Introduction
Glioma is the most common and lethal intracranial
tumor, posing a severe threat to human health and life (1); it is characterized by high morbidity
and relapse rates, and a poor survival rate. The annual morbidity
rate of cerebral glioma is ~6.1 per 100,000 individuals (2). Depending on its origin, glioma can be
classified into astrocytoma or oligodendroglioma. Glioma is divided
into grades I–IV by the World Health Organization (WHO); the median
survival rates of patients with grades III and IV glioma are 2 and
1 year, respectively (3). Despite
numerous advances being made in surgical excision, chemotherapy,
radiotherapy, targeted therapy and gene therapy, the currently
available therapeutic methods are not sufficient to completely
eliminate the glioma, and the therapeutic effect remains
unsatisfactory. Therefore, a more in-depth understanding of the
precise mechanisms responsible for the development of gliomas is
urgently required, which may be beneficial for the identification
of novel therapeutic targets.
Hepatocyte nuclear factor 4 (HNF4), which includes
HNF4A and HNF4G isoforms, belongs to the nuclear hormone receptor
superfamily (4). HNF4G is
considered to be a transcription factor. HNF4G is detected in the
human kidneys, stomach, pancreas, lungs, bladder and testicles
(5). Previous studies have
demonstrated that HNF4G is associated with glucose metabolism and
hyperuricemia (6,7). In addition, several studies have
observed that HNF4G functions as an oncogene, regulating cell
growth, apoptosis and invasion in certain types of human cancer,
including pancreatic, prostate, bladder, liver and lung cancer
(8–11). However, the function of HNF4G in
several other types of tumors, including glioma, has not yet been
fully elucidated. In particular, the possible molecular mechanisms
underlying the role of HNF4G in gliomas remain unclear.
Therefore, the present study measured the expression
of HNF4G in patients with glioma and investigated the function and
molecular mechanisms of HNF4G in glioma progression. The present
study compared HNF4G expression levels in glioma specimens and cell
lines with those in adjacent non-tumor tissues and normal cells,
respectively. The association of the mRNA expression of HNF4G and
the clinicopathological features of patients with glioma was
investigated. In addition, the effect of HNF4G on glioma cell
proliferation was examined in vitro and in vivo.
Furthermore, molecular mechanistic analyses were conducted to
investigate whether HNF4G affected glioma cell proliferation and
tumor growth via neuropilin-1 (NRP1).
Materials and methods
Human glioma samples
Human glioma specimens and adjacent non-tumor
tissues were obtained during glioma surgery in 59 patients (38 men
and 21 women; 33 cases ≥50 years old, 26 cases <50 years old;
mean age, 52 years old) between April 2019 and October 2020 at the
Department of Pathology, Xi'an Gaoxin Hospital (Xi'an, China).
Written informed consent was obtained from each patient. The
patients had not been treated with radiotherapy, chemotherapy or
other treatment prior to the surgery. Three small sections from
each tissue were promptly frozen and stored at −80°C for use in the
following experiments. The patient samples were divided into high
and low expression groups based on the median gene expression
levels (12). The present study was
approved by the Ethics Committee of Xi'an Gaoxin Hospital (Xi'an,
China; approval no. GXYY-XA-H-2022-068).
Cells and cell culture
The human glioma U87, LN229 and U251 cell lines were
purchased from Procell Life Science &Technology Co., Ltd. and
immortalized normal human astrocytes (NHAs) were purchased from
Shanghai BinsuiBio Co., Ltd. The U87 cell line is not the original
glioblastoma cell line established in 1968 at the University of
Uppsala; it is the U87 MG ATCC version (CVCL_UE09), which is most
probably a glioblastoma, but whose origin is unknown. The cells
were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) with
10% fetal calf serum (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at
37°C with 5% CO2.
Animals
A total of 4 male BALB/c nude mice (5 weeks old;
29.1±1.7 g) were purchased from Shanghai SLAC Laboratory Animal
Co., Ltd., and fed under pathogen-free conditions. Mice were
maintained at a temperature of 26°C, with 50% relative humidity,
ventilation 13 times/h and a 10-h light and 14-h dark cycle/day.
The food was autoclaved and the drinking water was sterile water
with a mixture of vitamins, and both were available ad libitum. The
Institutional Animal Care and Use Committee of Xi'an Gaoxin
Hospital approved the animal experiments (approval no.
GXYY-XA-A-2022-039).
Plasmid construction and
transfection
Full-length DNA sequences of HNF4G or NRP1 were
incorporated into the pCMV2-GV146 plasmid (Genechem Co. Ltd.),
respectively. A reporter plasmid (pGL3-NRP1-luc; Beijing AuGCT
DNA-SYN Biotechnology) was also constructed, which included the
490-bp DNA fragment 33484619-33485108 relative to the NRP1
promoter, which was located upstream of the firefly luciferase
reporter gene in pGL3-luc. The pGL3-luc and pGL3-NRP1-luc plasmids
were transfected into LN229 and U251 glioma cells at 37°C for 48 h
using Jet Prime (Polyplus-transfection SA).
Transfection with small interfering
RNA (siRNA)
siRNAs were purchased from Shanghai GenePharma Co.,
Ltd., for the knockdown of HNF4G and NRP1 gene expression. Human
HNF4G siRNA-1 (sense, 5′-CGGCACUACAUAAAUGUGATT-3′ and antisense,
5′-UCACAUUUAUGUAGUGCCGTT-3′), HNF4G siRNA-2 (sense,
5′-CGAGUGAGAGAAACACAUUTT-3′ and antisense,
5′-AAUGUGUUUCUCUCACUCGTT-3′), NRP1 siRNA-1 (sense,
5′-GGUUUCUCAGCAAACUACATT-3′ and antisense,
5′-UGUAGUUUGCUGAGAAACCTT-3′), NRP1 siRNA-2 (sense,
5′-CUGGCAUAUCUAUGAGAUUTT-3′ and antisense,
5′-AUCUCAUAGAUAUGCCAGTT-3′) and negative control siRNA (NC-siRNA
sense, 5′-UUCACCGACUUUGUCACGUTT-3′ and antisense,
5′-ACGUGACAAAGUCGGUGAATT-3′) were transiently transfected into
LN229 and U251 cells at the concentration of 50 nM at 37°C for 24,
48 or 72 h using Jet Prime (Polyplus-transfection SA).
Cell proliferation assay
The LN229 and U251 cells were independently
suspended at 20,000 cells/ml in DMEM with 10% fetal calf serum and
seeded in 96-well plates (200 µl/well). These cells were
transiently transfected with the control vector, HNF4G
overexpression vector, NRP1 overexpression vector, NC-siRNA (80
nM), HNF4G siRNA-1, HNF4G siRNA-2, NRP1 siRNA-1, NRP1 siRNA-2,
HNF4G siRNA-2 + control vector, or HNF4G siRNA-2 + NRP1
overexpression vector for 24, 48 or 72 h. Cell growth was measured
using an MTT assay (MilliporeSigma). MTT diluent (20 µl) was added
to each well followed by culturing for 4 h. Dimethylsulfoxide (150
µl) was then added to dissolve the formazan salt. The optical
density at 492 nm was examined using a microplate reader (BMG
Labtech GmbH).
Cell cycle assay
The LN229 and U251 glioma cells were harvested at 24
h following transfection. The cells were washed using PBS and
immobilized with 70% ethyl alcohol at 4°C. The cells were then
washed with PBS and prepared as a single cell suspension. Propidium
iodide (PI; 0.05 mg/ml; MilliporeSigma) containing RNase A (0.1
mg/ml) was added to each group of cells followed by incubation at
room temperature for 10 min for staining. Cell cycle analysis was
performed by a flow cytometer (EPICS XL; Beckman Coulter, Inc.)
using SYSTEM II Software (version 3.0; Beckman Coulter, Inc.).
