Introduction
Breast cancer is among the most common invasive
malignancies threatening the health of women worldwide (1). Despite the various breast cancer
treatment strategies that have been developed, breast cancer
remains the second leading cause of cancer-associated deaths in
women (2). The American Cancer
Society estimates that ~281,550 women were diagnosed with breast
cancer and ~43,600 women succumbed to breast cancer in 2021
(3). Currently, the primary
treatment for breast cancer is surgery combined with radiotherapy
or chemotherapy (1). Chemotherapy
is administered for the management of breast cancer prior to and
following surgery (4). Taxanes,
including Taxol (TAX), are used clinically as chemotherapeutic
agents for the treatment of early and advanced metastatic breast
cancer (5,6). However, long-term chemotherapy can
cause patients to develop resistance to chemotherapeutic drugs such
as TAX, which greatly limits the efficacy of chemotherapy and may
lead to systemic treatment failure (7,8).
Therefore, elucidation of the mechanism of resistance to TAX
chemotherapy is necessary to overcome these limitations and improve
the survival rate of patients with breast cancer.
Exosomes are carriers of proteins, lipids and
nucleic acids, including microRNAs (miRNAs), mRNAs and DNA, as well
other active metabolites (9), and
have been reported to play important roles in intercellular
communication (10). Crow et
al (11) demonstrated that
exosomes from platinum-resistant A2780 epithelial ovarian cancer
cells became resistant via the promotion of epithelial-mesenchymal
transition (EMT), which implies the importance of exosomes in
cancer drug resistance. miRNAs, a class of non-coding RNA with a
length of ~22 nucleotides, participate in multiple biological
processes, such as cell proliferation, metastasis, invasion,
apoptosis and aging, by regulating the expression of mRNAs
(12,13). Studies have indicated that exosomal
miRNAs are involved in tumorigenesis, metastasis, tumor response to
treatment and the pathogenesis of tumor drug resistance (14,15). A
study by Fu et al (16)
demonstrated that multidrug resistant Bel/5-FU cells delivered
miR-32-5p to sensitive Bel7402 cells via exosomes and activated the
PI3K/Akt pathway, which further induced multidrug resistance via
the regulation of angiogenesis and EMT. Another study showed that
miR-221 enrichment was closely associated with sorafenib
resistance, and hepatocellular carcinoma-associated exosomes
containing miR-221 induced sensitive cells to become sorafenib
resistant via the modulation of caspase-3 and suppression of
apoptosis (17). Additionally, Uhr
et al (18) suggested that
hsa-miR-187-5p and hsa-miR-106a-3p may be indicators of drug
resistance in breast cancer, and TAX sensitivity may be associated
with hsa-miR-556-5p. However, the roles of exosomal miRNAs and
their underlying mechanisms in the TAX resistance of breast cancer
require elucidation.
Therefore, in the present study, exosomes were
isolated from normal MCF-7 breast cancer cells and TAX-resistant
MCF-7 breast cancer cells, and then the miR-187-5p and miR-106a-3p
levels in the cells and exosomes were determined. As miR-187-5p was
significantly enriched in TAX-resistant cells and TAX-resistant
cell-derived exosomes, MCF-7 cells were treated with TAX for 48 h
and the effects of exosomal miR-187-5p on TAX resistance in breast
cancer cells and the associated underlying mechanisms were
investigated. The aim of the present study is to lay a foundation
for the discovery of novel therapeutic targets and pathways to
confront acquired resistance to TAX in breast cancer
chemotherapy.
Materials and methods
Cell culture
Breast cancer cells (MCF-7; drug sensitive, DS) and
TAX-resistant breast cancer cells (MCF-7/TAX; drug resistant, DR)
were purchased from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences and Procell Life Science &
Technology Co., Ltd., respectively. The MCF-7 and MCF-7/TAX cells
were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo
Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS;
Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin/100 µg/ml
streptomycin (Thermo Fisher Scientific, Inc.), and maintained in an
incubator containing 5% CO2 at 37°C.
Isolation and identification of
exosomes
The MCF-7 and MCF-7/TAX cell lines at 80–90%
confluence were washed with PBS three times. Then, the medium was
replaced with DMEM containing Systembio Exosome-Depleted FBS (cat.
no. EXO-FBS-50A-1; SBI System Biosciences). After being cultured
for 48 h, the cell supernatants of the MCF-7 and MCF-7/TAX cells
were collected and used for exosome isolation. Exosome isolation
from the cell supernatant was performed at 4°C as previously
described (19). Briefly, the cell
supernatant was centrifuged at 500 × g for 5 min and then at 2,000
× g for 30 min at 4°C. Subsequently, the supernatant was obtained
and mixed with an equal volume of pre-cooled 16% polyethylene
glycol. The mixture was incubated at 4°C overnight and centrifuged
at 10,000 × g for 60 min at 4°C. After removing the supernatant,
the sediments were resuspended in 1 ml PBS and centrifuged at
120,000 × g for 70 min at 4°C. After removing the supernatant, the
sediments were resuspended in 100 µl PBS to form an exosome
suspension.
A BCA protein assay kit (Thermo Fisher Scientific,
Inc.) was then used to determine the exosome concentration
according to the manufacturer's instructions. Based on the methods
of Lee et al (20),
transmission electron microscopy (TEM; JEOL, Ltd.) was used to
visualize the morphology of the isolated exosomes. Thereafter, the
particle size and distribution of the exosomes were examined by
nanoparticle tracking analysis (NTA) using a NanoSight NS300
particle size analyzer (Malvern Panalytical, Ltd.) as previously
described (21). Additionally, the
protein expression levels of heat shock protein 70 (HSP70), tumor
susceptibility gene 101 (TSG101) and CD9, which are
exosome-specific markers, were detected by western blotting using
their corresponding antibodies: Anti-HSP70 antibody (catalogue
number, 10995-1-AP; 1:1,000; ProteinTech Group, Inc.), anti-TSG101
antibody (catalogue number, 28283-1-AP; 1:1,000; ProteinTech Group,
Inc.) and anti-CD9 antibody (catalogue number, 20597-1-AP; 1:1,000;
Abcam). Anti-calnexin antibody (catalogue number, 10427-2-AP;
1:1,000; ProteinTech Group, Inc.) was used to detect calnexin as a
cell marker and anti-GAPDH antibody (catalogue number, 10494-1-AP;
1:2,000; ProteinTech Group, Inc.) was used to detect GAPDH as a
loading control (22).
Co-culture of MCF-7/TAX cell-derived
exosomes and MCF-7 cells
To determine the uptake of MCF-7/TAX cell-derived
exosomes by MCF-7 cells, a PKH67 staining kit (PKH67GL-1KT;
Sigma-Aldrich; Merck KGaA) was used to label the exosomes with a
green fluorescent dye, according to the manufacturer's
instructions. Briefly, 700 µl MCF-7/TAX cell-derived exosomes was
added to 1,300 µl Diluent C, and then 2 ml Diluent C with 16 µl
PKH67 was added. After mixing, the mixture was incubated at room
temperature for 5 min. Then, 4 ml 1% bovine serum albumin (BSA) was
added to eliminate the excess dye. After being centrifuged at
120,000 × g for 90 min at 4°C, the sediments were resuspended in
300 µl PBS, thereby forming a suspension of PKH67-labeled MCF-7/TAX
cell-derived exosomes.
MCF-7 cells were seeded into a 24-well plate at a
density of 3×104 cells/well and cultured overnight. The
next day, the medium was changed to serum-free medium and 10 µl
PKH67-labeled MCF-7/TAX cell-derived exosomes were added to the
cells. After co-culture for 48 h, the cells were washed with PBS
and fixed with 4% paraformaldehyde at room temperature for 20 min.
After washing with PBS, the cells were treated with 0.1% Triton
X-100 for 20 min at room temperature. After thrice washing with
PBS, 3% BSA blocking solution was added and the cells were
incubated at room temperature for 1 h. After washing, the
cytoskeleton was stained with red fluorescence by the addition of
actin red (Nanjing KeyGen Biotech Co., Ltd.) and incubation at room
temperature in the dark for 20 min. After washing, mounting medium
with DAPI (Thermo Fisher Scientific, Inc.) was added to seal the
slides, and images of the cells were captured under a laser
confocal scanning microscope (TCS SP8; Leica Microsystems,
Inc.).
