Introduction
Breast cancer is the most common cancer type
reported in women, and certain individuals have been shown to have
a genetic predisposition to its development (1). To elucidate the genetic contribution
to this disease, numerous genome-wide association studies (GWASs)
have been performed, and based on these studies, certain
susceptibility loci have been reported. Among these genetic
markers, rs1011970 in chromosome 9p21 has been reported to be
associated with breast cancer in multiple GWASs (2–4). That
is, the carrier of the risk allele at rs1011970 presents with a
markedly higher chance (10–20%) to develop breast cancer than
carriers of another allele (2–4).
However, it is important to note that this mutation may not be the
actual causal single nucleotide polymorphism (SNP) for breast
cancer. Indeed, usually only ~500,000 SNPs are utilized to
represent the variation of the whole genome out of a total of ~80
million SNPs (5). Therefore, it is
possible that the actual causal SNP for breast cancer may be an SNP
or SNPs that exist in strong linkage disequilibrium (LD) with
rs1011970, which has been frequently reported in recent functional
genomics studies based on GWAS results (6–8).
Moreover, as this SNP is located within the intron of
cyclin-dependent kinase inhibitor 2B (CDKN2B) antisense RNA 1
(CDKN2B-AS1) and the protein-coding gene closest to this SNP
is CDKN2B, it may be hypothesized that this SNP could be
associated with breast cancer through the function of these two
genes (2–4). However, to the best of our knowledge,
the study of this issue has not previously been reported. All these
factors hinder our understanding of the association between this
locus and breast cancer susceptibility.
Therefore, the present study used functional
genomics approaches to identify the identity of the actual causal
SNP at this locus and to elucidate the underlying mechanism
associated with this SNP in breast cancer. Through population
genetics analysis, rs77283072 was demonstrated to be in nearly
complete LD with rs1011970. Therefore, its function and role in
breast cancer were also evaluated.
Materials and methods
1000 Genomes Project (1KG) data
analysis
The nucleotide sequence for the 200 kb surrounding
the region of rs1011970 was downloaded from the 1KG project
(http://www.internationalgenome.org/)
for three representative populations, namely, the Utah Residents
with European Ancestry (CEU), Han Chinese in Beijing (CHB) and
Yoruba in Ibadan (YRI) populations. The LD pattern was assessed
using the Genome Variation Server 150 (http://gvs.gs.washington.edu/GVS150/).
Dual-luciferase assay
DNA was purified from the MCF-7 cell line using a
standard phenol-chloroform approach. The region proximal to
rs1011970 (~1.5 kb) was determined using PCR with
Phusion® High-Fidelity DNA Polymerase (Thermo Fisher
Scientific, Inc.) and the primer pair
5′-TATCTATTCAGCAAAGCCCCACTC-3′ and 5′-ATTAAGGGCTGCCAAGTCACAA-3′
(restriction enzyme cutting sites not shown) with the following
thermocycling conditions: 98°C for 30 sec; followed by 40 cycles of
98°C for 10 sec, 62°C for 30 sec and 72°C for 45 sec, and finally
72°C for 5 min. The PCR product was separated in 1.5% agarose gel
and visualized using SYBR gold (Beijing Solarbio Science &
Technology Co., Ltd.). After digestion with MluI and
XhoI (New England BioLabs, Inc.), PCR products were ligated
with a luciferase reporter, pGL3-promoter vector (Promega
Corporation). After Sanger sequencing with primers
5′-AAACAAAACGAAACAAAACAAACT-3′ and 5′-GCGGAGTTAGGGGCGGGATGG-3′
(Tsingke Biological Technology), the plasmid containing another
allele (T and A for rs1011970 and rs77283072, respectively) was
constructed using a Q5 Site-directed Mutagenesis Kit (New England
BioLabs, Inc.; primer sequences are presented in Table SI). Prior to transfection, all
plasmids were re-sequenced to verify their sequences and the
haplotype orientation.
The human breast cancer cell line, MCF-7, was
purchased from Conservation Genetics CAS Kunming Cell Bank
(http://www.kmcellbank.com/) and cultured
in HyClone® Dulbecco's modified Eagle's medium, high
glucose (Cytiva) with 10% fetal bovine serum (Biological
Industries) in 5% CO2 at 37°C. The constructed plasmids
(475 ng) were transfected by incubation with Lipofectamine™ 2000
(Thermo Fisher Scientific, Inc.) for 5 min at 37°C. Following
transfection, cells were cultured for an additional 36 h at 37°C,
washed with phosphate-buffered saline (Beijing Solarbio Science
& Technology Co., Ltd.) and lysed. The level of luciferase
expression was quantified using a Dual-Luciferase Reporter Assay
System (Promega Corporation) and normalized using the read of the
pRL-TK plasmid (25 ng; Promega Corporation). Six independent
repeats were performed for each transfection.
