Long noncoding RNAs (lncRNAs), sometimes known as
competitive endogenous RNA (ceRNA), are a class of noncoding RNAs
that are >200 nucleotides in length, which are involved in
multiple cellular processes, such as proliferation,
differentiation, maturation, apoptosis, metastasis, and metabolism
(1–3). By targeting complementary miRNAs,
ceRNAs can downregulate miRNA expression, indirectly regulating the
expression of the downstream mRNA and thus affecting the behavior
of a tumor cell (4). lncRNAs can
bind to DNA, RNA, and proteins to form complexes that regulate
transcription (5). A diseased state
is ultimately brought on via the regulation of gene expression via
various mechanisms, including mRNA stability and translation
(6–8). It has been shown that lncRNAs can be
categorized into different classes, which modulate tumor activity
through differing functions. For example, they act as molecular
sponges for miRNAs (9), modifiers
and decoys for ribosomal proteins or ribonucleic acids (10), catalysts for gene transcription
(11), gene methylation (12), protein kinase phosphorylation
(13), and stabilizers of protein
expression (14). lncRNAs can also
act as coactivators of signaling factors as agonists. Additionally,
they may alter the formation of tumors by sensitizing or
desensitizing the cancerous cells to chemotherapeutic drugs. For
example, the lncRNA LINC02525, which acts as a modifier of
ribosomal proteins, can function as an oncogene by interacting with
the ribosomal protein RPL35 to activate the translation of E2F1 and
subsequently enhance the transcription of the GTPase-activating
protein DEPDC1B to regulate the biological activity of
neuroblastoma (NB) cells (15).
Additionally, lincRNA-p21 acts as a gene transcription catalyst by
mediating the binding of hnRNPK to promote transcription of its
neighboring gene, cyclin-dependent kinase inhibitor 1A
(CDKN1A), which encodes p21, and mediates the binding of
hnRNPK to it, negatively regulating mRNA translation (10). lncRNAs can influence the
transcription of genes on several chromosomes and can also be used
as a decoy to trigger protein and RNA regulation (16). LINC00184 increases protein kinase
phosphorylation and gene methylation, which increases the level of
glycolysis in esophageal cancer (EC) cells by promoting PTEN
methylation and AKT phosphorylation (17). Through the
LINRIS/IGF2BP2/v-Myc avian myelocytomatosis viral oncogene
homolog (c-Myc) axis, the lncRNA LINRIS can stabilize
protein expression, prevent IGF2BP2 degradation, and
upregulate Myc expression to maintain glycolysis in
colorectal cancer (CRC) cells (12). It was found that the SNP
rs11672691 risk allele at the regulatory region 19q13 locus
upregulated the expression of transcription factors NK3 homeobox 1
(NKX3.1), Yin-Yang 1 (YY1), PCAT19-long through
promoter-enhancer switching bifunctional regulatory elements in the
SNP region, increased the expression of HNRNPAB-PCAT19-long
ribonucleoprotein complex and cell cycle gene expression (18). Increased levels of the transcription
factor homeobox A2 (HOXA2) and the carcinoembryonic
antigen-related cell adhesion molecule 21 (CEACAM21) are
also brought on by prostate cancer-associated transcript 19
(PCAT19) upregulation, which together promote prostate cancer (PCa)
invasion (13,18). As a desensitizer of tumor-resistant
cells, lncRNA differentiation antagonizing non-protein coding RNA
(DANCR) exerts a sponging effect through the miR-125b-5p/hexokinase
(HK)2 axis to downregulate miR-124b-5p expression and thus enhances
HK2 activity, desensitizing cisplatin-resistant CRC cells, while
silencing DANCR exhibits the opposite effect, decreasing the
resistance of CRC cells to cisplatin (9). LINC00504 functions as a coactivator of
c-Myc and stimulates CRC glucose metabolism levels by
increasing the recruitment of c-Myc to chromatin sites,
which is how chromatin modifiers are used to modulate gene
transcription (11). lncRNAs can
regulate tumor apoptosis by affecting chromosomal gene
transcription, post-transcription and the regulation of epigenetic
mechanisms such as gene methylation, which results in lncRNAs
acting as tumor suppressors or oncogenes, which provides new
potential targets for improving tumor prognosis and metastasis
(19).
The three primary metabolic pathways are glycolysis,
pentose phosphate, and oxidative phosphorylation, of which,
glycolysis is highly active in tumor cells (20). Otto Warburg, a German scientist,
discovered the Warburg effect in the 1950s, which states that when
oxygen levels are adequate, tumor cells do not undergo oxidative
phosphorylation like normal cells but instead produce the required
energy by aerobic glycolysis (21).
A complex process of glucose metabolism in neoplastic cells
contributes to the dysregulated lncRNA expression profiles in
neoplasms. Regulation via lncRNAs can alter gene transcription
levels, protein synthesis, and translation rates, affecting glucose
metabolism in tumor cells. Several studies have been conducted to
assess the specific molecular mechanisms of lncRNAs in tumor
glucose metabolism. In these studies, it was found that a single
lncRNA can exert a regulatory role in several types of cancer,
often have differing effects. For example, potassium voltage-gated
channel subfamily Q member 1 opposite strand/antisense transcript 1
(KCNQ1OT1) can promote the glycolysis of CRC cells through HK2
(22); in contrast, it can also
compete with miR-34c-5p to increase aldolase A (ALDOA) activity and
glycolysis levels in osteosarcoma (OS) cells (23). Similarly, LINC00504 increases
glycolysis in ovarian cancer (OC) (24) and CRC (11) by increasing the activity of the
glycolytic enzymes HK2, pyruvate dehydrogenase kinase 1 (PDK1),
pyruvate kinase 2 (PKM2), and the expression levels of c-Myc
in the corresponding tumor cells. In pituitary tumors (PT),
urothelial cancer associated 1 (UCA1) accelerates HK2 and lactate
dehydrogenase A (LDHA) activation to increase glycolysis (25). At the same time, in cervical
squamous cell carcinoma and endocervical adenocarcinoma (CESC)
(26), UCA1 and HK2 expression are
negatively correlated. It was found that several lncRNAs can
regulate a single gene to influence tumor progression. MYC
genes have a significant impact on the regulation of transcription.