Cell transfection
Cell transfection was carried out according to the
method described by Liao et al (23). In brief, MCF-7 cells were seeded in
a 24-well plate at a density of 5×104 cells/well and
cultured overnight. miR-187-5p mimics (sense:
5′-GGCTACAACACAGGACCCGGGC-3′, and antisense:
5′-CCGGGUCCUGUGUUGUAGCCTT-3′) and negative control (NC) mimics
(sense: 5′-UUCUCCGAACGUGUCACGUTT-3′, and antisense:
5′-ACGUGACACGUUCGGAGAATT-3′) were designed and provided by Yanzai
Biotechnology (Shanghai) Co., Ltd. After reaching 70% confluence,
the cells were transfected with 50 nM miR-187-5p mimics or NC
mimics using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.), following the manufacturer's protocol. After 6 h
of transfection, the medium was replaced with a complete medium and
the cells were cultured for another 48 h at room temperature. Total
RNA was extracted from the cells after transfection, and the cell
transfection efficiency was assessed by measuring the level of
miR-187-5p using reverse transcription-quantitative polymerase
chain reaction (RT-qPCR). The primers used for miR-187-5p are shown
in Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer | Sequence
(5′-3′) |
|---|
| hsa-miR-187-5p | RT:
GTCGTATCCAGTGCAGGGTC |
|
|
CGAGGTATTCGCACTGGATACGA |
|
| CGCCCGG |
|
| F:
GCGGCTACAACACAGGAC |
|
hsa-miR-106a-3p | RT:
GTCGTATCCAGTGCAGGGTC |
|
|
CGAGGTATTCGCACTGGATACGA |
|
| CGTAAGA |
|
| F:
GCCTGCAATGTAAGCACT |
| Downstream
universal primer |
GTGCAGGGTCCGAGGT |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| ABCD2 | F:
AATGGACCAGATCGAGTGCTG |
|
| R:
TGGGATAGAGGGTTTTCAGAGC |
| β-catenin | F:
AAAGCGGCTGTTAGTCACTGG |
|
| R:
CGAGTCATTGCATACTGTCCAT |
| c-Myc | F:
CCTGGTGCTCCATGAGGAGAC |
|
| R:
CAGACTCTGACCTTTTGCCAGG |
| Cyclin D1 | F:
GCTGCGAAGTGGAAACCATC |
|
| R:
CCTCCTTCTGCACACATTTGAA |
| GAPDH | F:
TGACAACTTTGGTATCGTGGA |
|
| AGG |
|
| R:
AGGCAGGGATGATGTTCTGGA |
|
| GAG |
Detection of cell viability using a
Cell Counting Kit-8 (CCK-8) assay in different treatment
groups
MCF-7 cells were seeded into a 96-well plate at a
density of 5×104 cells/well, and different
concentrations of TAX (0, 0.1, 0.2, 0.4, 0.8, 1.6, 3.2 and 6.4
µg/ml) were used to treat the cells for 48 h. The viability of the
MCF-7 cells was then determined to screen for the optimum
concentration of TAX using a CCK-8 assay (Beyotime Institute of
Biotechnology). Thereafter, the MCF-7 cells were divided into six
groups: Control, TAX, TAX + MCF-7/TAX cell-derived exosomes (TAX +
DR-Exo), TAX + MCF-7 cell-derived exosomes (TAX + DS-Exo), TAX + NC
mimics and TAX + miR-187-5p mimics. With the exception of those in
the control group, the cells were all treated with TAX for 48 h,
and then the cells in the TAX + DR-Exo, TAX + DS-Exo, TAX + NC
mimics and TAX + miR-187-5p mimics groups were treated with
MCF-7/TAX cell-derived exosomes (final concentration, 10 µg/ml) or
MCF-7 cell-derived exosomes (final concentration, 10 µg/ml), or
transfected with NC mimics or miR-185-5p mimics, respectively.
After culture for 24, 48 and 72 h, 10 µl CCK-8 reagent was added to
each well and the cells were incubated for 4 h. Finally, the
absorbance at 450 nm was measured using a microplate reader.
Cell apoptosis determined by flow
cytometry
The apoptosis of MCF-7 cells was examined using an
Annexin V-FITC/PI apoptosis assay kit (Beyotime Institute of
Biotechnology), according to the manufacturer's recommendations. In
brief, the cells in the various treatment groups were harvested and
resuspended in 1X binding buffer (100 µl). Next, 5 µl FITC-Annexin
V and 5 µl PI (50 µl/ml) were added to the cells, which were then
incubated in the dark for 15 min. Next, 400 µl 1X binding buffer
was added and a FACSCalibur flow cytometer (Becton-Dickinson and
Company) was used to analyze the cells. The total apoptotic rate
(early and late apoptosis) was calculated using CellQuest software
(version 4; Becton, Dickinson and Company).
Cell migration, invasion and colony
formation assays
Transwell assays were performed to determine the
migration and invasion of MCF-7 cells. Transwell chambers (pore
size, 8 µm; Guangzhou Jet Bio-Filtration Co., Ltd.) were used to
analyze cell migration, whereas Transwell chambers coated with
Matrigel at 37°C for 3 h were used for cell invasion. The upper
chamber contained 200 µl cell suspension (4×104 cells in
DMEM) and the lower chamber contained 500 µl DMEM containing 10%
FBS and 100 U/ml penicillin/100 µg/ml streptomycin. After 48 h of
incubation at 37°C, the chambers were removed and washed twice with
PBS. The cells were fixed with 4% paraformaldehyde at room
temperature for 20 min. After washing, the cells were stained with
crystal violet (Beyotime Institute of Biotechnology) at room
temperature for 20 min. After washing away the excess dye and
drying, the stained cells were observed and photographed under a
light microscope.
For colony formation, the cells were seeded into a
6-well plate at a density of 200 cells/well, and the cells with
different treatments were cultured in an incubator at 37°C for 14
days. When cell colonies were visible, the supernatant was
discarded and the cells were washed twice with PBS. Next, 4%
paraformaldehyde was added to fix the cells for 10 min at room
temperature. The excess paraformaldehyde was then removed and the
cells were stained with crystal violet at room temperature for 10
min. After washing and drying, images were captured and cell
colonies were counted using ImageJ software (version 1.47; National
Institutes of Health). A colony was defined as >50 cells.
Triplicate wells were analysed for each treatment group.
RT-qPCR
Total RNA was extracted from differently treated
cells using the RNAiso Plus kit (Takara Bio, Inc.) and twice the
volume of isopropanol. The purity and concentration of the
extracted total RNA were determined using a microplate reader.
Subsequently, the total RNA was reverse transcribed into cDNA using
a PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara Bio, Inc.)
following the manufacturer's instructions. For the analysis of
miRNA levels, the stem-loop method was used, as previously reported
(24), and U6 served as a reference
gene. GAPDH was used as the reference gene for mRNA expression.
RT-qPCR was performed with Power SYBR Green PCR Master Mix (Thermo
Fisher) and using the Applied Biosystems 7500 thermocycler (Applied
Biosystems). qPCR was initiated at 50°C for 2 min and proceeded
with 40 cycles at 95°C for 2 min, 95°C for 15 sec and 60°C for 60
sec. The sequences of all primers are listed in Table I, and the relative levels of
miR-187-5p, miR-106a-3p, ATP binding cassette subfamily D member 2
(ABCD2), β-catenin, c-Myc and cyclin D1 were calculated
using the 2−ΔΔCq method (25).
Western blotting
Total protein was isolated from differently treated
cells using RIPA lysis buffer (Beyotime Institute of
Biotechnology), and the total protein concentration was measured
using a BCA protein assay kit. Subsequently, the protein samples
(20 µg/lane) were separated using 10% SDS-PAGE, transferred to PVDF
membranes and blocked with 5% skimmed milk. After blocking at 37°C
for 1 h, the membranes were incubated with anti-β-catenin
(catalogue number, 17565-1-AP; 1:1,000), anti-c-Myc (catalogue
number, 10828-1-AP; 1:1,000), anti-cyclin D1 (catalogue number,
26939-1-AP; 1:1,000) and anti-GAPDH (catalogue number, 10494-1-AP;
1:2,000) antibodies, all from ProteinTech Group, Inc., or the
aforementioned anti-HSP70, anti-TSG101, anti-CD9 and anti-calnexin
antibodies at 4°C overnight. After washing, the membranes were
incubated with goat anti-rabbit/mouse IgG (H+L)-HRP secondary
antibody (catalogue number, 111-035-003/115-035-003; 1:5,000;
Jackson ImmunoResearch Laboratories, Inc.) at 37°C for 2 h. After
washing with PBST (1,000 ml 1X PBS + 1 ml Tween-20) five times, the
protein bands were visualized using a Millipore ECL system (Tanon
Science & Technology Co., Ltd.). The protein bands were
analyzed using Image-Pro Plus software (version 6.0; Media
Cybernetics Imaging Technologies Inc.).
Dual-luciferase reporter gene
assay
The TargetScan online tool (https://www.targetscan.org/vert_71/) was used to
predict the target of miR-187-5p, and a dual-luciferase reporter
gene assay was used to analyze the interaction between miR-187-5p
and ABCD2. The miR-187-5p mimic and ABCD2
3′-untranslated region (3′-UTR) were synthesized by and purchased
from Yanzai Biotechnology (Shanghai) Co., Ltd. The pGL3-basic
vector was provided by Yanzai Biotechnology (Shanghai) Co., Ltd.
and was used to construct reporter plasmids containing the
ABCD2 3′-UTR (pGL3-ABCD2-WT) and a mutated ABCD2
3′-UTR (pGL3-ABCD2-MUT). Afterwards, the pGL3-basic vector,
pGL3-ABCD2-WT or pGL3-ABCD2-MUT (500 ng) were co-transfected into
293T cells (National Collection of Authenticated Cell Cultures)
with miR-187-5p mimic (100 nM) or NC mimic (100 nM) using
Lipofectamine 2000, according to the manufacturer's instructions.
After culture for 48 h, a dual-luciferase reporter system (Promega
Corporation) was used to measure the luminescent signal in relative
light units. Renilla luciferase activity was normalized to
firefly luciferase activity.
Statistical analysis
Results are reported as the mean ± standard
deviation of at least three independent samples. GraphPad Prism 5
(GraphPad Software, Inc.) was used to perform the statistical
analyses. Comparisons among three or more groups were performed
using one-way analysis of variance followed by Tukey's post hoc
test to analyze the pairwise differences. P<0.05 was considered
to indicate a statistically significant difference.
