The use of an artificial intelligence algorithm for circulating tumor cell detection in patients with esophageal cancer
- Authors:
- Published online on: June 8, 2023 https://doi.org/10.3892/ol.2023.13906
- Article Number: 320
-
Copyright: © Akashi et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Esophageal squamous cell carcinoma (ESCC) remains a significant global challenge, having the 6th highest mortality worldwide and killing over 500,000 people in 2020 (1). Despite recent progress in multidisciplinary treatments against ESCC, many patients die from distant metastasis or recurrence after surgery (2). Circulating tumor cells (CTCs) are defined as cancer cells that depart from the primary tumor to enter the bloodstream (3) and are considered predictors of distant metastasis and cancer recurrence (4,5). In esophageal cancer, researchers associate CTC detection with advanced disease stage, poor therapeutic response, and prognosis (6,7).
Most CTC separation techniques are two-step: firstly, cell enrichment of the sample and secondly, CTC detection. Enrichment protocols for CTCs generally use cell surface markers or morphological features enabling CTC isolation via immunological assays, microfluidic devices, or density gradient centrifugation (6–8). Although subsequent detection methods include flow cytometry, biomechanical discrimination, and polymerase chain reaction (7,9), with marker-stained cell manual detection by microscope the most common method. Increased attention is being paid to these approaches thanks to recent reports exploring cancer cell heterogeneity in terms of malignant potential and stem cell properties. Accordingly, identifying heterogeneity and malignant subsets in CTCs is a priority, with the usefulness of various surface markers reported (4,10–13). However, the use of multiple markers makes CTC detection more complex and time-consuming. Therefore, an accurate, easy-to-use, and rapid detection method is required for clinical application.
Artificial intelligence (AI) is the simulation of human intelligence processes demonstrated by a computer program. AI can extract important information from large amounts of diverse data, classifying and summarizing common patterns. Potentially alleviating a significant quantity of human workload (14,15). Recently, attention has focused on a method called deep learning, which uses multiple layers of artificial neural networks and is modeled after the human cerebral cortex (16). Object recognition is a major application of deep learning, with convolutional neural networks (CNN) applied facilitate image diagnosis (17).
The aim of this study was to establish an accurate and rapid image processing algorithm based on CNN for CTC detection in patients with ESCC. We first investigated the AI algorithm's accuracy in distinguishing ESCC cell lines from peripheral blood mononuclear cells (PBMCs), then used the AI algorithm to detect CTCs in peripheral blood samples obtained from ESCC patients.
Materials and methods
Patients' eligibility and sampling
This study was approved by the ethics review board of the University of Toyama Hospital (R2021042) and written informed consent was obtained from all ESCC participants. Peripheral blood was collected from 10 newly diagnosed ESCC patients and 5 healthy volunteers. Patient samples were collected between January 2022 and October 2022. The eligibility criteria for patients were i) a confirmed diagnosis of ESCC, ii) undergoing treatment at the University of Toyama Hospital, and iii) no ESCC treatment prior to enrollment. All cases were diagnosed according to the 7th edition of the Union for International Cancer Control system (18).
For the four surgical patients, peripheral blood samples were extracted from each patient during general anesthesia via the arterial pressure line prior to the operation. From the six patients who underwent chemotherapy, blood samples were extracted via a median cubital vein. Peripheral blood samples were obtained from each healthy volunteer via a median cubital vein. Blood samples were collected in 3 ml ethylenediaminetetraacetic acid (EDTA) tubes. Samples were processed within 3 h of the collection as described below.
Cell lines and cell culture
Human ESCC cell lines (KYSE30, KYSE140, KYSE520, and KYSE1440) were purchased from the Japanese Collection of Research Bioresources (JCRB, Tokyo, Japan). These cell lines are authenticated using STR profiling in the JCRB. Cells were cultured in Dulbecco's Modified Eagle (DMEM) medium (Nacalai tesque, Kyoto, Japan), supplemented with 1% penicillin-streptomycin and 10% heat-inactivated fetal calf serum (FCS). The culture was grown in cell culture dishes in a humidified atmosphere containing 5% CO2 at 37°C. Cells were washed with phosphate-buffered saline without calcium and magnesium (PBS, FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) and harvested with Trypsin-EDTA (0.25%) (ThermoFisher, Massachusetts, USA). The harvested cells were processed immediately for imaging as described below.
Sample collection and processing
We collected 2.5 ml of peripheral blood samples from ESCC patients and healthy volunteers in EDTA tubes. Density gradient centrifugation was performed using the RosetteSep™ Human Circulating Epithelial Tumor Cell Enrichment Cocktail (StemCell™ Technologies, Vancouver, Canada) combined with Lymphoprep™ (StemCell™ Technologies, Vancouver, Canada). To the 2.5 ml blood sample was added 250 µl (50 µl/ml) of the RosetteSep™ cocktail and then incubated for 20 min at room temperature. Blood samples were diluted with equal volumes of PBS and carefully layered onto Lymphoprep™ then centrifuged at 3,600 rpm at room temperature for 20 min. After centrifugation, supernatants were transferred to another 15 ml conical tube with cells pelleted by centrifugation at 1,800 rpm for 20 min at room temperature. The enriched cells were collected, red blood cells were lysed by BD Pharm Lyse lysing solution (Becton, Dickinson and Company, New Jersey, USA), and washed in PBS.
Cell labeling
Cell fixation was performed using 4% paraformaldehyde. For staining, human monoclonal EpCAM-phycoerythrin (PE) (clone REA764; MACS Miltenyi Biotec, Cologne, Germany) antibodies were used. Antibodies were diluted 1:50 in 50 µl PBS containing 5% FBS. After incubation for 60 min, the cells were washed in PBS and pelleted by centrifuge at 1,200 rpm for 5 min at 4°C. SlowFade™ Diamond Antifade Mountant with DAPI (ThermoFisher, Massachusetts, USA) was added and deposited on a microscope slide to be prepared for imaging.
