Introduction
Osteosarcoma is a rare cancer that annually affects
3–5 men per million and 2–4 women per million worldwide (1,2). Due
to its complex and unclear pathogenesis, as well as uncertain
molecular biology, the etiology of osteosarcoma is still largely
unknown (3,4); however, it is currently considered
that age, sex and race are related to the occurrence of
osteosarcoma (5). Benefiting from
the advancement and broad application of neoadjuvant and adjuvant
chemotherapy, more patients with osteosarcoma undergo limb-sparing
radical surgery (6); however, a
proportion of patients still fail to respond well or may experience
early relapse, resulting in an unsatisfactory prognosis, with a
5-year event-free survival of 59% and overall survival (OS) of 71%
in patients with osteosarcoma in Europe and America (7). Thus, identifying markers that could
forecast disease progression, relapse and/or death is beneficial
for osteosarcoma management via improved stratification of patients
and optimization of treatments (8,9).
Chaperonin-containing tailless complex polypeptide 1
subunit 6A (CCT6A) is a recently discovered oncogene, which
promotes cancer progression by facilitating the proliferation,
invasiveness and stemness and of tumor cells (10–13).
In addition, it exhibits potency as a prognostic marker in numerous
types of cancer, such as lung cancer, gastric cancer and papillary
thyroid carcinoma (14–16). Notably, CCT6A interacts with cell
division cycle 20 (CDC20), as shown by the STRING database
(https://cn.string-db.org/) and further
validated by a previous study in papillary thyroid carcinoma cancer
tissue (16). CDC20 is a well-known
oncogene that facilitates the initiation and progression of several
types of cancer, including osteosarcoma (17–21).
Therefore, it was hypothesized that CCT6A may be involved in the
pathogenesis and prognosis of osteosarcoma. Furthermore, previous
studies have reported that CCT6A regulates osteosarcoma cell
proliferation via Akt activation (22) and that CCT6A is associated with
CDC20 in patients with papillary thyroid carcinoma (16). However, these studies did not
explore the association between CCT6A and clinical/pathological
features, neoadjuvant chemotherapy response and prognosis in
patients with osteosarcoma, nor did the assess the relationship
between CCT6A and CDC20 in patients with osteosarcoma. Therefore,
these are new explorational directions that the present study aimed
to evaluate.
The present study aimed to investigate the
relationship between CCT6A and CDC20, as well as their association
with clinical features, neoadjuvant chemotherapy response and
survival in patients with osteosarcoma. In addition, the study
aimed to further explore the effects of CCT6A and CDC20 knockdown
on the malignant behaviors of osteosarcoma cells.
Materials and methods
Patients
A total of 52 patients with osteosarcoma who were
treated with surgical resection at the Center Hospital Affiliated
to Shenyang Medical College (Shenyang, China) between July 2016 and
June 2020 were retrospectively screened in the present study. The
screening criteria were as follows: i) Primary osteosarcoma newly
diagnosed by pathology; ii) Enneking stage II (23); iii) lesion at the extremities; iv)
complete data on the tumor cell necrosis rate (TCNR) assessed after
neoadjuvant therapy; v) at least one follow-up record; and vi)
available tumor specimens frozen in liquid nitrogen. The exclusion
criteria were as follows: i) Secondary osteosarcoma; ii) distant
metastasis at initial diagnosis; and iii) a prior history of other
primary cancers or malignant diseases. The present study was
approved by the Ethics Committee of the Center Hospital Affiliated
to Shenyang Medical College (approval no. 2020-09-03). The patients
or their statutory guardians provided written informed consent.
Treatment
Patients with osteosarcoma received neoadjuvant
chemotherapy according to the Instituto Orthopedic Rizzoli-Section
of Osteosarcoma/Neoadjuvant chemotherapy-5 (IOR-OS/N-5) regimen
(24). Every 6 weeks was considered
a therapy cycle (3 weeks for treatment, 3 weeks for interval before
another cycle began). After two cycles (12 weeks) of neoadjuvant
chemotherapy, patients received surgical resection. After surgery,
patients were treated with the IOR-OS/N-5 regimen or the intensive
IOR-OS/N-5 regimen (increased dose) as adjuvant therapy; the
regimen was selected based on the TCNR assessed after neoadjuvant
chemotherapy (24). Adjuvant
therapy lasted for three or four cycles.
Sample processing
A total of 52 tumor specimens and 38 paired nontumor
samples (2 cm away from the tumor) were used. The different number
of samples was due to not all the nontumor tissues being routinely
stored. All samples were frozen in liquid nitrogen, and were
accessible and available for detecting the mRNA expression levels
of CCT6A and CDC20 by reverse transcription-quantitative PCR
(RT-qPCR) assay. The kits used were as follows: RNeasy Protect Mini
Kit (Qiagen GmbH) for RNA (tissues and cells) extraction; iScript™
cDNA Synthesis Kit (Bio-Rad Laboratories, Inc.) for RT, used
according to the manufacturer's protocol; and KOD SYBR®
qPCR Mix (Toyobo Life Science) for qPCR. The thermocycling
conditions included 1 cycle of 98°C for 2 min, followed by 40
cycles of 98°C for 10 sec and 61°C for 30 sec. The mRNA expression
levels of CCT6A and CDC20 were evaluated using the
2−ΔΔCq method (25), and
GAPDH was used as an internal reference gene. The primers used were
designed according to previous studies (15,26)
and are listed as follows: CCT6A forward,
5′-TGACGACCTAAGTCCTGACTG-3′ and reverse,
5′-ACAGAACGAGGGTTGTTACATTT-3′; CDC20 forward,
5′-GCAGACATTCACCCAGCATCA-3′ and reverse,
5′-CATCCACGGCACTCAGACAG-3′; and GAPDH forward,
5′-GAGTCCACTGGCGTCTTCAC-3′ and reverse,
5′-TGCTGATGATCTTGAGGCTGTT-3′.
Additionally, the tissue samples were fixed with 10%
formalin for 24 h at room temperature and embedded by paraffin,
then cut into 4-µm sections for the detection of CCT6A and CDC20
protein expression via immunohistochemistry (IHC). The IHC assay
was performed as described in a previous study (27). The following antibodies were used:
Rabbit anti-CCT6A antibody (cat. no. ab110905; 1:100; Abcam) and
rabbit anti-CDC20 antibody (cat. no. ab155921; 1:1,000; Abcam)
primary antibodies; and goat anti-rabbit HRP-conjugated secondary
antibody (cat. no. ab205718; 1:20,000; Abcam). The stained sections
were assessed using a light microscope. The staining intensity (0–3
score) and the staining density (0–4 score) were used as parameters
to semi-quantify protein expression (0–12 score), which was
calculated by multiplying the two aforementioned scores; high
protein expression (>3) and low protein expression (≤3) were
graded (27).
Data collection and follow-up
data
The demographics and disease characteristics of the
patients with osteosarcoma were collected from the medical records.
Biochemical indexes such as lactate dehydrogenase (LDH; normal
level, ≤300 U/l) and alkaline phosphatase (ALP; normal level
>500 U/l for patients <16 years old and >150 U/l for
patients ≥16 years old) were detected as routine hospital analyses.
In addition, Huvos grade (28) was
evaluated based on the efficacy of neoadjuvant chemotherapy
(assessed by TCNR). Disease-free survival (DFS) and OS time were
calculated from the follow-up data, with a final date of February
28, 2022. DFS was calculated from surgery to disease recurrence,
new primary osteosarcoma occurrence or death. OS was calculated
from neoadjuvant chemotherapy to death.
Transfection
Saos-2 and U-2 OS osteosarcoma cells (both from
American Type Culture Collection) were cultured at 37°C with 5%
CO2 in McCoy's 5a Modified Medium containing 10% fetal
bovine serum (FBS; both from Gibco; Thermo Fisher Scientific,
Inc.). CCT6A, CDC20 and negative control (NC) small interfering RNA
(siRNA) molecules (si-CCT6A, si-CDC20 and si-NC, respectively; 50
nM) were transfected into 5×105 Saos-2 and U-2 OS cells
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 8 h at 37°C. The siRNA sequences were as
follows: si-CCT6A, sense 5′-CCCTCGTTCTGTCACATTATT-3′, antisense
5′-TAATGTGACAGAACGAGGGTT-3′; si-CDC20, sense
5′-CGGCTTCGAAATATGACCATT-3′, antisense 5′-TGGTCATATTTCGAAGCCGTT-3′;
si-NC, sense 5′-TTCTCCGAACGTGTCACGTTT-3′, antisense
5′-ACGTGACACGTTCGGAGAATT-3′. Cells cultured normally were used as
normal controls. A total of 48 h post-transfection, the mRNA
expression levels of CCT6A and CDC20 were evaluated by RT-qPCR as
aforementioned.