Apoptosis assay
At 48 h post-transfection, the LN229 and U251 cells
were collected. The cells were washed twice with PBS. The cells
were then prepared as single cell suspensions with binding buffer
and stained using an Annexin-V-FITC/PI Apoptosis Detection kit
(Abcam). The number of apoptotic cells was detected and quantified
by a flow cytometer (EPICS XL; Beckman Coulter, Inc.) using SYSTEM
II Software (version 3.0; Beckman Coulter, Inc.).
Lentiviral construction
HNF4G short hairpin RNA (shRNA) was incorporated
into a lentiviral vector (GV118; Shanghai GeneChem Co., Ltd.) for
the silencing of HNF4G expression. The sequences were as follows:
Negative control (sh-Ctrl),
5′-AAAAGAGGCTTGCACAGTGCATTCAAGACGTGCACTGTGCAAGCCTCTTTT-3′; and
HNF4G shRNA,
5′-TCGAGTGAGAGAAACACATTTTCTCGAGAATGTGTTTCTCTCACTCGTTTTTTC-3′. The
U251 cells were cultured in 12-well plates. The solution of
lentiviral vector (0.5 ml 4×108 TU/ml) was then used to
infect the U251 cells with Polybrene 5 µg/ml (Shanghai GeneChem
Co., Ltd.) for 10 h at 37°C, after which the culture medium was
replaced with DMEM containing 10% fetal calf serum. At 2 days
post-infection, puromycin (cat. no. P9620; 25 µg/ml;
MilliporeSigma) was added in infected cells and the culture was
continued for 6 days for the tumor transplantation experiment.
Tumorigenicity assay
Following infection with sh-Ctrl or HNF4G shRNA, the
U251 cells were prepared as a single-cell suspension in DMEM.
Tumorigenicity was detected using 6-week-old BALB/c nude mice
(n=4/group). The infected U251 cells (2×106) were
resuspended in 100 µl DMEM and injected subcutaneously into the
posterior flanks of the mice. The transplanted tumors were measured
using a Vernier caliper every third day. The length (L) and width
(W) of the tumors were used to calculate the tumor volume (V) using
the following formula: V=(L × W2)/2. The nude mice were
euthanized by anesthesia with 3% isoflurane followed by the
injection of pentobarbital sodium (150 mg/kg) 31 days after the
injection of the cells. The cessation of breathing and heartbeat
were considered as confirmation of death. The tumors were then
isolated and weighed, after which the tumor tissues were frozen for
use in further assays.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the glioma samples,
mouse tumor tissues and glioma cells using an RNA Extraction Kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The RNA was reversed transcribed into
cDNA using the PrimeScript™ II 1st strand cDNA kit
(Takara Bio, Inc.) according to the manufacturer's protocol. qPCR
was then performed using SYBR Premix Ex Taq (Takara Biotechnology
Co., Ltd.). Reaction conditions were as follows: Initial
denaturation at 95°C for 5 min, followed by denaturation at 95°C
for 10 sec, annealing at 55°C for 25 sec and extension at 72°C for
10 sec, for 45 cycles. The primer sequences used were as follows:
HNF4G forward, 5′-ACAGAATAAGCACCAGAAG-3′ and reverse,
5′-TCACAGACATCACCAATAC-3′; NRP1 forward, 5′-CGTGGAAGTCTTCGATGGAG-3′
and reverse, 5′-AAGAAATGGCCCTGAAGACA-3′; and GAPDH forward,
5′-GCCGTATCGCTCAGACAC-3′ and reverse, 5′-GCCTAATACGACCAAATCC-3′.
The qPCR was performed using an IQ™5 Multicolor qRT-PCR
Detection System (Bio-Rad Laboratories, Inc.). The
2−ΔΔCq method (13) was
used to analyze the expression of the target genes using GAPDH as
the reference gene.
Western blot analysis
Protein was extracted from the glioma samples, mouse
tumor tissue and glioma cells using RIPA buffer (MilliporeSigma)
with protease inhibitors. Protein quantification was performed with
a BCA kit (Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. Equal amounts of protein samples (30 µg)
were subjected to sodium dodecyl sulfate-polyacrylamide (10%) gel
electrophoresis followed by transfer onto nitrocellulose membranes.
After blocking the membranes with skimmed milk (5%) at room
temperature for 2 h, primary antibodies were added followed by
incubation at 4°C for 10 h. The primary antibodies comprised HNF4G
antibody (cat. no. HPA005438; 1:1,000; MilliporeSigma), NRP1
antibody (cat. no. sc-5307; 1:1,000; Santa Cruz Biotechnology,
Inc.) and GAPDH antibody (cat. no. sc-47724; 1:1,000; Santa Cruz
Biotechnology, Inc.). The appropriate anti-rabbit (cat. no.
sc-2357) or anti-mouse (cat. no. sc-2005) horseradish
peroxidase-linked secondary antibody (both 1:2,000; Santa Cruz
Biotechnology, Inc.) was then added followed by incubation at room
temperature for 4 h. The membranes were finally incubated with
enhanced chemiluminescence reagent (GE Healthcare; Cytiva) for
chemiluminescence detection. The protein bands were scanned using
Syngene G:BOX Chemi XX6 system (Syngene) and quantified using
GeneTools software (version 3.06.02; Syngene). GAPDH was used to
normalize the protein expression data.
Chromatin immunoprecipitation
(ChIP)-RT-qPCR
After crosslinking the LN229 and U251 cells with
formaldehyde (1%) at room temperature for 18 min, glycine was added
for quenching. The cells were resuspended in SDS lysis buffer [50
mM Tris-HCl (pH 8.1), 10 mM EDTA and 1% SDS with freshly added PIC
at 1 µl PIC/800 µl total volume]. The LN229 and U251 cells were
then sonicated using an ultrasonic processor and nuclear lysates
were extracted, which contained chromatin that had been broken into
~200-bp DNA fragments. HNF4G or IgG (cat. nos. HPA005438 and I8765;
both 1:100; MilliporeSigma) antibodies were incubated with the
extracted DNA fragments for 13 h at 4°C. Dynabeads Protein A (200
µl; Thermo Fisher Scientific, Inc.) was added in 500 µl lysis
buffer and agitated in a shaker for 2 h. The beads were centrifuged
at 12,000 × g for 1 min at room temperature. The supernatant was
removed and transfered to a new 1.5-ml microfuge tube. The
DNA-protein complexes were washed in TE buffer at 65°C.
Subsequently, the crosslinking was reversed for 8 h at 65°C. The
QIAquick® PCR purification Kit (Qiagen GmbH) was used to
extract the binding DNA fragments. The predicted binding NRP1 DNA
fragments (UCSC Genome Browser; Human Feb. 2009, GRCh37/hg19)
(14) were verified using RT-qPCR
according to the aforementioned protocol. The primer sequences used
were as follows: Primer 1 forward, 5′-GCATTGACTTAGCCAGAAGGTGACA-3′
and reverse, 5′-GCTTTCCTGTTTCTCCATTGTCTGA-3′; primer 2 forward,
5′-ATGACTCAGACAATGGAGAAACAGG-3′ and reverse,
5′-ACAAGTTCAATCCAAACCACGCGGG-3′; primer 3 forward,
5′-GTAGACCCGCGTGGTTTGGATTGAA-3′ and reverse,
5′-GCCAGTCGGTCCTGTCACAGAGTCT-3′; primer 4 forward,
5′-CAGTAGACTCTGTGACAGGACCGAC-3′ and reverse,
5′-ATTGTCAGAGCAGGAGCGGTTTTGT-3′; and primer 5 forward,
5′-TAACAAAACCGCTCCTGCTCTGACA-3′ and reverse,
5′-GTTAAAAAAAATAAAAGTGAACAAC-3′. The target areas of the primers
are shown in Fig. S1.
