Introduction
According to Global Cancer Observatory data, lung
cancer is the second most common cancer worldwide (1). Although smoking is still a dominant
risk factor for lung cancer, other risk factors, including
environmental and occupational exposure, chronic lung disease, and
lifestyle factors, contribute to the risk of developing lung
cancer. Lung adenocarcinoma (LADC) is the most common type of lung
cancer, accounting for approximately 50% of all cases, and its
frequency is increasing. Even though the major cause of LADC is
smoking, it is also the most common type of lung cancer in
non-smokers (2).
In the past decade, advances have been achieved in
the epidemiology and prevention of lung cancer. Nevertheless, lung
cancer remains the leading cause of cancer death in both sexes and
in all ages. The lack of specific symptoms in the early stages and
practical biomarkers for screening and the prediction of patient
outcomes partially explain the poor survival rate and mortality of
these patients (3).
Oncogenesis is a highly complex, multi-step process
involving genetic and epigenetic changes. Lung cancer has a higher
tumor mutational burden than other cancer types, which may be
related to smoking and exposure to tobacco xenobiotics (4). A previous study examined the rates of
protein-altering mutations in 441 tumors. The findings obtained
showed that non-small cell lung cancer (NSCLC) had one of the
highest rates of protein-altering mutations, with rates in
adenocarcinomas and squamous cell carcinomas of 3.5 and 3.9 per
megabase, respectively (5). NSCLC
treatment strategies and prognoses are markedly affected by the
stage of the disease at diagnosis. Surgery is highly effective in
the early stages, but ineffective in patients with metastasis,
which accounts for the majority of cases at the time of diagnosis.
In a previous study, up to 69% of advanced NSCLC patients were
estimated to have targetable mutations in a number of genes,
including epidermal growth factor receptor (EGFR), anaplastic
lymphoma kinase, c-ROS oncogene 1, Kirsten rat sarcoma virus, V-RAF
murine sarcoma oncogene homolog B1, mesenchymal-epithelial
transition factor receptor, and human EGFR-related 2 (6).
Epigenetic alterations have been detected in all
human cancers, and these changes, including DNA methylation, play
an important role in the early development and progression of
diseases, particularly cancer (7).
The DNA methylation of intermittently distributed CpG sites
down-regulates the expression of tumor suppressor genes.
Furthermore, aberrant DNA methylation patterns have been identified
in many cases of lung cancer, with most being located in promoter
sequences (8,9). Moreover, a progressive CpG island
(CGI) methylation pattern was detected in atypical adenomatous
hyperplasia and adenocarcinoma in situ that was followed by
further methylation and the development of LADC (10,11).
These findings suggest that DNA methylation has potential in the
development of biomarker candidates for the early detection of
cancer and outcome predictions.
We previously performed the genome-wide screening of
aberrantly methylated CGIs using the paired tumorous and
non-tumorous tissues of 12 LADC samples. The top 10 significantly
methylated genes were noted and the gene showing the highest
methylation level was dipeptidyl peptidase-like 6 (DPP6).
Furthermore, the expression of DPP6 was down-regulated in
cancer cell lines (12).
DPP6 is a type II transmembrane protein and a member of the
prolyl oligopeptidase family of serine proteases (13,14).
DPP6 is mostly studied as an auxiliary subunit of
voltage-gated K+ channels of the Kv4 family. However, DPP6
hypermethylation has been identified in several cancer types
(15–19).
In the present study, we examined the DNA
methylation and gene expression levels of DPP6 in resected
LADC tissues to assess its potential as a prognostic marker for
LADC.
Materials and methods
Patients and tissue samples
Seventy-three LADC tissues, including 25 pairs of
tumor-matched normal tissues, were collected from patients who
underwent surgery at Tokushima University Hospital between April
1999 and November 2013. Tissue samples were snap-frozen and stored
at 80°C for the later isolation of DNA and RNA. A methylation
analysis and RT-PCR were performed on 73 tumor samples and 25
normal lung samples. Tumor staging was conducted based on the
seventh tumor-node-metastasis classification for lung cancer
(20). Tumors were classified
according to the predominant histological subtype proposed by the
2015 WHO classification (21). The
mean follow-up duration for 162 patients with LADC was 48 months
(range, 0.6-147 months), with 45 cases of recurrence in (27.8%) and
34 of death (21.0%).
The Ethics Committee of the University of Tokushima
approved the present study (Tokushima University Hospital, approval
no. 4071), and procedures were conducted according to the
Declaration of Helsinki. All patients provided written informed
consent.
Clinical characteristics
Clinical information on 73 LADC patients, including
survival data and basic characteristics, such as sex, age at
diagnosis, tumor stage, histology, smoking history, and EGFR
mutations, was obtained and shown in Table I.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
|
Characteristics | Value, n | % or IQR |
|---|
| Sex |
|
|
|
Male | 39 | 53% |
|
Female | 34 | 47% |
| Age | 66.8±9.8 | (43–84) |
| Non-smoker | 35 | 48% |
| Smoker | 38 | 48% |
| Brinkman index | 783±487 |
|
| Pathology |
|
|
|
Lepidic | 23 | 31.5% |
|
Papillary | 26 | 35.6% |
|
Solid | 3 | 4.1% |
|
Acinar | 5 | 6.8% |
|
Mixed or
others | 16 | 21.9% |
| Operation |
|
|
|
Pneumonectomy | 1 | 1.4% |
|
Lobectomy | 69 | 94.5% |
|
Segmentectomy | 1 | 1.4% |
|
Lobectomy +
segmentectomy | 3 | 4.1% |
| Complete
resection | 73 | 100% |
| Preoperative
chemotherapy | 1 | 1.4% |
| Postoperative
chemotherapy | 20 | 27.4% |
|
Cisplatin-based | 6 |
|
|
UFT | 13 |
|
|
Other | 1 |
|
| Pathological
stage |
|
|
|
IA | 38 | 52.1% |
|
IB | 20 | 27.4% |
|
IIA | 9 | 12.3% |
|
IIB | 4 | 5.5% |
|
III | 2 | 2.7% |
| pN factor |
|
|
|
0 | 65 | 89.0% |
|
1 | 6 | 8.2% |
|
2 | 2 | 2.7% |
| Pl factor
positive | 16 | 21.9% |
| v factor positive
(n=71) | 12 | 16.9% |
| ly factor
positive | 12 | 16.4% |
| EGFR mutation
(n=24) | 9 | 37.5% |
Global methylation analysis
We previously screened 12 paired
tumorous/non-tumorous LADC sample sets obtained from freshly frozen
specimens using Illumina HumanMethylation450 K Bead Chip to
identify aberrantly methylated CGIs in a genome-wide manner.