Chromosome conformation capture
(3C)
Spatial contacts between the enhancer and promoter
of nearby genes were evaluated using 3C and quantitative PCR
(qPCR). Briefly, MCF-7 cells (~1×108) were cross-linked
using 1% formaldehyde (Beijing Solarbio Science & Technology
Co., Ltd.) and lysed using lysis buffer (Solarbio) on ice for 10
min, and their chromatin was digested using EcoRI at 37°C
overnight. After ligation, the DNA was purified using a standard
phenol-chloroform method (9). The
bacterial artificial chromosome RP11-478M20 containing the genome
segment chr9: 21947303-22110179 (Genome Reference Consortium Human
Build 37) was cultured overnight at 37°C, isolated, digested,
ligated as a control and quantified using qPCR. qPCR was performed
using iQ™ SYBR® Green (Bio-Rad Laboratories, Inc.) with
following thermocycling conditions: 96°C for 10 min; followed by 40
cycles of 96°C for 10 sec and 60°C for 30 sec. The enrichment for
chromatin was evaluated using the 2−ΔΔCq approach
(10). The primer sequences are
presented in Table SII. Three
repeat experiments were performed for each unidirectional anchor
primer.
RNA-sequencing (RNA-seq) analysis
The RNA-seq data for lymphoblastoid cell lines (LCL)
from a previous study (11) were
obtained from the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra/) and aligned
with the CDKN2A mRNA sequence using bowtie2 2.4.4 (12). CDKN2A expression was
calculated using eXpress (13) with
default parameters and reported as fragments per kilo base of
transcript per million mapped fragments. The genotype for LCL was
obtained from the 1KG database and linear regression was performed
between genotype and CDKN2A expression using SPSS 20.0 (IBM
Corp.). Moreover, the association between rs77283072 and other
genes were searched in GTEx database (https://gtexportal.org).
Electrophoretic mobility-shift assay
(EMSA)
Nuclear extracts were isolated from MCF-7 cells
using a Nuclear and Cytoplasmic Protein Extraction kit (Beyotime
Institute of Biotechnology) and centrifuged at 14,000 × g for 10
min at 4°C. The concentration of nuclear protein was determined
using an Enhanced BCA Protein Assay Kit (Beyotime Institute of
Biotechnology). The probes for both alleles of rs77283072, which
were synthesized by Sangon Biotech Co., Ltd., and the sequences are
presented in Table SIII. The
probes were labeled with biotin using an EMSA Probe Biotin Labeling
Kit (Beyotime Institute of Biotechnology). Biotin-labeled probes
(10 fmol) and nuclear extracts (5 µg) were incubated in EMSA
binding buffer at room temperature for 20 min. The probe-protein
complexes were separated by electrophoresis using a 4.9%
non-denatured polyacrylamide gel and then transferred to nylon
membranes (Beyotime Institute of Biotechnology). For each allele,
probes labeled with biotin alone, and probe-protein complex
incubated with competitor oligonucleotides (non-labeled probes)
were also included as controls. The membranes were incubated with
streptavidin-horseradish peroxidase conjugate (Beyotime Institute
of Biotechnology) at room temperature for 5 min and then assessed
using an ECL chemiluminescence kit (MilliporeSigma).
Chromatin immunoprecipitation (ChIP)
assay
The histone modification surrounding rs77283072 was
searched in the ENCODE Portal database (https://www.encodeproject.org/) using its location.