Western blotting
The cells were lysed in RIPA buffer (Beyotime
Institute of Biotechnology) 48 h post-transfection. The
concentration of total protein was measured by BCA kit (Beyotime
Institute of Biotechnology). The thermally denatured proteins (20
µg) were then separated by SDS-PAGE on a 4–20% precast gel
(Beyotime Institute of Biotechnology). Subsequently, the proteins
were transferred to a nitrocellulose membrane (Pall Corporation),
which was incubated with CCT6A (cat. no. 19793-1-AP; 1:1,000; Wuhan
Sanying Biotechnology), CDC20 (cat. no. 10252-1-AP; 1:10,000; Wuhan
Sanying Biotechnology) and GAPDH (cat. no. AF7021; 1:2,000,
Affinity Biosciences) antibodies at 4°C overnight after being
blocked using 5% skimmed milk (Beyotime Institute of Biotechnology)
at 37°C for 1.5 h. The membrane was successively incubated with a
HRP-linked goat anti-rabbit secondary antibody (cat. no. S0001;
1:5,000; Affinity Biosciences) at 37°C for 1.5 h. An ECL kit
(Beyotime Institute of Biotechnology) was adopted to visualize the
protein bands. Finally, ImageJ (version 1.8; National Institutes of
Health) was used to semi-quantify protein expression.
Cell proliferation and apoptosis
To evaluate cell proliferation, 10 µl Cell Counting
Kit-8 reagent (Sangon Biotech Co., Ltd.) was added to the
3×103 cells at 0, 24, 48 and 72 h post-transfection. The
cells were then cultured at 37°C for 2 h, and the optical density
value at 450 nm was measured. Cell apoptosis was evaluated using a
TUNEL kit (Beyotime Institute of Biotechnology) at 48 h
post-transfection. The 6×104 cells were cultured with
TUNEL working solution at 37°C in the dark for 1 h after being
fixed in 4% paraformaldehyde (Sangon Biotech Co., Ltd.) at room
temperature for 0.5 h and permeabilized with 0.3% Triton X-100
(Sangon Biotech Co., Ltd.) for 5 min at room temperature. The cells
were stained with DAPI solution (Beyotime Institute of
Biotechnology) for 5 min at 37°C and antifade Mounting Medium
(Beyotime Institute of Biotechnology) before being sealed. The
images of 5 fields of view were captured by an inverted
fluorescence microscope (Motic Incorporation, Ltd.).
Transwell assay
Cell invasion was assessed by Transwell assay 48 h
post-transfection. The 8-µm 24-well Transwell insert (Corning,
Inc.) was precoated with Matrigel Matrix (Corning, Inc.) at 37°C
for 1 h. A total of 6×104 cells in McCoy's 5a Modified
Medium containing 1% FBS were seeded into the Transwell insert. The
lower chamber was filled with McCoy's 5a Modified Medium containing
10% FBS. The cells were then incubated at 37°C for 24 h and the
invasive cells were fixed with 4% paraformaldehyde for 20 min at
room temperature and stained with 0.1% crystal violet (Sangon
Biotech Co., Ltd.) for 10 min at room temperature. The images were
captured with an inverted fluorescence microscope.
Statistical analysis
Data are presented as the mean ± standard deviation,
median [interquartile range (IQR)] or count (%). SPSS v22.0 (IBM
Corp.) was used for statistical analysis and GraphPad Prism v9.0
(GraphPad Software; Dotmatics) was used to generate graphs.
Comparisons of parameters between tumor and adjacent tissues were
made using the Wilcoxon rank sum test. The correlation between
CCT6A and CDC20 expression was detected by Spearman's rank
correlation test. The expression levels of CCT6A and CDC20 in
patients with different characteristics were compared using the
Wilcoxon rank sum test or Kruskal-Wallis H rank sum test, as
appropriate. A receiver operating characteristic (ROC) curve was
constructed to display the ability of CCT6A and CDC20 expression to
identify patients with different Huvos grades (III–IV vs. I–II).
The association of CCT6A and CDC20 expression with DFS and OS was
evaluated by Kaplan-Meier curve analysis and log-rank test.
Independent factors related to pathological response, DFS and OS
were screened by multivariate logistic regression or Cox
proportional hazard regression models with a forward stepwise
method. The differences among groups in the cell experiments (in
triplicates) were analyzed by one-way ANOVA followed by Dunnett's
multiple comparisons test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Characteristics of patients with
osteosarcoma
The patients with osteosarcoma included in the
present study had a mean age of 20.2±8.0 years (range, 7–41 years),
and there were 21 (40.4%) female patients and 31 (59.6%) male
patients. Furthermore, there were 16 (30.8%) and 36 (69.2%)
patients at Enneking stages IIA and IIB, respectively. A total of
44 (84.6%) patients had no pathological fracture, whereas 8 (15.4%)
patients had a pathological fracture. Notably, all patients
received neoadjuvant chemotherapy according to the IOR-OS/N-5
protocol. Regarding Huvos grade, 9 (17.3%) patients had grade I, 21
(40.4%) patients had grade II, 21 (40.4%) patients had grade III
and 1 (1.9%) patient had grade IV osteosarcoma. Other specific
clinical information is listed in Table
I.
 | Table I.Clinical characteristics of patients
with osteosarcoma (n=52). |
Table I.
Clinical characteristics of patients
with osteosarcoma (n=52).
| Characteristic | Value |