Imaging, processing, and computational classification of cells using AI
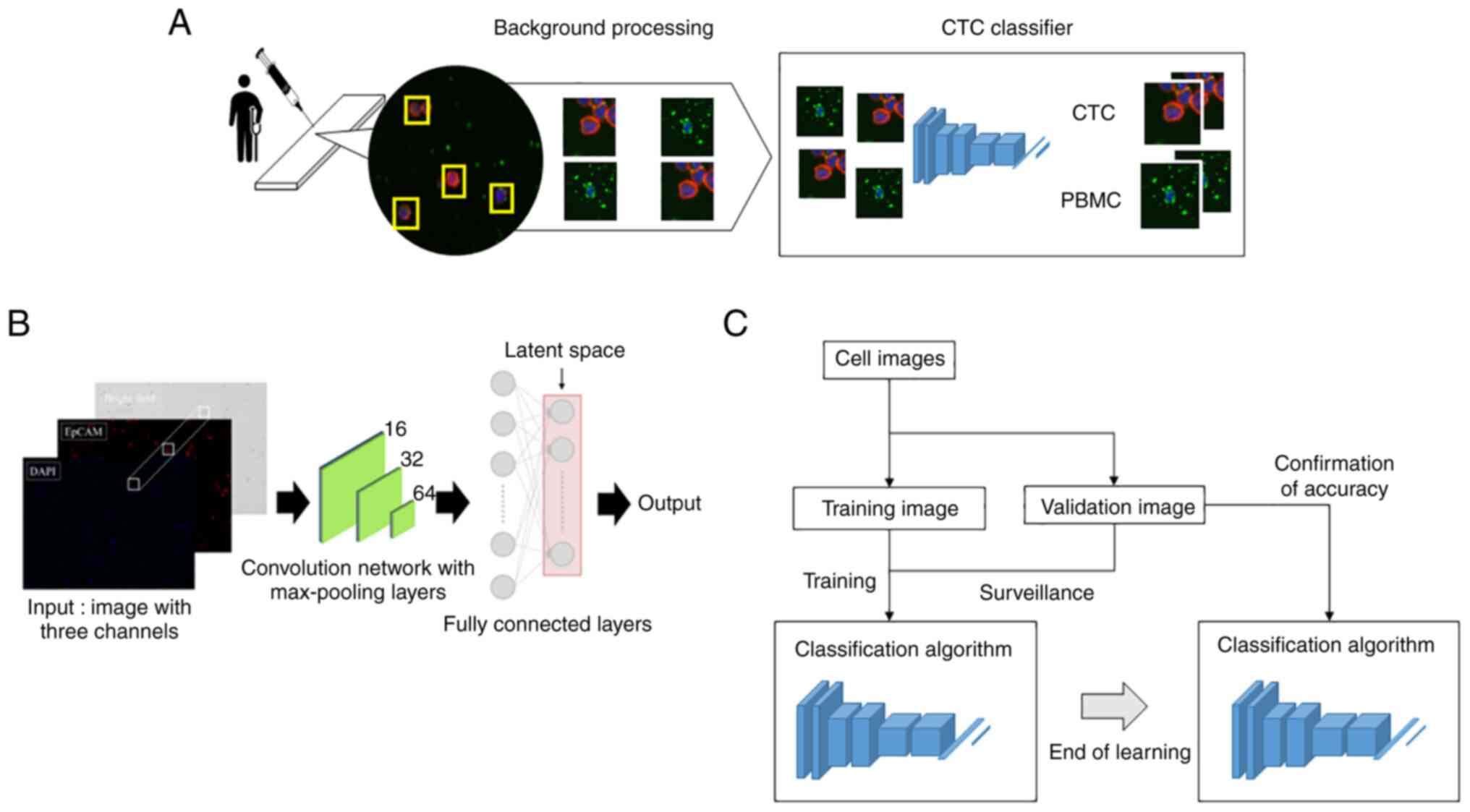
The cell classification process using the CNN-based algorithm is shown in Fig. 1A. Cellular regions were extracted from the microscopic image of KYSEs and blood samples. Luminance characteristic analysis was performed on these to control cell image background information. Then the brightness value components of each fluorescence were combined into a single image. Images were then fed into the CNN-based classifier for cancer cell evaluation.
Specifically, KYSEs and blood samples were prepared as above. Images were captured using the inverted microscope BZ-X800 (KEYENCE, Osaka, Japan). Images were taken at a 20× magnification through the objective lens. The acquired images were processed using an algorithm constructed in cooperation with the Department of Mechanical and Intellectual Systems Engineering, Faculty of Engineering, University of Toyama. Cell image cropping used the following morphological criteria: Extract the DAPI-positive site using Otsu's method (19). Narrowing down by requiring an EpCAM luminance of 20 or more. Furthermore, DAPI- and/or EpCAM-positive cells with an area greater than 700 pixels were excluded.
Fig. 1B shows the model with the classification network. The CNN consists of an input layer, hidden layer, and output layer. We input images of 64 pixels ×64 pixels to the input layer. The hidden layer includes many convolutional layers, pooling layers, and fully connected layers. The convolutional layer extracts various local features of the input layer through the convolution operation and normalizes the features for each channel image. The CNN performs feature extraction again at the pooling layer and semantically combines similar features to make the features robust to noise and deformation. The CNN samples these features, outputting them in a reduced processing size, this operation is repeated and continues with the fully connected layer. Each neuron in the fully connected layer is fully connected to all neurons in the previous layers. The fully connected layer integrates local information with class discrimination from the previous layers by the rectified linear unit (ReLU) function. Finally, the output value of the fully connected layer is passed to the output layer.
A schematic diagram of the evaluation method for the cell classifying AI algorithm accuracy is shown in Fig. 1C. The cell identification algorithm is trained using training images of cancer cells and PBMCs. Cell identification accuracy is then confirmed using validation images. By repeating this process, the AI algorithm accuracy is evaluated with regard to the training data variability.
The hardware environment used for computation was; CPU:Intel(r) Core(TM) i9-10980XE CPU @ 3.00GHz, Memory: 96GB, GPU:NVIDIA GeForce RTX 3090, V-RAM:24GB. The software environments used for computation were Python Ver.3.6.13, CUDA Ver.10.1, opencv Ver.4.5.3.56, cuDNN Ver.7.6.5, TensorFlow Ver.2.6.0, Keras Ver.2.6.0, NumPy Ver.1.19.5, pandas Ver.1.1.5, openpyel Ver.3.0.9, matplotlib Ver.3.3.4, scikit-learn Ver.0.24.2, seaborn Ver.0.11.2, shap Ver.0.40.0.