The online programs Match (http://www.gene-regulation.com/cgi-bin/pub/programs/match/bin/match.cgi)
and JASPAR (http://jaspar.genereg.net/) were used to predict
potential transcription factors (TFs) by inputting the surrounding
sequence of rs77283072. ChIP was performed using an EZ ChIP Kit
(MilliporeSigma). Briefly, MCF-7 cells (~1×107 cells)
were cross-linked using formaldehyde at 37°C for 10 min. After
washing, the cells were scraped, lysed using lysis buffer
(MilliporeSigma) on ice for 10 min, sonicated on ice for 20 cycles
(10 sec each) into small fragments (400–800 bp) and pre-cleared
with 60 µl protein A beads. The protein-chromatin complex (1 ml)
was immunoprecipitated using 2 µg mouse antibody for predicted TFs
or normal mouse IgG (Santa Cruz Biotechnology, Inc.) as a control
at 4°C overnight. The antibodies for the following TFs were
utilized: REL proto-oncogene (cat. no. sc-373713X), ETS
transcription factor ELK1 (cat. no. sc-65986), POU class 2 homeobox
1 (cat. no. sc-8024) or paired box 6 (cat. no. sc-53106; Santa Cruz
Biotechnology Inc.). After washing with low salt, high salt, LiCl
and TE buffer (MilliporeSigma), the captured protein-chromatin
complex was dissolved using elution buffer (MilliporeSigma) and the
cross-link was broken by adding 20 µl 5M NaCl and heating at 65°C
for 4 h. Protein was removed by digestion using proteinase K (Roche
Diagnostics) at 45°C for 1 h and the DNA was purified using the
column supplied with the EZ ChIP Kit. Finally, qPCR was performed
to evaluate the enrichment of the DNA obtained using iQ™
SYBR® Green (Bio-Rad Laboratories, Inc.) and the primers
as follows: 5′-TCATGTGGCAGTGGCAAGAGTAAAA-3′ and
5′-AGGGTGAGGTAACTGAATCCCGAG-3′. This primer pair was designed based
on the genome segment surrounding rs77283072. The thermocycling
conditions were as follows: 96°C for 10 min; followed by 40 cycles
of 96°C for 10 sec and 60°C for 30 sec. Since a standard curve
approach and multiple calibrations and controls were used in qPCR,
a reference gene is not necessary for this experiment (14). For each ChIP assay, a positive
control segment (one genome region that was known to bind TF)
(15–18) was included and ChIP-qPCR was
performed using the primers in Table
SIV.
ChIP-seq data were obtained from the Sequence Read
Archive database and aligned with human genome sequence using
bowtie2 2.4.4 (12). The enrichment
for TFs was evaluated by software MACS 1.4.4 (Model-based analysis
of ChIP-seq) (19).
Statistical analysis
Independent Student's t-tests were used to compare
the luciferase activities and ChIP results. Analysis of variance
(ANOVA) and Bonferroni's post hoc test were used to evaluate the 3C
enrichment. Linear regression with additive genetic model was used
to evaluate the association between genotype and gene expression.
All statistical analyses were performed using SPSS 20.0 (IBM
Corp.). P<0.05 was considered to indicate a statistically
significant difference.
Results
LD pattern surrounding rs1011970
Within the 200-kb region surrounding rs1011970,
genetic variations were identified in the CEU (n=792), CHB (n=793)
and YRI (n=1,324) populations. Among these, only one non-coding SNP
located 1.3 kb upstream, rs77283072, was in almost complete LD with
rs1011970 (r2=1.000, 0.951 and 1.000 for CEU, CHB and
YRI, respectively; Fig. S1) and
was therefore expected to present a similar signal to that of
rs1011970 in GWAS. All other SNPs exhibited relatively low LD with
rs1011970 (all r2<0.64). Therefore, it was possible
to hypothesize that these two SNPs were likely to be the causal
SNPs in breast cancer.
Function of rs1011970 and
rs77283072
To elucidate the function of these two SNPs,
plasmids with different alleles of rs1011970 and rs77283072 were
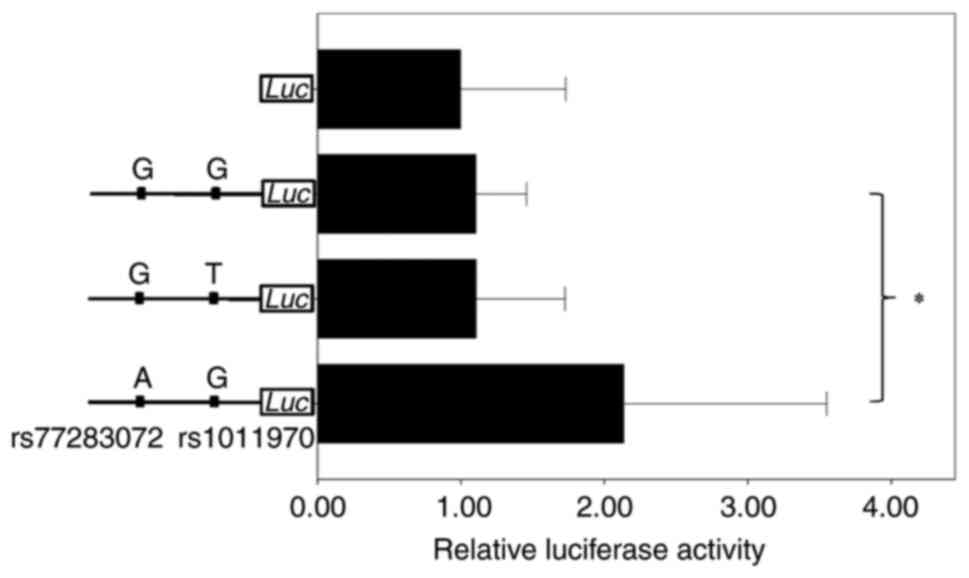
generated (Fig S2), and
transfection and luciferase assessment were performed. As presented
in Fig. 1, the rs1011970 alleles
did not demonstrate any significant difference in relative
luciferase activity levels (P=0.988). However, the luciferase
activity levels of the A allele of rs77283072 was ~93.4% higher
compared with that of the G allele (P=0.000068), which indicated
that rs77283072 was the causal SNP in breast cancer.