Results
Characterization of MCF-7 and
MCF-7/TAX cell-derived exosomes
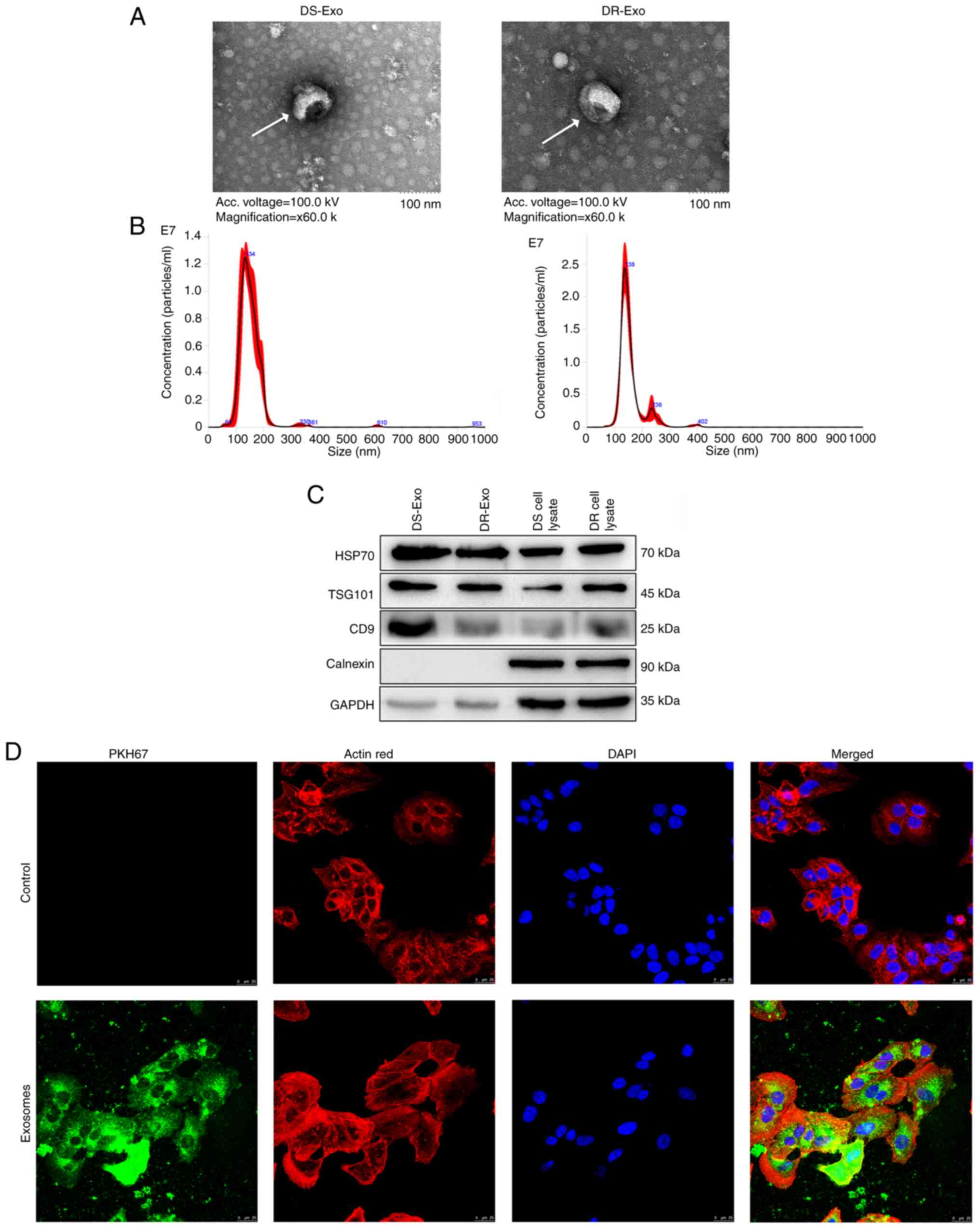
Exosomes isolated from MCF-7 and MCF-7/TAX cells
were characterized using TEM, NTA and western blotting. The TEM
results revealed that the morphology of the exosomes isolated from
both types of cells was cup-shaped and approximately round with a
diameter of ~100 nm (Figs. 1A and
S1). NTA showed that the major
peaks of the substances from the MCF-7 and MCF-7/TAX cells occurred
at sizes of ~134 and 138 nm, respectively (Fig. 1B), which was in accordance with the
size distribution of exosomes reported previously (26). Additionally, western blotting
revealed that HSP70, TSG101 and CD9, which are exosome-specific
markers, were all expressed in the exosomes (Fig. 1C). However, calnexin, which is
specific for cells, was only expressed in the cell lysate and not
expressed in the exosomes (Fig.
1C). These results indicate that exosomes were successfully
isolated from the MCF-7 and MCF-7/TAX cells.
To observe the uptake of the exosomes isolated from
MCF/TAX cells in MCF-7 cells, PKH67 was used to stain the exosomes
green (Fig. S2), actin red was
used to stain the cytoskeleton red and the MCF-7 cell nuclei were
stained blue using DAPI. Following co-culture for 48 h, green
fluorescence was observed in the MCF-7 cells (Fig. 1D), indicating that PKH67-labeled
MCF-7/TAX cell-derived exosomes were taken up by MCF-7 cells after
the co-culture.
Selection of miRNAs and cell
transfection efficiency
With the aim of selecting miRNAs for subsequent
study, a literature search was conducted, which indicated that
miR-187-5p and miR-106a-3p are closely associated with TAX
resistance (18). Therefore,
miR-187-5p and miR-106a-3p were chosen for further validation. The
relative level of miR-187-5p was significantly higher in MCF-7/TAX
(DR) cells compared with MCF-7 (DS_ cells (P<0.05), and its
level was also significantly higher in MCF-7/TAX cell-derived
exosomes compared with MCF-7 cell-derived exosomes (P<0.05;
Fig. 2A). However, miR-106a-3p was
not detected in cells or exosomes (data not shown), which may be
due to low abundance in this cell line. Therefore, miR-187-5p was
selected for evaluation in subsequent experiments.
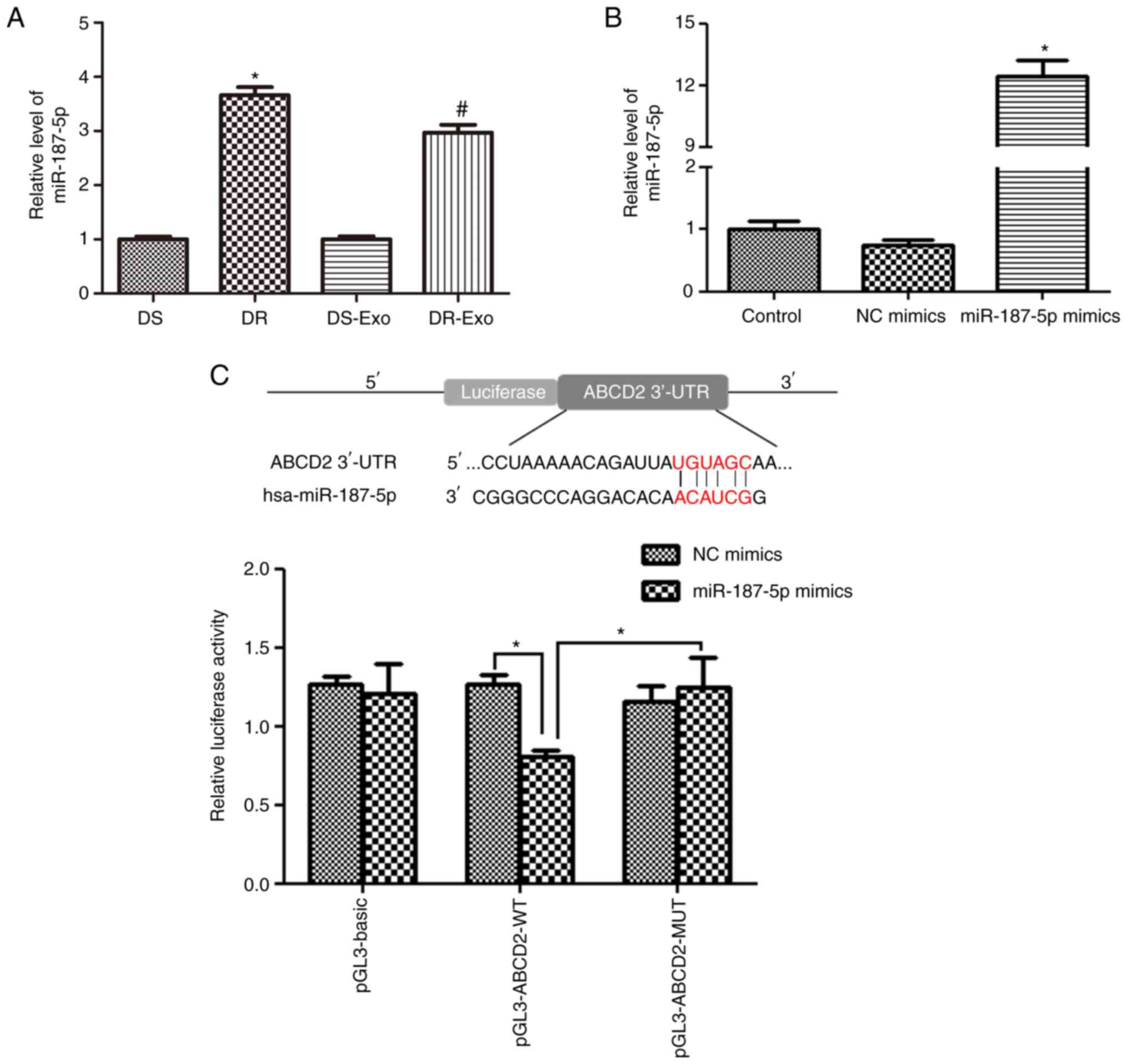 | Figure 2.Cell transfection efficiency and the
association between miR-187-5p and ABCD2. (A) Relative
levels of miR-187-5p in cells and exosomes. *P<0.05 vs. the DS
group; #P<0.05 vs. the DS-Exo group. (B) Cell
transfection efficiency of MCF-7 cells evaluated by measuring the
relative level of miR-187-5p. *P<0.05 vs. the control group. (C)
ABCD2 was confirmed to directly bind with miR-187-5p by a
dual-luciferase reporter gene assay. *P<0.05 as indicated. DS,
drug sensitive MCF-7 cells; DR, drug resistant MCF/TAX cells; Exo,
exosomes; control, untransfected MCF-7 cells; NC, negative control;
miR, microRNA; pGL3-basic, empty vector; pGL3-ABCD2-WT, wild-type
3′-UTR ABCD2 reporter plasmid; pGL3-ABCD2-MUT, mutated
3′-UTR ABCD2 reporter plasmid; UTR, untranslated region;
ABCD2, ATP binding cassette subfamily D member 2. |
MCF-7 cells with miR-187-5p enrichment were
constructed by transfection with miR-187-5p mimics, and the level
of miR-187-5p was determined by RT-qPCR to evaluate the
transfection efficiency. No significant difference in miR-187-5p
levels was detected between the control and NC mimics groups
(P>0.05; Fig. 2B). Compared with
the control group, the relative level of miR-187-5p in the
miR-187-5p mimics group was increased to 12.46±0.78, which was
significantly elevated compared with that in the control and NC
mimics groups (P<0.05; Fig. 2B).