LncRNA GLCC1 (27) and LINC00504
(11) can regulate c-Myc to
promote glycolysis of tumor cells and accelerate tumor growth,
whereas LINC00261 (28) is
negatively correlated with c-Myc. With MIR17HG being
significantly upregulated in CRC (29) while acting as a tumor growth
suppressor in OC (30), the effect
of lncRNA expression varies in different tumors. As shown in
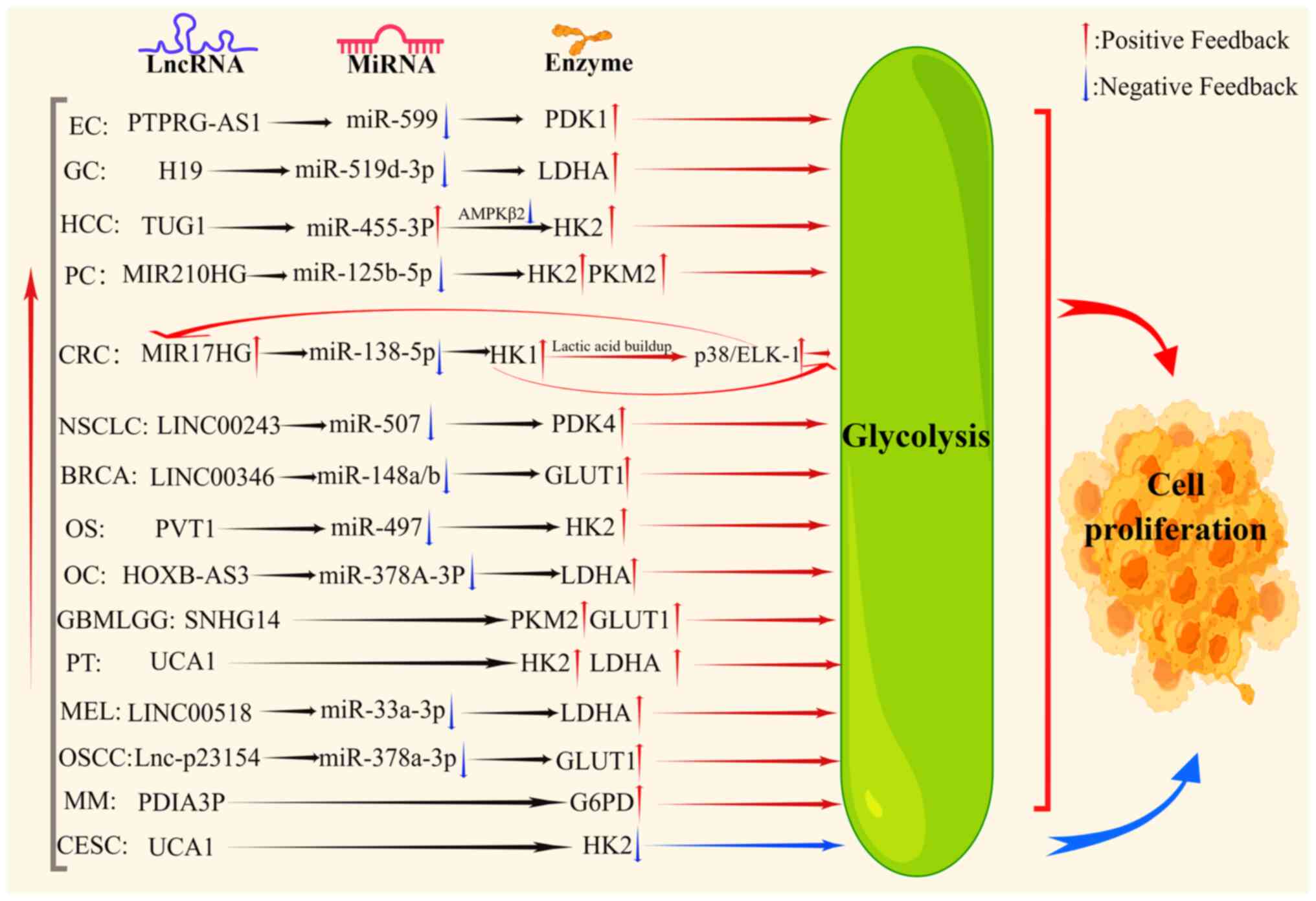
Fig. 1, it was found that lncRNAs,
including protein tyrosine phosphatase receptor type G antisense
RNA 1 (PTPRG-AS1) (31), H19
(32), MIR210 host gene (MIR210HG)
(33), MIR17HG (29), LINC00243 (34), and LINC00346 (35), promote the growth of tumor cells,
altering the expression of key gluconeogenesis enzymes through
sponging of miRNAs. Additionally, several signaling molecules are
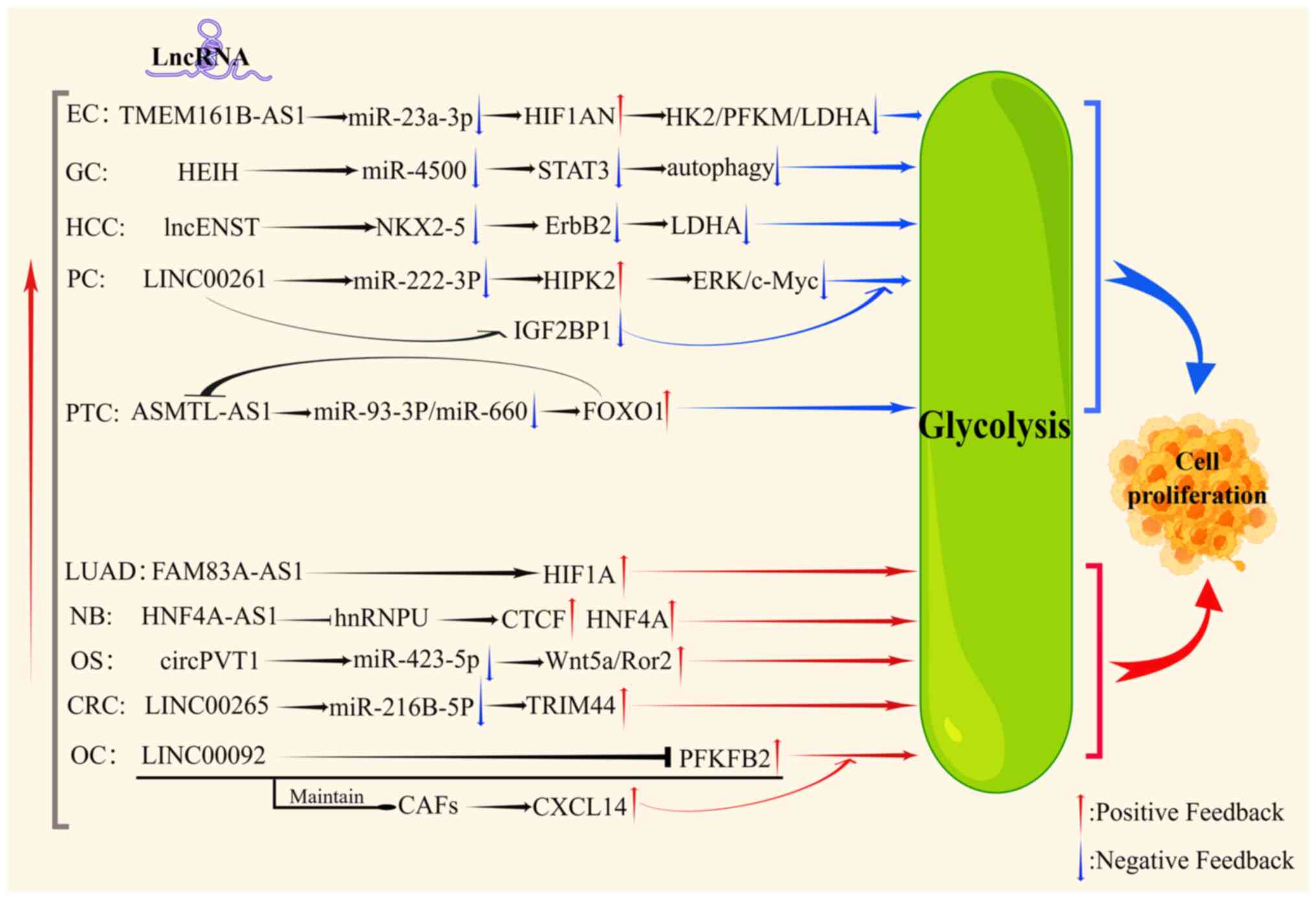
regulated by lncRNAs, such as transmembrane protein 161B antisense
RNA 1 (TMEM161B-AS1) (36), high
expression in hepatocellular carcinoma (HEIH) (37), lncENST (38), and LINC00261 (28), that can affect glycolysis and tumor
cell activity (Fig. 2). The effects
of lncRNAs and glycometabolism in cancer are summarized in this
review.
