Q, Ma
CM, Zhuo L, Guo D, Zheng K, Zhou C, et al: PAK4 phosphorylates
fumarase and blocks TGFβ-induced cell growth arrest in lung cancer
cells. Cancer Res. 79:1383–1397. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang H, Dong S, Liu Y, Ma F, Fang J, Zhang
W, Shao S, Shen H and Jin J: DAB2 suppresses gastric cancer
migration by regulating the Wnt/β-catenin and Hippo-YAP signaling
pathways. Transl Cancer Res. 9:1174–1184. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tian X and Zhang Z: miR-191/DAB2 axis
regulates the tumorigenicity of estrogen receptor-positive breast
cancer. IUBMB Life. 70:71–80. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hocevar BA: Loss of disabled-2 expression
in pancreatic cancer progression. Sci Rep. 9:75322019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu HT, Yang LH, Li QC, Liu SL, Liu D, Xie
XM and Wang EH: Disabled-2 and Axin are concurrently colocalized
and underexpressed in lung cancers. Hum Pathol. 42:1491–1498. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie XM, Zhang ZY, Yang LH, Yang DL, Tang
N, Zhao HY, Xu HT, Li QC and Wang EH: Aberrant hypermethylation and
reduced expression of disabled-2 promote the development of lung
cancers. Int J Oncol. 43:1636–1642. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li C, Chen J, Chen T, Xu Z, Xu C, Ding C,
Wang Y, Lei Z, Zhang HT and Zhao J: Aberrant hypermethylation at
sites-86 to 226 of DAB2 gene in non-small cell lung cancer. Am J
Med Sci. 349:425–431. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Du L, Zhao Z, Ma X, Hsiao TH, Chen Y,
Young E, Suraokar M, Wistuba I, Minna JD and Pertsemlidis A:
miR-93-directed downregulation of DAB2 defines a novel oncogenic
pathway in lung cancer. Oncogene. 33:4307–4315. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang L, Huang P, Li Q, Wang D and Xu CX:
miR-134-5p promotes stage I lung adenocarcinoma metastasis and
chemoresistance by targeting DAB2. Mol Ther Nucleic Acids.
18:627–637. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hardie DG: AMPK-sensing energy while
talking to other signaling pathways. Cell Metab. 20:939–952. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kahn BB, Alquier T, Carling D and Hardie
DG: AMP-activated protein kinase: Ancient energy gauge provides
clues to modern understanding of metabolism. Cell Metab. 1:15–25.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ashrafizadeh M, Mirzaei S, Hushmandi K,
Rahmanian V, Zabolian A, Raei M, Farahani MV, Goharrizi MASB, Khan
H, Zarrabi A and Samarghandian S: Therapeutic potential of AMPK
signaling targeting in lung cancer: Advances, challenges and future
prospects. Life Sci. 278:1196492021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tong WH, Sourbier C, Kovtunovych G, Jeong
SY, Vira M, Ghosh M, Romero VV, Sougrat R, Vaulont S, Viollet B, et
al: The glycolytic shift in fumarate-hydratase-deficient kidney
cancer lowers AMPK levels, increases anabolic propensities and
lowers cellular iron levels. Cancer Cell. 20:315–327. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuan SSF, Hou MF, Hsieh YC, Huang CY, Lee
YC, Chen YJ and Lo S: Role of MRE11 in cell proliferation, tumor
invasion, and DNA repair in breast cancer. J Natl Cancer Inst.
104:1485–1502. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang YY, Chen YK, Lo S, Chi TC, Chen YH,
Hu SC, Chen YW, Jiang SS, Tsai FY, Liu W, et al: MRE11 promotes
oral cancer progression through RUNX2/CXCR4/AKT/FOXA2 signaling in
a nuclease-independent manner. Oncogene. 40:3510–3532. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang CH, Wang PJ, Hsieh YC, Lo S, Lee YC,
Chen YC, Tsai CH, Chiu WC, Chu-Sung Hu S, Lu CW, et al: Resistin
facilitates breast cancer progression via TLR4-mediated induction
of mesenchymal phenotypes and stemness properties. Oncogene.
37:589–600. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang JY, Wang YY, Lo S, Tseng LM, Chen
DR, Wu YC, Hou MF and Yuan SF: Visfatin mediates malignant
behaviors through adipose-derived stem cells intermediary in breast
cancer. Cancers (Basel). 12:292019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang YY, Chen HD, Lo S, Chen YK, Huang YC,
Hu SC, Hsieh YC, Hung AC, Hou MF and Yuan SF: Visfatin enhances
breast cancer progression through CXCL1 induction in
tumor-associated macrophages. Cancers (Basel). 12:35262020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Udupa S, Nguyen S, Hoang G, Nguyen T,
Quinones A, Pham K, Asaka R, Nguyen K, Zhang C, Elgogary A, et al:
Upregulation of the glutaminase II pathway contributes to glutamate
production upon glutaminase 1 inhibition in pancreatic cancer.