Nineteen CGIs were identified as differentially hypermethylated in
the DPP6 gene, with a false discovery rate of <0.05 and β
difference (tumor-non-tumorous tissue) of >0.25. CGI in
DPP6 was ranked as the most hypermethylated CGI with a high
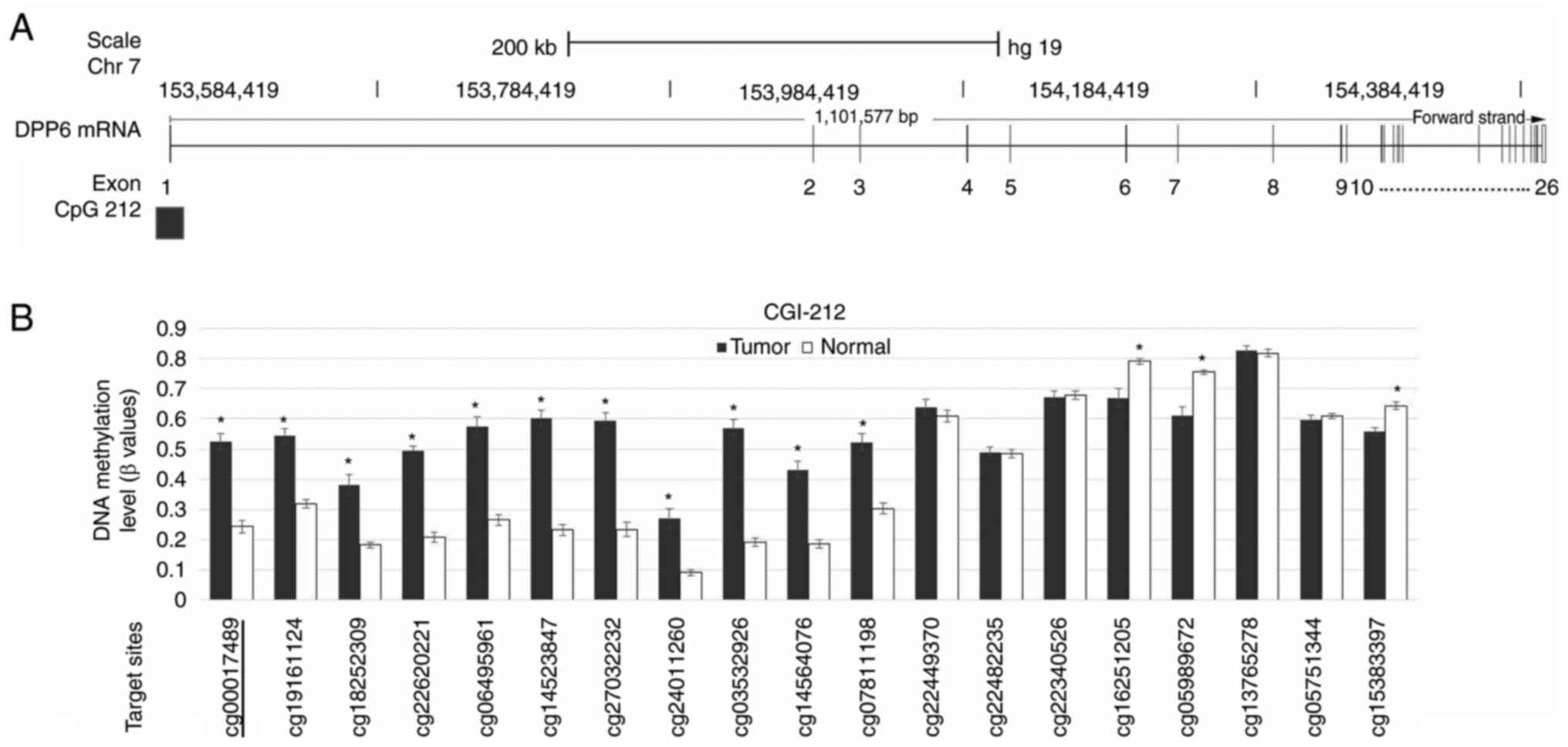
P-value. A schematic diagram of the DPP6 mRNA structure is
shown in Fig. 1A. Among the 26
exons of DPP6 mRNA, the CGI was located around exon 1.
Fig. 1B shows the results of the
array-based methylation status of each CpG site within DPP6.
We examined the DNA methylation of 4 CpG sites near cg00017489 of
the DPP6 gene.
Nucleic acid isolation
DNA and RNA were extracted from frozen tissue using
the QIAamp DNA Mini Kit 50 and RNeasy Mini kit (Qiagen GmbH)
according to the manufacturer's instructions.
Bisulfite pyrosequencing
Genomic DNA was converted to bisulfite using the
manufacturer's specified EpiTect Bisulfite Kit 48 (Qiagen).
Transformed DNA was amplified by PCR with Pyromark PCR-Kit 200
(Qiagen) before pyrosequencing. Sequencing primers were developed
using PyroMark Assay Design 2.0: forward,
TTTGGGTGGGTTTGTATATGAATTTG; reverse,
(biotin)-TACCCCTAAACCCTATTCCAT-CAATCATC. Pyrosequencing was
performed according to published protocols using PSQ 96MA (Qiagen)
to obtain the methylation rate at the mean of four CpG sites. The
methylation rate at each CpG was calculated using PyroQ-CpG
1.0.9.
Real-time quantitative PCR
Reverse transcription was performed with iScript
Reverse transcription Supermix. SsoAdvanced Universal
SYBR® Green Supermix (Bio-Rad) and the DPP6
Taqman Gene expression assay were used for quantitative PCR
employing the following primers: DPP6 forward
5′-ATGCAGGGGAACGTGATG-3′, DPP6 reverse
5′-GCAGTGCAATTGCTATTCCTT-3. GAPDH forward
5′-AGCCACATCGCTCAGACAC-3′, GAPDH reverse
5′-GCCCAATACGACCAAATCC-3′ GAPDH was used as the internal
control gene.
Statistical analysis
The paired t-test and Wilcoxon signed-rank
test were used for comparisons between paired samples when data
were and were not normally distributed, respectively. The
relationships between methylation and mRNA expression levels and
clinical characteristics, including tumor stage, histological
patterns, smoking history, blood vessel invasion, lymph vessel
invasion, pleural invasion, and lymph node metastasis, were
examined using the unpaired t-test for normally distributed
data and the Mann-Whitney test for not normally distributed data.
Multiple group comparisons were conducted using a one-way analysis
of variance followed by Tukey's multiple comparison test for
normally distributed data and the Kruskal-Wallis test followed by
Dunn's multiple comparison test for not normally distributed data.