|---|
| Mean age ± SD,
years | 20.2±8.0 |
| Sex, n (%) |
|
|
Female | 21 (40.4) |
|
Male | 31 (59.6) |
| Location, n
(%) |
|
|
Femur | 29 (55.8) |
|
Tibia | 18 (34.6) |
|
Others | 5 (9.6) |
| Enneking stage, n
(%) |
|
|
IIA | 16 (30.8) |
|
IIB | 36 (69.2) |
| Pathological
fracture, n (%) |
|
|
No | 44 (84.6) |
|
Yes | 8 (15.4) |
| Median serum ALP at
baseline (IQR), U/l | 157.8
(91.5-233.9) |
| Median serum LDH at
baseline (IQR), U/l | 232.5
(198.1-312.1) |
| Neoadjuvant
regimen, n (%) |
|
|
IOR-OS/N-5 | 52 (100.0) |
| Median TCNR (IQR),
% | 81.0
(54.3-93.0) |
| Huvos grade, n
(%) |
|
|
I | 9 (17.3) |
|
II | 21 (40.4) |
|
III | 21 (40.4) |
|
IV | 1 (1.9) |
| Surgery type, n
(%) |
|
|
Limb salvage | 45 (86.5) |
|
Amputation | 7 (13.5) |
CCT6A and CDC20 expression and their
intercorrelation
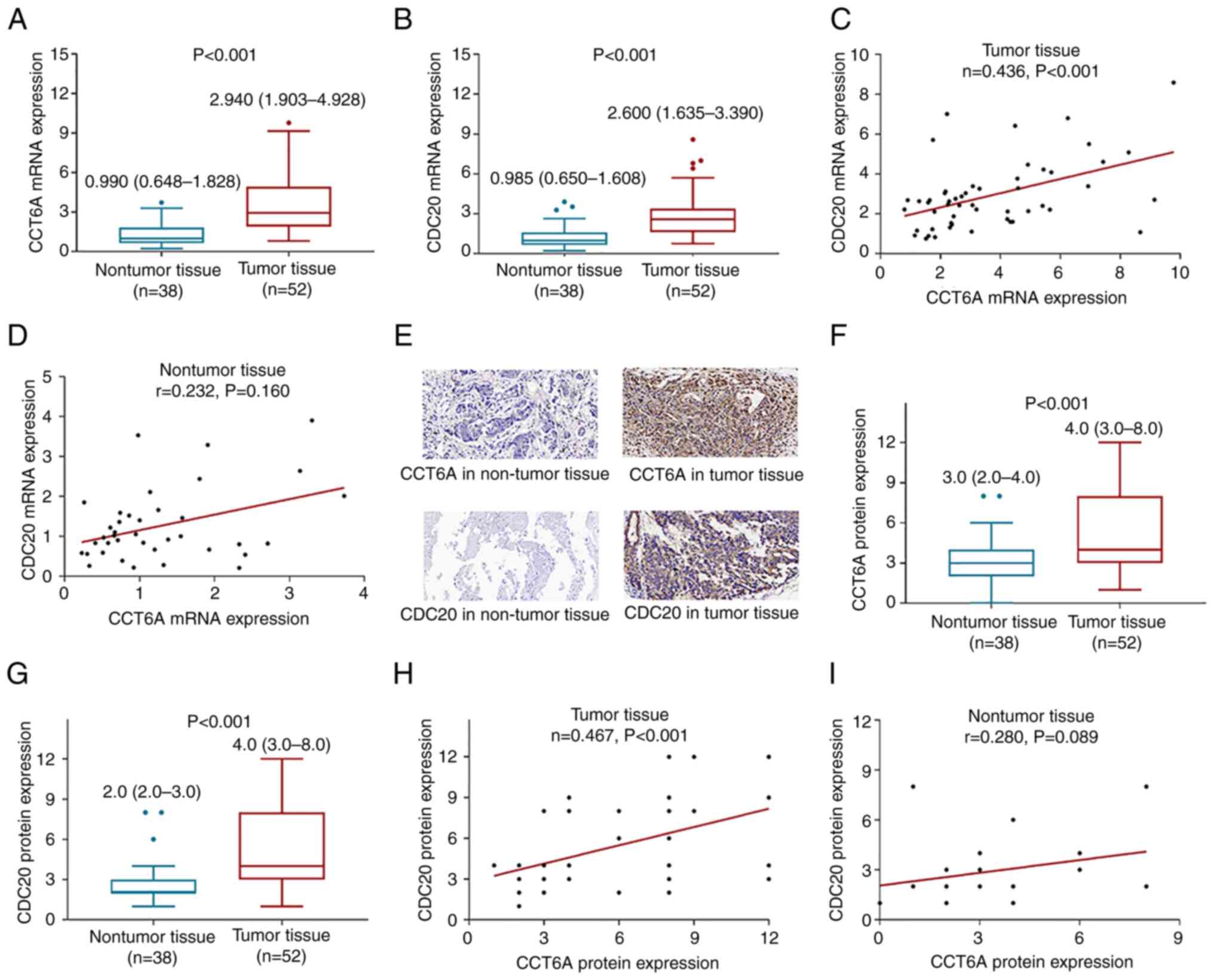
The mRNA expression levels of CCT6A were increased
in tumor tissues [median (IQR): 2.940 (1.903-4.928)] compared with
those in nontumor tissues [median (IQR): 0.990 (0.648-1.828)]
(P<0.001; Fig. 1A). Meanwhile,
CDC20 mRNA expression was also elevated in tumor tissues
(P<0.001; Fig. 1B). In addition,
CCT6A mRNA expression was positively correlated with CDC20 mRNA
expression in tumor tissues (r=0.436; P<0.001; Fig. 1C). However, no correlation was
discovered between CCT6A and CDC20 mRNA expression in nontumor
tissues from patients with osteosarcoma (r=0.232; P=0.160; Fig. 1D).
Representative images of IHC staining of CCT6A and
CDC20 in tumor tissues and nontumor tissues are shown in Fig. 1E. The protein expression levels of
CCT6A were elevated in tumor tissues (IHC score: 5.4±3.2) compared
with those in nontumor tissues (IHC score: 3.0±1.7) (P<0.001;
Fig. 1F). Furthermore, CDC20
protein expression was also increased in tumor tissues (P<0.001;
Fig. 1G). In addition, CCT6A
protein expression was positively correlated with CDC20 protein
expression in tumor tissues (r=0.467; P<0.001; Fig. 1H), but CCT6A protein expression was
not correlated with CDC20 protein expression in nontumor tissues
(r=0.280; P=0.089; Fig. 1I).
Association of tumor CCT6A and CDC20
with clinical features
Increased tumor CCT6A mRNA expression was associated
with a higher Enneking stage (IIB vs. IIA) (P=0.039) and abnormal
(>300 U/l) (vs. normal, ≤300 U/l) serum LDH at baseline
(P=0.048) in patients with osteosarcoma (Table II). Notably, elevated tumor CDC20
mRNA expression was only related to higher Enneking stage (IIB vs.
IIA) (P=0.044) in patients with osteosarcoma (Table II). Tumor CCT6A and CDC20 mRNA
expression was not associated with other clinical features in
patients with osteosarcoma (all P>0.05; Table II).
 | Table II.Association of tumor CCT6A and CDC20
expression with clinical characteristics. |
Table II.
Association of tumor CCT6A and CDC20
expression with clinical characteristics.
|
| RT-qPCR for mRNA
expression evaluation | IHC for protein
expression evaluation |
|---|
|
|
|
|
|---|
| Characteristic | Tumor CCT6A mRNA
expression, median (IQR) | P-value | Tumor CDC20 mRNA
expression, median (IQR) | P-value | Tumor CCT6A protein
expression, median (IQR) | P-value | Tumor CDC20 protein
expression, median (IQR) | P-value |
|---|
| Age |
| 0.111 |
| 0.399 |
| 0.406 |
| 0.677 |
|
≤20 years | 2.670
(1.760–4.430) |
| 2.505
(1.845–3.060) |
| 4.0 (3.0–8.0) |
| 4.0 (3.0–8.0) |
|
|
>20 years | 3.815
(2.250–6.420) |
| 2.735
(1.523–4.625) |
| 4.0 (3.0–8.3) |
| 4.0 (3.0–8.0) |
|
| Sex |
| 0.634 |
| 0.589 |
| 0.704 |
| 0.834 |
|
Female | 3.070
(1.975–5.315) |
| 3.040
(1.605–4.590) |
| 4.0 (3.0–8.0) |
| 4.0 (2.5–8.0) |
|
|
Male | 2.630
(1.820–4.920) |
| 2.570
(1.600–3.260) |
| 4.0 (3.0–8.0) |
| 4.0 (3.0–8.0) |
|
| Location |
| 0.681 |
| 0.166 |
| 0.602 |
| 0.155 |
|
Femur | 3.080
(1.985–5.545) |
| 3.040
(2.160–4.160) |
| 4.0 (3.0–8.0) |
| 6.0 (3.0–8.0) |
|
|
Tibia | 2.770
(2.038–4.350) |
| 2.330
(1.585–2.723) |
| 4.0 (3.0–8.0) |
| 4.0 (2.8–4.0) |
|
|
Others | 2.320
(1.405–5.590) |
| 2.140
(1.035–4.720) |
| 3.0 (2.5–6.5) |
| 4.0 (2.5–8.0) |
|
| Enneking stage |
| 0.039 |
| 0.044 |
| 0.004 |
| 0.047 |
|
IIA | 2.325
(1.348–4.078) |
| 2.180
(1.290–2.730) |
| 3.0 (2.3–3.8) |
| 4.0 (2.0–4.0) |
|
|
IIB | 3.250
(2.230–5.598) |
| 2.700
(2.113–4.195) |
| 6.0 (3.3–8.0) |
| 4.0 (3.0–8.0) |
|
| Pathological
fracture |
| 0.447 |
| 0.723 |
| 0.948 |
| 0.623 |
|
No | 2.940
(2.183–5.290) |
| 2.530
(1.635–3.365) |
| 4.0 (3.0–8.0) |
| 4.0 (3.0–8.0) |
|
|
Yes | 3.105
(1.368–4.550) |
| 2.665
(1.728–5.068) |
| 6.0 (2.0–8.0) |
| 4.0 (3.0–11.3) |
|
| Serum ALP at
baselinea |
| 0.335 |
| 0.369 |
| 0.559 |
| 0.277 |
|
Normal | 2.670
(2.083–4.515) |
| 2.435
(1.485–3.315) |
| 4.0 (3.0–8.0) |
| 4.0 (2.8–8.0) |
|
|
Abnormal | 3.815
(1.760–6.938) |
| 2.700
(1.810–3.885) |
| 5.0 (3.0–8.3) |
| 4.0 (3.0–8.3) |
|
| Serum LDH at
baselineb |
| 0.048 |
| 0.187 |
| 0.016 |
| 0.203 |
|
Normal | 2.500
(1.800–4.330) |
| 2.440
(1.600–3.120) |
| 4.0 (3.0–8.0) |
| 4.0 (3.0–8.0) |
|
|
Abnormal | 4.570
(2.740–6.840) |
| 3.290
(1.740–4.850) |
| 8.0 (4.0–9.0) |
| 4.0 (3.0–10.5) |
|
In terms of protein expression, tumor CCT6A protein
expression was associated with increased Enneking Stage (IIB vs.