Proteomics. 19:18004512019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bardella C, Olivero M, Lorenzato A, Geuna
M, Adam J, O'Flaherty L, Rustin P, Tomlinson I, Pollard PJ and Di
Renzo MF: Cells lacking the fumarase tumor suppressor are protected
from apoptosis through a hypoxia-inducible factor-independent,
AMPK-dependent mechanism. Mol Cell Biol. 32:3081–3094. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barker KT, Bevan S, Wang R, Lu YJ,
Flanagan AM, Bridge JA, Fisher C, Finlayson CJ, Shipley J and
Houlston RS: Low frequency of somatic mutations in the FH/multiple
cutaneous leiomyomatosis gene in sporadic leiomyosarcomas and
uterine leiomyomas. Br J Cancer. 87:446–448. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kiuru M, Lehtonen R, Arola J, Salovaara R,
Järvinen H, Aittomäki K, Sjöberg J, Visakorpi T, Knuutila S, Isola
J, et al: Few FH mutations in sporadic counterparts of tumor types
observed in hereditary leiomyomatosis and renal cell cancer
families. Cancer Res. 62:4554–4557. 2002.PubMed/NCBI
|
|
30
|
Sciacovelli M, Gonçalves E, Johnson TI,
Zecchini VR, da Costa AS, Gaude E, Drubbel AV, Theobald SJ, Abbo
SR, Tran MG, et al: Fumarate is an epigenetic modifier that elicits
epithelial-to-mesenchymal transition. Nature. 537:544–547. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schmidt C, Sciacovelli M and Frezza C:
Fumarate hydratase in cancer: A multifaceted tumour suppressor.
Semin Cell Dev Biol. 98:15–25. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ge X, Li M, Yin J, Shi Z, Fu Y, Zhao N,
Chen H, Meng L, Li X, Hu Z, et al: Fumarate inhibits PTEN to
promote tumorigenesis and therapeutic resistance of type2 papillary
renal cell carcinoma. Mol Cell. 82:1249–1260.e7. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Johnson TI, Costa AS, Ferguson AN and
Frezza C: Fumarate hydratase loss promotes mitotic entry in the
presence of DNA damage after ionising radiation. Cell Death Dis.
9:9132018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Isaacs JS, Jung YJ, Mole DR, Lee S,
Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, et
al: HIF overexpression correlates with biallelic loss of fumarate
hydratase in renal cancer: Novel role of fumarate in regulation of
HIF stability. Cancer Cell. 8:143–153. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sullivan LB, Martinez-Garcia E, Nguyen H,
Mullen AR, Dufour E, Sudarshan S, Licht JD, Deberardinis RJ and
Chandel NS: The proto-oncometabolite fumarate binds glutathione to
amplify ROS-dependent signaling. Mol Cell. 51:236–248. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gonçalves E, Sciacovelli M, Costa ASH,
Tran MGB, Johnson TI, Machado D, Frezza C and Saez-Rodriguez J:
Post-translational regulation of metabolism in fumarate hydratase
deficient cancer cells. Metab Eng. 45:149–157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chang YJ, Huang JY, Lin CH and Wang BY:
Survival and treatment of lung cancer in Taiwan between 2010 and
2016. J Clin Med. 10:46752021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang BY, Huang JY, Cheng CY, Lin CH, Ko J
and Liaw YP: Lung cancer and prognosis in Taiwan: A
population-based cancer registry. J Thorac Oncol. 8:1128–1135.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sciacovelli M and Frezza C: Fumarate
drives EMT in renal cancer. Cell Death Differ. 24:1–2. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hou C, Ishi Y, Motegi H, Okamoto M, Ou Y,
Chen J and Yamaguchi S: Overexpression of CD44 is associated with a
poor prognosis in grade II/III gliomas. J Neurooncol. 145:201–210.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang JG, Xu C, Zhang L, Zhu W, Shen H and
Deng HW: Identify gene expression pattern change at transcriptional
and post-transcriptional levels. Transcription. 10:137–146. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Martin J, Herbert B and Hocevar B:
Disabled-2 downregulation promotes epithelial-to-mesenchymal
transition. Br J Cancer. 103:1716–1723. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun C, Yao X, Jiang Q and Sun X: miR-106b
targets DAB2 to promote hepatocellular carcinoma cell proliferation
and metastasis. Oncol Lett. 16:3063–3069. 2018.PubMed/NCBI
|
|
44
|
Yang YF, Wang YY, Hsiao M, Lo S, Chang YC,
Jan YH, Lai TC, Lee YC, Hsieh YC and Yuan SF: IMPAD1 functions as
mitochondrial electron transport inhibitor that prevents ROS
production and promotes lung cancer metastasis through the
AMPK-Notch1-HEY1 pathway. Cancer Lett. 485:27–37. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
He K, Guo X, Liu Y, Li J, Hu Y, Wang D and
Song J: TUFM downregulation induces epithelial-mesenchymal
transition and invasion in lung cancer cells via a mechanism
involving AMPK-GSK3β signaling. Cell Mol Life Sci. 73:2105–2121.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Han SY, Jeong YJ, Choi Y, Hwang SK, Bae YS
and Chang YC: Mitochondrial dysfunction induces the invasive
phenotype, and cell migration and invasion, through the induction
of AKT and AMPK pathways in lung cancer cells. Int J Mol Med.
42:1644–1652. 2018.PubMed/NCBI
|
|
47
|
Wu SB, Wu YT, Wu TP and Wei YH: Role of
AMPK-mediated adaptive responses in human cells with mitochondrial
dysfunction to oxidative stress. Biochim Biophys Acta.
1840:1331–1344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Godlewski J, Nowicki MO, Bronisz A, Nuovo
G, Palatini J, De Lay M, Van Brocklyn J, Ostrowski MC, Chiocca EA
and Lawler SE: MicroRNA-451 regulates LKB1/AMPK signaling and
allows adaptation to metabolic stress in glioma cells. Mol Cell.
37:620–632. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gongol B, Sari I, Bryant T, Rosete G and
Marin T: AMPK: An epigenetic landscape modulator. Int J Mol Sci.
19:32382018. View Article : Google Scholar : PubMed/NCBI
|