The majority of malignant tumors occur in the
digestive system, which includes the esophagus, the stomach, the
liver, the pancreas, and the colon (39). Their characteristics of slow
diagnosis, challenging treatment, poor prognosis, and high
mortality rates have become significant clinical issues that must
be resolved. Numerous studies have shown that lncRNAs can target
key enzymes and various signaling factors that affect glycolysis
and are crucial for regulating digestive system tumor
expression.
EC is a common, aggressive, malignant tumor with a
poor prognosis worldwide. An ever-increasing number lncRNAs are
being reported to be involved in the progression of EC. However,
more research is required on lncRNAs and their potential molecular
mechanisms for regulating EC glucose metabolism.
A recent study found that PTPRG-AS1 upregulated PDK1
expression through a PTPRG-AS1/miR-599/PDK1 axis via sponging
miR-599 (31), thereby promoting EC
cell proliferation, migration, and glycolysis. The lncRNA actin γ 1
pseudogene (AGPG) is a novel stimulator of glycolysis and
tumorigenesis. It regulates
6-phosphofructose-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3),
negatively regulated by the P53 oncogene to enhance sugar
metabolism and promote the progression of the cell cycle. Future
treatments for EC may be tailored using AGPG, which targets an
AGPG/P53/PFKFB3 axis (40).
GC is ranked the fifth most common type of cancer
worldwide and the third leading cause of death in patients with
high cancer risk. Most patients with GC have a poor survival rate,
with a tendency for their 5-year survival rate to be <50%
(41).
HCC is among the most prevalent malignancies
worldwide, particularly in East Asia and Africa. The high
prevalence of these diseases is caused by the lack of monitoring
and treatment options (54). Below
is a summary of recent research on the role of lncRNA in regulating
aerobic glycolysis in HCC.
PKM2 was found to be essential, and E2F1-activated
SNHG1 regulates the miR-326/PKM2 axis to promote PKM2 expression,
glycolysis, and HCC cell proliferation (55). The orphan nuclear receptor Nur77
regulates the Nur77-WFDC21P-PFKP/PKM2 axis by transcriptionally
activating the lncRNA WAP four-disulfide core domain 21 pseudogenes
(WFDC21P), which suppresses PFKP and PKM2 and impedes HCC
glycolysis (56). LDHA and PKM2 are
more readily bound to the fibroblast growth factor receptor type 1
by the lncRNA highly upregulated in liver cancer (HULC), resulting
in their phosphorylation and facilitating glycolysis (57). By inhibiting forkhead box M1
(FOXM1) transactivation through STAT3 and regulating
glucose transporter Type 1 (GLUT1) by competing with STAT3,
lncRNA SLC2A1-AS1 deactivates HCC cells by interfering with the
FOXM1/GLUT1 axis and inhibits aerobic glycolysis in HCC
cells (58). AMPKβ2, a
downstream marker gene of miR-455-3P, was found to be downregulated
to stimulate the enzymatic protein activity of HK2, and TUG1 was
found to activate HK2 and promote cell proliferation, metastasis,
and glycolysis by upregulating the expression of miR-455-3p through
the TUG1/miR-455-3p/AMPKβ2 axis (59). The inactivation of GLUT1 and HK2,
caused by genistein-induced downregulation of HIF1A,
promotes the apoptosis of HCC aerobic glycolytic cells and the
antitumor effect of sorafenib on HCC cells (60). According to a study, aspirin can
target the inhibition of aerobic glycolysis and PFKFB3
overexpression in HCC cells, sensitizing sorafenib-resistant HCC
cells. The findings provide a strong theoretical basis for the
targeted treatment of sorafenib-resistant HCC cells and the
eradication of HCC cells with aspirin and sorafenib (61).
A previous study reported that the Warburg effect is
mediated by human epidermal growth factor receptor (ErbB) 2,
which activates heat shock factor 1 before upregulating LDHA
(62). Its promoter may interact
negatively with the transcription factor Nkx2-5. Inhibiting
the Warburg effect in HCC cells, lncENST directly binds to and
activates the transcription factor Nkx2-5 through an
Nkx2-5/ErbB2 axis. This inhibits ErbB2 gene
transcription and downregulates ErbB2 protein expression
(38). HOX transcript antisense RNA
(HOTAIR) regulates a miR-130a-3p/HIF1A axis on miR-130a-3p
uptake, resulting in high expression of HIF1A, which
inhibits glycolysis in hypoxia-induced HCC cells (63). Regulating GSK3β and Wnt/β catenin
signaling and promoting HCC, SNHG5 promotors (64). In HCC cells, lncRNA cyclin-dependent
kinase inhibitor 2B antisense RNA 1 (CDKN2B-AS1) sponge let-7c-5p
promotes upregulation of nucleosomal assembly protein 1 like
1SS-mediated PI3K/Akt/mTOR axis-stimulated
glycolysis (65). While NEAT1 is
the primary transcriptional target of mTOR, it can also
control the lncRNA transcriptome in HCC. When activated in cancer,
mTORC1 inhibits NEAT1_2 expression while facilitating mRNA
splicing and the expression of key glycolytic enzymes, which
promotes HCC glucose metabolism and cell proliferation (66). By controlling the
MAPK/extracellular regulated protein kinases (ERK)
pathway, hypoxia-inducible lncRNA neuropeptide S receptor 1
antisense RNA 1 was also found to promote the proliferation and
glycolysis of HCC cells (67). The
target gene of the lncRNA Fiveprime to Xist (FTX) is the peroxisome
proliferator-activated receptor γ (PPARγ), and FTX functions
downstream of PPAR. Through overexpression of the FTX/PPARγ
axis, FTX promotes activation of the PPARγ pathway, which
attenuates the expression of tumor necrosis factor-α (TNFα),
leptin, and PDK1, regulating GLUT4, PDK1, and PFKL in HCC cells,
and maintains increased glycolysis in HCC (68). Additionally, the progression of HCC
may be aided by the regulation of Bcl-2-related protein A1
(BCL2A1) expression by the lncRNA POU3F3 adjacent noncoding
transcript 1 (PANTR1) via sponging of miR-587 (69).