Luciferase reporter gene assay
The LN229 and U251 cells were plated into 96-well
plates. Each group was established in six parallel wells. The LN229
and U251 cells were transfected with pGL3-luc or pGL3-NRP1-luc, and
co-transfected with pGL3-NRP1-luc and HNF4G siRNAs or HNF4G
overexpression vector at 37°C for 2 days. The luciferase activity
was detected using a Dual-Luciferase Reporter Assay System (Promega
Corporation). Luciferase activity was normalized to Renilla
luciferase activity.
The cancer genome atlas (TCGA) data
analysis
To verify that HNF4G regulates NRP1, microarray data
were collected from the public database, TCGA (http://gepia.cancer-pku.cn/detail.php?gene=HNF4G), and
the correlation between HNF4G and NRP1 expression was analyzed by
using simple linear regression.
Statistical analysis
All statistical data were analyzed using SPSS 25.0
software (IBM Corp.). All experiments were performed with at least
three independent assays. Data are presented as the mean ± standard
deviation. Unpaired Student's t-test or one-way ANOVA was performed
to analyze the differences between two or multiple independent
groups, respectively. The ANOVA test followed by the Bonferroni
post hoc test was used for pairwise comparisons. Paired Student's
t-test was used to analyze differences in HNF4G and NRP1 mRNA
levels between glioma tissue and normal tissue. Fisher's exact test
was used to analyze the association between HNF4G mRNA expression
and clinicopathological features. Pearson's correlation analysis
was conducted to analyze the correlation between HNF4G and NRP1
expression. P<0.05 was considered to indicate a statistically
significant difference.
Results
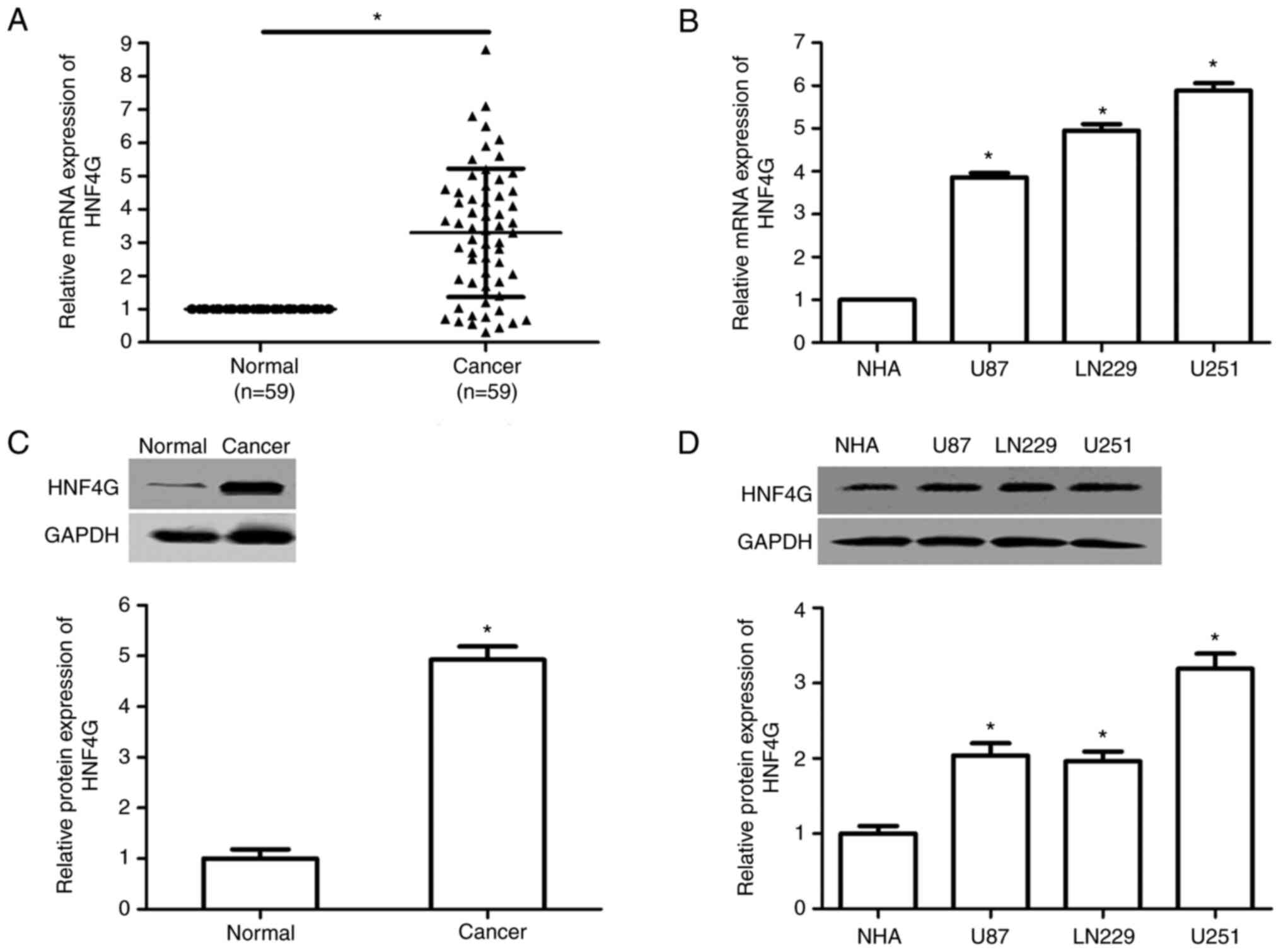
HNF4G expression is upregulated in
human glioma samples and cell lines
To explore the role of HNF4G in glioma, the
expression of HNF4G in glioma specimens and cell lines was
evaluated using RT-qPCR and western blot analysis. The results
revealed that the mRNA and protein expression levels of HNF4G were
significantly upregulated in human glioma samples compared with
those in adjacent normal tissues (P<0.001; Fig. 1A and C). Moreover, the high mRNA
expression of HNF4G was associated with the WHO pathological grade,
isocitrate dehydrogenase [NADP(+)] 1 (IDH1) mutation, tumor size
and the Karnofsky Performance Scale (KPS) score (all P<0.001;
Table I). The HNF4G mRNA and
protein expression levels were also significantly higher in the
U87, LN229 and U251 human glioma cell lines than in the NHAs
(P<0.001; Fig. 1B and D).
 | Table I.Association between HNF4G mRNA
expression and clinicopathological features in patients with
glioma. |
Table I.
Association between HNF4G mRNA
expression and clinicopathological features in patients with
glioma.