The relationships between the methylation and mRNA expression
levels of DPP6 and basic clinical characteristics were
examined by the chi-square test. Survival data analyzed by the
Kaplan-Meier analysis with the Log-rank test were used to compare
overall survival (OS) and disease-free survival (DFS) rates across
high/low methylation and high/low mRNA expression levels. The
cut-off values were made from ROC curves. The multivariate survival
analyses were calculated using the likelihood ratio test of the
stratified Cox's proportional hazard regression analysis. All
statistical analyses were performed using GraphPad Prism, version
5.00; and SPSS (version 24.0; IBM Corp.).
Results
Methylation and mRNA expression levels
of DPP6 in LADC samples and paired adjacent normal tissues
The methylation and mRNA expression levels of the
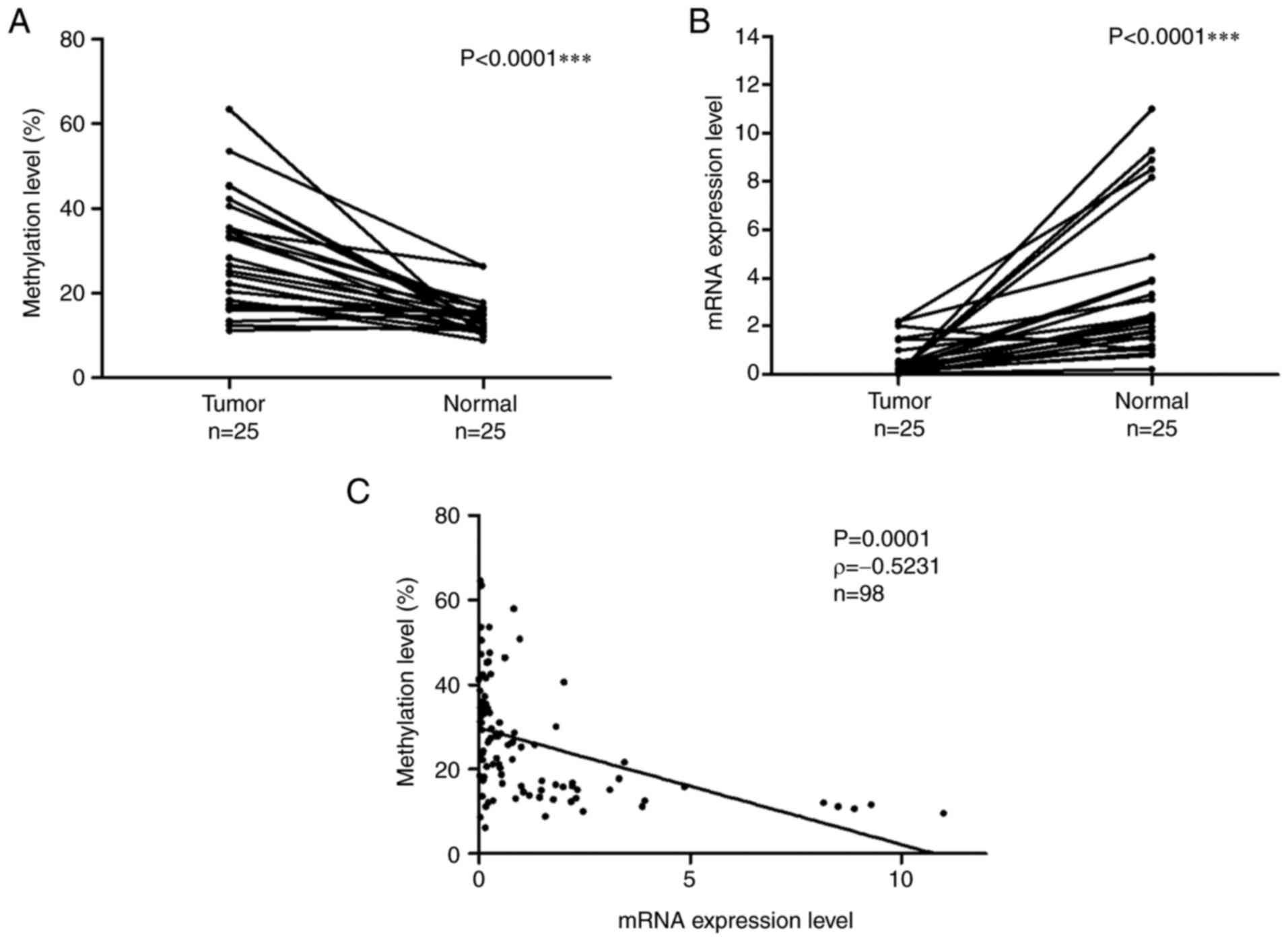
DPP6 gene were examined in 25 matched samples. Fig. 2A shows the methylation levels of the
DPP6 gene in LADC and normal lung tissues. DNA methylation
levels were significantly higher in tumor samples than in normal
samples (the Wilcoxon signed-rank test, P<0.0001). Fig. 2B shows the mRNA expression levels of
DPP6 in LADC and normal lung tissues. mRNA expression levels
were significantly lower in tumor samples than in normal samples
(the Wilcoxon signed-rank test, P<0.0001).
The relationship between the DNA methylation and
mRNA expression levels of DPP6 in 98 samples /73 tumor and
25 normal/ was examined with Spearman's rank correlation (Fig. 2C). The obtained results revealed a
negative relationship between methylation and mRNA expression
levels (P=0.0001, ρ=−0.5231).
Relationships between methylation and
mRNA expression levels of DPP6 and the malignant behavior of
LADC
To examine the effects of the hypermethylation of
DPP6 on the malignancy and carcinogenesis of LADC, we
measured methylation and mRNA expression levels in 73 samples and
compared the results obtained with histology patterns and stage
grading.
DPP6 methylation levels were lower in the IA
group than in the other groups. Furthermore, slightly higher mRNA
expression levels were observed in the IA group (Fig. 3A and 3B). No significant differences were
observed in methylation levels in samples with the lepidic and
other histological patterns (Fig.
3C). However, DPP6 mRNA expression levels were higher in
samples with the lepidic pattern than in those with other
histological patterns (Fig. 3D)
(the Mann-Whitney test P=0.0393). The relationships between the
methylation and mRNA expression levels of DPP6 and basic
clinical characteristics are shown in Table II. Data were analyzed by
cross-tabulation with the chi-square test. Methylation levels were
significantly higher in LADC with than in that without pleural
invasion (Table II). mRNA levels
were significantly lower in LADC with than in that without pleural
invasion (Table II). LADC with
vascular invasion had higher methylation and lower mRNA expression
levels of DPP6. The relationships between the mRNA
expression levels of DPP6 and blood and lymph vessel invasions are
shown in Fig S1.