Classification of ESCC cell lines
To validate AI image recognition accuracy in distinguishing ESCC cells from PBMCs, we used images of ESCC cell lines stained with DAPI and EpCAM (KYSE30: 640 images, KYSE140: 194 images, KYSE520: 1037 images, KYSE1440: 347 images) and PBMCs from healthy volunteers (400 images). Specifically, we trained the AI using images of a KYSE and PBMC, shown a pair at a time and in order. Then the AI evaluated other image sets of KYSEs and PBMCs (KYSE30 vs. PBMCs, KYSE140 vs. PBMCs, KYSE520 vs. PBMCs, KYSE1440 vs. PBMCs), withholding the answers, to identify cell image as KYSE or PBMC.
Comparison of cell detection between AI image processing and manual cell count
To compare cell-detecting speeds between AI and humans, the AI and three researchers (TA, YN, TY) counted a total cell number in three identical images of KYSE140 and PBMCs each. Specifically, KYSE140 (1.0×105 cells) and PBMCs (1.0×107 cells), stained with DAPI as above, with 2×2 view images taken and merged to create 3 images for each KYSE140 and PBMCs from healthy volunteers. The AI and three researchers then counted the DAPI-positive areas recognized as cells, recording the time required to count.
Comparing AI and human image recognition accuracy
To compare image recognition accuracy between AI and humans, four researchers and pre-trained AI were tested to distinguish between KYSE140 and PBMCs. As described in the previous section, the AI was pre-trained using segmented 194 and 400 single cell images of KYSE140 and PBMCs, respectively, both stained with DAPI and EpCAM. Three sets of 100 images (50 images each were randomly selected from the 194 and 400 images of KYSE140 and PBMCs, respectively) were presented to the AI and four researchers separately (TA, YN, TY, NM), to identify the cell images as either KYSE140 or PBMC. To each image was assigned a hidden answer as to whether it was a KYSE or a PBMC. The researchers classified cells as KYSE140 and PBMCs based on the detection of EpCAM-positive/DAPI-stained cells and EpCAM-negative/DAPI-stained cells, respectively. The analysis time required for the 100 images was also noted.
Statistical analysis
All analyses were carried out with JMP 16.0 software (SAS Institute Inc., Cary, NC, USA). A confusion matrix was used to observe specificity, sensitivity, and accuracy. Difference between the AI and manual accuracy using image sets of KYSE140 and PBMCs was determined using the Wilcoxon rank-sum test P<0.05 was considered to indicate a statistically significant difference.
Results
Validation of the image recognition accuracy of AI
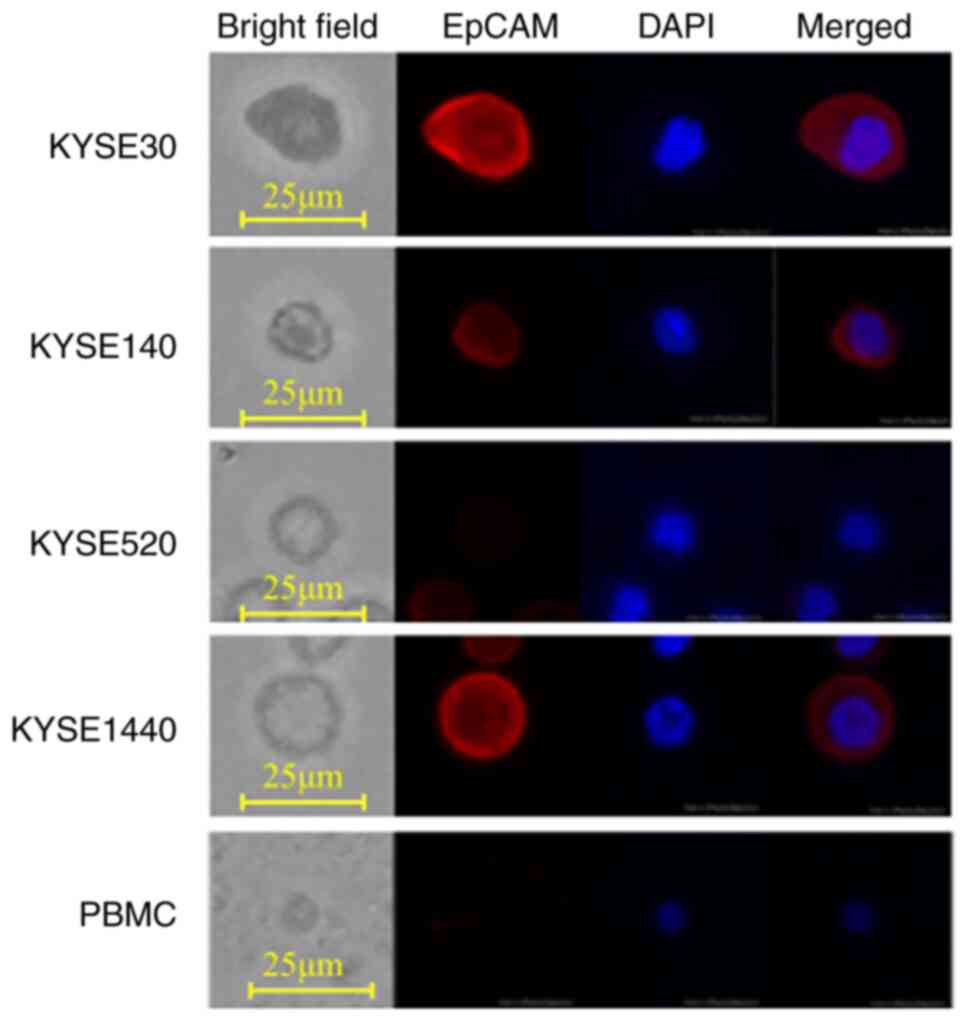
Firstly, the image recognition accuracy of the trained AI in distinguishing ESCC cell lines from PBMCs was evaluated. The AI was trained using identified paired images of single cells from ESCC cell lines and PBMC. Then the AI was shown paired images of a KYSE and PBMC, with the answer hidden, and tasked to identify which was the KYSE. Representative images of four ESCC cell lines (KYSE30, KYSE140, KYSE520, and KYSE1440) and PBMCs are shown in Fig. 2. PBMCs had no EpCAM expression and were small in both cell size and nucleus. KYSE520 did not express EpCAM, both KYSE30 and KYSE1440 strongly expressed EpCAM, while KYSE140 weakly expressed EpCAM. The AI differentiated KYSE30, KYSE140, KYSE520, and KYSE1440 from PBMCs with an accuracy of 99.9, 99.8, 99.8, and 100%, respectively, when trained using the same cell lines (Table I). Interestingly, even using KYSEs not used for training, the specificity in distinguishing KYSEs from PBMCs was greater than 99.6%, regardless of the KYSE combination used in training and examination. On the other hand, sensitivity varied from 20.4 to 100% depending on the KYSE combination used in training and examination (Table I). Among these four ESCC cell lines, we further validated the efficiency of the AI trained with KYSE140 by comparing it to human manual CTC detection.