|
|
| HNF4G mRNA
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Patients (n) | High (n=49) | Low (n=10) | aP-value |
|---|
| Sex |
|
|
| 0.907 |
|
Male | 38 | 31 | 7 |
|
|
Female | 21 | 18 | 3 |
|
| Age, years |
|
|
| 0.839 |
|
≥50 | 33 | 27 | 6 |
|
|
<50 | 26 | 22 | 4 |
|
| WHO grade |
|
|
| <0.001 |
| I +
II | 20 | 12 | 8 |
|
| III +
IV | 39 | 37 | 2 |
|
| IDH1 |
|
|
| <0.001 |
|
Mutation | 27 (Nod, 13; Cod,
16) | 25 | 2 |
|
|
Wild-type | 32 (Nod, 28; Cod,
6) | 24 | 8 |
|
| 1p/19q |
|
|
| 0.065 |
| No
deletion | 38 | 31 | 7 |
|
|
Codeletion | 21 | 18 | 3 |
|
| Tumor size, cm |
|
|
| <0.001 |
| ≥5 | 35 | 32 | 3 |
|
|
<5 | 24 | 17 | 7 |
|
| KPS score |
|
|
| <0.001 |
|
<80 | 23 | 17 | 6 |
|
|
≥80 | 36 | 32 | 4 |
|
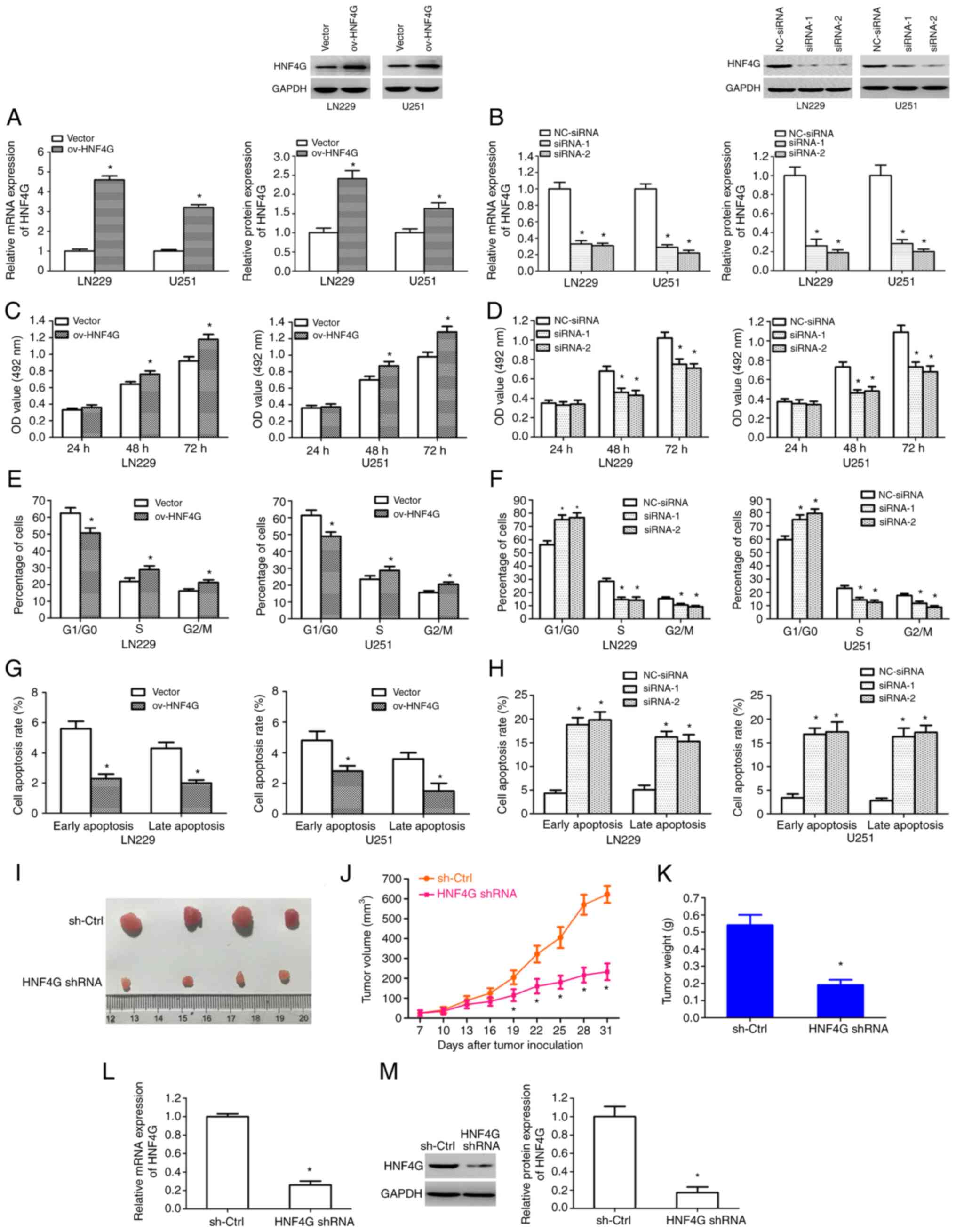
HNF4G promotes glioma cell
proliferation in vitro and in vivo
To investigate the biological function of HNF4G in
human glioma, LN229 and U251 cells were stably transfected with
HNF4G overexpression plasmid, control empty plasmid, HNF4G siRNAs
or NC-siRNA, independently. The HNF4G overexpression plasmid
significantly upregulated the mRNA and protein expression levels of
HNF4G in the LN229 and U251 cells; by contrast, the HNF4G siRNAs
significantly downregulated the mRNA and protein expression levels
of HNF4G (P<0.001; Fig. 2A and
B). The results of MTT assays revealed that HNF4G
overexpression significantly promoted glioma cell proliferation
(P<0.001; Fig. 2C) after 48 and
72 h, whereas the silencing of HNF4G significantly suppressed
glioma cell proliferation at these time points (P<0.001;
Fig. 2D). Cell cycle analysis
revealed that HNF4G overexpression led to a significant reduction
in the number of cells in the G0/G1 phase and
significant increases in the number of cells in the S and
G2/M phases (P<0.001; Figs. 2E and S2A); by contrast, the silencing of HNF4G
resulted in the significant accumulation of cells in the
G0/G1 phase and a corresponding reduction in
the number of cells in the S and G2/M phases
(P<0.001; Figs. 2F and S2B). Moreover, analysis of cell apoptosis
using flow cytometry revealed that HNF4G overexpression reduced the
rates of early and late apoptosis (Figs. 2G and S2C), whereas the silencing of HNF4G
enhanced the rate of early- and late-apoptotic cells to a
significant extent (P<0.001; Figs.
2H and S2D). Subsequently, an
HNF4G shRNA lentiviral vector was constructed and stably
transfected U251 gliomas were generated. Nude mice were
subcutaneously injected in the posterior flanks with HNF4G
shRNA-infected or sh-Ctrl-infected U251 cells and tumor growth was
monitored for 31 days. Observation of the excised tumors revealed
that HNF4G shRNA markedly inhibited tumor growth compared with that
in the sh-Ctrl group, in which the largest tumor diameter was 9 mm
(Fig. 2I). The volumes and weights
of the tumors derived from HNF4G shRNA-transfected cells were
significantly lower than those derived from sh-Ctrl-transfected
cells (P<0.001; Fig. 2J and K).
The results also revealed that HNF4G shRNA significantly suppressed
the mRNA and protein expression levels of HNF4G in the xenograft
tumors (P<0.001; Fig. 2L and M).
These results suggest that HNF4G facilitated glioma cell
proliferation and tumor growth, and suppressed cell apoptosis.
HNF4G activates NRP1 transcription by
binding to its promoter
To determine the mechanisms underlying the effect of
HNF4G in the modulation of glioma development, the UCSC Genome
Browser was used to predict and select the downstream target gene
of HNF4G. The results indicated that HNF4G binds to the promoter
region of the NRP1 gene (Fig. 3A).
ChIP-RT-qPCR further confirmed that HNF4G bound to the promoter
regions of NRP1 in LN229 and U251 cells, and primarily bound to the
segments of primers 3–5 (P<0.001; Fig. 3B). Promoter reporter assays were
used to verify whether HNF4G regulates NRP1 transcription by
binding to its promoter. The binding sequence of NRP1 was inserted
downstream of the luciferase gene. The LN22 and U251 cells were
transfected with the constructed plasmids for 2 days and the
luciferase activity was detected. The results revealed that the
luciferase activity in the cells transfected with pGL3-NRP1-luc was
significantly higher than that in the cells transfected with
pGL3-luc (P<0.001; Fig. 3C).
When the glioma cells were co-transfected with pGL3-NRP1-luc and
the HNF4G overexpression vector, the luciferase activity was
significantly enhanced compared with that in the control vector
group (P<0.001; Fig. 3D). A
significant reduction in luciferase activity was observed in the
cells co-transfected with pGL3-NRP1-luc and HNF4G siRNA compared
with that in the cells co-transfected with pGL3-NRP1-luc and
NC-siRNA (P<0.001; Fig. 3E).