These results indicate that MCF-7 cells with miR-187-5p enrichment
were successfully established and were suitable for further
experiments.
ABCD2 directly binds with
miR-187-5p
To explore the downstream regulatory mechanism of
miR-187-5p, the online tool TargetScan was used to predict the
target gene of miR-187-5p. ABCD2 was identified as a
potential target of miR-187-5p owing to the 3′-UTR of ABCD2
including a binding site for miR-187-5p (Fig. 2C). A dual-luciferase reporter gene
assay was then performed to confirm the relationship between
ABCD2 and miR-187-5p. In the cells transfected with
pGL3-basic vector, no significant difference was found in the
relative luciferase activity between the NC mimics and miR-187-5p
mimics (P>0.05; Fig. 2C).
However, in the cells transfected with pGL3-ABCD2-WT, the relative
luciferase activity was significantly reduced after co-transfection
with miR-187-5p mimics compared with co-transfection with NC mimics
(P<0.05). When ABCD2 was mutated (pGL3-ABCD2-MUT
plasmid), the relative luciferase activity after co-transfection
with miR-187-5p mimics increased significantly compared with that
observed for pGL3-ABCD2-WT and miR-187-5p mimics co-transfection
(P<0.05), and was restored to a level similar to that achieved
with NC mimics (P>0.05; Fig.
2C). These results suggest that ABCD2 directly binds to
miR-187-5p.
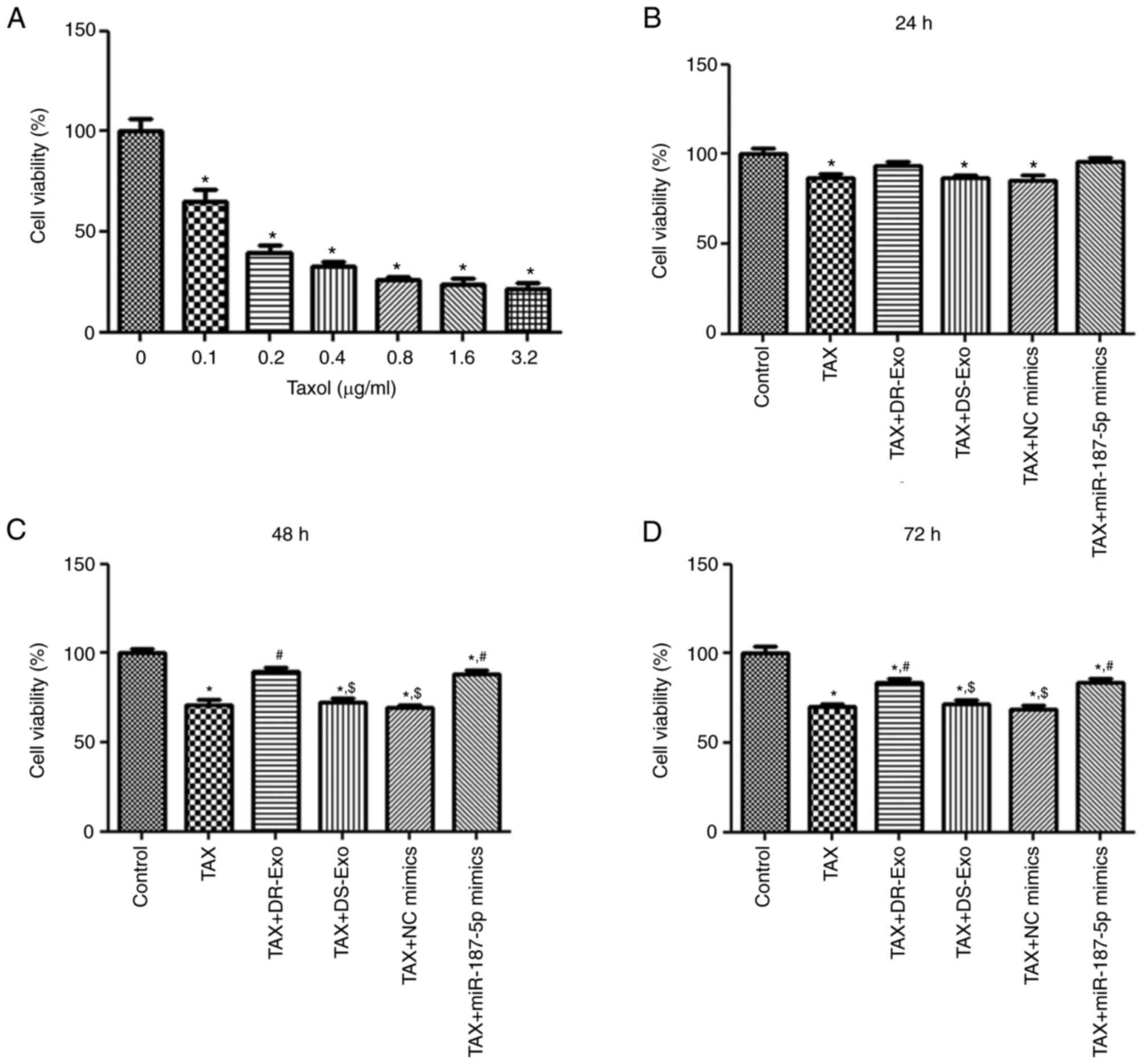
Screening for the optimal TAX
concentration and cell viability analysis
To determine the optimal TAX concentration,
different concentrations of TAX were used to treat MCF-7 cells and
the viability of the cells was measured using a CCK-8 assay. The
viability of cells treated with TAX concentrations of 0.1, 0.2,
0.4, 0.8, 1.6 and 3.2 µg/ml was significantly lower than that of
cells that did not receive TAX treatment (P<0.05; Fig. 3A). On the premise that MCF-7 cells
treated with 0.1 µg/ml TAX had the highest viability among all the
treatment groups, this concentration was selected for
administration to the MCF-7 cells in subsequent experiments.
To understand the role of MCF-7/TAX cell-derived
exosomal miR-187-5p in the viability of TAX-induced MCF-7 cells,
the viability of TAX-induced MCF-7 cells that were treated with
MCF-7/TAX cell-derived (DR) exosomes or transfected with miR-187-5p
mimics was determined. After culture for 24 h, the viability of
MCF-7 cells in the TAX, TAX + DS-Exo and TAX + NC mimics groups was
significantly lower than that in the control group (P<0.05),
while there was no significant difference in cell viability among
the TAX, TAX + DR-Exo, TAX + DS-Exo and TAX + miR-187-5p mimics
groups (P>0.05; Fig. 3B). After
culture for 48 and 72 h, the cell viability of the TAX + DR-Exo and
TAX + miR-187-5p mimics groups was significantly higher compared
with that of the TAX group (P<0.05), and no significant
differences were observed between the TAX + DR-Exo and TAX +
miR-187-5p mimics, as well as among the TAX, TAX + DS-Exo and TAX +
NC mimics groups (P>0.05; Fig. 3C
and D). These results indicate that TAX inhibited the viability
of MCF-7 cells, and that MCF-7/TAX cell-derived exosomes and
miR-187-5p enrichment reversed the TAX-induced loss in viability of
the MCF-7 cells.