The efficiency of AI image processing compared to manual counting
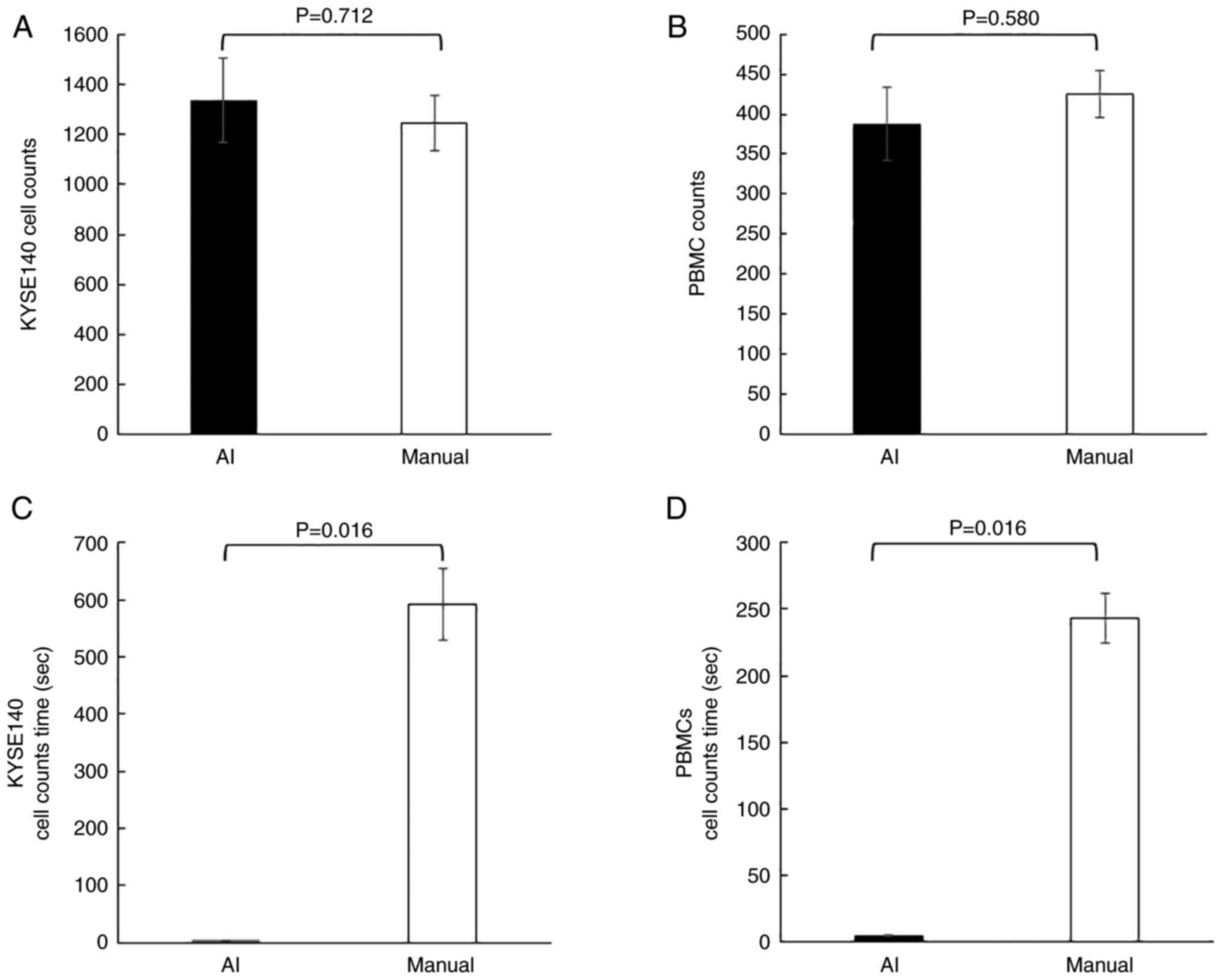
Secondly, the AI and three researchers each counted the number of DAPI-stained cells in three identical images of KYSE140 and PBMCs. AI image processing and manual counting detected the same number of KYSE140 cells and PBMCs (n=1,335.3±168.6 and 1,246.1±113.0 for KYSE140 cells by AI and manual detection, respectively, P=0.71 Fig. 3A; n=387.7±45.6 and 425.6±29.1 for PBMCs, P=0.58, Fig. 3B).
Whereas, using KYSE140, AI image processing and manual counting took 4.9±0.3 and 591.4±62.4 sec, respectively, with a significant difference (P=0.016, Fig. 3C). Using PBMCs, AI image processing and manual counting took 4.9±0.3 and 243.3±18.8 sec, respectively, with a significant difference (P=0.016, Fig. 3D).
These results showed no significant difference in the number of cells detected between AI and humans, but yielded a significantly shorter AI analysis time.