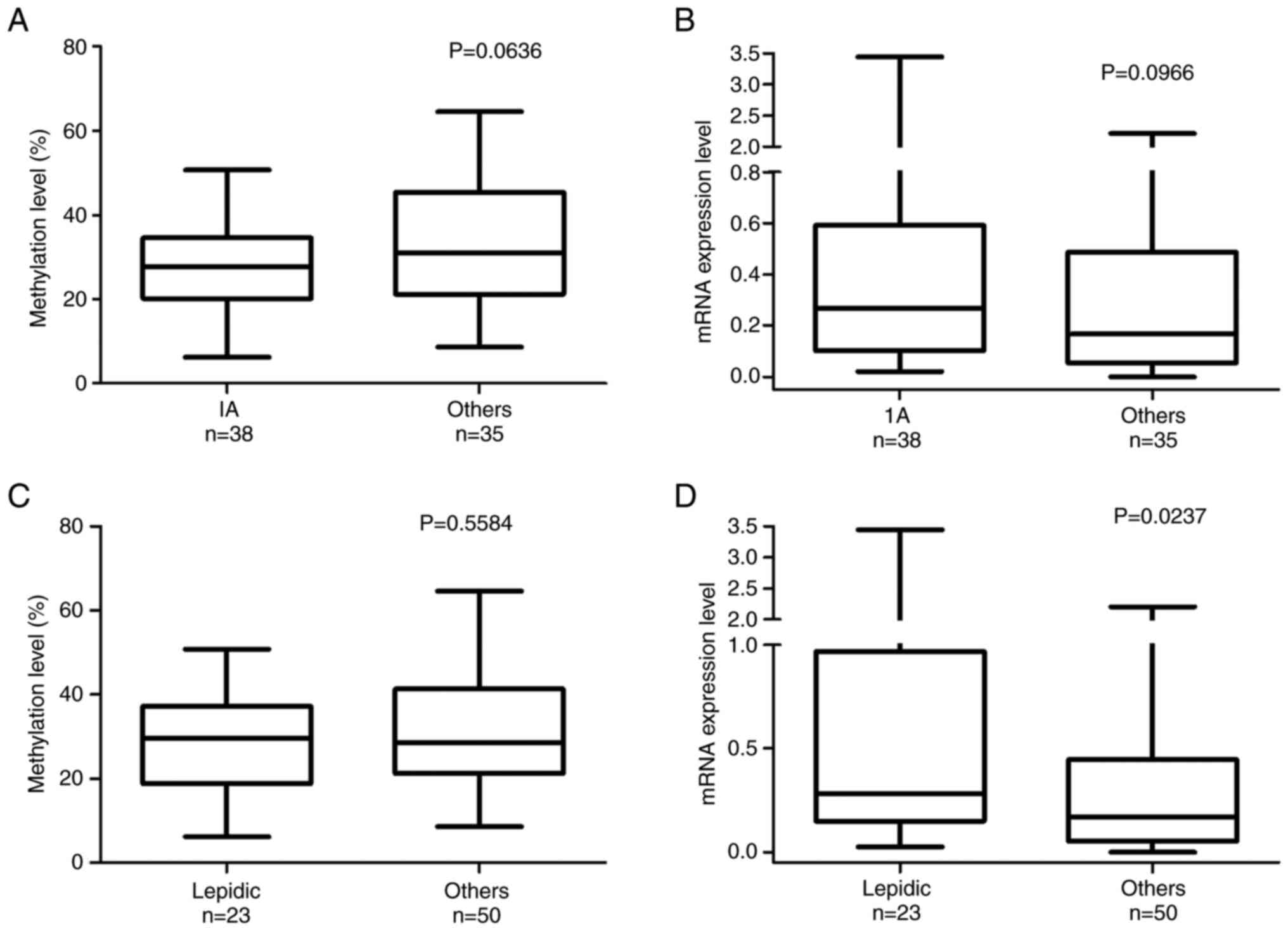 | Figure 3.DNA methylation and mRNA expression
levels of DPP6 in lung cancer stages and histological patterns. The
methylation and mRNA expression levels of DPP6 in LADC tumor
samples (n=73) were analyzed using pyrosequencing and reverse
transcription-quantitative PCR, respectively. According to the
World Health Organisation histological classification, the (A)
methylation (%) and (B) mRNA expression levels of DPP6 are shown in
lung cancer stages (IA, n=38; others: IB, n=20; IIA, n=9; IIb, n=4;
IIIa, n=1). The (C) methylation (%) and (D) mRNA expression levels
of DPP6 are shown in lung cancer histological patterns (lepidic,
n=23; others: papillary, n=26; acinar, n=5; solid, n=3; and mixed
n=16). DPP6, dipeptidyl peptidase-like 6. |
 | Table II.Characteristics of patients grouped
by the median value of DNA methylation or mRNA expression. |
Table II.
Characteristics of patients grouped
by the median value of DNA methylation or mRNA expression.