Regulatory target gene of the enhancer
containing rs77283072
As presented in Fig.
S3, a search using the ENCODE Portal demonstrated that there
were H3K4m1 and H3K27ac histone modifications close to rs77283072.
As these two types of histone modification were previously reported
to be the markers for active enhancers (20), it was possible to hypothesize that
this SNP might have the ability to alter enhancer activity.
Meanwhile, there were also other kinds of histone modifications
with unknown function, such as H3K36m3 and H4K20m1 (Fig. S3). Although rs77283072 is located
within the intron region of CDKN2B-AS1, the actual
regulatory target of this enhancer remains unknown. To identify the
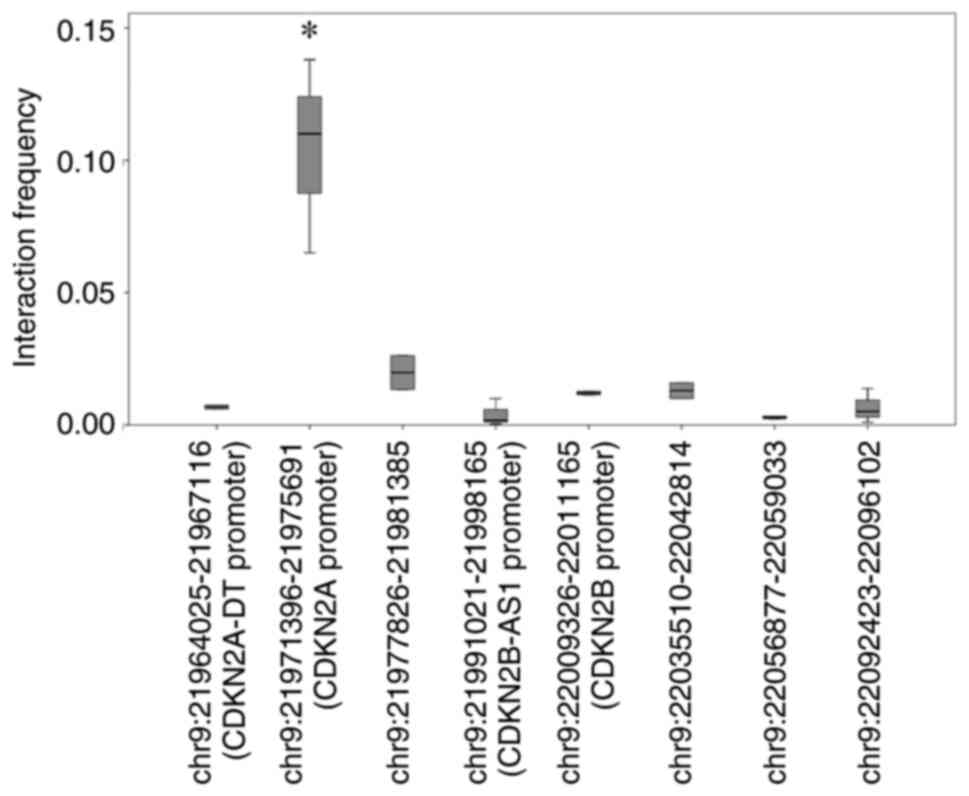
potential target, 3C-qPCR was used to assess the interaction
between this enhancer and proximal gene promoters. The anchor
promoter was intended to bind the enhancer region, whereas the
target primers were designed to bind with the promoter of CDKN2A
divergent transcript (CDKN2A-DT), CDKN2A, CDKN2B-AS1
and CDKN2B, and four random genome regions. The amounts of
3C products were used to represent the interaction frequency
between the enhancer and target gene promoter, and were compared
using ANOVA. As presented in Fig.
2, no increases in interaction were demonstrated in the
promoters of CDKN2A-DT, CDKN2B-AS1 and CDKN2B.