Comparison of image recognition accuracy between AI and humans
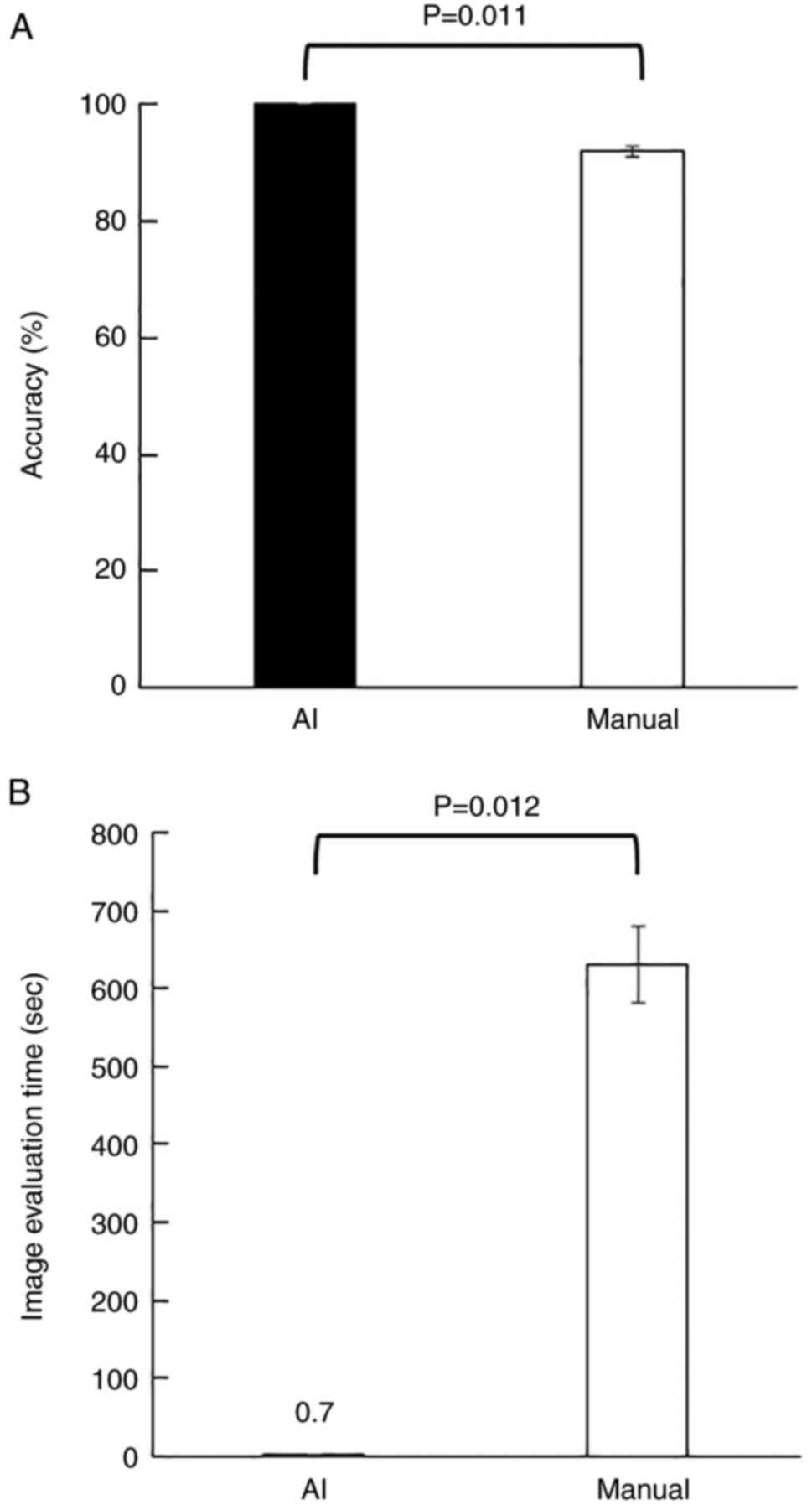
To compare AI and human image recognition accuracy in distinguishing cancer cells from PBMCs, the trained AI and four researchers were tasked to identify KYSE140 from PBMCs using images of 100 EpCAM/DAPI stained cells (50 of KYSE140 and 50 of PBMCs) with the answers withheld. After evaluating the three sets of 100 images, the AI completely distinguished KYSE140 from PBMCs with both a sensitivity and specificity of 100%, while the researchers distinguished them with a sensitivity and specificity of 86 and 97.5%, respectively (Table II). The average accuracies of the AI and researchers were 100 and 91.8% with a significant difference (P=0.011, Fig. 4A). The average times taken to classify 100 images for the AI and researchers were 0.7±0.01 and 630.4±49.5 sec, with a significant difference (P=0.012, Fig. 4B).
Table II.Comparison of image recognition accuracy between manual and AI methods, using KYSE140 and PBMC images. |
Detection of CTCs in blood samples of ESCC patients using the AI algorithm
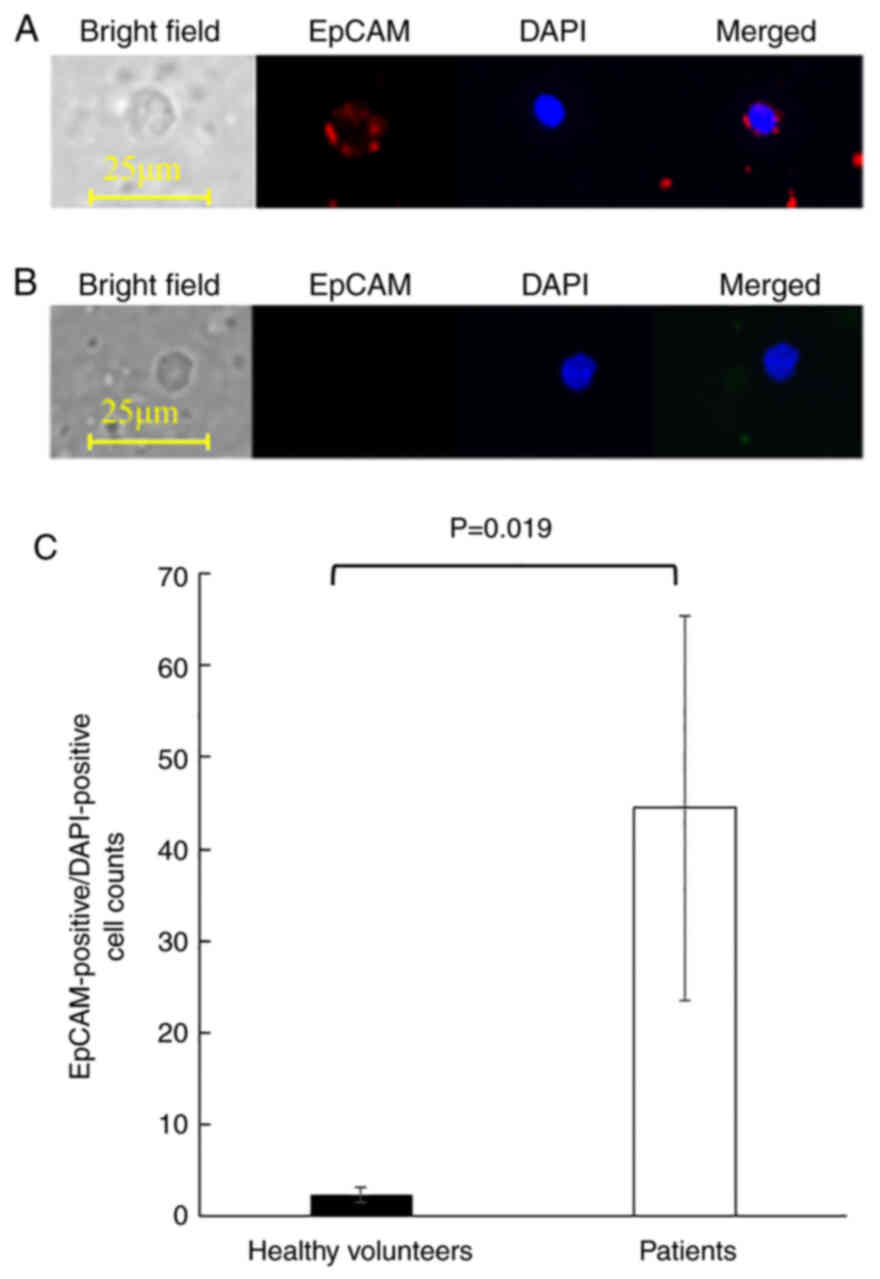
Finally, CTCs from the peripheral blood of 10 ESCC patients were enriched and processed using the image recognition AI algorithm to evaluate its clinical application. The clinicopathological characteristics of the patients are summarized in Table III. The patient population consisted of 5 men and 5 women, with a median age of 71.9 years (range, 54–79 years). Two patients presented with stage I disease and 8 patients with stage III. Blood samples from 5 healthy volunteers with a median age of 35.3 years (range, 30–39 years) were used as negative controls. Representative images of EpCAM-positive/DAPI-positive cells detected from patients were shown in Fig. 5A. The combination of nuclear DAPI staining and cell surface expression of EpCAM indicated that the cells were mononuclear cells of epithelial origin. On the other hand, PBMCs detected from healthy volunteers were small, had round nuclei, and did not express EpCAM, indicating that they were lymphocytes (Fig. 5B). Although EpCAM-positive/DAPI-positive cells were detected in all examined samples, ESCC patients yielded significantly more EpCAM-positive/DAPI-positive cells than the healthy volunteers (mean cell counts of 2.4±0.8 and 44.5±20.9, respectively, P=0.019, Fig. 5C).
Discussion
Though the presence of CTCs in ESCC patients is widely accepted, methods of CTC identification with high accuracy and efficiency are still under investigation. The performance of recent CNN-based diagnostic support tools is reaching a level comparable to experts in various medical fields (20–22).
In this study, we established a CNN-based image processing algorithm and validated its performance with ESCC cell lines and blood from ESCC patients. These results demonstrated that AI distinguished cancer cells from PMBC by factors other than EpCAM expression, a reliable clinical marker. This AI algorithm distinguished each type of ESCC cell line from PBMCs with an accuracy of more than 99.8% when the AI was trained with the same KYSE. Regardless of the combination of KYSEs used for training and examination, specificity in distinguishing KYSEs from PBMCs was more than 99.6%. On the other hand, sensitivity in distinguishing KYSEs from