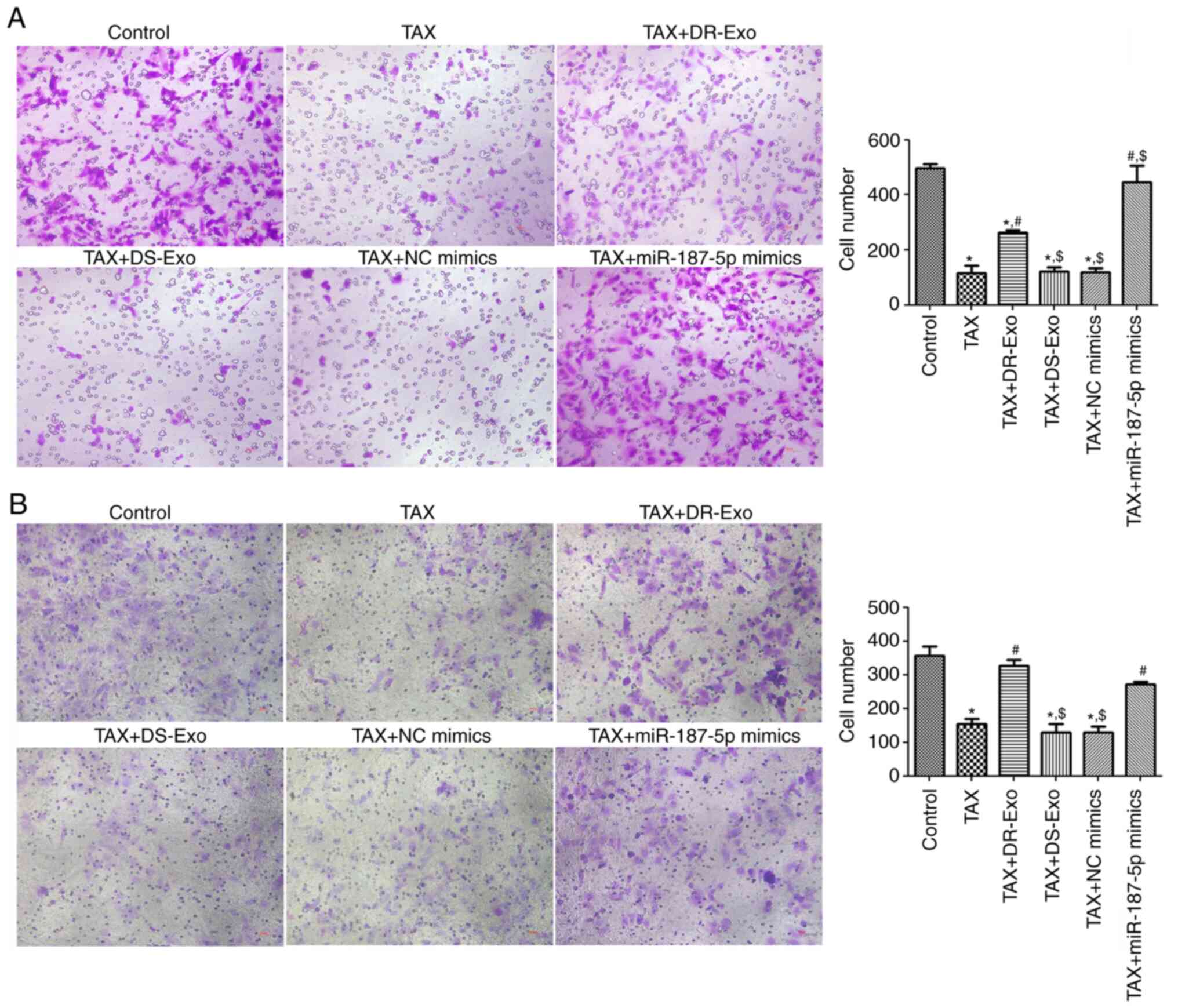
Cell migration and invasion
analyses
To investigate further the effects of MCF-7/TAX
cell-derived exosomal miR-187-5p on the migration and invasion of
TAX-induced MCF-7 cells, Transwell assays were performed to test
the migration and invasion of MCF-7 cells induced by TAX. The
numbers of migrated and invaded cells in the TAX group were
significantly lower compared with those in the control group
(P<0.05; Fig. 4). Compared with
cells treated only with TAX, the numbers of migrated and invaded
cells treated with MCF-7/TAX cell-derived exosomes or transfected
with miR-187-5p mimics were significantly higher after TAX
treatment (P<0.05), whereas the numbers of migrated and invaded
cells did not change significantly following treatment with MCF-7
cell-derived exosomes or transfection with NC mimics (P>0.05;
Fig. 4). In terms of migrated
cells, the cell number in the TAX + miR-187-5p mimics group was
significantly higher than that in the TAX + DR-Exo group
(P<0.05; Fig. 4A). In terms of
invaded cells, no significant difference in the cell number was
detected between these two groups (P>0.05; Fig. 4B). These results imply that TAX
suppressed the migration and invasion of MCF-7 cells, while
MCF-7/TAX cell-derived exosomes and miR-187-5p enrichment
attenuated the effects of TAX. In addition, the effects of
miRNA-187-5p and DR-Exo of MCF-7 cell growth may contribute to the
difference in migration and invasion.
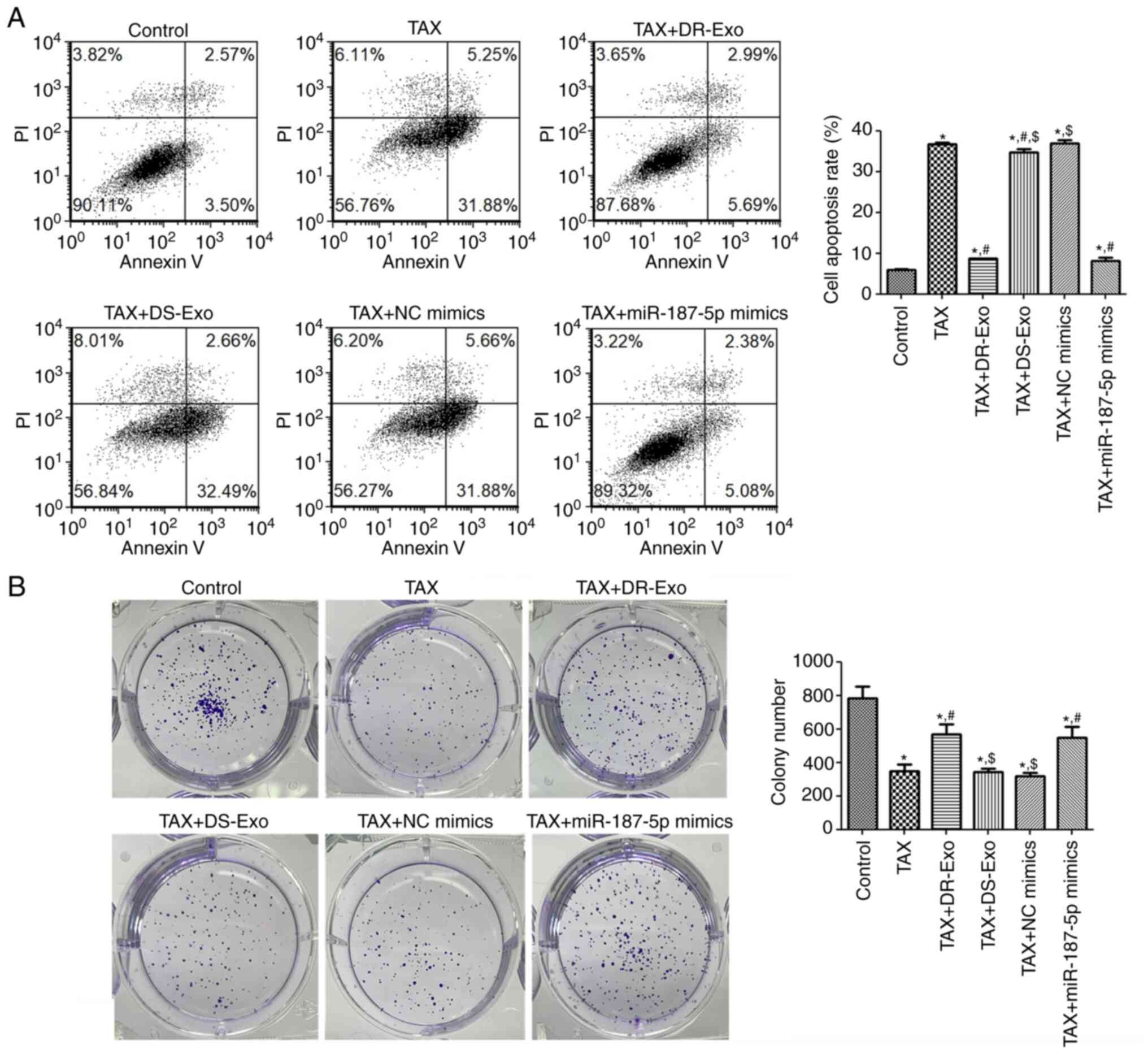
Cell apoptosis and colony formation
analyses
The apoptosis of MCF-7 cells with different
treatments was determined using flow cytometry to investigate the
actions of MCF-7/TAX cell-derived exosomal miR-187-5p on the
apoptosis of TAX-induced MCF-7 cells. The cell apoptosis rate was
significantly enhanced after TAX treatment compared with that in
the control group (P<0.05), and no significant difference was
found in the cell apoptosis rate between the TAX and TAX + NC
mimics groups (P>0.05; Fig. 5A).