However, if we compared the ligation frequency between
CDKN2A promoter and other 7 segments involved in the assay,
a significant increase could be observed at the CDKN2A
promoter (~86.0 kb between the CDKN2A promoter and the
enhancer; P=0.0038), which indicated that CDKN2A was the
regulatory target of this enhancer.
Expression quantitative trait locus
(eQTL) analysis
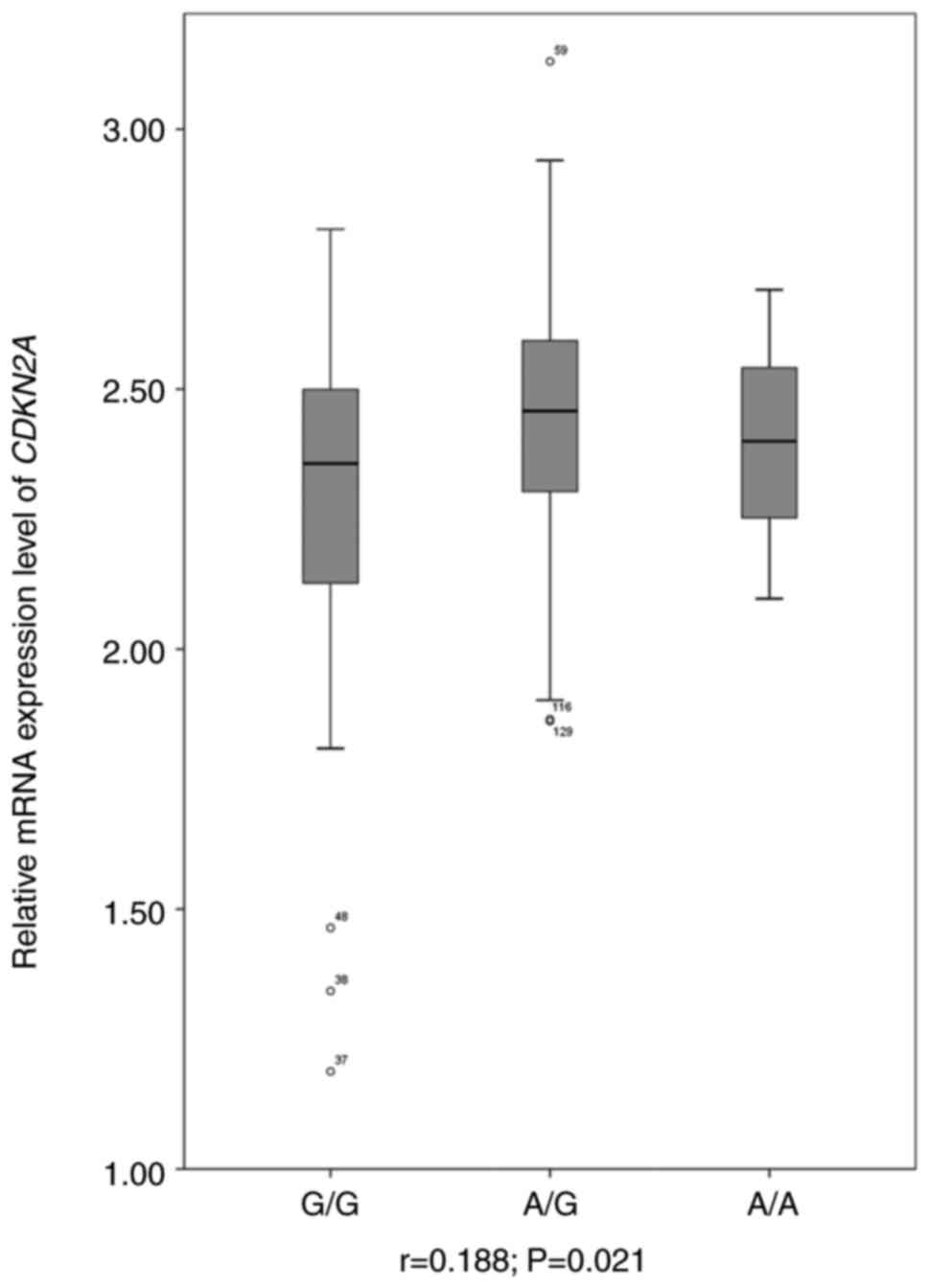
If rs77283072 was indeed capable of regulating
CDKN2A expression, this SNP would be an eQTL for this gene.
To evaluate this hypothesis, RNA-seq data for a widely used model,
LCL, was downloaded and CDKN2A mRNA expression levels were
assessed. Linear regression was performed to evaluate the
association between genotype and gene expression. As presented in
Fig. 3, CDKN2A mRNA
expression levels were demonstrated to be significantly associated
with the rs77283072 genotype (r=0.188; P=0.021). The A allele of
rs77283072 was associated with a higher mRNA expression level of
CDKN2A, a finding that was consistent with the results of
the luciferase assay.