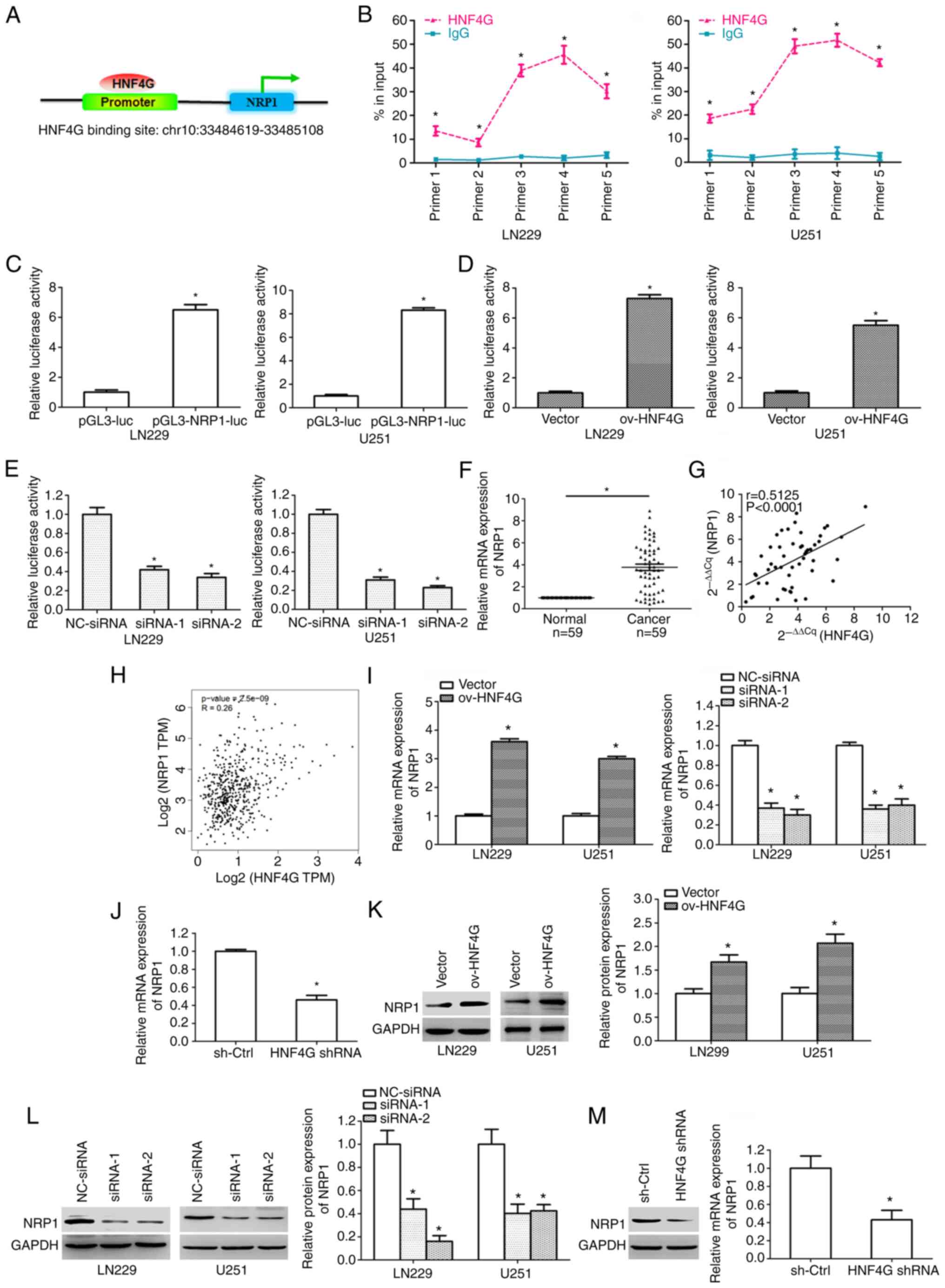 | Figure 3.HNF4G promotes NRP1 transcription by
binding its promoter region in glioma cells. (A) The HNF4G binding
site in the NRP1 promoter was analyzed using the UCSC Genome
Browser. (B) Chromatin immunoprecipitation-reverse
transcription-quantitative PCR confirmed that HNF4G bound to the
promoter region of NRP1 in LN229 and U251 cells. (C) Luciferase
activity in LN229 and U251 cells 2 days after transfection with
pGL3-NRP1-luc. Luciferase activity in LN229 and U251 cells after
co-transfection with (D) pGL3-NRP1-luc and HNF4G overexpression
vector, and (E) pGL3-NRP1-luc and HNF4G siRNA. (F) NRP1 mRNA
expression was significantly upregulated in glioma samples compared
with that in normal tissues. (G) A notable positive association was
detected between HNF4G and NRP1 mRNA expression in glioma tissues.
(H) The Cancer Genome Atlas data revealed that HNF4G expression was
positively associated with NRP1 expression in glioma. (I) NRP1 mRNA
expression in glioma cells transfected with HNF4G overexpression
vector or siRNA. (J) NRP1 mRNA expression in xenograft tumors. NRP1
protein expression in glioma cells transfected with (K) HNF4G
overexpression vector and (L) HNF4G siRNA. (M) NRP1 protein
expression in xenograft tumors. *P<0.001 vs. respective control,
n=3. HNF4G, hepatocyte nuclear factor 4γ; NRP1, neuropilin-1; luc,
luciferase; ov, overexpression; siRNA, small interfering RNA; NC,
negative control; sh-Ctrl, short hairpin control; shRNA, short
hairpin RNA. |
The analysis of patient tissues showed that the mRNA
expression of NRP1 was also significantly upregulated in glioma
samples compared with that in the adjacent normal tissues
(P<0.001; Fig. 3F). In addition,
a significant positive association between HNF4G and NRP1 mRNA
expression was detected in the glioma tissues (P<0.001; Fig. 3G). TCGA data also revealed that
HNF4G expression was positively associated with NRP1 expression in
glioma (P<0.001; Fig. 3H). HNF4G
overexpression significantly upregulated the mRNA and protein
expression levels of NRP1 in LN229 and U251 cells, while HNF4G
siRNA significantly downregulated them (P<0.001; Fig. 3I, K and L). Furthermore, HNF4G shRNA
significantly suppressed the mRNA and protein expression levels of
NRP1 in xenograft tumors (P<0.001; Fig. 3J and M). Together, these data
demonstrate that HNF4G promoted NRP1 transcription by binding to
its promoter in glioma cells.