The MCF-7/TAX cell-derived exosomes, MCF-7 cell-derived exosomes
and miR-187-5p mimics significantly reduced the TAX-induced cell
apoptosis rate compared with that in the TAX group (P<0.05),
with the reduction induced by the MCF-7/TAX cell-derived exosomes
and miR-187-5p mimics being particularly evident (P<0.05;
Fig. 5A). These results indicate
that TAX induced the apoptosis of MCF-7 cells, whereas MCF-7/TAX
cell-derived exosomes and miR-187-5p enrichment attenuated the
apoptosis caused by TAX.
Thereafter, the colony formation of MCF-7 cells
subjected to different treatments was examined. The number of MCF-7
cell colonies formed after treatment with TAX decreased
significantly compared with that in the control group, (P<0.05;
Fig. 5B). No significant
differences in colony numbers were detected among the TAX, TAX +
DS-Exo and TAX + NC mimics groups (P>0.05; Fig. 5B). In addition, the colony numbers
in the TAX + DR-Exo and TAX + miR-187-5p mimics groups were
significantly higher than those in the TAX group (P<0.05), and
significantly lower than those in the control group (P<0.05;
Fig. 5B). These results show that
TAX inhibited the colony formation of MCF-7 cells, whereas
MCF-7/TAX cell-derived exosomal miR-187-5p promoted TAX-induced
MCF-7 colony formation.
Effects of exosomal miR-187-5p on the
expression of ABCD2, β-catenin, c-Myc and cyclin D1
To investigate the molecular mechanisms of exosomal
miR-187-5p involvement in the TAX-induced effects on MCF-7 cells,
the expression levels of ABCD2, β-catenin, c-Myc and cyclin D1 were
determined using RT-qPCR and western blotting. RT-qPCR showed that
TAX significantly upregulated ABCD2 mRNA expression compared
with that in the control group (P<0.05), and MCF-7 cell-derived
exosomes and NC mimics did not significantly alter the increase in
ABCD2 mRNA expression caused by TAX (P>0.05; Fig. 6A). However, the ABCD2 mRNA
expression following TAX treatment was significantly downregulated
by MCF-7/TAX cell-derived exosomes and miR-187-5p mimics compared
with TAX alone (P<0.05), and was reduced to a level similar to
that of the control group (P>0.05; Fig. 6A). The trend in β-catenin mRNA
expression in the different groups was opposite to that of
ABCD2 mRNA expression (Fig.
6B). The mRNA expression of c-Myc was significantly
downregulated after TAX treatment compared with that in the control
group (P<0.05) and did not significantly change after treatment
with MCF-7 cell-derived exosomes or NC mimics transfection compared
with that in the TAX group (P>0.05; Fig. 6C). However, the mRNA expression of
c-Myc was significantly upregulated by MCF-7/TAX cell-derived
exosomes and miR-187-5p mimics compared with that in the TAX group
(P<0.05), and the action of miR-187-5p mimics was significantly
greater compared with that of the TAX + DR-Exo group (P<0.05;
Fig. 6C). Cyclin D1 mRNA expression
was significantly downregulated in the cells treated with TAX
compared with that in the control group (P<0.05) and was also
significantly downregulated in the TAX-induced cells treated with
MCF-7 cell-derived exosomes and NC mimics (P<0.05); no
significant differences were detected among the TAX, TAX + DS-Exo
and TAX + NC mimics groups (P>0.05; Fig. 6D). However, when the TAX-induced
cells were treated with MCF-7/TAX cell-derived exosomes or
transfected with miR-187-5p mimics, cyclin D1 mRNA expression was
significantly upregulated compared with that in cells only treated
with TAX (P<0.05; Fig. 6D).
Additionally, the protein levels of β-catenin, c-Myc and cyclin D1
were detected using western blotting, and representative protein
bands are shown in Fig. 6E. The
quantitative analysis showed that the trends in β-catenin, c-Myc
and cyclin D1 protein expression in the different groups detected
by western blotting were similar to those of β-catenin, c-Myc and
cyclin D1 mRNA expression measured by RT-qPCR (Fig. 6F-H).
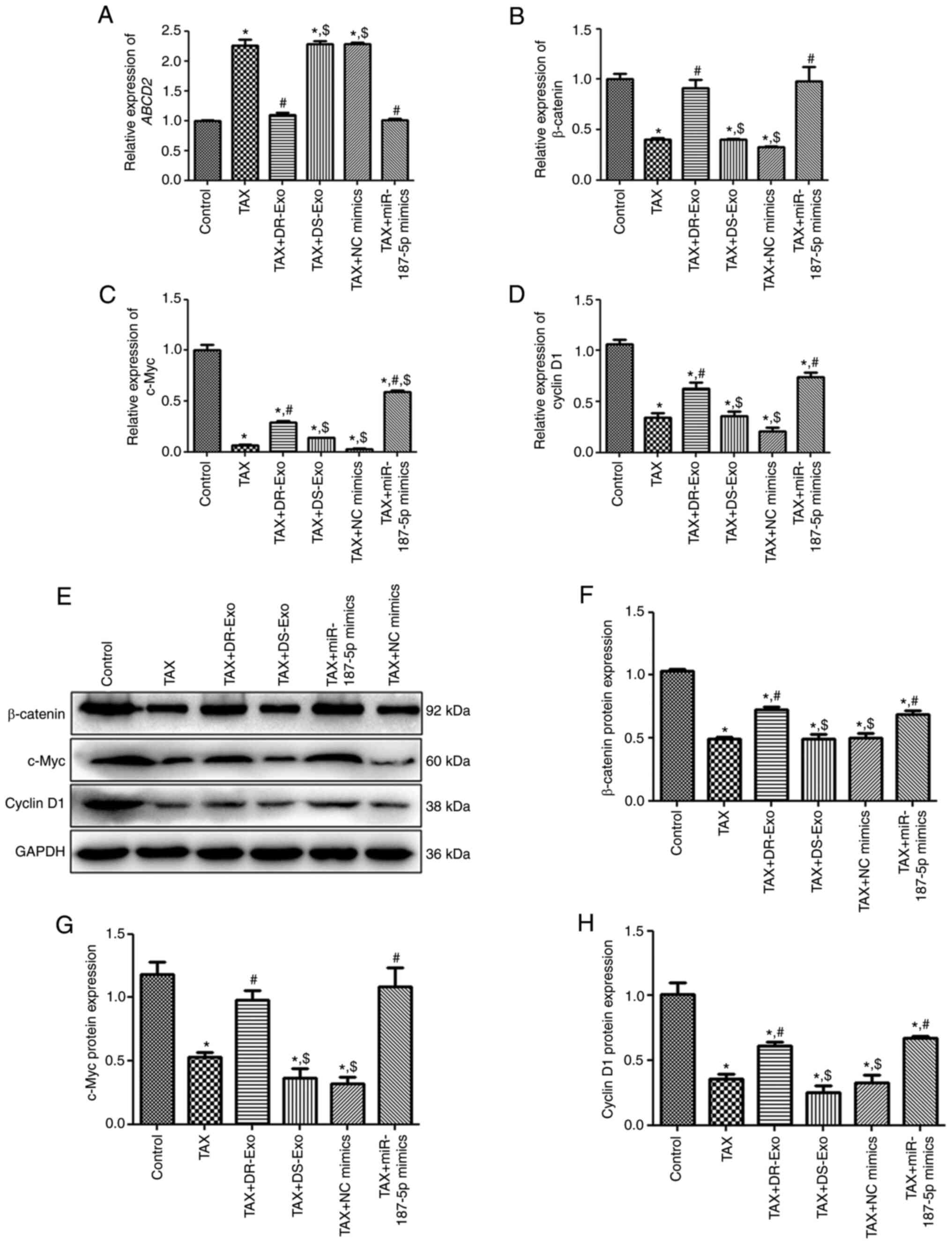 | Figure 6.Effects of exosomal miR-187-5p on the
expression of ABCD2, β-catenin, c-Myc and cyclin D1 in MCF-7 cells.