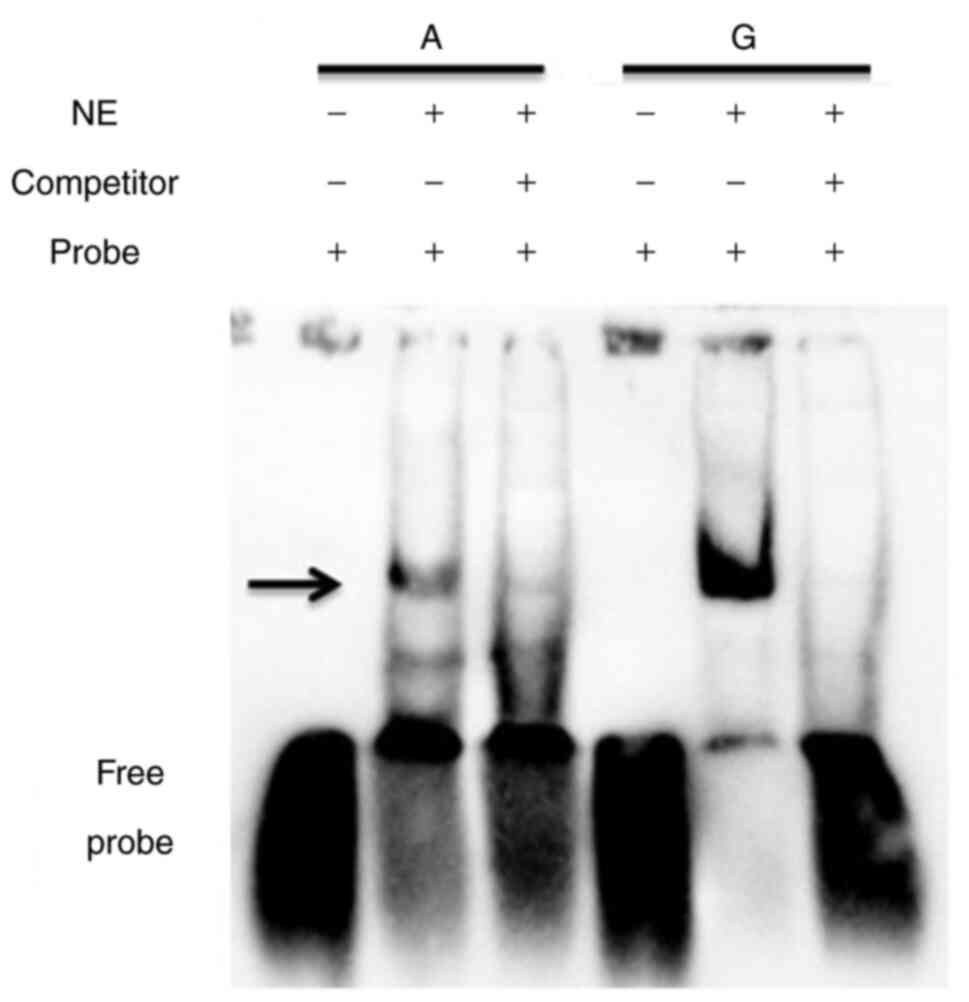
Differences in the TF binding affinity
between the rs77283072 alleles
Considering the function and location of rs77283072,
it was hypothesized that this SNP could have the ability to alter
the binding affinity of TFs. To evaluate this, EMSA was performed.
As presented in Fig. 4, a marked
difference was demonstrated in the binding affinity between the
rs77283072 alleles. The interaction of probe and nuclear proteins
was abolished by competitor oligonucleotides, which indicated that
the binding was specific. Furthermore, the more highly expressed
allele of rs77283072, the A allele, demonstrated a markedly lower
affinity with nuclear proteins, which suggested that the protein
interacting with the enhancer containing rs77283072 exerted a
negative regulatory effect on gene expression.
These results indicated that rs77283072 may have
been located within the binding sites of the REL proto-oncogene,
ETS transcription factor ELK1, POU class 2 homeobox 1 or paired box
6 TFs. To identify the actual TF, ChIP was performed. For each
antibody, a positive control segment was included and significant
enrichment was shown, as presented in Fig. S4, which demonstrated that the ChIP
assay could be utilized to identify the TFs surrounding rs77283072.