| A, Characteristics
of patients grouped by the median value of DNA methylation |
|---|
|
|---|
|
|
| DPP6 methylation
level |
|
|---|
|
|
|
|
|
|---|
| Factors | Group | High | Low | P-value |
|---|
| Sex | Female/male | 20/17 | 14/22 | 0.194a |
| Age±SD |
| 67.56±10.29 | 66.25±9.506 | 0.778b |
| Smoking |
Non-smoker/smoker | 15/21 | 20/17 | 0.290a |
| Stage (ver.7) | IA/others | 15/21 | 23/14 | 0.080a |
| IP | -/+ | 35/2 | 32/4 | 1.000a |
| Emphysema | -/+ | 35/2 | 34/2 | 1.000a |
| Tumor size
(mm±SD) |
| 28.72±13.17 | 25.44±14.31 | 0.202b |
| Metastasis | -/+ | 33/3 | 32/5 | 0.711a |
| Pleural | -/+ | 23/13 | 34/3 | 0.004a |
| Vascular | -/+ | 26/9 | 33/3 | 0.050a |
| Lymph vessel | -/+ | 30/6 | 31/6 | 0.959a |
| Pathology |
Lep/others | 12/24 | 11/26 | 0.740a |
|
| B,
Characteristics of patients grouped by the median value of mRNA
expression. |
|
|
|
| DPP6 methylation
level |
|
|
|
|
|
|
| Factors | Group | High | Low | P-value |
|
| Sex | Female/male | 15/21 | 19/18 | 0.407a |
| Age±SD |
| 67.38±8.597 | 66.25±11.02 | 0.620b |
| Smoking |
Non-smoker/smoker | 20/17 | 15/21 | 0.290a |
| Stage (ver.7) | IA/others | 21/16 | 17/19 | 0.415a |
| IP | -/+ | 34/3 | 33/3 | 1.000a |
| Emphysema | -/+ | 36/1 | 33/3 | 0.358a |
| Tumor size
(mm±SD) |
| 25.35±10.44 | 29±16.29 | 0.690c |
| Metastasis | -/+ | 33/3 | 32/5 | 0.711a |
| Pleural | -/+ | 21/6 | 26/10 | 0.233a |
| Vascular | -/+ | 34/2 | 25/10 | 0.010a |
| Lymph vessel | -/+ | 34/3 | 27/9 | 0.050a |
| Pathology | Lep/others | 14/23 | 9/27 | 0.238a |
Prognostic value of DPP6 mRNA
expression and methylation levels in LADC
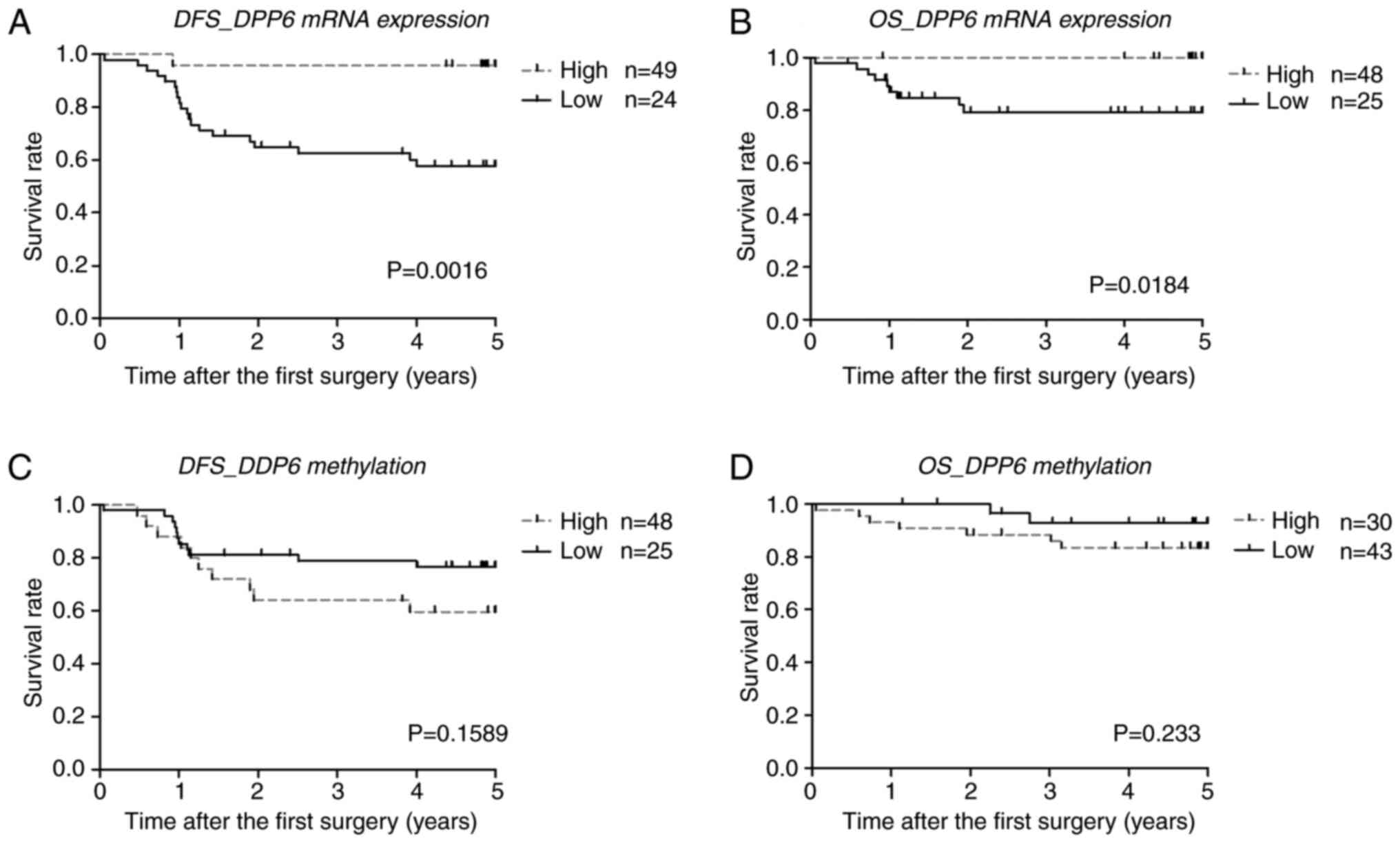
We performed DFS and OS analyses of the mRNA
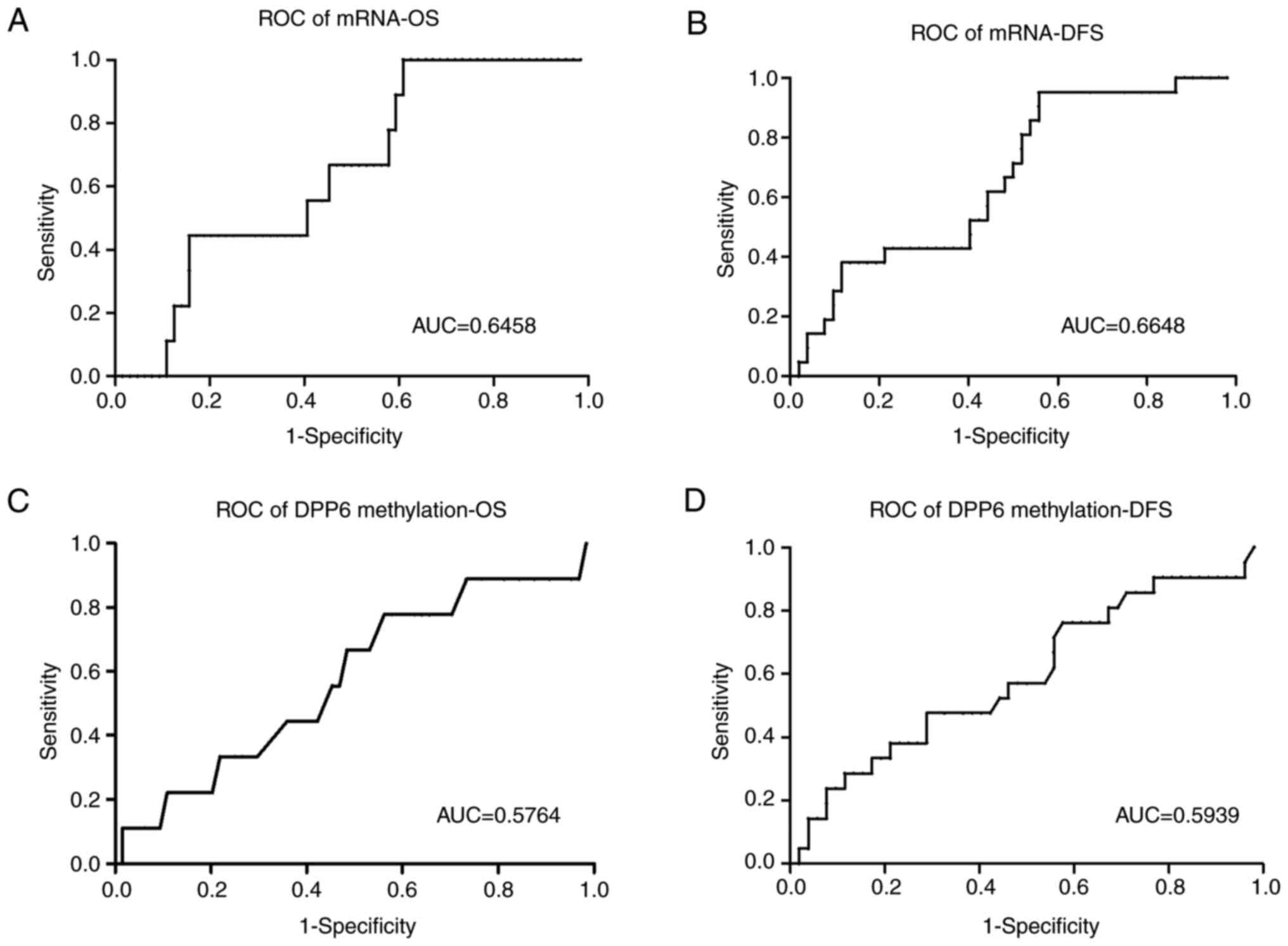
expression and methylation levels of DPP6 (Fig. 4). Samples were divided into high/low
expression and high/low methylation levels using cut-off values
generated from the ROC/Receiver-Operating characteristic/curves
(Fig. 5). DFS and OS rates were
significantly higher in samples with high mRNA expression levels of
DPP6 than in those with low levels (the Kaplan-Meier
analysis, DFS P=0.0016 cut-off value 0.4 and OS P=0.0184 cut-off
value 0.3).
However, no significant differences were observed in
DFS or OS between samples with high and low DPP6 methylation
levels (the Kaplan-Meier analysis, DFS P=0.1589 cut-off value 27
and OS P=0.233 cut-off value 34.5). The ROC curves of DFS and OS
are shown in (Fig. 5).
The multivariate Cox's regression analysis revealed
that the tumor staging (HR; 0.289, 95% CI; 0.094-0.885 P=0.030) was
an independent prognostic factor for DFS (Table III) and the patients' age (HR;
5.377, 95% CI; 1.086-26.623 P=0.039) for OS (Table III). According to the multivariate