IIA; P=0.004) and abnormal (vs. normal) serum LDH at baseline
(P=0.016) (Table II). Furthermore,
tumor CDC20 protein expression was only related to elevated
Enneking stage (IIB vs. IIA; P=0.047; Table II). Tumor CCT6A and CDC20 protein
expression was not associated with other clinical characteristics
in patients with (all P>0.05; Table
II).
Association of tumor CCT6A and CDC20
with pathological response
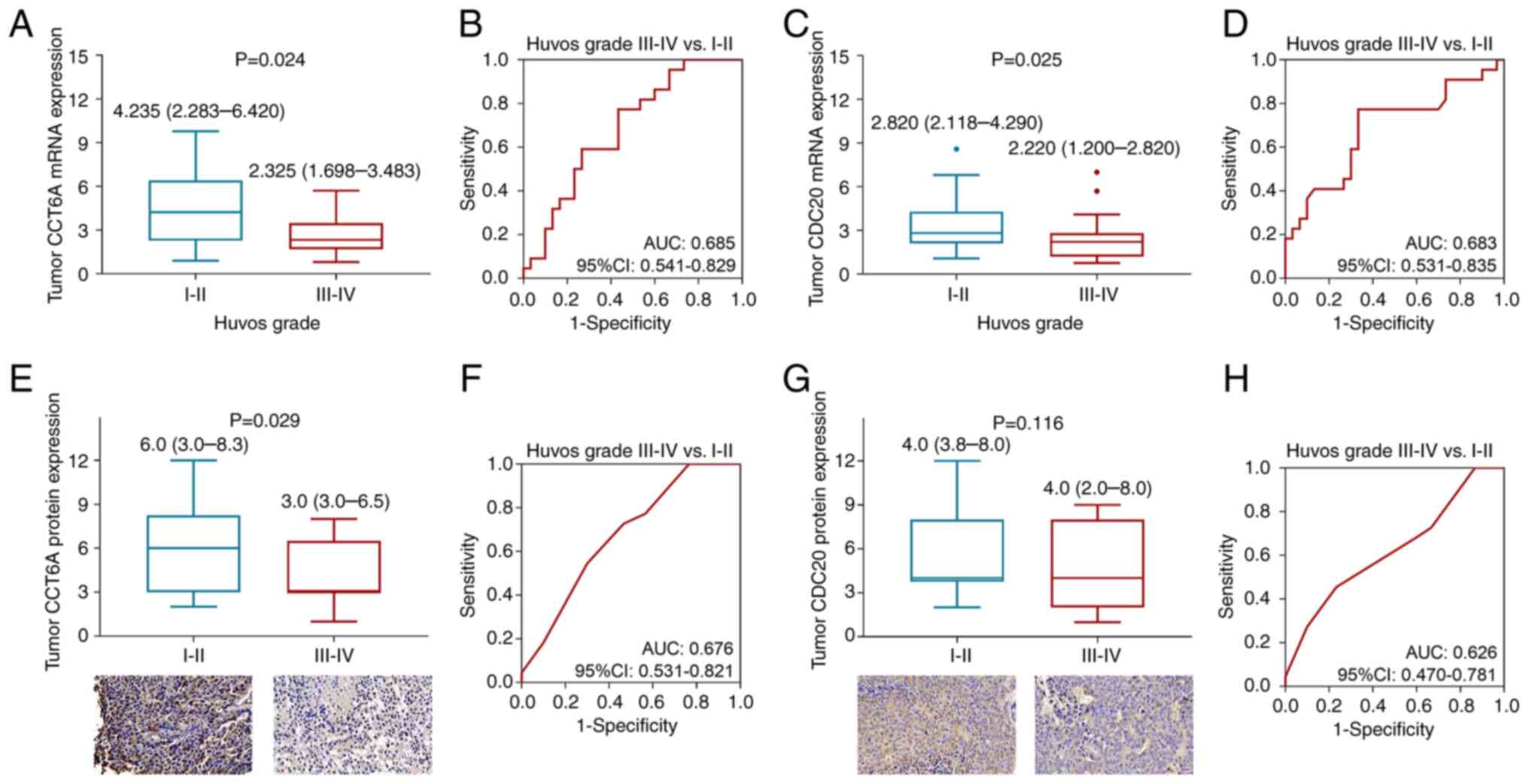
Tumor CCT6A mRNA expression was increased in
patients with Huvos grade I–II osteosarcoma compared with in those
with Huvos grade III–IV osteosarcoma [median (IQR): 4.235
(2.283-6.420) vs. 2.325 (1.698-3.483); P=0.024; Fig. 2A]. Notably, tumor CCT6A mRNA
expression possessed an acceptable capacity for discriminating
patients with Huvos grade I–II from those with Huvos grade III–IV
[area under the curve (AUC): 0.685; 95% confidence interval (CI):
0.541-0.829; Fig. 2B]. In addition,
higher tumor CDC20 mRNA expression was related to a lower Huvos
grade (P=0.025; Fig. 2C),and CDC20
mRNA expression also showed an acceptable capacity for
discriminating patients with Huvos grade I–II from those with Huvos
grade III–IV (AUC: 0.683; 95% CI: 0.531-0.835; Fig. 2D).
Regarding protein expression, tumor CCT6A protein
expression was elevated in patients with Huvos grade I–II
osteosarcoma compared with those with Huvos grade III–IV
osteosarcoma (IHC score: 6.3±3.4 vs. 4.2±2.3; P=0.014; Fig. 2E). Furthermore, tumor CCT6A protein
expression had a good capacity to discriminate patients with Huvos
grade I–II from those with Huvos grade III–IV (AUC: 0.676; 95% CI:
0.531-0.821; Fig. 2F). By contrast,
tumor CDC20 protein expression was not related to Huvos grade
(P=0.149; Fig. 2G), and could not
discriminate patients with Huvos grade I–II from those with Huvos
grade III–IV (AUC: 0.626; 95% CI: 0.470-0.781; Fig. 2H).
Independent factors related to
pathological response
Univariate analysis revealed that higher tumor CCT6A
mRNA expression (cut off by median value) [odds ratio (OR)=0.674;
P=0.018], higher tumor CCT6A protein expression (IHC=3 as cut off)
(OR=0.786; P=0.027) and abnormal serum ALP (>500 U/l for
patients <16 years old and >150 U/l for patients ≥16 years
old) at baseline (OR=0.225; P=0.018) were related to the reduced
possibility of realizing a good pathological response (Huvos grade
III–IV) (Table III). Further
multivariate logistic regression analysis revealed that higher
tumor CCT6A mRNA expression (OR=0.677; P=0.033) and abnormal serum
ALP at baseline (OR=0.248; P=0.037) were independently related to a
lower probability of achieving a good pathological response (Huvos
grade III–IV) (Table III).
 | Table III.Logistic regression analysis for
Huvos grade III–IV. |
Table III.
Logistic regression analysis for
Huvos grade III–IV.
| A, Univariate
logistic regression analysis |
|---|
|
|---|
|
|
|
| 95% CI |
|---|
|
|
|
|
|
|---|
| Items | P-value | OR | Lower | Upper |
|---|
| Higher tumor CCT6A
mRNA expression | 0.018 | 0.674 | 0.486 | 0.935 |
| Higher tumor CDC20
mRNA expression | 0.085 | 0.707 | 0.477 | 1.049 |
| Higher tumor CCT6A
protein expression | 0.027 | 0.786 | 0.635 | 0.973 |
| Higher tumor CDC20
protein expression | 0.151 | 0.864 | 0.707 | 1.055 |
| Age (>20 years
vs. ≤20 years) | 0.338 | 1.727 | 0.565 | 5.283 |
| Sex (male vs.