As presented in Fig. S5, none of
the antibodies demonstrated significant enrichment of the chromatin
surrounding rs77283072, which indicated that none of the proposed
TFs could, in fact, bind this region. Multiple ChIP-seq data was
also analyzed for TFs. However, no significant enrichment was
identified for the rs77283072 surrounding region (all P>0.05;
data not shown). Therefore, the identity of the TF that interacted
with this enhancer remained unclear.
Discussion
In the present study, the LD pattern was utilized to
search for potential causal SNPs for breast cancer. Through a
functional genomics approach, both the actual causal SNP and the
underlying mechanism were identified. The results of the present
study indicated that it was not rs1011970 but rs77283072 that could
influence individual breast cancer risk by the regulation of
CDKN2A expression levels. The present study demonstrated the
connection between the genetic variation in this locus and breast
cancer susceptibility.
Owing to the relatively short distance between this
locus and CDKN2B, it has previously been proposed that the
SNPs in this locus are associated with breast cancer through
regulation of CDKN2B expression (3). Moreover, in the GTEx database,
rs77283072 was demonstrated to be significantly associated with
CDKN2B expression levels in whole blood
(P=1.1×10−10). However, the G allele of rs77283072 was
the high-expression allele according to the GTEx data, which was in
contrast to the results of the luciferase assay in the present
study. Moreover, no interaction was demonstrated between the
enhancer and CDKN2B promoter. Taken together, these findings
indicated that CDKN2B was not likely to be involved in the
association between this locus and breast cancer
susceptibility.
However, the 3C results from the present study
clearly demonstrated that an interaction did occur between this
enhancer and the CDKN2A promoter. CDKN2A is an
important tumor suppressor gene, which mainly encodes two proteins
through alternative splicing, p16INK4A and p14ARF, both
of which have been reported to inhibit the cell cycle (21). p16INK4A is able to
prevent the activity of cyclin-dependent kinase 4/6 and
retinoblastoma protein (22,23).
However, p14ARF has been reported to stabilize the tumor suppressor
protein p53 through interaction with murine double minute 2
(24,25). It has been proposed that the
inactivation of CDKN2A either by mutation or through DNA
methylation occurs in ~20% of patients with breast cancer (26). Therefore, the observation that
rs77283072 was able to influence the risk of breast cancer by
regulating CDKN2A mRNA expression levels was not unexpected.
Considering the widespread and universal functions of the proteins
p16INK4A and p14ARF, rs77283072 might also contribute to
the onset of carcinogenesis in other tissues. Moreover, rs77283072
has been reported to be significantly associated with prostate
cancer, according to one GWAS (P=6.4×10−7) (27). Furthermore, the association between
rs1011970 and multiple cancer types (4) could also be attributed to the function
of rs77283072.
It would have been ideal to identify the potential
TFs which interacted with rs77283072 in the present study. ChIP
assays were performed for all predictions using common
bioinformatics approaches but failed to identify the TF binding
rs77283072. Multiple ChIP-sequencing datasets were also used and
the reads were aligned with the human genome using bowtie2
(10), and the enrichment was
analyzed using software MACS (19).
However, no significant peaks for any TFs in the rs77283072
surrounding region were identified (data not shown). As there are
hundreds of TFs within the human cell, it is difficult to directly
perform ChIP assays without bioinformatics work and this requires
further investigation.
In the luciferase experiment, the relative enhancer
activity of the A/T combination for rs77283702/rs1011970 locus was
not investigated, which constitutes a potential limit for the
study. However, since SNPs usually executes their role on gene
expression relatively independently, this might not alter the
conclusion that rs1011970 does not have the ability to alter
CDKN2A expression and further influence breast cancer
risk.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 31370129) and the Fundamental
Research Funds for the Central Universities (grant no.
GK202001004).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available at https://www.jianguoyun.com/p/DYEJH4wQ_cv3BRizpOkEIAA
and from the corresponding author on reasonable request.
Authors' contributions
CS conceived and designed the study, and wrote the
manuscript. GHH, SDL, XQS, YC, LS and QNS performed the
experiments. GHH analyzed the data. GHH and CS confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Michailidou K, Beesley J, Lindstrom S,
Canisius S, Dennis J, Lush MJ, Maranian MJ, Bolla MK, Wang Q and
Shah M: Genome-wide association analysis of more than 120,000
individuals identifies 15 new susceptibility loci for breast
cancer. Nat Genet. 47:373–380. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Turnbull C, Ahmed S, Morrison J, Pernet D,
Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS, et
al: Genome-wide association study identifies five new breast cancer
susceptibility loci. Nat Genet. 42:504–507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fehringer G, Kraft P, Pharoah PD, Eeles
RA, Chatterjee N, Schumacher FR, Schildkraut JM, Lindström S,
Brennan P, Bickeböller H, et al: Cross-cancer genome-wide analysis
of lung, ovary, breast, prostate, and colorectal cancer reveals
novel pleiotropic associations. Cancer Res. 76:5103–5114. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
The 1000 Genomes Project Consortium, . A
global reference for human genetic variation. Nature. 526:68–74.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li YK, Zhang XX, Yang Y, Gao J, Shi Q, Liu
SD, Fu WP and Sun C: Convergent evidence supports TH2LCRR as a
novel asthma susceptibility gene. Am J Respir Cell Mol Biol.