PC is a highly lethal malignancy that is often
diagnosed in the first instance as advanced PC, and thus has a poor
prognosis, with an overall 5-year survival rate of <6% (70,71).
By 2030, PDAC is predicted to be the second leading cause of
cancer-related death, and is refractory to most current treatment
strategies (72). There are few
functional targets identified for the management of PC and their
underlying mechanisms remain to be determined. The following is a
summary of the known findings.
The lncRNA MIR210HG was found to be upregulated in
PC and is a key oncogenic regulator of PC invasiveness and
glycolysis. Through the MIR210HG/miR-125b-5p/HK2/PKM2 axis,
MIR210HG and miR-125b-5p bind competitively to target HK2/PKM2
expression, enhancing PC cell invasion and glycolysis (33).
CRC is one of the most prevalent cancers of the
digestive system worldwide, the fourth most common form of cancer
in the United States, after breast cancer, lung cancer, and PCa,
and the second leading cause of cancer-related death, after lung
cancer, in terms of cancer mortality worldwide (74,75).
Meanwhile, in the 21st century, CRC is rising in the cancer
spectrum and is in the top five of the most common cancers in China
(76). Patients with CRC have a
5-year overall survival rate of ~64% (74). Several studies have shown that
lncRNAs and glucose metabolism significantly impact colon cancer
development, progression, and outcomes.
lncRNAs are crucial for regulating glucose
metabolism in several types of cancer, including thyroid cancer
(TC), lung cancer (LC), breast invasive carcinoma (BRCa), NB, OS,
CESC, OC, glioma, PT, melanoma, oral squamous cell carcinoma, and
multiple myeloma, amongst others (25,91–98).
Table I lists certain lncRNAs that
regulate tumor glycolysis and their identified roles.
It has been found that acetylserotonin
O-methyltransferase like antisense RNA 1 (ASMTL-AS1) upregulates
the expression of forkhead box O1 (FOXO1) and inhibits
glycolysis in papillary thyroid carcinoma (PTC) through an
ASMTL-AS1/miR-93-3P/miR-660/FOXO1 axis. A positive feedback
loop created by the recombination of FOXO1 and ASMTL-AS1 can
further inhibit the occurrence and progression of PTC (105). It was also shown that the lncRNA
sequence similarity family 83 member A-antisense ribonucleic acid 1
(FAM83A-AS1) upregulated HIF1A expression through a
HIF1A/glycolytic axis to accelerate the progression of lung
adenocarcinoma (106). Hepatocyte
nuclear factor 4 α antisense RNA 1 (HNF4A-AS1) binds to
heterogeneous nuclear ribonucleoprotein U (hnRNPU) through an
HNF4A-AS1/hnRNPU/CCCTC-binding factor (CTCF) axis, which
transactivates CTCF upregulates HNF4A expression, and promotes
aerobic glycolysis and NB cell invasion (107). To activate Wnt family member 5A
(Wnt5a)/receptor tyrosine kinase-like orphan receptor 2
(Ror2) signaling and promote OS glycolysis and cell
proliferation, circPVT1 binds and inhibits miR-423-5p through the
circPVT1-miR-423-5p-Wnt5a/Ror2 axis (108). It was found that LINC00092 and
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (PFKFB2)
together promoted glycolysis in OC metastasis, which in turn
maintained the characteristics of cancer-associated fibroblasts in
the tumor microenvironment. In addition, the chemokine CXCL14
promoted OC metastasis in vivo and in vitro, forming
a positive feedback loop that regulated the biological activity of
OC (109). The HIPPO pathway
regulates cell proliferation and cell invasion. LINC00857
upregulates the expression of the oncogene YAPA1 through a
LINC00857/miR-486-5p/YAP1 axis, which binds to the
miR-486-5p pathway, a pathway that inactivates the HIPPO pathway to
enhance OC cell proliferation and glycolysis (110). Another study found that LINC00504
promoted the activation of glycolytic enzymes PDK1, PKM2, and HK2
by sponging miR-1244. This enhanced OC cell proliferation and
aerobic glycolysis (24).
RNA-binding signal transduction-associated protein 3 (KHDRBS3) can
promote glycolysis through the gene claudin (CLDN6), which
is associated with chemoresistance and a key player in several
types of cancer. Studies have shown that MIR17HG can inhibit
paclitaxel by downregulating the expression of KHDRBS3 and
CLDN6 by targeting the KHDRBS3/MIR17HG/CLDN6 axis.
Paclitaxel cell resistance and glycolysis are novel approaches to
treating OC-resistant cells (30).
lncRNAs participate in transport, in the activity
of key enzymes, in the regulation of miRNAs, in the regulation of
various signaling pathways, and in the regulation of tumor energy
metabolism, thus having a broad impact on the malignant properties
of cancerous cells. Tumor occurrence and progression are highly
correlated with glucose metabolism. Blocking glycolysis or the
pentose phosphate pathway can effectively inhibit the proliferation
of tumor cells and even result in their death. Therefore,
glycolysis-related enzymes may be a novel therapeutic target.
However, additional research is required to determine the
underlying mechanism of lncRNA regulation of energy metabolism in
different types of tumors. For example, it is unknown whether the
identified transcription factors and their derived lncRNAs can
target and coordinate the regulation of tumor progression. Further
research is required to determine how different transcription
factors, via lncRNAs, regulate tumor glucose metabolism directly,
indirectly, cooperatively, or antagonistically and whether lncRNAs
can establish a specific relationship to regulate the expression of
tumors jointly. The mechanism of targeted and precise regulation of
tumor drug resistance via the interaction between lncRNAs and drugs
also needs to be studied. Further research is required to fully
understand how different transcription factors regulate tumor
expression in the microenvironment under both hypoxic and normoxic
conditions. lncRNAs can act as sponges that bind to miRNAs to
inhibit the regulatory effect of said miRNAs on their target mRNAs.
The discovery of the ceRNA mechanism, which reduces the inhibitory
effect of miRNAs on target genes, opens novel avenues for further
investigation into the lncRNAs. This review may assist in
understanding the molecular mechanisms of tumor energy metabolism
regulation and offer a fresh perspective for future studies on
tumor diagnosis and targeted therapy.
Not applicable.
The work was supported by the Natural Science Foundation Project
of Xizang Autonomous Region (grant no. XZ202101ZR0074G), the Major
Cultivation Project of Xizang Minzu University (grant no. 20MDT02),
and the National Innovation and Entrepreneurship Training Program
for College Students in 2022 (grant no. 202210695033).
Not applicable.