female) | 0.524 | 0.695 | 0.227 | 2.130 |
| Location |
|
|
|
|
|
Femur | Reference |
|
|
|
|
Tibia | 0.417 | 1.636 | 0.498 | 5.379 |
|
Others | 0.930 | 1.091 | 0.157 | 7.592 |
| Enneking Stage (IIB
vs. IIA) | 0.054 | 0.300 | 0.088 | 1.023 |
| Pathological
fracture (yes vs. no) | 0.293 | 0.400 | 0.073 | 2.204 |
| Serum ALP at
baselinea (abnormal
vs. normal) | 0.018 | 0.225 | 0.066 | 0.770 |
| Serum LDH at
baselineb (abnormal
vs. normal) | 0.115 | 0.316 | 0.075 | 1.326 |
|
| B, Multivariate
logistic regression analysis |
|
|
|
|
| 95% CI |
|
|
|
|
|
| Items | P-value | OR | Lower | Upper |
|
| Higher tumor mRNA
CCT6A expression | 0.033 | 0.677 | 0.473 | 0.969 |
| Serum ALP at
baselinea (abnormal
vs. normal) | 0.037 | 0.248 | 0.067 | 0.918 |
Association of tumor CCT6A and CDC20
expression with DFS and OS
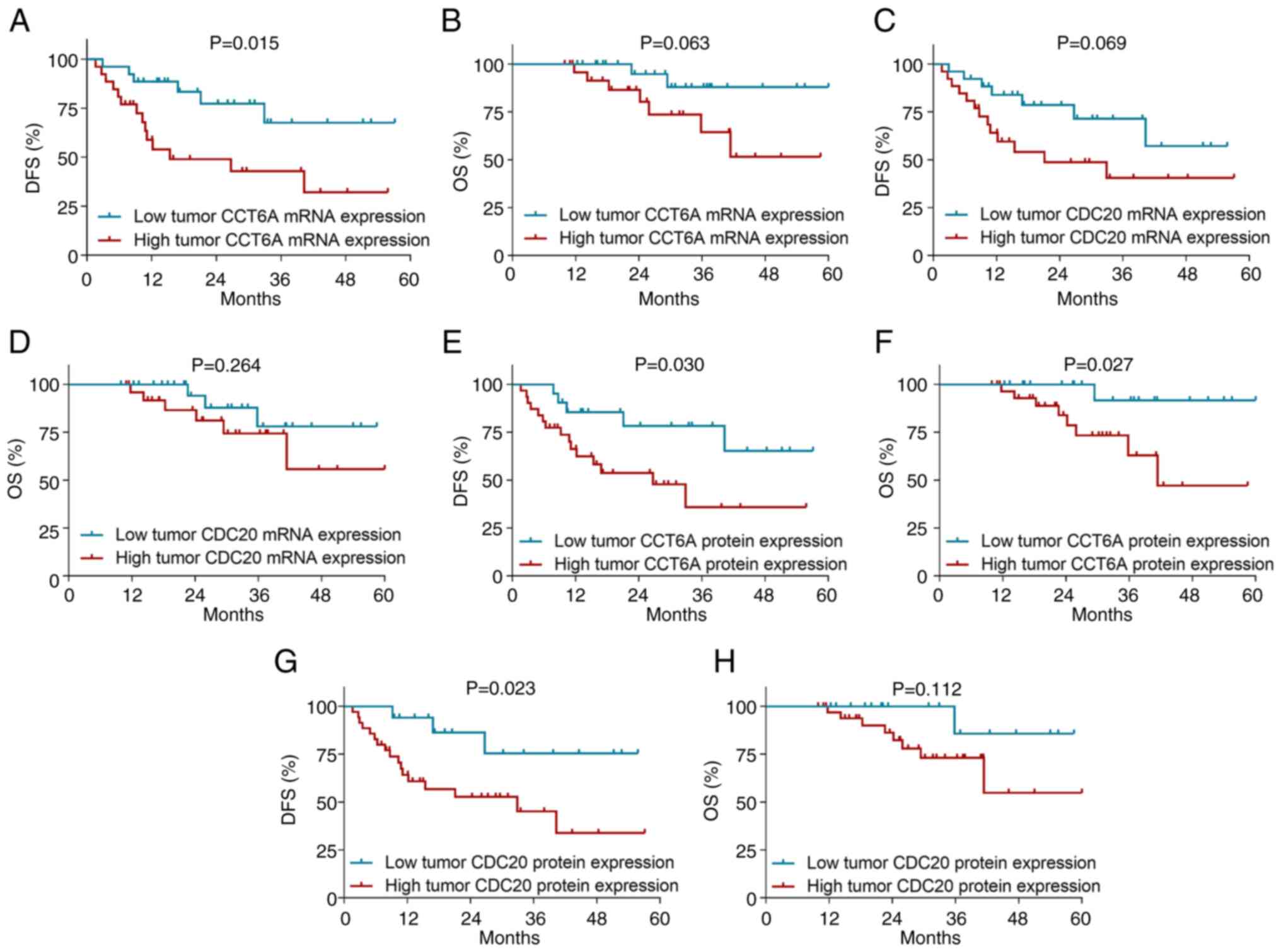
High tumor CCT6A high mRNA expression (cut off by
median value) was associated with poor DFS in patients with
osteosarcoma (P=0.015; Fig. 3A),
but it was not associated with OS (P=0.063; Fig. 3B). Furthermore, tumor CDC20 mRNA
expression was not associated with DFS (P=0.069; Fig. 3C) or OS (P=0.264; Fig. 3D) in patients with osteosarcoma.
High tumor CCT6A protein expression was associated
to worse DFS (P=0.030; Fig. 3E) and
OS (P=0.027; Fig. 3F) in patients
with osteosarcoma. Moreover, high tumor CDC20 protein was only
associated with poor DFS (P=0.023; Fig.
3G), but not OS (P=0.112; Fig.
3H) in patients with osteosarcoma.
Independent factors predicting DFS and
OS
Univariate analysis revealed that high tumor CCT6A
mRNA expression (vs. low) [hazard ratio (HR)=3.096; P=0.021], high
tumor CCT6A protein expression (vs. low) (HR=2.974; P=0.038), high
tumor CDC20 protein expression (vs. low) (HR=3.768; P=0.035),
higher Enneking stage (IIB vs. IIA) (HR=4.052; P=0.027), and
abnormal serum LDH at baseline (vs. normal) (HR=2.983; P=0.018)
were related to worse DFS, whereas tibia lesion (vs. femur)
(HR=0.244; P=0.027) and good pathological response (Huvos grade
III–IV vs. I–II) (HR=0.237; P=0.013) were linked to better DFS in
patients with osteosarcoma (Table
IV). Further multivariate analysis suggested that high tumor
CCT6A mRNA expression (vs. low) (HR: 2.960; P=0.028) was
independently associated with shorter DFS, whereas a good
pathological response (Huvos grade III–IV vs. I–II) (HR=0.241;
P=0.016) was independently related to prolonged DFS (Table IV).
 | Table IV.Cox's proportional hazard regression
analysis for DFS. |
Table IV.
Cox's proportional hazard regression
analysis for DFS.
| A, Univariate Cox's
proportional hazard regression analysis |
|---|
|
|---|
|
|
|
| 95% CI |
|---|
|
|
|
|
|
|---|
| Items | P-value | HR | Lower | Upper |
|---|
| Tumor CCT6A mRNA
expression (high vs. low) | 0.021 | 3.096 | 1.185 | 8.087 |
| Tumor CDC20 mRNA
expression (high vs. low) | 0.077 | 2.298 | 0.915 | 5.772 |
| Tumor CCT6A protein
expression (high vs. low) | 0.038 | 2.974 | 1.063 | 8.319 |
| Tumor CDC20 protein
expression (high vs. low) | 0.035 | 3.768 | 1.101 | 12.898 |
| Age (>20 vs. ≤20
years) | 0.559 | 0.764 | 0.310 | 1.882 |
| Sex (male vs.
female) | 0.289 | 1.656 | 0.652 | 4.206 |
| Location |
|
|
|
|
|
Femur | Reference |
|
|
|
|
Tibia | 0.027 | 0.244 | 0.070 | 0.850 |
|
Others | 0.503 | 0.603 | 0.137 | 2.649 |
| Enneking stage (IIB
vs. IIA) | 0.027 | 4.052 | 1.175 | 13.978 |
| Pathological
fracture (yes vs. no) | 0.178 | 2.165 | 0.704 | 6.656 |
| Serum ALP at
baselinea (abnormal
vs. normal) | 0.186 | 1.830 | 0.748 | 4.477 |
| Serum LDH at
baselineb (abnormal
vs. normal) | 0.018 | 2.983 | 1.211 | 7.350 |
| Huvos grade (III–IV
vs. I–II) | 0.013 | 0.237 | 0.076 | 0.733 |
| Surgery type
(amputation vs. limb salvage) | 0.725 | 0.768 | 0.177 | 3.340 |
|
| B, Multivariate
Cox's proportional hazard regression analysis |
|
|
|
|
| 95% CI |
|
|
|
|
|
| Items | P-value | HR | Lower | Upper |
|
| Tumor CCT6A mRNA
expression (high vs. low) | 0.028 | 2.960 | 1.126 | 7.786 |
| Huvos grade (III–IV
vs. I–II) | 0.016 | 0.241 | 0.076 | 0.769 |
In addition, univariate analysis revealed that only
abnormal serum LDH at baseline (vs. normal) (HR=4.100; P=0.036) was
associated with poor OS, whereas good pathological response (Huvos
grade III–IV vs. I–II) (HR=0.077; P=0.022) was associated with a
satisfactory OS in patients with osteosarcoma (Table V). Further multivariate analysis
revealed that only good pathological response (Huvos grade III–IV
vs. I–II) was independently associated with a better OS in patients
with osteosarcoma (HR=0.077; P=0.022; Table V).
 | Table V.Cox's proportional hazard regression
analysis for OS. |
Table V.