MCF-7 cells were treated with TAX for 48 h and various treatments
or transfections were applied. Relative mRNA expression of (A)
ABCD2, (B) β-catenin, (C) c-Myc and (D) cyclin D1 in MCF-7
cells with different treatments determined by
reverse-transcription-quantitative PCR. (E) Representative images
of β-catenin, c-Myc and cyclin D1 western blots. Quantified protein
levels of (F) β-catenin, (G) c-Myc and (H) cyclin D1 in MCF-7 cells
with different treatments. *P<0.05 vs. the control group;
#P<0.05 vs. the TAX group; $P<0.05 vs.
the TAX + DR-Exo group. miR, microRNA; ABCD2, ATP binding cassette
subfamily D member 2; TAX, Taxol; control, untreated MCF-7 cells;
DS-Exo, drug sensitive MCF-7 cell-derived exosomes; DR-Exo, drug
resistant MCF/TAX cell-derived exosomes; NC, negative control. |
Discussion
Breast cancer is the most frequently occurring
cancer in women and the second most common cancer worldwide
(27). TAX is a first-line
chemotherapeutic agent for the treatment of patients with breast
cancer (28). However, long-term
TAX chemotherapy causes patients with breast cancer to acquire TAX
resistance (29), which contributes
to clinical treatment failure and markedly reduces the survival
rate. Exosomes, particularly tumor-derived exosomes, have been
reported to deliver miRNAs that can confer drug resistance to
target cells and play essential roles in the drug resistance of
tumors (11,30). Therefore, in the present study,
exosomes were extracted from MCF-7 and MCF-7/TAX cells so that
their effects could be investigated. Western blotting showed that
exosome-specific markers HSP70, TSG101 and CD9 were all expressed
in the exosomes and cell lysate, which indicated that a large
number of exosomes were present. The difference in the expression
levels of CD9 observed between the exosomes and cell lysate may be
due to the source of cells, and the presence of TAX. Based on the
presence of these markers combined with the results of NTA and TEM,
it can be inferred that exosomes were successfully isolated from
MCF-7 and MCF-7/TAX cells. After that, the levels of miR-187-5p and
miR-106a-3p were measured in the exosomes and cells. The results
revealed that miR-187-5p was significantly enriched in MCF-7/TAX
cells and MCF-7/TAX cell-derived exosomes, which indicated that the
resistance MCF-7 cells to TAX was closely associated with
miR-187-5p enrichment.
There is evidence that the exosome-mediated transfer
of miRNAs alters the drug resistance of cancer cells, and is a
potential biomarker for the progression and treatment of cancer
(30). A study by Santos et
al (31) showed that resistant
cell-derived exosomes were enriched in miR-155, which inhibited the
migration ability of breast cancer cells and promoted
chemoresistance in sensitive cells by exosomal transfer. Another
study indicated that tumor-associated macrophage-derived exosomes
transmitted miR-21 to gastric cancer cells, which increased the
cell viability and colony formation of the cancer cells and
restrained their apoptosis, thus endowing them with cisplatin
resistance (32). To explore the
roles of MCF-7/TAX cell-derived exosomes in the delivery of
miR-187-5p during the treatment of MCF-7 cells with TAX, MCF-7
cells were treated with TAX and then either treated with MCF-7/TAX
cell-derived exosomes or transfected with miR-187-5p mimics.
Treatment with TAX significantly inhibited the viability,
migration, invasion and colony formation of MCF-7 cells and
promoted their apoptosis. However, MCF-7/TAX cell-derived exosomes
and miR-187-5p mimics reversed the TAX-induced changes in cell
viability, apoptosis, migration, invasion and colony formation.
Furthermore, the growth-inducing effects of miRNA-187-5p and
MCF-7/TAX cells-derived exosomes on MCF-7 cells may increase their
migration and invasion. TAX has been shown to be effective against
various cancers via the repression of cancer cell viability and
induction of apoptosis (33,34).
Additionally, a study reported that miR-187-5p is an oncogene that
increased the risk of recurrence in bladder cancer, while its
overexpression promoted the proliferation and mobility of bladder
cancer cells and repressed their apoptosis (35), implying that miR-187-5p may play an
important role in the development of cancers. Based on the results
of the present study, it may be speculated that TAX-resistant MCF-7
cell-derived exosomes regulate the growth of TAX-induced MCF-7
cells via the delivery of miR-187-5p, thus affecting the drug
resistance of breast cancer cells.
A study by Hu et al (36) showed that exosomes secreted by
colorectal cancer cells activated the Wnt/β-catenin signaling
pathway by promoting the stabilization and nuclear translocation of
β-catenin, resulting in resistance to 5-fluorouracil and
oxaliplatin. Another study demonstrated that exosomes released from
cisplatin-resistant A549 human lung cancer cells induced
therapeutic resistance by upregulating mTOR expression (37). These reports suggest that exosomes
affect various signal transduction pathways and thereby regulate
drug resistance (30). Moreover,
the Wnt/β-catenin signaling pathway, which induces EMT, invasion
and metastasis, has been reported to serve an important role in the
development and progression of breast cancer (38). Therefore, in the present study, the
effects of the Wnt/β-catenin signaling pathway on TAX resistance in
breast cancer were investigated. The results showed that TAX
significantly upregulated ABCD2 and downregulated β-catenin, c-Myc
and cyclin D1 expression compared with that in untreated MCF-7
cells, whereas the MCF-7/TAX cell-derived exosomes and miR-187-5p
mimics significantly attenuated the effects of TAX. A
dual-luciferase reporter gene assay confirmed that miR-187-5p
directly bound to the ABCD2 UTR. ATP-binding box (ABC)
transporters promote the development of anticancer drug resistance
via ATP-dependent drug efflux. ABCD2 is a type of ABC transporter
protein that is overexpressed in patients with breast cancer
receiving neoadjuvant chemotherapy (39) and is associated with the progression
and prognosis of breast cancer (40). Overall, it can be inferred that
TAX-resistant MCF-7 cell-derived exosomal miR-187-5p mediates TAX
resistance in breast cancer cells via the direct targeting of
ABCD2.
Park et al (41) reported that ABCD2 is a direct target
of β-catenin, indicating that ABCD2 may be involved in the
Wnt/β-catenin signaling pathway. β-catenin is the core
transcription factor of the Wnt/β-catenin signaling pathway; it is
a key factor in the transmission of signals to the nucleus and the
initiation of Wnt-specific gene transcription, which may contribute
to the specificity of various cells and tissues (42). Cyclin D1 is a target gene of the
Wnt/β-catenin signaling pathway that drives cancer cell
proliferation, and its activation is closely associated with poor
clinical prognosis in breast cancer (43). Cyclin D1 is necessary for the
proliferation of tamoxifen-resistant breast cancer cells, and its
overexpression is associated with poor outcomes in patients with
breast cancer treated with tamoxifen (44). The protein product of the oncogene
c-Myc is an essential downstream effector of cell proliferation
induced by the Wnt/β-catenin signaling pathway, and c-Myc is
usually upregulated in various cancers (45). Xu et al (46) demonstrated that the
LINC01094/miR-577 axis regulates the expression of β-catenin, c-Myc
and cyclin D1 in ovarian cancer cells, thereby affecting the
proliferation, migration, invasion and EMT of these cells. Another
study showed that aspirin overcame tamoxifen resistance in estrogen
receptor-positive breast cancer by downregulating c-Myc and cyclin
D1 proteins (47). These reports,
together with the results of the present study, support the
hypothesis that TAX-resistant MCF-7 cell-derived exosome-delivered
miR-187-5p regulates the viability, apoptosis, migration, invasion
and colony formation of TAX-induced cells through the
c-Myc/Wnt/β-catenin signaling pathway, thereby contributing to TAX
resistance in breast cancer cells.
In conclusion, TAX-resistant cell-secreted
exosome-delivered miR-187-5p promoted the viability, migration,
invasion and colony formation of TAX-induced breast cancer cells
while suppressing their apoptosis, thus conferring TAX resistance.
The TAX resistance mechanism of exosomal miR-187-5p in breast
cancer may be associated with the direct targeting of ABCD2
expression and the c-Myc/Wnt/β-catenin signaling pathway.
Additional experiments using TAX-resistant breast cancer cells as
positive controls will be conducted in the future to further
validate the experimental results. The findings of the present
study reveal the underlying mechanism of TAX resistance in breast
cancer and suggest that exosomal miR-187-5p/ABCD2 and Wnt/β-catenin
signaling are new potential targets and pathways for the treatment
of patients with breast cancer who are resistant to TAX-based
chemotherapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TW and QZ designed the experiments; TW, DZ, XW and
NZ performed the experiments and analyzed the experimental results;
QZ supervised the experiments; TW drafted the paper and QZ revised
the manuscript. QZ and TW confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
McDonald ES, Clark AS, Tchou J, Zhang P
and Freedman GM: Clinical diagnosis and management of breast
cancer. J Nucl Med. 57 (Suppl 1):9S–16S. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gupta N, Gupta P and Srivastava SK:
Penfluridol overcomes paclitaxel resistance in metastatic breast
cancer. Sci Rep. 9:50662019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hassan MS, Ansari J, Spooner D and Hussain
SA: Chemotherapy for breast cancer (Review). Oncol Rep.
24:1121–1131. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ghersi D, Willson ML, Chan MM, Simes J,
Donoghue E and Wilcken N: Taxane-containing regimens for metastatic
breast cancer. Cochrane Database Syst Rev.
10:CD0033662015.PubMed/NCBI
|
|
6
|
Megerdichian C, Olimpiadi Y and Hurvitz
SA: nab-Paclitaxel in combination with biologically targeted agents
for early and metastatic breast cancer. Cancer Treat Rev.