XH contributed to drafting of the manuscript. ZZ
and XC were involved in writing the paper and revising it
critically for important intellectual content. XW reviewed and
edited the manuscript. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Ma Y, Yang Y, Wang F, Moyer MP, Wei Q,
Zhang P, Yang Z, Liu W, Zhang H, Chen N, et al: Long non-coding RNA
CCAL regulates colorectal cancer progression by activating
Wnt/β-catenin signalling pathway via suppression of activator
protein 2α. Gut. 65:1494–1504. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang X, Xie Z, Lei X and Gan R: Long
non-coding RNA GAS5 in human cancer. Oncol Lett. 20:2587–2594.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang Z, Zhou JK, Peng Y, He W and Huang
C: The role of long noncoding RNAs in hepatocellular carcinoma. Mol
Cancer. 19:772020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen B, Dragomir MP, Fabris L, Bayraktar
R, Knutsen E, Liu X, Tang C, Li Y, Shimura T, Ivkovic TC, et al:
The long noncoding RNA CCAT2 induces chromosomal instability
through BOP1-AURKB signaling. Gastroenterology. 159:2146–2162.e33.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ji D, Wang Y, Li H, Sun B and Luo X: Long
non-coding RNA LINC00461/miR-149-5p/LRIG2 axis regulates
hepatocellular carcinoma progression. Biochem Biophys Res Commun.
512:176–181. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gil N and Ulitsky I: Regulation of gene
expression by cis-acting long non-coding RNAs. Nat Rev Genet.
21:102–117. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi H, Li K, Feng J, Liu G, Feng Y and
Zhang X: LncRNA-DANCR Interferes With miR-125b-5p/HK2 axis to
desensitize colon cancer cells to cisplatin vis activating
anaerobic glycolysis. Front Oncol. 10:10342020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dimitrova N, Zamudio JR, Jong RM, Soukup
D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA and Jacks
T: LincRNA-p21 activates p21 in cis to promote Polycomb target gene
expression and to enforce the G1/S checkpoint. Mol Cell.
54:777–790. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feng J, Ma J, Liu S, Wang J and Chen Y: A
noncoding RNA LINC00504 interacts with c-Myc to regulate tumor
metabolism in colon cancer. J Cell Biochem. 120:14725–14734. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen
YX, Liu J, Luo XJ, Meng Q, Pu HY, et al: LncRNA LINRIS stabilizes
IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer.
Mol Cancer. 18:1742019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao P, Xia JH, Sipeky C, Dong XM, Zhang Q,
Yang Y, Zhang P, Cruz SP, Zhang K, Zhu J, et al: Biology and
clinical implications of the 19q13 aggressive prostate cancer
susceptibility locus. Cell. 174:576–589.e18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bian Z, Zhang J, Li M, Feng Y, Wang X,
Zhang J, Yao S, Jin G, Du J, Han W, et al: LncRNA-FEZF1-AS1
promotes tumor proliferation and metastasis in colorectal cancer by
regulating PKM2 Signaling. Clin Cancer Res. 24:4808–4819. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu PY, Tee AE, Milazzo G, Hannan KM, Maag
J, Mondal S, Atmadibrata B, Bartonicek N, Peng H, Ho N, et al: The
long noncoding RNA lncNB1 promotes tumorigenesis by interacting
with ribosomal protein RPL35. Nat Commun. 10:50262019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Goodall GJ and Wickramasinghe VO: RNA in
cancer. Nat Rev Cancer. 21:22–36. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li W, Huang K, Wen F, Cui G, Guo H, He Z
and Zhao S: LINC00184 silencing inhibits glycolysis and restores
mitochondrial oxidative phosphorylation in esophageal cancer
through demethylation of PTEN. EBioMedicine. 44:298–310. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hua JT, Ahmed M, Guo H, Zhang Y, Chen S,
Soares F, Lu J, Zhou S, Wang M, Li H, et al: Risk SNP-Mediated
promoter-enhancer switching drives prostate cancer through lncRNA
PCAT19. Cell. 174:564–575.e18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo W, Dong Z, Liu S, Qiao Y, Kuang G, Guo
Y, Shen S and Liang J: Promoter hypermethylation-mediated
downregulation of miR-770 and its host gene MEG3, a long non-coding
RNA, in the development of gastric cardia adenocarcinoma. Mol
Carcinog. 56:1924–1934. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ganapathy-Kanniappan S and Geschwind JF:
Tumor glycolysis as a target for cancer therapy: Progress and
prospects. Mol Cancer. 12:1522013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen C, Wei M, Wang C, Sun D, Liu P, Zhong
X and Yu W: Long noncoding RNA KCNQ1OT1 promotes colorectal
carcinogenesis by enhancing aerobic glycolysis via hexokinase-2.
Aging (Albany NY). 12:11685–11697. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv
L, Liu J, Xu Y, Shen Y and Yang M: Noncoding RNAs in gastric
cancer: Implications for drug resistance. Mol Cancer. 19:622020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Y, He X, Chen Y and Cao D: Long
non-coding RNA LINC00504 regulates the Warburg effect in ovarian
cancer through inhibition of miR-1244. Mol Cell Biochem. 464:39–50.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu G, Wang L and Li Y: Inhibition of
lncRNA-UCA1 suppresses pituitary cancer cell growth and prolactin
(PRL) secretion via attenuating glycolysis pathway. In Vitro Cell
Dev Biol Anim. 56:642–649. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fan L, Huang C, Li J, Gao T, Lin Z and Yao
T: Long non-coding RNA urothelial cancer associated 1 regulates
radioresistance via the hexokinase 2/glycolytic pathway in cervical
cancer. Int J Mol Med. 42:2247–2259. 2018.PubMed/NCBI
|
|
27
|
Tang J, Yan T, Bao Y, Shen C, Yu C, Zhu X,
Tian X, Guo F, Liang Q, Liu Q, et al: LncRNA GLCC1 promotes
colorectal carcinogenesis and glucose metabolism by stabilizing
c-Myc. Nat Commun. 10:34992019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhai S, Xu Z, Xie J, Zhang J, Wang X, Peng
C, Li H, Chen H, Shen B and Deng X: Epigenetic silencing of LncRNA
LINC00261 promotes c-myc-mediated aerobic glycolysis by regulating
miR-222-3p/HIPK2/ERK axis and sequestering IGF2BP1. Oncogene.
40:277–291. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao S, Guan B, Mi Y, Shi D, Wei P, Gu Y,
Cai S, Xu Y, Li X, Yan D, et al: LncRNA MIR17HG promotes colorectal
cancer liver metastasis by mediating a glycolysis-associated
positive feedback circuit. Oncogene. 40:4709–4724. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu X, Qiu L, Feng H, Zhang H, Yu H, Du Y,
Wu H, Zhu S, Ruan Y and Jiang H: KHDRBS3 promotes paclitaxel
resistance and induces glycolysis through modulated MIR17HG/CLDN6
signaling in epithelial ovarian cancer. Life Sci. 293:1203282022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu K, Wang Z, Huang Y, Yao L, Kang N, Ge
W, Zhang R and He W: LncRNA PTPRG-AS1 facilitates glycolysis and
stemness properties of esophageal squamous cell carcinoma cells
through miR-599/PDK1 axis. J Gastroenterol Hepatol. 37:507–517.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sun L, Li J, Yan W, Yao Z, Wang R, Zhou X,
Wu H, Zhang G, Shi T and Chen W: H19 promotes aerobic glycolysis,
proliferation, and immune escape of gastric cancer cells through
the microRNA-519d-3p/lactate dehydrogenase A axis. Cancer Sci.