Cox's proportional hazard regression
analysis for OS.
| A, Univariate Cox's
proportional hazard regression analysis |
|---|
|
|---|
|
|
|
| 95% CI |
|---|
|
|
|
|
|
|---|
| Items | P-value | HR | Lower | Upper |
|---|
| Tumor CCT6A mRNA
expression (high vs. low) | 0.085 | 3.984 | 0.826 | 19.227 |
| Tumor CDC20 mRNA
expression (high vs. low) | 0.276 | 2.163 | 0.540 | 8.659 |
| Tumor CCT6A protein
expression (high vs. low) | 0.059 | 7.455 | 0.927 | 59.973 |
| Tumor CDC20 protein
expression (high vs. low) | 0.147 | 4.705 | 0.580 | 38.195 |
| Age (>20 vs. ≤20
years) | 0.443 | 0.581 | 0.145 | 2.326 |
| Sex (male vs.
female) | 0.428 | 1.753 | 0.437 | 7.036 |
| Location |
|
|
|
|
|
Femur | Reference |
|
|
|
|
Tibia | 0.672 | 0.729 | 0.169 | 3.151 |
|
Others | 0.873 | 1.193 | 0.138 | 10.348 |
| Enneking stage (IIB
vs. IIA) | 0.295 | 2.324 | 0.480 | 11.262 |
| Pathological
fracture (yes vs. no) | 0.137 | 3.398 | 0.677 | 17.048 |
| Serum ALP at
baselinea (abnormal
vs. normal) | 0.212 | 2.336 | 0.616 | 8.855 |
| Serum LDH at
baselineb (abnormal
vs. normal) | 0.036 | 4.100 | 1.099 | 15.288 |
| Huvos grade (III–IV
vs. I–II) | 0.022 | 0.077 | 0.009 | 0.693 |
| Surgery type
(amputation vs. limb salvage) | 0.930 | 0.911 | 0.114 | 7.308 |
|
| B, Multivariate
Cox's proportional hazard regression analysis |
|
|
|
|
| 95% CI |
|
|
|
|
|
| Items | P-value | HR | Lower | Upper |
|
| Huvos grade (III–IV
vs. I–II) | 0.022 | 0.077 | 0.009 | 0.693 |
CCT6A and CDC20 in osteosarcoma cell
lines
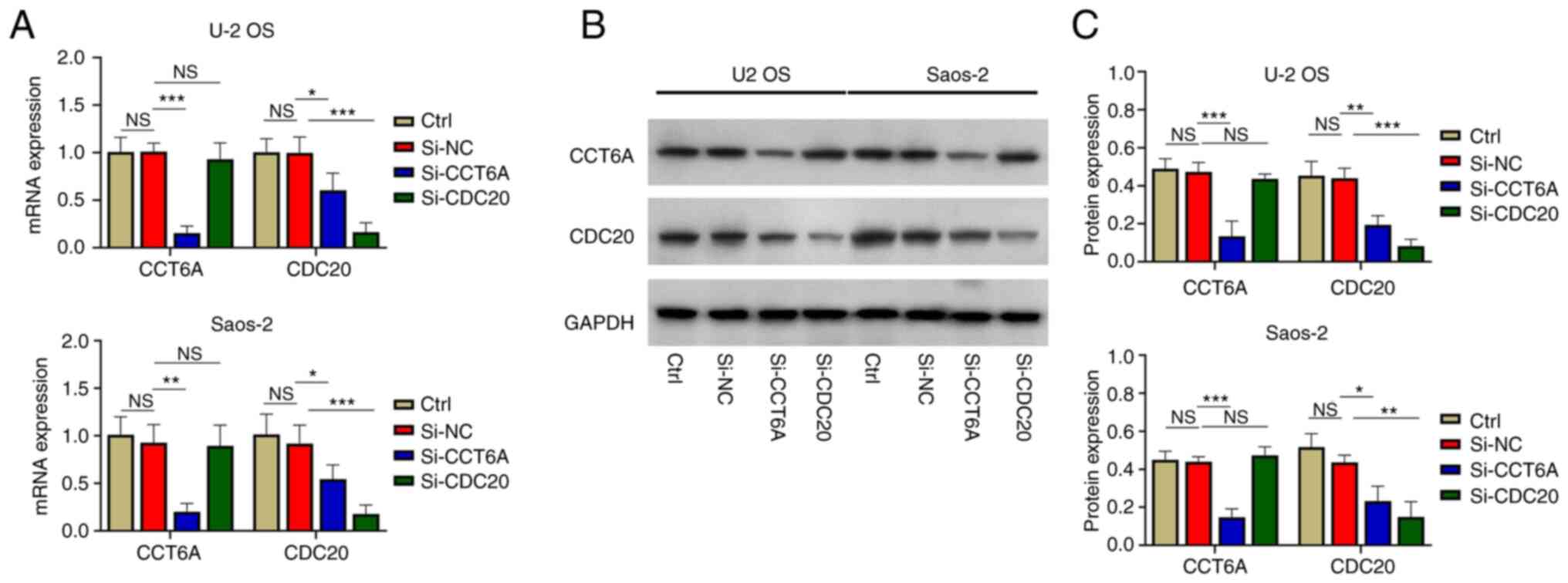
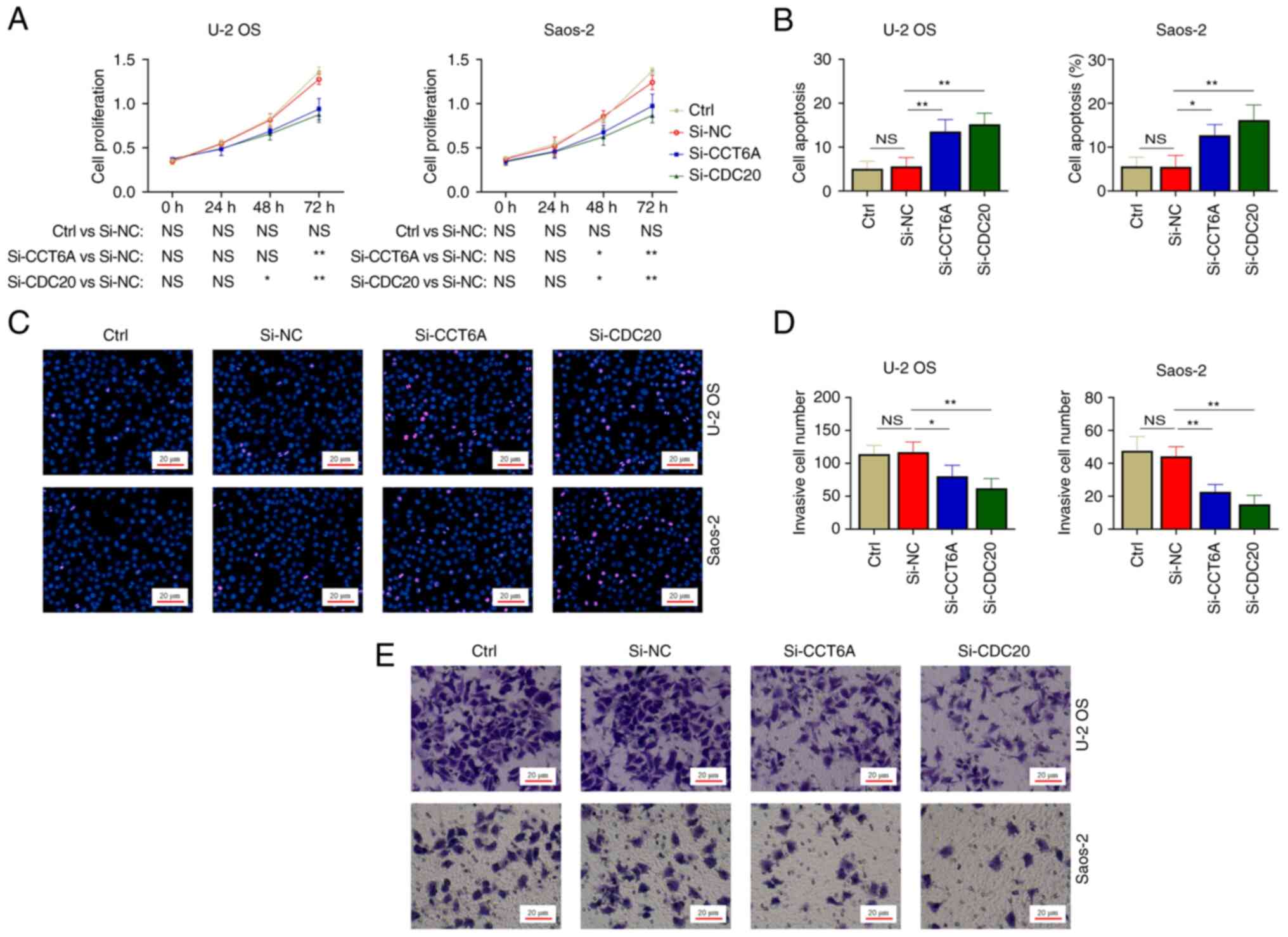
U-2 OS and Saos-2 cells were transfected with
si-CCT6A, si-CDC20 or si-NC. Firstly, it was revealed that the mRNA
(Fig. 4A) and protein expression
levels (Fig. 4B and C) of CCT6A
were significantly reduced in the cells transfected with si-CCT6A
siRNA compared with in those transfected with si-NC (all
P<0.01). Furthermore, the mRNA (Fig.