analysis the DNA methylation and mRNA expression level of DPP6 were
not significant to predict DFS and OS.
 | Table III.Cox proportional hazard regression
analysis of disease-free and overall survival in 73 lung
adenocarcinoma samples. |
Table III.
Cox proportional hazard regression
analysis of disease-free and overall survival in 73 lung
adenocarcinoma samples.
| A, Cox proportional
hazard regression analysis of disease-free survival in 73 lung
adenocarcinoma samples. |
|---|
|
|---|
|
| Multivariate
analysis |
|---|
|
|
|
|---|
| Factors | Hazard ratio | 95% CI | P-value |
|---|
| Sex, male (n=39)
vs. female (n=34) | 1.075 | 0.149–7.734 | 0.943 |
| Age, ≥67 (n=36) vs.
<67 (n=37) | 2.475 | 0.967–6.334 | 0.059 |
| Smoking, non-smoker
(n=35) vs. smoker (n=38) | 0.344 | 0.049–2.432 | 0.285 |
| Stage, (ver.7) IA
(n=38) vs. others (n=35) | 0.289 | 0.094–0.885 | 0.030 |
| Metastasis,
negative (n=65) vs. positive (n=8) | 2.994 | 0.880–10.183 | 0.079 |
| Lymph vessel,
negative (n=61) vs. positive (n=12) | 2.048 | 0.591–7.101 | 0.259 |
| Pathology, lepidic
(n=23) vs. others (n=50) | 0.611 | 0.165–2.262 | 0.461 |
| mRNA expression,
low (n=37) vs. high (n=36) | 1.991 | 0.731–5.425 | 0.178 |
| DNA methylation,
low (n=36) vs. high (n=37) | 0.695 | 0.271–1.782 | 0.449 |
|
| B, Cox
proportional hazard regression analysis of overall survival in 73
lung adenocarcinoma samples. |
|
|
| Multivariate
analysis |
|
|
|
| Factors | Hazard
ratio | 95% CI | P-value |
|
| Sex, male (n=39)
vs. female (n=34) | 0.900 | 0.012–68.553 | 0.962 |
| Age, ≥67 (n=36) vs.
<67 (n=37) | 5.377 | 1.086–26.623 | 0.039 |
| Smoking, non-smoker
(n=35) vs. smoker (n=38) | 0.505 | 0.006–39.686 | 0.759 |
| Stage, (ver.7) IA
(n=38) vs. others (n=35) | 0.389 | 0.076–1.983 | 0.256 |
| Metastasis,
negative (n=65) vs. positive (n=8) | 0.351 | 0.025–4.898 | 0.436 |
| Lymph vessel,
negative (n=61) vs. positive (n=12) | 1.673 | 0.220–12.719 | 0.619 |
| Pathology, lepidic
(n=23) vs. others (n=50) | 0.293 | 0.029–2.915 | 0.295 |
| mRNA expression,
low (n=37) vs. high (n=36) | 1.748 | 0.398–7.682 | 0.460 |
| DNA methylation,
low (n=36) vs. high (n=37) | 0.562 | 0.127–2.480 | 0.447 |
Discussion
In addition to its role in the modulation of A-type
potassium channels in neurons and also in neuronal development and
synaptic plasticity (13),
DPP6 is involved in tumorigenesis (22,23).
Furthermore, the hypermethylation of DPP6 has been
identified in several cancer types and is associated with survival
rates, which will be discussed in the following paragraphs.
We examined the methylation and gene expression
levels of the DPP6 gene. Low methylation and high mRNA
expression levels of DPP6 were observed in normal tissues.