PBMCs varied between 20.4 and 100%. This indicates that some cancer cells are misidentified as PBMCs depending on the combination of KYSE used for training and examination. The lower differentiation sensitivity in the identification of KYSE520 after training on KYSE30, as well as in the identification of KYSE520 after training on KYSE1440, is partly explained by differences in EpCAM expression levels. However, KYSE30 was interestingly well distinguished after training on KYSE520 with an accuracy of 99.8%, despite marked differences in EpCAM expression, indicating that the AI algorithm distinguishes cells using factors other than EpCAM expression, such as cell morphology and nuclear staining. One strength of a diagnostic system that uses deep learning is that the AI can discover previously unknown features that are invisible to the human eye, such as minute differences in nucleus structure (16,23).
AI differentiated ESCC cell lines from PBMCs better than humans. Our AI algorithm was both faster and more accurate than humans. This may be due to as-yet unidentified hierarchical features that help AI distinguish cancer cells from PMBCs. The AI algorithm counted almost the same number of cells but was significantly faster than humans. Additionally, the AI algorithm distinguished KYSE from PBMC perfectly, unlike humans. Sensitivity was also lower in humans compared to AI. Researchers recognized KYSE140 and PBMCs based solely on EpCAM-positive/DAPI-stained cells and EpCAM-negative/DAPI-stained cells, respectively. Therefore, EpCAM expression heterogeneity in individual KYSE140 cells, as well as non-specific PBMC staining, may contribute to errors in the determination of EpCAM positivity by researchers. Also, it is possible that the AI algorithm accurately recognized the EpCAM expression cut-off value through pre-training, or that features were recognized that were independent of EpCAM expression (24). It is of interest to evaluate whether diagnosis by a human will approach that of the AI when manual recognition includes additional cytological details, such as cell size, shape, and nucleus-to-cytoplasmic ratio alongside EpCAM expression (25). Nevertheless, recognition accuracy among researchers varies, with trained pathologists continuing to use subjective criteria in cytology (25). It is possible that humans are unable to match AI's recognition capabilities.
AI counted and classified cells up to 850 times faster than humans. A full range search (X, Y, and Z axis) is required for humans to recognize a cell as slides have three-dimensional structure, despite their flat, two-dimensional appearance. In fact, this step requires the most time during the CTC detection process. However, AI performs rapid image acquisition and analysis. Reducing analysis time greatly improves efficiency, enabling accelerated AI algorithm evolution through training with a large library of images. In this study, the AI algorithm was preliminarily applied to detect EpCAM-positive/DAPI-positive cells in ESCC patients. EpCAM-positive/DAPI-positive cells were detected in blood samples from ESCC patients using the AI algorithm, suggesting potential clinical applications. The average number of EpCAM-positive cells in the patients was 44.5 cells while in healthy volunteers it was 2.4 cells, agreeing with previous reports (6,10).