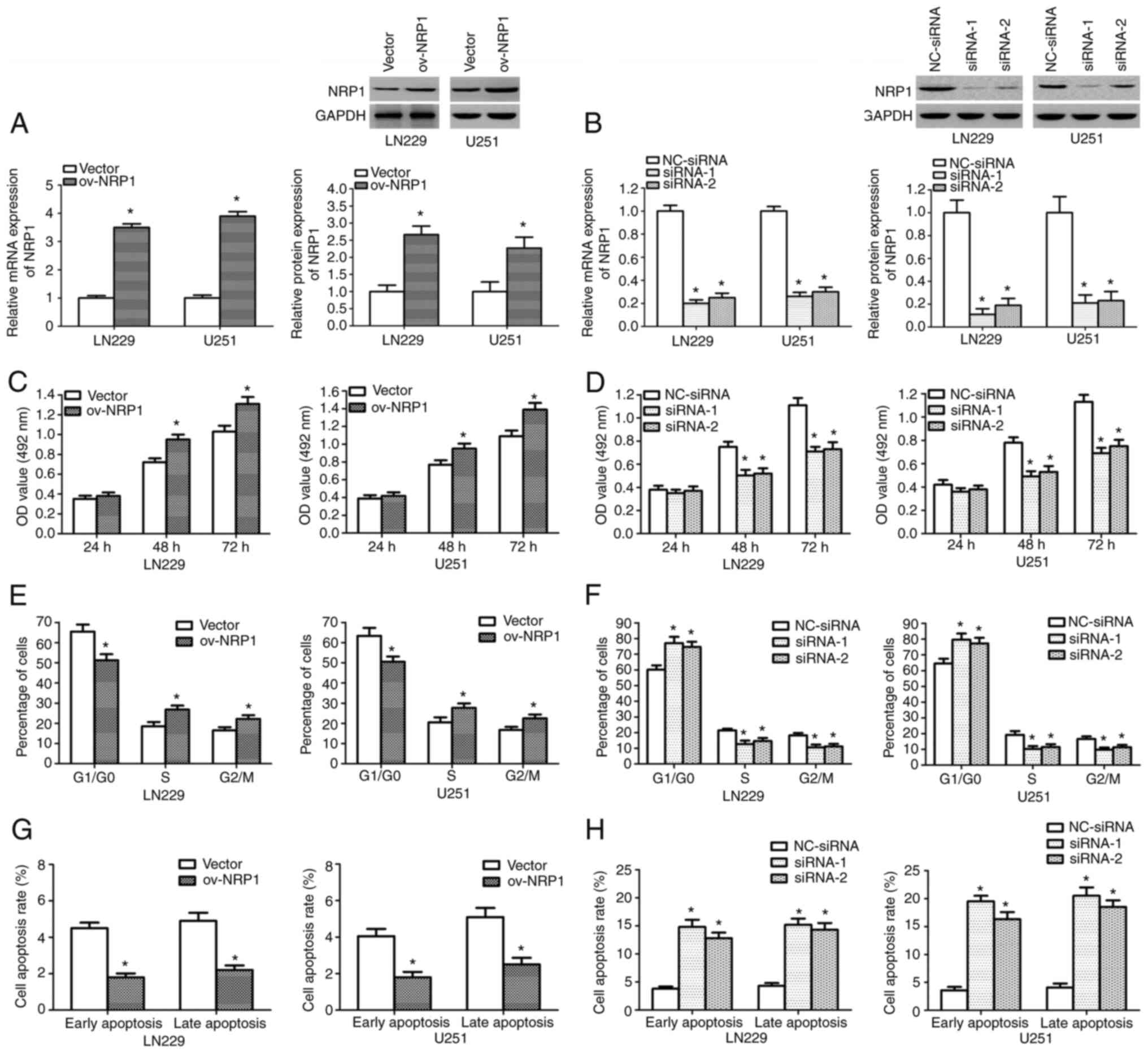
NRP1 promotes glioma cell
proliferation
To explore the role of NRP1 in human glioma, the
LN229 and U251 cells were stably transfected with NRP1
overexpression plasmid, control empty plasmid, NRP1 siRNAs or
NC-siRNA, independently. NRP1 overexpression significantly
increased the mRNA and protein expression levels of NRP1 in the
cells, whereas the knockdown of NRP1 significantly suppressed the
mRNA and protein expression levels of NRP1 (P<0.001; Fig. 4A and B). The results of MTT assays
revealed that NRP1 overexpression markedly promoted glioma cell
proliferation after 48 and 72 h (P<0.001; Fig. 4C), while the knockdown of NRP1
substantially inhibited glioma cell growth at these time points
(P<0.001; Fig. 4D). Flow
cytometry revealed that NRP1 overexpression induced significant
changes in the cell cycle, resulting in a marked significant
reduction in the proportion of cells in the
G0/G1 phase and an increase in the proportion
of cells in the S and G2/M phases (P<0.001; Figs. 4E and S3A). However, the knockdown of NRP1 led
to the significant accumulation of cells in the
G0/G1 phase and a reduction in the proportion
of cells in the S and G2/M phases (P<0.001; Figs. 4F and S3B). In addition, flow cytometry revealed
that NRP1 overexpression significantly decreased the rates of early
and late apoptosis (P<0.001; Figs.
4G and S3C), whereas the
knockdown of NRP1 significantly increased the rates of early and
late apoptosis (P<0.001; Figs.
4H and S3D). These results
demonstrate that NRP1 had oncogenic effects.
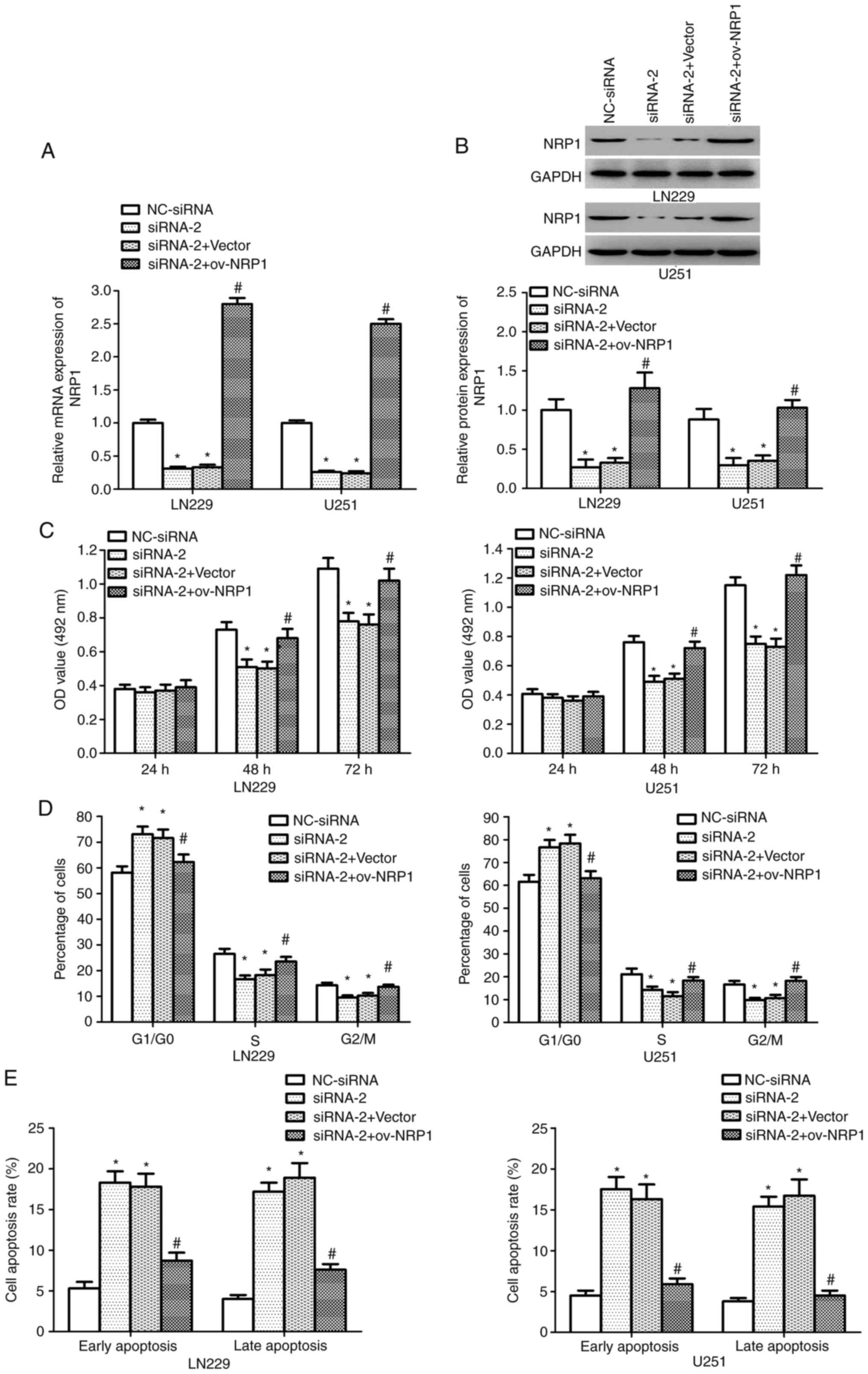
HNF4G facilitates glioma cell growth
by upregulating NRP1 expression
To validate the previous findings which indicated
that HNF4G enhances glioma cell proliferation by modulating NRP1
transcription, HNF4G siRNA-2 and NRP1 overexpression vector were
co-transfected into LN22 and U251 cells. The knockdown of HNF4G
decreased the NRP1 mRNA and protein expression levels in LN229 and
U251 cells, whereas co-transfection with the NRP1 overexpression
vector reversed this effect (P<0.001; Fig. 5A and B). Furthermore, MTT assay
results revealed that NRP1 overexpression reversed the effects of
HNF4G knockdown on cell proliferation (P<0.001; Fig. 5C). Cell cycle analysis confirmed
that HNF4G knockdown led to a significant accumulation of cells in
the G0/G1 phase and a reduction in the
proportion of cells in the S and G2/M phase, while
co-transfection with NRP1 overexpression vector counteracted the
effects of HNF4G knockdown on the cell cycle (P<0.001; Figs. 5D and S4A). Furthermore, apoptosis analysis
showed that while the knockdown of HNF4G significantly increased
the proportions of early- and late-apoptotic cells, co-transfection
with NRP1 overexpression vector eliminated the effects of HNF4G
siRNA-2 on apoptosis (P<0.001; Figs.