On the other hand, high methylation and low mRNA expression levels
of DPP6 were detected in tumor tissues. These results
indicate an inverse relationship between the methylation and mRNA
expression levels of DPP6 in LADC. The relationship between
the DNA methylation and mRNA expression levels of DPP6 was
analyzed by Spearman's rank correlation (P=0.001).
DPP6 is primarily a tumor suppressor gene. A
previous study on pancreatic ductal adenocarcinoma reported that
the β-value of the DPP6 promoter was significantly higher in
tumor samples than in normal samples, while the methylation levels
of the DPP6 promoter negatively correlated with mRNA
expression levels in 72 paired tissues (15). Another study detected the
hypermethylation of DPP6 in esophageal adenocarcinoma, which
showed the down-regulated mRNA expression of DPP6 in 78
paired samples (16). Furthermore,
DPP6 was shown to be significantly methylated in acute
myeloid leukemia, and analyses of its gene expression revealed its
down-regulation (17).
Collectively, these findings indicate that DPP6 is tightly
regulated during oncogenic development. Moreover, the
hypermethylation of DPP6 was reported in clear cell renal
cell carcinoma (ccRCC) and was associated with advanced and
high-grade tumors (18).
DPP6 was also identified as one of the differentially
methylated genes in LADC.
This study used data obtained from UCSC Xena, which
included 29 paired samples. The methylation levels of DPP6
were higher in tumor samples than in normal samples (19). These findings indicate that the
hypermethylation of DPP6 occurs in several cancer types and
contributes to the low mRNA expression levels of DPP6.
In the present study, the methylation and expression
levels of DPP6 were associated with lung cancer stages and
histological patterns. Low methylation and high expression levels
of DPP6 were detected in the early stage of LADC (IA). In
contrast, high methylation and low expression levels of DPP6
were observed in the advanced stages of LADC (including IB, II, and
III). The methylation levels of DPP6 were lower in samples
with the lepidic pattern than in those with other histological
patterns. Furthermore, the mRNA expression levels of DPP6
were significantly higher in the former than in the latter. LADC
with a lepidic histology pattern is less malignant than that with
other histological patterns.
DPP6 was identified as one of the genetically
altered genes that regulate vascular invasion in human pancreatic
cancer (24). In the present study,
positive blood and lymph vessel invasion was associated with low
mRNA expression levels of DPP6 (Fig. S1). Collectively, these findings and
the present results suggest that high methylation and low
expression levels of DPP6 have a negative impact on the
prognosis of patients with several types of cancers, including
LADC.
The survival rate of patients with lung cancer is
low due to the lack of specific symptoms in the early stages and
practical biomarkers for screening and outcome predictions.
Therefore, we analyzed OS and DFS based on a receiver operating
characteristic curve. The methylation level of DPP6 did not
significantly differ between the groups examined; however, the
survival rate was higher in the group with low methylation levels
than in that with high methylation levels. On the other hand, the
survival rate was significantly higher in the group with high mRNA
expression levels than in that with low expression levels.
A previous study examined the survival rates of
patients with breast cancer based on the expression of DPP6.
According to Kaplan-Meier plots, high expression levels of
DPP6 were associated with higher DFS rates (n=2898)
(25). Furthermore, a Kaplan-Meier
survival analysis of candidate biomarkers and 12 genes, including
DPP6, revealed their significant effects on the prognosis of
pancreatic cancer patients. The OS rate was higher in the group
with high mRNA expression levels of DPP6 (n=150) (26). Therefore, high methylation and low
mRNA expression levels of DPP6 may have a negative impact on
the prognosis of patients with cancer.
Limited information is currently available on the
oncogenic properties of the DPP6 gene. In patients with
esophageal carcinoma, high expression levels of DPP6 were
associated with shorter survival, and DPP6 was negatively
regulated by activating enhancer-binding protein 2 (AP-2α) and
positively by AP-2γ (27).
Furthermore, in patients with esophageal squamous cell carcinoma,
DPP6 was up-regulated by ARID3A and down-regulated by ZNF354C
(28). DPP6 is also involved
in the regulation of cell-specific phenotypes and its deregulation
may result in carcinogenesis. Low methylation and high expression
levels of the DPP6 gene have been observed in colon cancer
(29). Based on these findings,
DPP6 may act as a tumor suppressor gene in LADC; however,
the methylation and expression levels of DPP6 may depend on
the cancer type.
We performed immunohistochemistry to detect the
expression of DPP6. However, we were unable to evaluate the
results obtained because the protein expression of DPP6 in
healthy lungs was generally low. Therefore, we need a more
sensitive assay to assess tissue protein expression. Furthermore,
DPP6 was stained in the interstitial tissues, but not tumor
cells of tumor tissues, indicating non-specific staining. The
function of DPP6 in the regulation of tumor progression
currently remains unknown. Therefore, further studies are needed to
obtain more detailed insights into this novel biomarker.
In conclusion, the methylation levels of DDP6
differed between lung cancer tissues and normal tissues. The low
mRNA expression level of DPP6 was associated with the malignant
features of lung cancer samples and poor survival rates. The
present results indicate the potential of DPP6 as a biomarker for
lung cancer screening and outcome predictions.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
BM, KK, SS, and BT analyzed and interpreted patient
data on DNA methylation using pyrosequencing and mRNA expression by
RT-qPCR. KK, HTo, and HTa designed and conducted the present study.
KK, CT and NK performed histological examinations of lung cancer
and combined the clinical data of patients. BM and KK were major
contributors to the writing of the manuscript. BM and KK confirm
the authenticity of raw data. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
The Ethics Committee of the University of Tokushima
approved the present study (Tokushima University Hospital; approval
no. 4071), and all procedures were conducted according to the
Declaration of Helsinki. All patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CGIs
|
CpG islands
|
|
LADC
|
lung adenocarcinomas
|
|
DPP6
|
dipeptidyl peptidase-like 6
|
|
NSCLC
|
non-small cell lung cancer
|
|
EGFR
|
epidermal growth factor receptor
|
|
OS
|
overall survival
|
|
DFS
|
disease-free survival
|
References
|
1
|
Sheikh MA, Malik YS, Yu H, Lai M, Wang X
and Zhu X: Epigenetic regulation of Dpp6 expression by Dnmt3b and
its novel role in the inhibition of RA induced neuronal
differentiation of P19 cells. PLoS One. 8:e558262013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Succony L, Rassl DM, Barker AP, McCaughan
FM and Rintoul RC: Adenocarcinoma spectrum lesions of the lung:
Detection, pathology and treatment strategies. Cancer Treat Rev.