112:2245–2259. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu T, Li G, Wang C, Gong G, Wang L, Li C,
Chen Y and Wang X: MIR210HG regulates glycolysis, cell
proliferation, and metastasis of pancreatic cancer cells through
miR-125b-5p/HK2/PKM2 axis. RNA Biol. 18:2513–2530. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feng X and Yang S: Long non-coding RNA
LINC00243 promotes proliferation and glycolysis in non-small cell
lung cancer cells by positively regulating PDK4 through sponging
miR-507. Mol Cell Biochem. 463:127–136. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Y, Li H, Wang W, Yu X and Xu Q:
LINC00346 regulates glycolysis by modulation of glucose transporter
1 in breast cancer cells. Mol Cell Probes. 54:1016672020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shi Z, Li G, Li Z, Liu J and Tang Y:
TMEM161B-AS1 suppresses proliferation, invasion and glycolysis by
targeting miR-23a-3p/HIF1AN signal axis in oesophageal squamous
cell carcinoma. J Cell Mol Med. 25:6535–6549. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang H, Shen X, Xiong S, Peng L, Mai W
and Xin L: HEIH promotes malignant progression of gastric cancer by
regulating STAT3-Mediated autophagy and glycolysis. Dis Markers.
2022:26345262022.PubMed/NCBI
|
|
38
|
Chen G, Jiang J, Wang X, Feng K and Ma K:
lncENST suppress the warburg effect regulating the tumor progress
by the Nkx2-5/ErbB2 Axis in hepatocellular carcinoma. Comput Math
Methods Med. 2021:69595572021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cao W, Chen HD, Yu YW, Li N and Chen WQ:
Changing profiles of cancer burden worldwide and in China: A
secondary analysis of the global cancer statistics 2020. Chin Med J
(Engl). 134:783–791. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vargas RE and Wang W: Significance of long
non-coding RNA AGPG for the metabolism of esophageal cancer. Cancer
Commun (Lond). 40:313–315. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pei LJ, Sun PJ, Ma K, Guo YY, Wang LY and
Liu FD: LncRNA-SNHG7 interferes with miR-34a to de-sensitize
gastric cancer cells to cisplatin. Cancer Biomark. 30:127–137.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hu J, Huang L, Ding Q, Lv J and Chen Z:
Long noncoding RNA HAGLR sponges miR-338-3p to promote 5-Fu
resistance in gastric cancer through targeting the LDHA-glycolysis
pathway. Cell Biol Int. 46:173–184. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qian Y, Song W, Wu X, Hou G, Wang H, Hang
X and Xia T: DLX6 Antisense RNA 1 modulates glucose metabolism and
cell growth in gastric cancer by targeting microRNA-4290. Dig Dis
Sci. 66:460–473. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Deng P, Li K, Gu F, Zhang T, Zhao W, Sun M
and Hou B: LINC00242/miR-1-3p/G6PD axis regulates Warburg effect
and affects gastric cancer proliferation and apoptosis. Mol Med.
27:92021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dai T, Zhang X, Zhou X, Hu X, Huang X,
Xing F, Tian H and Li Y: Long non-coding RNA VAL facilitates PKM2
enzymatic activity to promote glycolysis and malignancy of gastric
cancer. Clin Transl Med. 12:e10882022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jin X, Qiao L, Fan H, Liao C, Zheng J,
Wang W, Ma X, Yang M, Sun X and Zhao W: Long non-coding RNA MSC-AS1
facilitates the proliferation and glycolysis of gastric cancer
cells by regulating PFKFB3 expression. Int J Med Sci. 18:546–554.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xu Z, Lv H, Wang Y, Hu C, Chen S, Du Y and
Shi Can Cheng X: HAND2-AS1 inhibits gastric adenocarcinoma cells
proliferation and aerobic glycolysis via miRNAs Sponge. Cancer
Manag Res. 12:3053–3068. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Qian C, Xu Z, Chen L, Wang Y and Yao J:
Long noncoding RNA LINC01391 restrained gastric cancer aerobic
glycolysis and tumorigenesis via targeting miR-12116/CMTM2 axis. J
Cancer. 11:6264–6276. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yan B, Ren Z, Sun J, Ding C and Yang D:
IGF2-AS knockdown inhibits glycolysis and accelerates apoptosis of
gastric cancer cells through targeting miR-195/CREB1 axis. Biomed
Pharmacother. 130:1106002020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tan HY, Wang C, Liu G and Zhou X: Long
noncoding RNA NEAT1-modulated miR-506 regulates gastric cancer
development through targeting STAT3. J Cell Biochem. 120:4827–4836.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sun K, Hu P and Xu F: LINC00152/miR-139-5p
regulates gastric cancer cell aerobic glycolysis by targeting
PRKAA1. Biomed Pharmacother. 97:1296–1302. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li M, Cai O, Yu Y and Tan S: Paeonol
inhibits the malignancy of Apatinib-resistant gastric cancer cells
via LINC00665/miR-665/MAPK1 axis. Phytomedicine. 96:1539032022.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bertuccio P, Turati F, Carioli G,
Rodriguez T, La Vecchia C, Malvezzi M and Negri E: Global trends
and predictions in hepatocellular carcinoma mortality. J Hepatol.
67:302–309. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang Y, Yang F, Peng Q, Mei K, He H and
Yang Q: Long non-coding RNA SNHG1 activates glycolysis to promote
hepatocellular cancer progression through the miR-326/PKM2 axis. J
Gene Med. 24:e34402022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Guan YF, Huang QL, Ai YL, Chen QT, Zhao
WX, Wang XM, Wu Q and Chen HZ: Nur77-activated lncRNA WFDC21P
attenuates hepatocarcinogenesis via modulating glycolysis.
Oncogene. 39:2408–2423. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang C, Li Y, Yan S, Wang H, Shao X, Xiao
M, Yang B, Qin G, Kong R, Chen R and Zhang N: Interactome analysis
reveals that lncRNA HULC promotes aerobic glycolysis through LDHA
and PKM2. Nat Commun. 11:31622020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shang R, Wang M, Dai B, Du J, Wang J, Liu
Z, Qu S, Yang X, Liu J, Xia C, et al: Long noncoding RNA SLC2A1-AS1
regulates aerobic glycolysis and progression in hepatocellular
carcinoma via inhibiting the STAT3/FOXM1/GLUT1 pathway. Mol Oncol.