4A) and protein expression levels (Fig. 4B and C) of CDC20 were reduced in U-2
OS and Saos-2 cells transfected with si-CDC20 siRNA compared with
in those transfected with si-NC (all P<0.01). These findings
indicated that transfection was successful.
Effect of CCT6A and CDC20 knockdown on
cell proliferation, apoptosis and invasion in osteosarcoma cell
lines
The cell proliferation assay illustrated that CCT6A
knockdown reduced cell proliferation at 72 h in U-2 OS cells
(P<0.01), and at 48 and 72 h in Saos-2 cells (both P<0.05),
whereas CDC20 knockdown decreased cell proliferation at 48 and 72 h
in both U-2 OS and Saos-2 cells (all P<0.05) (Fig. 5A). Furthermore, the cell apoptosis
assay showed that CCT6A and CDC20 knockdown increased the apoptotic
rate in U-2 OS and Saos-2 cells (all P<0.05; Fig. 5B and C). Moreover, the Transwell
assay revealed that the number of invasive cells was reduced by
CCT6A and CDC20 knockdown in U-2 OS and Saos-2 cells (all
P<0.05; Fig. 5D and E).
Discussion
CCT6A and CDC20 are overexpressed in various types
of cancer (16,27,29–32),
and a previous study reported that CCT6A is positively associated
with CDC20 in patients with papillary thyroid carcinoma (16). However, the dysregulation and
intercorrelation of CCT6A and CDC20 in patients with osteosarcoma
still needs to be explored. The present study discovered that the
expression levels of CCT6A and CDC20 were increased in tumor
tissues compared with those in nontumor tissues in patients with
osteosarcoma. The potential reason could be that increased CCT6A
and CDC20 may inhibit cell cycle arrest, thereby facilitating
malignant cell changes (10,21),
thus CCT6A and CDC20 are highly expressed in tumor tissues compared
with nontumor tissues.
The present study also revealed that increased tumor
CCT6A expression was associated with higher Enneking stage (IIB vs.
IIA) and abnormal serum LDH (vs. normal), whereas elevated tumor
CDC20 expression was associated with higher Enneking stage in
patients with osteosarcoma. The probable reasons for this may be:
i) Increased tumor CCT6A and CDC20 expression could facilitate the
extra-periosteal invasion of tumor cells; therefore, increased
tumor CCT6A and CDC20 are linked with increased Enneking stage in
patients with osteosarcoma (11,33);
and ii) elevated tumor CCT6A could promote tumor cell migration
(11); thus, an increase in tumor
CCT6A is associated with abnormal serum LDH in osteosarcoma
patients. Therefore, increased tumor CCT6A and CDC20 expression may
be associated with a worse tumor burden in patients with
osteosarcoma.
Notably, tumor CCT6A and CDC20 expression was
increased in patients with osteosarcoma with poor pathological
response compared with in those with good pathological response
(reflected by Huvos grade). The possible reason would be that
increased tumor CCT6A and CDC20 expression could enhance drug
resistance, thereby decreasing the efficacy of neoadjuvant
chemotherapy (13,21). Therefore, tumor CCT6A and CDC20
expression could reflect the outcome of neoadjuvant chemotherapy.
Further multivariate regression analysis confirmed that higher
tumor CCT6A was independently linked with a lower possibility of
achieving a good pathological response (Huvos grade III–IV vs.
I–II).
Furthermore, the present study revealed that
elevated tumor CCT6A expression was associated with poor DFS and OS
in patients with osteosarcoma. The potential reason would be that
as aforementioned, increased tumor CCT6A represented higher
Enneking stage and abnormal serum LDH, as well as poor neoadjuvant
chemotherapy response. Therefore, tumor CCT6A is negatively
associated with DFS and OS in patients with osteosarcoma. Notably,
multivariate Cox proportional hazard regression analysis revealed
that high tumor CCT6A expression was independently associated with
poor DFS, which further confirmed the prognostic value of tumor
CCT6A in patients osteosarcoma. However, high tumor CCT6A
expression could not independently predict OS; the potential
reasons are as follows: i) The incidence of OS events was
relatively lower than that of DFS events, and the sample size was
insufficient, which led to low statistical power; and ii) OS may be
affected by other factors, such as treatment after tumor recurrence
(34); therefore, the prognostic
values of tumor CCT6A in patients with osteosarcoma would be
less.
The present study also conducted in vitro
experiments, and observed that CCT6A and CDC20 knockdown inhibited
cell proliferation and invasion in two osteosarcoma cell lines.
These data further explain the relationship between CCT6A and CDC20
and tumor stages or markers in patients with osteosarcoma.
Meanwhile, it was observed that CCT6A knockdown also reduced CDC20
expression, indicating that CCT6A might regulate CDC20 secretion,
although further validation is needed.
A previous study reported on the co-expression
relationship between CCT6A and CDC20 proteins in patients with
papillary thyroid carcinoma (16).
The present study also observed the co-expression relationship
between CCT6A and CDC20 protein/mRNA in patients with osteosarcoma,
which was in line with previous study in patients with papillary
thyroid carcinoma (16).
Despite the interesting findings of the present
study, some limitations should be noted. First, the sample size was
not large enough to provide a sound conclusion, which could be
validated in further studies; however, osteosarcoma is a rare form
of cancer. Second, the IOR-OS/N-5 regimen was recommended as
neoadjuvant chemotherapy for osteosarcoma at our institution; the
present study assessed the relationship between CCT6A and CDC20 and
the response and survival benefit of the neoadjuvant IOR-OS/N-5
regimen followed by surgery, whereas other neoadjuvant regimens
should be further explored. Third, only patients with newly
diagnosed osteosarcoma were analyzed in the present study;
therefore, further investigations should be carried out in patients
with recurrent osteosarcoma. Finally, the follow-up duration of the
current study was not long enough to make a long-term prognostic
evaluation of CCT6A and CDC20 for osteosarcoma.
In conclusion, CCT6A is associated with CDC20,
Enneking stage and prognosis, and its knockdown inhibits
osteosarcoma cell proliferation and invasion. These data indicated
the close relationship between CCT6A and the pathogenesis and
prognosis of osteosarcoma; however, further validation is
needed.
Acknowledgements
Not applicable.
Funding
This study was supported by osteosarcoma the Education Science
Foundation of Liaoning Province (grant no. SYYX202015).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NW contributed to the study conception and design.
Material preparation, data collection and analysis were performed
by YC and JW. The first draft of the manuscript was written by JW
and all authors commented on previous versions of the manuscript.