66:283–292. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li X, Xu X, Fang J, Wang L, Mu Y, Zhang P,
Yao Z, Ma Z and Liu Z: Rs2853677 modulates snail1 binding to the
TERT enhancer and affects lung adenocarcinoma susceptibility.
Oncotarget. 7:37825–37838. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Almeida R, Ricano-Ponce I, Kumar V, Deelen
P, Szperl A, Trynka G, Gutierrez-Achury J, Kanterakis A, Westra HJ,
Franke L, et al: Fine mapping of the celiac disease-associated LPP
locus reveals a potential functional variant. Hum Mol Genet.
23:2481–2489. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hagege H, Klous P, Braem C, Splinter E,
Dekker J, Cathala G, de Laat W and Forné T: Quantitative analysis
of chromosome conformation capture assays (3C-qPCR). Nat Protoc.
2:1722–1733. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pickrell JK, Marioni JC, Pai AA, Degner
JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y and
Pritchard JK: Understanding mechanisms underlying human gene
expression variation with RNA sequencing. Nature. 464:768–772.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Roberts A and Pachter L: Streaming
fragment assignment for real-time analysis of sequencing
experiments. Nat Methods. 10:71–73. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nie L, Vazquez AE and Yamoah EN:
Identification of transcription factor-DNA interactions using
chromatin immunoprecipitation assays. Methods Mol Biol.
493:311–321. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li XX, Peng T, Gao J, Feng JG, Wu DD, Yang
T, Zhong L, Fu WP and Sun C: Allele-specific expression identified
rs2509956 as a novel long-distance cis-regulatory SNP for SCGB1A1,
an important gene for multiple pulmonary diseases. Am J Physiol
Lung Cell Mol Physiol. 317:L456–L463. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang X, Gamble MJ, Stadler S, Cherrington
BD, Causey CP, Thompson PR, Roberson MS, Kraus WL and Coonrod SA:
Genome-wide analysis reveals PADI4 cooperates with Elk-1 to
activate c-Fos expression in breast cancer cells. PLoS Genet.
7:e10021122011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun C, Southard C, Witonsky DB, Kittler R
and Di Rienzo A: Allele-specific down-regulation of RPTOR
expression induced by retinoids contributes to climate adaptations.
PLoS Genet. 6:e10011782010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi Q, Yao XY, Wang HY, Li YJ, Zhang XX
and Sun C: Breast cancer-associated SNP rs72755295 is a
cis-regulatory variation for human EXO1. Genet Mol Biol.
45:e202104202022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feng J, Liu T, Qin B, Zhang Y and Liu XS:
Identifying ChIP-seq enrichment using MACS. Nat Protoc.
7:1728–1740. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Calo E and Wysocka J: Modification of
enhancer chromatin: What, how, and why? Mol Cell. 49:825–837. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Serra S and Chetty R: p16. J Clin Pathol.
71:853–858. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liggett WH Jr and Sidransky D: Role of the
p16 tumor suppressor gene in cancer. J Clin Oncol. 16:1197–1206.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Serrano M: The tumor suppressor protein
p16INK4a. Exp Cell Res. 237:7–13. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Xiong Y and Yarbrough WG: ARF
promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus
deletion impairs both the Rb and p53 tumor suppression pathways.
Cell. 92:725–734. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pomerantz J, Schreiber-Agus N, Liegeois
NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee
HW, et al: The Ink4a tumor suppressor gene product, p19Arf,
interacts with MDM2 and neutralizes MDM2′s inhibition of p53. Cell.
92:713–723. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Poi MJ and Tsai MD: Regulatory
mechanisms of tumor suppressor P16(INK4A) and their relevance to
cancer. Biochemistry. 50:5566–5582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schumacher FR, Al Olama AA, Berndt SI,
Benlloch S, Ahmed M, Saunders EJ, Dadaev T, Leongamornlert D,
Anokian E, Cieza-Borrella C, et al: Association analyses of more
than 140,000 men identify 63 new prostate cancer susceptibility
loci. Nat Genet. 50:928–936. 2018. View Article : Google Scholar : PubMed/NCBI
|