14:1381–1396. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lin YH, Wu MH, Huang YH, Yeh CT, Cheng ML,
Chi HC, Tsai CY, Chung IH, Chen CY and Lin KH: Taurine up-regulated
gene 1 functions as a master regulator to coordinate glycolysis and
metastasis in hepatocellular carcinoma. Hepatology. 67:188–203.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li S, Li J, Dai W, Zhang Q, Feng J, Wu L,
Liu T, Yu Q, Xu S, Wang W, et al: Genistein suppresses aerobic
glycolysis and induces hepatocellular carcinoma cell death. Br J
Cancer. 117:1518–1528. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li S, Dai W, Mo W, Li J, Feng J, Wu L, Liu
T, Yu Q, Xu S, Wang W, et al: By inhibiting PFKFB3, aspirin
overcomes sorafenib resistance in hepatocellular carcinoma. Int J
Cancer. 141:2571–2584. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhao YH, Zhou M, Liu H, Ding Y, Khong HT,
Yu D, Fodstad O and Tan M: Upregulation of lactate dehydrogenase A
by ErbB2 through heat shock factor 1 promotes breast cancer cell
glycolysis and growth. Oncogene. 28:3689–3701. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hu M, Fu Q, Jing C, Zhang X, Qin T and Pan
Y: LncRNA HOTAIR knockdown inhibits glycolysis by regulating
miR-130a-3p/HIF1A in hepatocellular carcinoma under hypoxia. Biomed
Pharmacother. 125:1097032020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li Y, Guo D, Zhao Y, Ren M, Lu G, Wang Y,
Zhang J, Mi C, He S and Lu X: Long non-coding RNA SNHG5 promotes
human hepatocellular carcinoma progression by regulating
miR-26a-5p/GSK3β signal pathway. Cell Death Dis. 9:8882018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Huang Y, Xiang B, Liu Y, Wang Y and Kan H:
LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human
hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer
Lett. 437:56–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang H, Su X, Burley SK and Zheng XFS:
mTOR regulates aerobic glycolysis through NEAT1 and nuclear
paraspeckle-mediated mechanism in hepatocellular carcinoma.
Theranostics. 12:3518–3533. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
He H, Chen T, Mo H, Chen S, Liu Q and Guo
C: Hypoxia-inducible long noncoding RNA NPSR1-AS1 promotes the
proliferation and glycolysis of hepatocellular carcinoma cells by
regulating the MAPK/ERK pathway. Biochem Biophys Res Commun.
533:886–892. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li X, Zhao Q, Qi J, Wang W, Zhang D, Li Z
and Qin C: lncRNA Ftx promotes aerobic glycolysis and tumor
progression through the PPARγ pathway in hepatocellular carcinoma.
Int J Oncol. 53:551–566. 2018.PubMed/NCBI
|
|
69
|
Ma X, Mao Z, Zhu J, Liu H and Chen F:
lncRNA PANTR1 Upregulates BCL2A1 expression to promote
tumorigenesis and warburg effect of hepatocellular carcinoma
through restraining miR-587. J Immunol Res. 2021:17368192021.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Neoptolemos JP, Kleeff J, Michl P,
Costello E, Greenhalf W and Palmer DH: Therapeutic developments in
pancreatic cancer: Current and future perspectives. Nat Rev
Gastroenterol Hepatol. 15:333–348. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Rahib L, Smith BD, Aizenberg R, Rosenzweig
AB, Fleshman JM and Matrisian LM: Projecting cancer incidence and
deaths to 2030: The unexpected burden of thyroid, liver, and
pancreas cancers in the United States. Cancer Res. 74:2913–2921.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xu F, Huang M, Chen Q, Niu Y, Hu Y, Hu P,
Chen D, He C, Huang K, Zeng Z, et al: LncRNA HIF1A-AS1 promotes
gemcitabine resistance of pancreatic cancer by enhancing glycolysis
through modulating the AKT/YB1/HIF1α pathway. Cancer Res.
81:5678–5691. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Miller KD, Nogueira L, Devasia T, Mariotto
AB, Yabroff KR, Jemal A, Kramer J and Siegel RL: Cancer treatment
and survivorship statistics, 2022. CA Cancer J Clin. 72:409–436.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wu C, Li M, Meng H, Liu Y, Niu W, Zhou Y,
Zhao R, Duan Y, Zeng Z, Li X, et al: Analysis of status and
countermeasures of cancer incidence and mortality in China. Sci
China Life Sci. 62:640–647. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yan T, Shen C, Jiang P, Yu C, Guo F, Tian
X, Zhu X, Lu S, Han B, Zhong M, et al: Risk SNP-induced
lncRNA-SLCC1 drives colorectal cancer through activating glycolysis
signaling. Signal Transduct Target Ther. 6:702021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Huang JZ, Chen M, Chen D, Gao XC, Zhu S,
Huang H, Hu M, Zhu H and Yan GR: A peptide encoded by a putative
lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell.
68:171–184.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Li S, Zhu K, Liu L, Gu J, Niu H and Guo J:
lncARSR sponges miR-34a-5p to promote colorectal cancer invasion
and metastasis via hexokinase-1-mediated glycolysis. Cancer Sci.
111:3938–3952. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lan Z, Yao X, Sun K, Li A, Liu S and Wang
X: The interaction between lncRNA SNHG6 and hnRNPA1 contributes to
the growth of colorectal cancer by enhancing aerobic glycolysis
through the regulation of alternative splicing of PKM. Front Oncol.
10:3632020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zheng H, Zhang M, Ke X, Deng X, Li D and
Wang Q, Yan S, Xue Y and Wang Q: LncRNA XIST/miR-137 axis
strengthens chemo-resistance and glycolysis of colorectal cancer
cells by hindering transformation from PKM2 to PKM1. Cancer
Biomark. 30:395–406. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Cui K, Wu H, Fan J, Zhang L, Li H, Guo H,
Yang R and Li Z: The mixture of ferulic acid and P-Coumaric acid
suppresses colorectal cancer through lncRNA 495810/PKM2 mediated
aerobic glycolysis. Int J Mol Sci. 23:121062022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wang Y, Yu G, Liu Y, Xie L, Ge J, Zhao G
and Lin J: Hypoxia-induced PTTG3P contributes to colorectal cancer
glycolysis and M2 phenotype of macrophage. Biosci Rep.
41:BSR202107642021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Sun S, Li W, Ma X and Luan H: Long
Noncoding RNA LINC00265 promotes glycolysis and lactate production
of colorectal cancer through regulating of miR-216b-5p/TRIM44 Axis.
Digestion. 101:391–400. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yu Z, Wang Y, Deng J, Liu D, Zhang L, Shao
H, Wang Z, Zhu W, Zhao C and Ke Q: Long non-coding RNA COL4A2-AS1
facilitates cell proliferation and glycolysis of colorectal cancer
cells via miR-20b-5p/hypoxia inducible factor 1 alpha subunit axis.