NW and YC confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Center Hospital Affiliated to Shenyang Medical College. The
patients or their statutory guardians provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Prater S and McKeon B: Osteosarcoma.
StatPearls. StatPearls Publishing; Treasure Island, FL: 2022
|
|
2
|
Sadykova LR, Ntekim AI, Muyangwa-Semenova
M, Rutland CS, Jeyapalan JN, Blatt N and Rizvanov AA: Epidemiology
and risk factors of osteosarcoma. Cancer Invest. 38:259–269. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Azevedo JWV, de Medeiros Fernandes TAA,
Fernandes JV Jr, de Azevedo JCV, Lanza DCF, Bezerra CM, Andrade VS,
de Araújo JMG and Fernandes JV: Biology and pathogenesis of human
osteosarcoma. Oncol Lett. 19:1099–1116. 2020.PubMed/NCBI
|
|
4
|
Czarnecka AM, Synoradzki K, Firlej W,
Bartnik E, Sobczuk P, Fiedorowicz M, Grieb P and Rutkowski P:
Molecular biology of osteosarcoma. Cancers (Basel). 12:21302020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen B, Zeng Y, Liu B, Lu G, Xiang Z, Chen
J, Yu Y, Zuo Z, Lin Y and Ma J: Risk factors, prognostic factors,
and nomograms for distant metastasis in patients with newly
diagnosed osteosarcoma: A population-based study. Front Endocrinol
(Lausanne). 12:6720242021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jafari F, Javdansirat S, Sanaie S, Naseri
A, Shamekh A, Rostamzadeh D and Dolati S: Osteosarcoma: A
comprehensive review of management and treatment strategies. Ann
Diagn Pathol. 49:1516542020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smeland S, Bielack SS, Whelan J, Bernstein
M, Hogendoorn P, Krailo MD, Gorlick R, Janeway KA, Ingleby FC,
Anninga J, et al: Survival and prognosis with osteosarcoma:
Outcomes in more than 2000 patients in the EURAMOS-1 (European and
American Osteosarcoma Study) cohort. Eur J Cancer. 109:36–50. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xiao B, Liu L, Li A, Xiang C, Wang P, Li H
and Xiao T: Identification and verification of immune-related gene
prognostic signature based on ssGSEA for osteosarcoma. Front Oncol.
10:6076222020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Smrke A, Anderson PM, Gulia A, Gennatas S,
Huang PH and Jones RL: Future directions in the treatment of
osteosarcoma. Cells. 10:1722021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zeng G, Wang J and Huang Y, Lian Y, Chen
D, Wei H, Lin C and Huang Y: Overexpressing CCT6A contributes to
cancer cell growth by affecting the G1-To-S phase transition and
predicts A negative prognosis in hepatocellular carcinoma. Onco
Targets Ther. 12:10427–10439. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang X, Tong Y, Ye W and Chen L: HOXB2
increases the proliferation and invasiveness of colon cancer cells
through the upregulation of CCT6A. Mol Med Rep. 25:1742022.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li B, Lu X, Ma C, Sun S, Shu X, Wang Z and
Sun W: Long non-coding RNA NEAT1 promotes human glioma tumor
progression via miR-152-3p/CCT6A pathway. Neurosci Lett.
732:1350862020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peng X, Chen G, Lv B and Lv J:
MicroRNA-148a/152 cluster restrains tumor stem cell phenotype of
colon cancer via modulating CCT6A. Anticancer Drugs. 33:e610–e621.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Wang X, Xu L, Lin Y and Zhang J:
CCT6A and CHCHD2 are coamplified with EGFR and associated with the
unfavorable clinical outcomes of lung adenocarcinoma. Dis Markers.
2022:15601992022.PubMed/NCBI
|
|
15
|
He T, Yu D, Wang Z, Guo C, Chang Y and
Wang D: Chaperonin-containing tailless complex polypeptide 1
subunit 6A links with aggravating tumor features and disease-free
survival in surgical gastric cancer patients: A long-term follow-up
study. Clin Res Hepatol Gastroenterol. 46:1019132022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng W, Li W, Zhang X, Cen W and Liu Y:
The intercorrelation among CCT6A, CDC20, CCNB1, and PLK1
expressions and their clinical value in papillary thyroid carcinoma
prognostication. J Clin Lab Anal. 36:e246092022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Volonte D, Sedorovitz M and Galbiati F:
Impaired Cdc20 signaling promotes senescence in normal cells and
apoptosis in non-small cell lung cancer cells. J Biol Chem.
298:1024052022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Greil C, Engelhardt M and Wäsch R: The
role of the APC/C and its coactivators Cdh1 and Cdc20 in cancer
development and therapy. Front Genet. 13:9415652022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ni K and Hong L: Current progress and
perspectives of CDC20 in female reproductive cancers. Curr Mol Med.
23:193–199. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang H, Liu Z, Wu P, Wang H and Ren W:
NUSAP1 Accelerates osteosarcoma cell proliferation and cell cycle
progression via upregulating CDC20 and cyclin A2. Onco Targets
Ther. 14:3443–3454. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao Y, Guo C, Fu S, Cheng Y and Song C:
Downregulation of CDC20 suppressed cell proliferation, induced
apoptosis, triggered cell cycle arrest in osteosarcoma cells, and
enhanced chemosensitivity to cisplatin. Neoplasma. 68:382–390.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zeng W, Wu M, Cheng Y, Liu L, Han Y, Xie
Q, Li J, Wei L, Fang Y, Chen Y, et al: CCT6A knockdown suppresses
osteosarcoma cell growth and Akt pathway activation in vitro. PLoS
One. 17:e02798512022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Enneking WF, Spanier SS and Goodman MA: A
system for the surgical staging of musculoskeletal sarcoma. Clin
Orthop Relat Res. 106–120. 1980.PubMed/NCBI
|
|
24
|
Qiao S, Qi K, Liu C, Xu C, Ma J, Xu X, Li
C and Wang Z: Long intergenic non-coding RNA 511 correlates with
improved prognosis, and hinders osteosarcoma progression both in
vitro and in vivo. J Clin Lab Anal. 34:e231642020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu MS, Ma QY, Liu DD, Li XJ, Deng LJ, Li
N, Shen J, Zhao Z and Chen JX: CDC20 and its downstream genes:
Potential prognosis factors of osteosarcoma. Int J Clin Oncol.
24:1479–1489. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma J, Yang L, Feng H, Zheng L, Meng H and
Li X: CCT6A may act as a potential biomarker reflecting tumor size,
lymphatic metastasis, FIGO stage, and prognosis in cervical cancer
patients. J Clin Lab Anal. 35:e237932021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huvos AG and Higinbotham NL: Primary
fibrosarcoma of bone. A clinicopathologic study of 130 patients.
Cancer. 35:837–847. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cai Y, Wu D and Zhan L: CCT6A expression
in hepatocellular carcinoma and its correlation with clinical
characteristics, liver function indexes, tumor markers and
prognosis. Clin Res Hepatol Gastroenterol. 46:1017962022.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang J, Liu C, Xu G, Liang T, Yu C, Liao
S, Zhang Z, Lu Z, Wang Z, Chen J, et al: CCT6A, a novel prognostic
biomarker for Ewing sarcoma. Medicine (Baltimore). 100:e244842021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang X, Zhang X, Li X, Bao H, Li G, Li N,
Li H and Dou J: Connection between CDC20 expression and
hepatocellular carcinoma prognosis. Med Sci Monit.
27:e9267602021.PubMed/NCBI
|
|
32
|
Gayyed MF, El-Maqsoud NM, Tawfiek ER, El
Gelany SA and Rahman MF: A comprehensive analysis of CDC20
overexpression in common malignant tumors from multiple organs: Its
correlation with tumor grade and stage. Tumour Biol. 37:749–762.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao Y, Zhang B, Wang Y and Shang G: Cdc20
inhibitor apcin inhibits the growth and invasion of osteosarcoma
cells. Oncol Rep. 40:841–848. 2018.PubMed/NCBI
|
|
34
|
Evenhuis RE, Acem I, Rueten-Budde AJ,
Karis DSA, Fiocco M, Dorleijn DMJ, Speetjens FM, Anninga J,
Gelderblom H and van de Sande MAJ: Survival analysis of 3 different
age groups and prognostic factors among 402 patients with skeletal
high-grade osteosarcoma. Real world data from a single tertiary
sarcoma center. Cancers (Basel). 13:4862021. View Article : Google Scholar : PubMed/NCBI
|