5E and S4B). These findings
confirm that HNF4G promoted glioma cell proliferation and
suppressed cell apoptosis by promoting NRP1 expression.
Discussion
As a major transcription factor, HNF4G is frequently
upregulated in several types of cancer and functions as a pivotal
oncogene (8,11). Previous studies have demonstrated
that HNF4G facilitates the proliferation and invasion of bladder
cancer cells (10,15). Wang et al (8) found that the overexpression of HNF4G
promoted the progression and metastasis of pancreatic ductal
adenocarcinoma. In addition, Shukla et al (9) observed that the HNF4G/HNF1A
transcription loop accelerated prostate cancer cell proliferation
and oncogenesis. Furthermore, Wang et al (11) demonstrated that the expression of
HNF4G was significantly upregulated in lung cancer tissues, and
that the overexpression of HNF4G facilitated lung cancer cell
proliferation and tumorigenesis. In the present study, it was
observed that the expression of HNF4G was upregulated in human
glioma tissues and cell lines. The high mRNA expression of HNF4G
was associated with the WHO pathological grade, IDH1 mutation,
tumor size and KPS score. HNF4G promoted glioma cell proliferation
in vitro, as well as cell cycle G1-S phase
transition and tumor growth. Moreover, experiments in which HNF4G
was overexpressed or knocked down revealed that HNF4G markedly
suppressed apoptosis. The previous studies found that HNF4G
facilitated the proliferation, invasion and metastasis in certain
cancer types, and the present study revealed similar findings for
glioma; specifically, HNF4G promoted glioma cell proliferation and
cell cycle transition, and suppressed apoptosis. However,
metastasis was not evaluated. These findings indicate that HNF4G
functions as an oncogene in human glioma, and thus has potential
for use as a novel therapeutic target for glioma.
HNF4G promotes bladder cancer progression by
facilitating the transcription of hyaluronan synthase 2 (10). It has been reported that HNF4G and
HNF1A activate enhancer chromatin and upregulate the expression of
gastrointestinal transcriptome genes (9,16). In
the present study, to investigate the potential molecular mechanism
of HNF4G in glioma, NRP1 was predicted as a downstream target gene
of HNF4G by bioinformatics analysis. ChIP-RT-qPCR and promoter
reporter assays verified that HNF4G upregulated NRP1 transcription
by binding to its promoter. Furthermore, HNF4G expression
positively associated with NRP1 expression in glioma. The
overexpression and silencing of HNF4G revealed that HNF4G promoted
NRP1 expression in glioma. These findings confirm the prediction
that HNF4G upregulates NRP1 expression by binding to its promoter
region in glioma cells.
NRP1 is a transmembrane glycoprotein and a
multi-functional co-receptor for several signaling pathways, such
as hepatocyte growth factor (HGF), semaphorins, platelet-derived
growth factor and vascular endothelial growth factor (VEGF)
(17–19). NRP1 has a pivotal role in embryonic
angiogenesis and neurogenesis (20,21).
Moreover, NRP1 can regulate cell mitogenesis, migration, motility,
proliferation, survival and apoptosis by binding to VEGF and HGF
(22–26). There is evidence to indicate that
NRP1 is involved in tumorigenesis and progression. For example,
studies have shown that the upregulation of NRP1 promotes
pancreatic cancer progression by modulating the HGF/c-Met pathway
(27), VEGF-A/NRP1 signaling
promotes breast cancer metastasis (28), and NRP1 facilitates oral squamous
cell carcinoma cell growth and invasion by interacting with CMTM6
(29). In addition, the VEGFR-NRP1
axis has been demonstrated to promote the angiogenesis, growth and
metastasis of gastric cancer (30).
Furthermore, NRP1 has been shown to facilitate nasopharyngeal
carcinoma cell migration and invasion (31). In glioma, previous studies have
established that NRP1 promotes glioma cell proliferation, invasion,
migration and tumor growth, and suppresses cell apoptosis (32–34).
In the present study, it was confirmed that NRP1 promoted glioma
cell proliferation and inhibited cell apoptosis. Additionally, NRP1
overexpression reversed the effects induced by HNF4G knockdown on
cell proliferation, cell cycle progression and apoptosis. These
findings demonstrate that HNF4G promoted glioma cell proliferation
and suppressed cell apoptosis by promoting NRP1 transcription.
In conclusion, HNF4G is considered as an oncogene in
glioma. In the present study, the results demonstrated that HNF4G
expression was upregulated in glioma. HNF4G facilitated cell
proliferation and cell cycle progression, and inhibited apoptosis
by promoting NRP1 transcription. These results indicate that HNF4G
may be an effective therapeutic target for glioma.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Shaanxi Province Natural Science
Foundation (grant nos. 2015JM8419 and 2021JM-582) and the Key
Research and Development Program of Shaanxi (grant no.
2019SF-217).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HC and LZ designed the experiments. HC, QZ and LZ
conducted the experiments. ZL provided research materials and
analyzed data. LZ, HC and LZ wrote the manuscript. All authors read
and approved the final version of the manuscript. HC and LZ confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Xi'an Gaoxin Hospital (approval no. GXYY-XA-2022-052).
All patients provided written informed consent. The Institutional
Animal Care and Use Committee of Xi'an Gaoxin Hospital approved the
animal experiments (approval no. GXYY-XA-A-2022-039).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen J, Wu X, Xing Z, Ma C, Xiong W, Zhu X
and He X: FOXG1 expression is elevated in glioma and inhibits
glioma cell apoptosis. J Cancer. 9:778–783. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ostrom QT, Cote DJ, Ascha M, Kruchko C and
Barnholtz-Sloan JS: Adult glioma incidence and survival by race or
ethnicity in the United States from 2000 to 2014. JAMA Oncol.
4:1254–1262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Komori T, Sasaki H and Yoshida K: Revised
WHO classification of tumours of the central nervous system:
Summary of the revision and perspective. No Shinkei Geka.
44:625–635. 2016.PubMed/NCBI
|
|
4
|
Bertrand S, Brunet FG, Escriva H,
Parmentier G, Laudet V and Robinson-Rechavi M: Evolutionary
genomics of nuclear receptors: From twenty-five ancestral genes to
derived endocrine systems. Mol Biol Evol. 21:1923–1937. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Drewes T, Senkel S, Holewa B and Ryffel
GU: Human hepatocyte nuclear factor 4 isoforms are encoded by
distinct and differentially expressed genes. Mol Cell Biol.