Bioengineered. 12:6251–6263. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Li C, Wang P, Du J, Chen J, Liu W and Ye
K: LncRNA RAD51-AS1/miR-29b/c-3p/NDRG2 crosstalk repressed
proliferation, invasion and glycolysis of colorectal cancer. IUBMB
Life. 73:286–298. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang Z, Yang W, Li N, Chen X, Ma F, Yang
J, Zhang Y, Chai X, Zhang B, Hou X, et al: LncRNA MCF2L-AS1
aggravates proliferation, invasion and glycolysis of colorectal
cancer cells via the crosstalk with miR-874-3p/FOXM1 signaling
axis. Carcinogenesis. 42:263–271. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Guo X, Zhang Y, Liu L, Yang W and Zhang Q:
HNF1A-AS1 regulates cell migration, invasion and glycolysis via
modulating miR-124/MYO6 in colorectal cancer cells. Onco Targets
Ther. 13:1507–1518. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Li YL, Zhang XX, Yao JN, Gao B, Gao SL,
Wang CF, Zhou HN and Zhang LF: ZEB2-AS1 regulates the expression of
TAB3 and promotes the development of colon cancer by adsorbing
microRNA-188. Eur Rev Med Pharmacol Sci. 24:4180–4189.
2020.PubMed/NCBI
|
|
90
|
Wang X, Zhang H, Yin S, Yang Y, Yang H,
Yang J, Zhou Z, Li S, Ying G and Ba Y: lncRNA-encoded pep-AP
attenuates the pentose phosphate pathway and sensitizes colorectal
cancer cells to Oxaliplatin. EMBO Rep. 23:e531402022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Hong J, Guo F, Lu SY, Shen C, Ma D, Zhang
X, Xie Y, Yan T, Yu T, Sun T, et al: F. nucleatum targets lncRNA
ENO1-IT1 to promote glycolysis and oncogenesis in colorectal
cancer. Gut. 70:2123–2137. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhang H, Qin D, Jiang Z and Zhang J:
SNHG9/miR-199a-5p/Wnt2 axis regulates cell growth and aerobic
glycolysis in glioblastoma. J Neuropathol Exp Neurol. 78:939–948.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lu J, Liu X, Zheng J, Song J, Liu Y, Ruan
X, Shen S, Shao L, Yang C, Wang D, et al: Lin28A promotes
IRF6-regulated aerobic glycolysis in glioma cells by stabilizing
SNHG14. Cell Death Dis. 11:4472020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Liu X, Zhu Q, Guo Y, Xiao Z, Hu L and Xu
Q: LncRNA LINC00689 promotes the growth, metastasis and glycolysis
of glioma cells by targeting miR-338-3p/PKM2 axis. Biomed
Pharmacother. 117:1090692019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
He Z, You C and Zhao D: Long non-coding
RNA UCA1/miR-182/PFKFB2 axis modulates glioblastoma-associated
stromal cells-mediated glycolysis and invasion of glioma cells.
Biochem Biophys Res Commun. 500:569–576. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Liu Y, He D, Xiao M, Zhu Y, Zhou J and Cao
K: Long noncoding RNA LINC00518 induces radioresistance by
regulating glycolysis through an miR-33a-3p/HIF-1α negative
feedback loop in melanoma. Cell Death Dis. 12:2452021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wang Y, Zhang X, Wang Z, Hu Q, Wu J, Li Y,
Ren X, Wu T, Tao X, Chen X, et al: LncRNA-p23154 promotes the
invasion-metastasis potential of oral squamous cell carcinoma by
regulating Glut1-mediated glycolysis. Cancer Lett. 434:172–183.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yang X, Ye H, He M, Zhou X, Sun N, Guo W,
Lin X, Huang H, Lin Y, Yao R and Wang H: LncRNA PDIA3P interacts
with c-Myc to regulate cell proliferation via induction of pentose
phosphate pathway in multiple myeloma. Biochem Biophys Res Commun.
498:207–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Huo N, Cong R, Sun ZJ, Li WC, Zhu X, Xue
CY, Chen Z, Ma LY, Chu Z, Han YC, et al: STAT3/LINC00671 axis
regulates papillary thyroid tumor growth and metastasis via
LDHA-mediated glycolysis. Cell Death Dis. 12:7992021. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Xue L, Li J, Lin Y, Liu D, Yang Q, Jian J
and Peng J: m6 A transferase METTL3-induced lncRNA ABHD11-AS1
promotes the Warburg effect of non-small-cell lung cancer. J Cell
Physiol. 236:2649–2658. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Song J, Wu X, Liu F, Li M, Sun Y, Wang Y,
Wang C, Zhu K, Jia X, Wang B and Ma X: Long non-coding RNA PVT1
promotes glycolysis and tumor progression by regulating miR-497/HK2
axis in osteosarcoma. Biochem Biophys Res Commun. 490:217–224.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Han X and Yang Y, Sun Y, Qin L and Yang Y:
LncRNA TUG1 affects cell viability by regulating glycolysis in
osteosarcoma cells. Gene. 674:87–92. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Shen Y, Xu J, Pan X, Zhang Y, Weng Y, Zhou
D and He S: LncRNA KCNQ1OT1 sponges miR-34c-5p to promote
osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell
Death Dis. 11:2782020. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Xu S, Jia G, Zhang H, Wang L, Cong Y, Lv
M, Xu J, Ruan H, Jia X, Xu P and Wang Y: LncRNA HOXB-AS3 promotes
growth, invasion and migration of epithelial ovarian cancer by
altering glycolysis. Life Sci. 264:1186362021. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Feng Z, Chen R, Huang N and Luo C: Long
non-coding RNA ASMTL-AS1 inhibits tumor growth and glycolysis by
regulating the miR-93-3p/miR-660/FOXO1 axis in papillary thyroid
carcinoma. Life Sci. 244:1172982020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Chen Z, Hu Z, Sui Q, Huang Y, Zhao M, Li
M, Liang J, Lu T, Zhan C, Lin Z, et al: LncRNA FAM83A-AS1
facilitates tumor proliferation and the migration via the
HIF-1α/glycolysis axis in lung adenocarcinoma. Int J Biol Sci.
18:522–535. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Song H, Li D, Wang X, Fang E, Yang F, Hu
A, Wang J, Guo Y, Liu Y, Li H, et al: HNF4A-AS1/hnRNPU/CTCF axis as
a therapeutic target for aerobic glycolysis and neuroblastoma
progression. J Hematol Oncol. 13:242020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Wan J, Liu Y, Long F, Tian J and Zhang C:
circPVT1 promotes osteosarcoma glycolysis and metastasis by
sponging miR-423-5p to activate Wnt5a/Ror2 signaling. Cancer Sci.
112:1707–1722. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhao L, Ji G, Le X, Wang C, Xu L, Feng M,
Zhang Y, Yang H, Xuan Y, Yang Y, et al: Long Noncoding RNA
LINC00092 acts in cancer-associated fibroblasts to drive glycolysis
and progression of ovarian cancer. Cancer Res. 77:1369–1382. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Lin X, Feng D, Li P and Lv Y: LncRNA
LINC00857 regulates the progression and glycolysis in ovarian
cancer by modulating the Hippo signaling pathway. Cancer Med.
9:8122–8132. 2020. View Article : Google Scholar : PubMed/NCBI
|