In a recent report on CNN-based detection of CTC in cancer patients, Guo et al processed the enriched CTC fraction for immunofluorescence in situ hybridization against chromosome 8 centromere, considering a cell as a CTC if it were CD45-/DAPI+/with more than two centromeres (26). After pre-training with segmented images of 555 CTCs and 10777 non-CTCs, their CNN model identified CTCs with a sensitivity and specificity of 97.2 and 94.0%, respectively (26). With a similar number of cell images used for pre-training, the sensitivity and specificity on the test set were comparable to our results. This demonstrates the usefulness of the CNN-based algorithm for CTC detection. Further research is required to determine optimal markers, in terms of accuracy and convenience, to define CTCs for pre-training the AI algorithms.
Immunological detection of EpCAM expression is a robust method for CTC identification. However, certain limitations are being identified. Epithelial-to-mesenchymal transition (EMT) is reported during CTC detachment from the primary tumor, along with transformation to mesenchymal and stem-like properties (27–29). As a result of EMT, downregulation of epithelial markers such as EpCAM and upregulation of interstitial markers such as cell surface vimentin (CSV) are observed (27–32). Previous evaluations of EpCAM-based positive enrichment reported CTC detection rates in the range of 18–50% (33). Given reports on the involvement of EMT in treatment resistance (33) and tumor stem cell maintenance (34), the clinical significance of EpCAM-negative CTCs is suggested (33,34). Therefore, methods based on the combination of epithelial and mesenchymal markers may improve the clinical relevance of CTC detection.
In addition, using higher resolution images and setting cutoff values over the number of cases also improved detection rates. Taking advantage of AI's ability to autonomously identify hierarchical features (23), it is possible to establish an AI algorithm upon accumulated cases which identify via currently unknown marker-independent features. Further improvements in efficiency and system evolution automation may provide quick and accurate diagnoses based on simple sample preparation, ideally requiring only bright field image acquisition.
In this study, a small number of ESCC patients and healthy volunteers were compared by CTCs detection methods to assess the potential of our AI algorithm. CTC detection impacts prognostic value in ESCC patients, as indicated by several reports (32). The prognostic significance of AI-based CTC detection compared to conventional CTC detection remains to be evaluated in large prospective studies.
In addition, molecular mechanisms regulating the malignant potential of CTCs are still being elucidated (35,36). Future investigation of the correlation between the unknown features referenced by the AI algorithm in CTC detection and the molecular characteristics of CTCs may provide a basis for the development of novel diagnostic and therapeutic strategies against ESCC.
This study has certain limitations. First, a small number of EpCAM positive cells were detected in PBMCs prepared from healthy volunteers, meaning that not all the EpCAM positive cells were CTCs in ESCC patients. EpCAM positive cells may correspond to contamination of skin cells or immature blood cells (5,37). However, when many EpCAM positive cells are detected in ESCC patients, the majority are considered CTCs. Detection improvements include using high-resolution images, understanding EpCAM positive cells in healthy volunteers, and setting appropriate cutoff values over several cases. Second, the criteria that AI applied to distinguish ESCC cells from PBMC are a black box. This makes it unclear as to whether the present conditions are applicable to other cancers. Finally, CTCs were detected in ESCC patients by image processing under the conditions used in the cell lines to establish a prototype AI and preliminarily applied to the patient's samples. To establish better AI, supervised learning algorithms are best performed with many ESCC cell lines. Also, our future goal is to establish AI trained by peripheral blood samples from ESCC patients. However, this AI training faces the problem of setting a positive standard. Preparation of a true CTC to effectively and robustly train an AI for the correct answer is not trivial.
In conclusion, our results demonstrated that the CNN-based image processing algorithm for CTC detection provides higher reproducibility and a shorter analysis time compared to manual detection by the human eye. In addition, the AI algorithm appears to distinguish CTCs based on unknown features, independent of marker expressions.
Acknowledgements
The authors would like to thank Mr. Yusuke Kishi and Mr. Kisuke Tanaka (Department of Mechanical and Intellectual Systems Engineering, Faculty of Engineering, University of Toyama) for their assistance with the experiment.
Funding
This work was partly supported by JSPS KAKENHI (grant nos. 21K08729, 22K07185 and 22K08730).
Availability of data and materials
The datasets used and/or analyzed during the current study are not publicly available due to individual participants' privacy but are available from the corresponding author on reasonable request.
Authors' contributions
TA, TO, HT TY, YN, TaF, TMa, HB and TsF were involved in the conception and design, experiments, data analysis, data interpretation and manuscript writing. KT, YY, AY and TS were involved in the conception and design, data analysis using machine learning classifiers, data interpretation and manuscript writing. TMi, TW, KH, TI, SS, IH, KS, SH, IY and KM were involved in the conception and design, case enrollment, informed consent, blood sample collection, data interpretation and manuscript writing. TA, TO and KT confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The protocol of this study was approved by the ethics review board of University of Toyama Hospital (approval no. R2021042) and written informed consent was obtained from all participants.
Patient consent for publication
The patients have provided written informed consent for publication of the data in this manuscript.
Competing interests
The authors declare that they have no competing interests.