99:1022372021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bade BC and Dela Cruz CS: Lung cancer
2020: Epidemiology, etiology, and prevention. Clin Chest Med.
41:1–24. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zito Marino F, Bianco R, Accardo M, Ronchi
A, Cozzolino I, Morgillo F, Rossi G and Franco R: Molecular
heterogeneity in lung cancer: From mechanisms of origin to clinical
implications. Int J Med Sci. 16:981–989. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kan Z, Jaiswal BS, Stinson J, Janakiraman
V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, et al:
Diverse somatic mutation patterns and pathway alterations in human
cancers. Nature. 466:869–873. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wheeler DA and Wang L: From human genome
to cancer genome: the first decade. Genome Res. 23:1054–1062. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jha G, Azhar S, Rashid U, Khalaf H,
Alhalabi N, Ravindran D and Ahmad R: Epigenetics: The key to future
diagnostics and therapeutics of lung cancer. Cureus.
13:e197702021.PubMed/NCBI
|
|
8
|
Rauch TA, Wang Z, Wu X, Kernstine KH,
Riggs AD and Pfeifer GP: DNA methylation biomarkers for lung
cancer. Tumour Biol. 33:287–296. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chao YL and Pecot CV: Targeting
epigenetics in lung cancer. Cold Spring Harb Perspect Med.
11:a0380002021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Selamat SA, Galler JS, Joshi AD, Fyfe MN,
Campan M, Siegmund KD, Kerr KM and Laird-Offringa IA: DNA
methylation changes in atypical adenomatous hyperplasia,
adenocarcinoma in situ, and lung adenocarcinoma. PLoS One.
6:e214432011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chung JH, Lee HJ, Kim BH, Cho NY and Kang
GH: DNA methylation profile during multistage progression of
pulmonary adenocarcinomas. Virchows Arch. 459:201–211. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kajiura K, Masuda K, Naruto T, Kohmoto T,
Watabnabe M, Tsuboi M, Takizawa H, Kondo K, Tangoku A and Imoto I:
Frequent silencing of the candidate tumor suppressor TRIM58 by
promoter methylation in early-stage lung adenocarcinoma.
Oncotarget. 8:2890–2905. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Strop P, Bankovich AJ, Hansen KC, Garcia
KC and Brunger AT: Structure of a human A-type potassium channel
interacting protein DPPX, a member of the dipeptidyl aminopeptidase
family. J Mol Biol. 343:1055–1065. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nadal MS, Ozaita A, Amarillo Y, Vega-Saenz
de Miera E, Ma Y, Mo W, Goldberg EM, Misumi Y, Ikehara Y, Neubert
TA and Rudy B: The CD26-related dipeptidyl aminopeptidase-like
protein DPPX is a critical component of neuronal A-type K+
channels. Neuron. 37:449–461. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao X, Cao D, Ren Z, Liu Z, Lv S, Zhu J,
Li L, Lang R and He Q: Dipeptidyl peptidase like 6 promoter
methylation is a potential prognostic biomarker for pancreatic
ductal adenocarcinoma. Biosci Rep. 40:BSR202002142020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xi T and Zhang G: Epigenetic regulation on
the gene expression signature in esophagus adenocarcinoma. Pathol
Res Pract. 213:83–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Saied MH, Marzec J, Khalid S, Smith P,
Down TA, Rakyan VK, Molloy G, Raghavan M, Debernardi S and Young
BD: Genome wide analysis of acute myeloid leukemia reveal leukemia
specific methylome and subtype specific hypomethylation of repeats.
PLoS One. 7:e332132012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kang HW, Park H, Seo SP, Byun YJ, Piao XM,
Kim SM, Kim WT, Yun SJ, Jang W, Shon HS, et al: Methylation
signature for prediction of progression free survival in surgically
treated clear cell renal cell carcinoma. J Korean Med Sci.
34:e1442019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li M, Zhang C, Zhou L, Li S, Cao YJ, Wang
L, Xiang R, Shi Y and Piao Y: Identification and validation of
novel DNA methylation markers for early diagnosis of lung
adenocarcinoma. Mol Oncol. 14:2744–2758. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goldstraw P, Crowley J, Chansky K, Giroux
DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V and Sobin L;
International Association for the Study of Lung Cancer
International Staging Committee and Participating Institutions, .
The IASLC lung cancer staging project: proposals for the revision
of the TNM stage groupings in the forthcoming (seventh) edition of
the TNM Classification of malignant tumours. J Thorac Oncol.
2:706–714. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Travis WD, Brambilla E, Burke AP, Marx A
and Nicholson AG: Introduction to the 2015 world health
organization classification of tumors of the lung, pleura, thymus,
and heart. J Thorac Oncol. 10:1240–1242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sedo A and Kraml J: Dipeptidyl peptidase
IV in cell proliferation and differentiation. Sb Lek. 95:285–288.
1994.PubMed/NCBI
|
|
23
|
Kotacková L, Baláziová E and Sedo A:
Expression pattern of dipeptidyl peptidase IV activity and/or
structure homologues in cancer. Folia Biol (Praha). 55:77–84.
2009.PubMed/NCBI
|
|
24
|
Jones S, Zhang X, Parsons DW, Lin JC,
Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et
al: Core signaling pathways in human pancreatic cancers revealed by
global genomic analyses. Science. 321:1801–1806. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Choy TK, Wang CY, Phan NN, Khoa Ta HD,
Anuraga G, Liu YH, Wu YF, Lee KH, Chuang JY and Kao TJ:
Identification of Dipeptidyl Peptidase (DPP) family genes in
clinical breast cancer patients via an integrated bioinformatics
approach. Diagnostics (Basel). 11:12042021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jia Y, Shen M, Zhou Y and Liu H:
Development of a 12-biomarkers-based prognostic model for
pancreatic cancer using multi-omics integrated analysis. Acta
Biochim Pol. 67:501–508. 2020.PubMed/NCBI
|
|
27
|
Kolat D, Kaluzinska Z, Bednarek AK and
Pluciennik E: Prognostic significance of AP-2α/γ targets as cancer
therapeutics. Sci Rep. 12:54972022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Xu Y, Li Z, Zhu Y, Wen S, Wang M,
Lv H, Zhang F and Tian Z: Identification of the key transcription
factors in esophageal squamous cell carcinoma. J Thorac Dis.
10:148–161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Irizarry RA, Ladd-Acosta C, Wen B, Wu Z,
Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al:
The human colon cancer methylome shows similar hypo- and
hypermethylation at conserved tissue-specific CpG island shores.
Nat Genet. 41:178–186. 2009. View
Article : Google Scholar : PubMed/NCBI
|