16:925–931. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baraille F, Ayari S, Carrière V, Osinski
C, Garbin K, Blondeau B, Guillemain G, Serradas P, Rousset M,
Lacasa M, et al: Glucose tolerance is improved in mice invalidated
for the nuclear receptor HNF-4γ: A critical role for
enteroendocrine cell lineage. Diabetes. 64:2744–2756. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen BD, Chen XC, Pan S, Yang YN, He CH,
Liu F, Ma X, Gai MT and Ma YT: TT genotype of rs2941484 in the
human HNF4G gene is associated with hyperuricemia in Chinese Han
men. Oncotarget. 8:26918–26926. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang C, Zhang T, Liao Q, Dai M, Guo J,
Yang X, Tan W, Lin D, Wu C and Zhao Y: Metformin inhibits
pancreatic cancer metastasis caused by SMAD4 deficiency and
consequent HNF4G upregulation. Protein Cell. 12:128–144. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shukla S, Cyrta J, Murphy DA, Walczak EG,
Ran L, Agrawal P, Xie Y, Chen Y, Wang S, Zhan Y, et al: Aberrant
activation of a gastrointestinal transcriptional circuit in
prostate cancer mediates castration resistance. Cancer Cell.
32:792–806.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Okegawa T, Ushio K, Imai M, Morimoto M and
Hara T: Orphan nuclear receptor HNF4G promotes bladder cancer
growth and invasion through the regulation of the hyaluronan
synthase 2 gene. Oncogenesis. 2:e582013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Zhang J, Xu L, Zheng Y, Ling D and
Yang Z: Expression of HNF4G and its potential functions in lung
cancer. Oncotarget. 9:18018–18028. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Z, Ni F, Yu F, Cui Z, Zhu X and Chen
J: Prognostic significance of mRNA expression of CASPs in gastric
cancer. Oncol Lett. 18:4535–4554. 2019.PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun H, Tian J, Xian W, Xie T and Yang X:
miR-34a inhibits proliferation and invasion of bladder cancer cells
by targeting orphan nuclear receptor HNF4G. Dis Markers.
2015:8792542015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen L, Toke NH, Luo S, Vasoya RP, Fullem
RL, Parthasarathy A, Perekatt AO and Verzi MP: A reinforcing
HNF4-SMAD4 feed-forward module stabilizes enterocyte identity. Nat
Genet. 51:777–785. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hong TM, Chen YL, Wu YY, Yuan A, Chao YC,
Chung YC, Wu MH, Yang SC, Pan SH, Shih JY, et al: Targeting
neuropilin 1 as an antitumor strategy in lung cancer. Clin Cancer
Res. 13:4759–4768. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Karjalainen K, Jaalouk DE, Bueso-Ramos CE,
Zurita AJ, Kuniyasu A, Eckhardt BL, Marini FC, Lichtiger B, O'Brien
S, Kantarjian HM, et al: Targeting neuropilin-1 in human leukemia
and lymphoma. Blood. 117:920–927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gelfand MV, Hagan N, Tata A, Oh WJ,
Lacoste B, Kang KT, Kopycinska J, Bischoff J, Wang JH and Gu C:
Neuropilin-1 functions as a VEGFR2 co-receptor to guide
developmental angiogenesis independent of ligand binding. Elife.
3:e037202014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gu C, Rodriguez ER, Reimert DV, Shu T,
Fritzsch B, Richards LJ, Kolodkin AL and Ginty DD: Neuropilin-1
conveys semaphorin and VEGF signaling during neural and
cardiovascular development. Dev Cell. 5:45–57. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bagri A and Tessier-Lavigne M: Neuropilins
as semaphorin receptors: In vivo functions in neuronal cell
migration and axon guidance. Adv Exp Med Biol. 515:13–31. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pan Q, Chanthery Y, Liang WC, Stawicki S,
Mak J, Rathore N, Tong RK, Kowalski J, Yee SF, Pacheco G, et al:
Blocking neuropilin-1 function has an additive effect with
anti-VEGF to inhibit tumor growth. Cancer Cell. 11:53–67. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sulpice E, Plouët J, Bergé M, Allanic D,
Tobelem G and Merkulova-Rainon T: Neuropilin-1 and neuropilin-2 act
as coreceptors, potentiating proangiogenic activity. Blood.
111:2036–2045. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jia H, Cheng L, Tickner M, Bagherzadeh A,
Selwood D and Zachary I: Neuropilin-1 antagonism in human carcinoma
cells inhibits migration and enhances chemosensitivity. Brit J
Cancer. 102:541–552. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee P, Goishi K, Davidson AJ, Mannix R,
Zon L and Klagsbrun M: Neuropilin-1 is required for vascular
development and is a mediator of VEGF-dependent angiogenesis in
zebrafish. Proc Natl Acad Sci USA. 99:10470–10475. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Raskopf E, Vogt A, Standop J, Sauerbruch T
and Schmitz V: Inhibition of neuropilin-1 by RNA-interference and
its angiostatic potential in the treatment of hepatocellular
carcinoma. Z Gastroenterol. 48:21–27. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsushita A, Götze T and Korc M:
Hepatocyte growth factor-mediated cell invasion in pancreatic
cancer cells is dependent on neuropilin-1. Cancer Res.
67:10309–10316. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song Y, Zeng S, Zheng G, Chen D, Li P,
Yang M, Luo K, Yin J, Gu Y, Zhang Z, et al: FOXO3a-driven miRNA
signatures suppresses VEGF-A/NRP1 signaling and breast cancer
metastasis. Oncogene. 40:777–790. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng Y, Wang C, Song A, Jiang F, Zhou J,
Li G, Zhang W, Ye J, Ding X, Zhang W, et al: CMTM6 promotes cell
proliferation and invasion in oral squamous cell carcinoma by
interacting with NRP1. Am J Cancer Res. 10:1691–1709.
2020.PubMed/NCBI
|
|
30
|
Mei B, Chen J, Yang N and Peng Y: The
regulatory mechanism and biological significance of the
Snail-miR590-VEGFR-NRP1 axis in the angiogenesis, growth and
metastasis of gastric cancer. Cell Death Dis. 11:2412020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang Z, Cheng C, Xiong H, Wang Y, Chen
KK, Yang J, Xiao B, Zhang R, Li S and Sang Y: NRP1 promotes cell
migration and invasion and serves as a therapeutic target in
nasopharyngeal carcinoma. Int J Clin Exp Pathol. 11:2460–2469.
2018.PubMed/NCBI
|
|
32
|
Evans IM, Yamaji M, Britton G, Pellet-Many
C, Lockie C, Zachary IC and Frankel P: Neuropilin-1 signaling
through p130Cas tyrosine phosphorylation is essential for growth
factor-dependent migration of glioma and endothelial cells. Mol
Cell Biol. 31:1174–1185. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Higgins DMO, Caliva M, Schroeder M,
Carlson B, Upadhyayula PS, Milligan BD, Cheshier SH, Weissman IL,
Sarkaria JN, Meyer FB and Henley JR: Semaphorin 3A mediated brain
tumor stem cell proliferation and invasion in EGFRviii mutant
gliomas. BMC Cancer. 20:12132020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hamerlik P, Lathia JD, Rasmussen R, Wu Q,
Bartkova J, Lee M, Moudry P, Bartek J Jr, Fischer W, Lukas J, et
al: Autocrine VEGF-VEGFR2-neuropilin-1 signaling promotes glioma
stem-like cell viability and tumor growth. J Exp Med. 209:507–520.
2012. View Article : Google Scholar : PubMed/NCBI
|