References
|
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Watanabe M, Otake R, Kozuki R, Toihata T, Takahashi K, Okamura A and Imamura Y: Recent progress in multidisciplinary treatment for patients with esophageal cancer. Surg Today. 50:12–20. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Castro-Giner F and Aceto N: Tracking cancer progression: From circulating tumor cells to metastasis. Genome Med. 12:312020. View Article : Google Scholar : PubMed/NCBI | |
|
Ganesh K and Massague J: Targeting metastatic cancer. Nat Med. 27:34–44. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Rodrigues P and Vanharanta S: Circulating tumor cells: Come together, right now, over metastasis. Cancer Discov. 9:22–24. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Ujiie D, Matsumoto T, Endo E, Okayama H, Fujita S, Kanke Y, Watanabe Y, Hanayama H, Hayase S, Saze Z, et al: Circulating tumor cells after neoadjuvant chemotherapy are related with recurrence in esophageal squamous cell carcinoma. Esophagus. 18:566–573. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Xu HT, Miao J, Liu JW, Zhang LG and Zhang QG: Prognostic value of circulating tumor cells in esophageal cancer. World J Gastroenterol. 23:1310–1318. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Ohnaga T, Shimada Y, Takata K, Obata T, Okumura T, Nagata T, Kishi H, Muraguchi A and Tsukada K: Capture of esophageal and breast cancer cells with polymeric microfluidic devices for CTC isolation. Mol Clin Oncol. 4:599–602. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Watanabe T, Okumura T, Hirano K, Yamaguchi T, Sekine S, Nagata T and Tsukada K: Circulating tumor cells expressing cancer stem cell marker CD44 as a diagnostic biomarker in patients with gastric cancer. Oncol Lett. 13:281–288. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Yamaguchi T, Okumura T, Hirano K, Watanabe T, Nagata T, Shimada Y and Tsukada K: Detection of circulating tumor cells by p75NTR expression in patients with esophageal cancer. World J Surg Oncol. 14:402016. View Article : Google Scholar : PubMed/NCBI | |
|
Kojima H, Okumura T, Yamaguchi T, Miwa T, Shimada Y and Nagata T: Enhanced cancer stem cell properties of a mitotically quiescent subpopulation of p75NTR-positive cells in esophageal squamous cell carcinoma. Int J Oncol. 51:49–62. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Correnti M and Raggi C: Stem-like plasticity and heterogeneity of circulating tumor cells: Current status and prospect challenges in liver cancer. Oncotarget. 8:7094–7115. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Semaan A, Bernard V, Kim DU, Lee JJ, Huang J, Kamyabi N, Stephens BM, Qiao W, Varadhachary GR, Katz MH, et al: Characterisation of circulating tumour cell phenotypes identifies a partial-EMT sub-population for clinical stratification of pancreatic cancer. Br J Cancer. 124:1970–1977. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Shimizu H and Nakayama KI: Artificial intelligence in oncology. Cancer Sci. 111:1452–1460. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Elemento O, Leslie C, Lundin J and Tourassi G: Artificial intelligence in cancer research, diagnosis and therapy. Nat Rev Cancer. 21:747–752. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Zeune LL, Boink YE, van Dalum G, Nanou A, de Wit S, Andree KC, Swennenhuis JF, van Gils SA, Terstappen LWMM and Brune C: Deep learning of circulating tumour cells. Nat Machine Intelligence. 2:124–133. 2020. View Article : Google Scholar | |
|
Russakovsky O, Deng J, Su H, Krause J, Bernstein M, Berg A and Fei-Fei L: ImageNet large scale visual recognition challenge. Int J Computer Vision. 115:211–252. 2015. View Article : Google Scholar | |
|
Sobin LH, Gospodarowicz M and Wittekind C: TNM Classification of Malignant Tumours, 7th edition, UICC International Union Against Cancer 2010. Hoboken, NJ: Wiley-Blackwell; 2010 | |
|
Nobuyuki O: A Threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man, and Cybernetics. 9:pp62–66. 1979. View Article : Google Scholar : PubMed/NCBI | |
|
Liu X, Faes L, Kale AU, Wagner SK, Fu DJ, Bruynseels A, Mahendiran T, Moraes G, Shamdas M, Kern C, et al: A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit Health. 1:e271–e297. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Kamba S, Tamai N, Saitoh I, Matsui H, Horiuchi H, Kobayashi M, Sakamoto T, Ego M, Fukuda A, Tonouchi A, et al: Reducing adenoma miss rate of colonoscopy assisted by artificial intelligence: A multicenter randomized controlled trial. J Gastroenterol. 56:746–757. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM and Thrun S: Dermatologist-level classification of skin cancer with deep neural networks. Nature. 542:115–118. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou LQ, Wang JY, Yu SY, Wu GG, Wei Q, Deng YB, Wu XL, Cui XW and Dietrich CF: Artificial intelligence in medical imaging of the liver. World J Gastroenterol. 25:672–682. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Zheng S, Lin HK, Lu B, Williams A, Datar R, Cote RJ and Tai YC: 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices. 13:203–213. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Moore MJ, Sebastian JA and Kolios MC: Determination of cell nucleus-to-cytoplasmic ratio using imaging flow cytometry and a combined ultrasound and photoacoustic technique: A comparison study. J Biomed Opt. 24:1–10. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Guo Z, Lin X, Hui Y, Wang J, Zhang Q and Kong F: Circulating tumor cell identification based on deep learning. Front Oncol. 12:8438792022. View Article : Google Scholar : PubMed/NCBI | |
|
Habli Z, AlChamaa W, Saab R, Kadara H and Khraiche ML: Circulating tumor cell detection technologies and clinical utility: Challenges and opportunities. Cancers (Basel). 12:19302020. View Article : Google Scholar : PubMed/NCBI | |
|
Miki Y, Yashiro M, Kuroda K, Okuno T, Togano S, Masuda G, Kasashima H and Ohira M: Circulating CEA-positive and EpCAM-negative tumor cells might be a predictive biomarker for recurrence in patients with gastric cancer. Cancer Med. 10:521–528. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Han D, Chen K, Che J, Hang J and Li H: Detection of epithelial-mesenchymal transition status of circulating tumor cells in patients with esophageal squamous carcinoma. Biomed Res Int. 2018:76101542018. View Article : Google Scholar : PubMed/NCBI | |
|
Gao Y, Fan WH, Song Z, Lou H and Kang X: Comparison of circulating tumor cell (CTC) detection rates with epithelial cell adhesion molecule (EpCAM) and cell surface vimentin (CSV) antibodies in different solid tumors: A retrospective study. PeerJ. 9:e107772021. View Article : Google Scholar : PubMed/NCBI | |
|
Chaw SY, Abdul Majeed A, Dalley AJ, Chan A, Stein S and Farah CS: Epithelial to mesenchymal transition (EMT) biomarkers-E-cadherin, beta-catenin, APC and Vimentin-in oral squamous cell carcinogenesis and transformation. Oral Oncol. 48:997–1006. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Shi Y, Ge X, Ju M, Zhang Y, Di X and Liang L: Circulating tumor cells in esophageal squamous cell carcinoma-Mini review. Cancer Manag Res. 13:8355–8365. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al: Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 527:472–476. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Deng Z, Wu S, Wang Y and Shi D: Circulating tumor cell isolation for cancer diagnosis and prognosis. EBioMedicine. 83:1042372022. View Article : Google Scholar : PubMed/NCBI | |
|
Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, Krol I, Scheidmann MC, Beisel C, Stirnimann CU, et al: Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 176:98–112. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW and Terstappen LW: Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 10:6897–6904. 2004. View Article : Google Scholar : PubMed/NCBI |