40:614–625. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bukowski K, Kciuk M and Kontek R:
Mechanisms of multidrug resistance in cancer chemotherapy. Int J
Mol Sci. 21:32332020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Koual M, Tomkiewicz C, Cano-Sancho G,
Antignac JP, Bats AS and Coumoul X: Environmental chemicals, breast
cancer progression and drug resistance. Environ Health. 19:1172020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang X, Li Z, Cui Y, Cui X, Chen C and
Wang Z: Exosomes isolated from bone marrow mesenchymal stem cells
exert a protective effect on osteoarthritis via lncRNA
LYRM4-AS1-GRPR-miR-6515-5p. Front Cell Dev Biol. 9:6443802021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang L and Yu D: Exosomes in cancer
development, metastasis, and immunity. Biochim Biophys Acta Rev
Cancer. 1871:455–468. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Crow J, Atay S, Banskota S, Artale B,
Schmitt S and Godwin AK: Exosomes as mediators of platinum
resistance in ovarian cancer. Oncotarget. 8:11917–11936. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang F, Ning Z, Ma L, Liu W, Shao C, Shu Y
and Shen H: Exosomal miRNAs and miRNA dysregulation in
cancer-associated fibroblasts. Mol Cancer. 16:1482017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Li S, Li L, Li M, Guo C, Yao J
and Mi S: Exosome and exosomal microRNA: Trafficking, sorting, and
function. Genomics Proteomics Bioinformatics. 13:17–24. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Geretto M, Pulliero A, Rosano C, Zhabayeva
D, Bersimbaev R and Izzotti A: Resistance to cancer
chemotherapeutic drugs is determined by pivotal microRNA
regulators. Am J Cancer Res. 7:1350–1371. 2017.PubMed/NCBI
|
|
16
|
Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T,
Wen H, Yang Y, Wang S, Wang J, et al: Exosomal microRNA-32-5p
induces multidrug resistance in hepatocellular carcinoma via the
PI3K/Akt pathway. J Exp Clin Cancer Res. 37:522018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fornari F, Pollutri D, Patrizi C, Bella
TL, Marinelli S, Gardini AS, Marisi G, Toaldo MB, Baglioni M,
Salvatore V, et al: In hepatocellular carcinoma miR-221 modulates
sorafenib resistance through inhibition of caspase-3-mediated
apoptosis. Clin Cancer Res. 23:3953–3965. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Uhr K, Prager-van der Smissen WJC, Heine
AAJ, Ozturk B, van Jaarsveld MTM, Boersma AWM, Jager A, Wiemer EAC,
Smid M, Foekens JA and Martens JWM: MicroRNAs as possible
indicators of drug sensitivity in breast cancer cell lines. PLoS
One. 14:e02164002019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thery C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
supernatants and biological fluids. Curr Protoc Cell Biol Chapter
3. Unit 3.22. 2006.PubMed/NCBI
|
|
20
|
Lee YS, Kim SY, Ko E, Lee JH, Yi HS, Yoo
YJ, Je J, Suh SJ, Jung YK, Kim JH, et al: Exosomes derived from
palmitic acid-treated hepatocytes induce fibrotic activation of
hepatic stellate cells. Sci Rep. 7:37102017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martins TS, Catita J, Rosa IK, Silva OA
and Henriques AG: Exosome isolation from distinct biofluids using
precipitation and column-based approaches. PLoS One.
13:e01988202018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH,
Chen C, Li H, Li P, Quinn D, Dao M, et al: Isolation of exosomes
from whole blood by integrating acoustics and microfluidics. Proc
Natl Acad Sci USA. 114:10584–10589. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liao Z, Chen Y, Duan C, Zhu K, Huang R,
Zhao H, Hintze M, Pu Q, Yuan Z, Lv L, et al: Cardiac telocytes
inhibit cardiac microvascular endothelial cell apoptosis through
exosomal miRNA-21-5p-targeted cdip1 silencing to improve
angiogenesis following myocardial infarction. Theranostics.
11:268–291. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang C, Liu X, Zhao K, Zhu Y, Hu B, Zhou
Y, Wang M, Wu Y, Zhang C, Xu J, et al: miRNA-21 promotes
osteogenesis via the PTEN/PI3K/Akt/HIF-1alpha pathway and enhances
bone regeneration in critical size defects. Stem Cell Res Ther.
10:652019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Park ST and Kim J: Trends in
next-generation sequencing and a new era for whole genome
sequencing. Int Neurourol J. 20:S76–S83. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kolak A, Kaminska M, Sygit K, Budny A,
Surdyka D, Kukiełka-Budny B and Burdan F: Primary and secondary
prevention of breast cancer. Ann Agric Environ Med. 24:549–553.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Denkert C, Liedtke C, Tutt A and von
Minckwitz G: Molecular alterations in triple-negative breast
cancer-the road to new treatment strategies. Lancet. 389:2430–2442.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang S, Zhang H, Ghia EM, Huang J, Wu L,
Zhang J, Lam S, Lei Y, He J, Cui B, et al: Inhibition of
chemotherapy resistant breast cancer stem cells by a ROR1 specific
antibody. Proc Natl Acad Sci U S A. 116:1370–1377. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mashouri L, Yousefi H, Aref AR, Ahadi AM,
Molaei F and Alahari SK: Exosomes: composition, biogenesis, and
mechanisms in cancer metastasis and drug resistance. Mol Cancer.
18:752019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Santos JC, Lima NDS, Sarian LO, Matheu A,
Ribeiro ML and Derchain SFM: Exosome-mediated breast cancer
chemoresistance via miR-155 transfer. Sci Rep. 8:8292018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie
G, Ma Y and Shen L: Exosomal transfer of tumor-associated
macrophage-derived miR-21 confers cisplatin resistance in gastric
cancer cells. J Exp Clin Cancer Res. 36:532017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kampan NC, Madondo MT, McNally OM, Quinn M
and Plebanski M: Paclitaxel and its evolving role in the management
of ovarian cancer. Biomed Res Int. 2015:4130762015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Weaver BA: How Taxol/paclitaxel kills
cancer cells. Mol Biol Cell. 25:2677–2681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Z, Lin C, Zhao L, Zhou L, Pan X, Quan
J, Peng X, Li W, Li H, Xu J, et al: Oncogene miR-187-5p is
associated with cellular proliferation, migration, invasion,
apoptosis and an increased risk of recurrence in bladder cancer.
Biomed Pharmacother. 105:461–469. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hu YB, Yan C, Mu L, Mi YL, Zhao H, Hu H,
Li XL, Tao DD, Wu YQ, Gong JP and Qin JC: Exosomal Wnt-induced
dedifferentiation of colorectal cancer cells contributes to
chemotherapy resistance. Oncogene. 38:1951–1965. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qin X, Yu S, Zhou L, Shi M, Hu Y, Xu X,
Shen B, Liu S, Yan D and Feng J: Cisplatin-resistant lung cancer
cell-derived exosomes increase cisplatin resistance of recipient
cells in exosomal miR-100-5p-dependent manner. Int J Nanomedicine.
12:3721–3733. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dey N, Barwick BG, Moreno CS,
Ordanic-Kodani M, Chen Z, Oprea-Ilies G, Tang W, Catzavelos C,
Kerstann KF, Sledge GW Jr, et al: Wnt signaling in triple negative
breast cancer is associated with metastasis. BMC Cancer.
13:5372013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hlavac V, Brynychova V, Vaclavikova R,
Ehrlichová M, Vrána D, Pecha V, Koževnikovová R, Trnková M, Gatěk
J, Kopperová D, et al: The expression profile of ATP-binding
cassette transporter genes in breast carcinoma. Pharmacogenomics.
14:515–529. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Soucek P, Hlavac V, Elsnerova K,
Vaclavikova R, Kozevnikovova R and Raus K: Whole exome sequencing
analysis of ABCC8 and ABCD2 genes associating with clinical course
of breast carcinoma. Physiol Res. 64:S549–S557. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Park CY, Kim HS, Jang J, Lee H, Lee JS,
Yoo JE, Lee DR and Kim DW: ABCD2 is a direct target of beta-catenin
and TCF-4: Implications for X-linked adrenoleukodystrophy therapy.
PLoS One. 8:e562422013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jia XX, Zhu TT, Huang Y, Zeng XX, Zhang H
and Zhang WX: Wnt/β-catenin signaling pathway regulates asthma
airway remodeling by influencing the expression of c-Myc and cyclin
D1 via the p38 MAPK-dependent pathway. Exp Ther Med. 18:3431–3438.
2019.PubMed/NCBI
|
|
43
|
Lin SY, Xia W, Wang JC, Kwong KY, Spohn B,
Wen Y, Pestell RG and Hung MC: Beta-catenin, a novel prognostic
marker for breast cancer: Its roles in cyclin D1 expression and
cancer progression. Proc Natl Acad Sci USA. 97:4262–4266. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shi Q, Li Y, Li S, Jin L, Lai H, Wu Y, Cai
Z, Zhu M, Li Q, Li Y, et al: LncRNA DILA1 inhibits Cyclin D1
degradation and contributes to tamoxifen resistance in breast
cancer. Nat Commun. 11:55132020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gosens R, Baarsma HA, Heijink IH, Oenema
TA, Halayko AJ, Meurs H and Schmidt M: De novo synthesis of
{beta}-catenin via H-Ras and MEK regulates airway smooth muscle
growth. FASEB J. 24:757–768. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu J, Zhang P, Sun H and Liu Y:
LINC01094/miR-577 axis regulates the progression of ovarian cancer.
J Ovarian Res. 13:1222020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cheng R, Liu YJ, Cui JW, Yang M, Liu XL,
Li P, Wang Z, Zhu LZ, Lu SY, Zou L, et al: Aspirin regulation of
c-myc and cyclinD1 proteins to overcome tamoxifen resistance in
estrogen receptor-positive breast cancer cells. Oncotarget.
8:30252–30264. 2017. View Article : Google Scholar : PubMed/NCBI
|