Introduction
Opioids are still the most commonly used drugs for
cancer pain relief, with oral morphine that is recommended as the
first-line drug for moderate-to-severe cancer pain in the
international guidelines (1,2), and
oxycodone and hydromorphone, both of immediate-release (IR) and of
modified-release oral formulations as well as oral methadone that
are effective alternatives. Hydromorphone was unavailable until
2017, and methadone requires a special license for use; thus,
morphine and oxycodone are the most frequently used opioids in
Japan.
Because of its many formulations, such as tablet,
liquid medicine and suppository, morphine is superior in terms of
its ease of administration, convenience of use in patients
undergoing home care, and effectiveness in patients with
respiratory distress (3).
Furthermore, immediate-release (IR) morphine reaches a high serum
concentration more rapidly (Tmax=0·9 h) as compared with oxycodone
(Tmax=1·9 h) (4). On the other
hand, since the oral bioavailability ratio of oxycodone is higher
than that of morphine, it can be safely used in patients with
chronic kidney diseases and may also be useful for patients with
neuropathic pain (5–7), making selection of the most
appropriate opioid more difficult. There is lack of consensus
regarding the choice of drug (e.g., morphine or oxycodone) and the
dose required to provide quick and potent pain relief in individual
patients (3), especially since
opioid sensitivity and the associated side effects vary widely
among patients.
Numerous studies have proposed that the
catechol-O-methyltransferase (COMT) 472G→A (rs4680,
pVal158Met) single nucleotide polymorphism (SNP) may be a
predictive biomarker of the response to morphine treatment
(8). In these studies, patients
with the GG genotype received the highest dose of morphine
(9,10), while those with the AA genotype
received the lowest dose (11).
Based on these studies, we performed a randomized
controlled trial-the RELIEF study (Trial registration number:
UMIN000015579), in which we compared the efficacy of morphine and
oxycodone using the COMT rs4680 SNP as a biomarker. We
randomized total of 140 patients (1:1) into a morphine group (Group
M) or an oxycodone group (Group O) and evaluated the patients to
determine the ability of pain controllability of the drugs, and
found that in Group M, not only patients with the rs4680 GG
genotype, but also those with the non-GG (GA/AA) genotype required
higher doses of opioids, as compared with Group O (12). The results of this study, together
with those of others, imply that the analysis of the COMT
genotype alone may not be sufficient to adequately individualize
opioid therapy (4,13). To explore SNPs other than the
COMT rs4680 SNP, which may aid in individualizing treatment
with morphine or oxycodone, we focused on a few other SNPs that
have previously been suggested to be linked to pain sensitivity
and/or opioid efficacy. The investigation consisted of two parts: a
development study of 94 cases to screen for the candidate SNPs and
a validation study including an additional 135 cases from the
RELIEF study to validate these SNPs.
Patients and methods
Patients and samples
Two subsets of patients with advanced malignancies
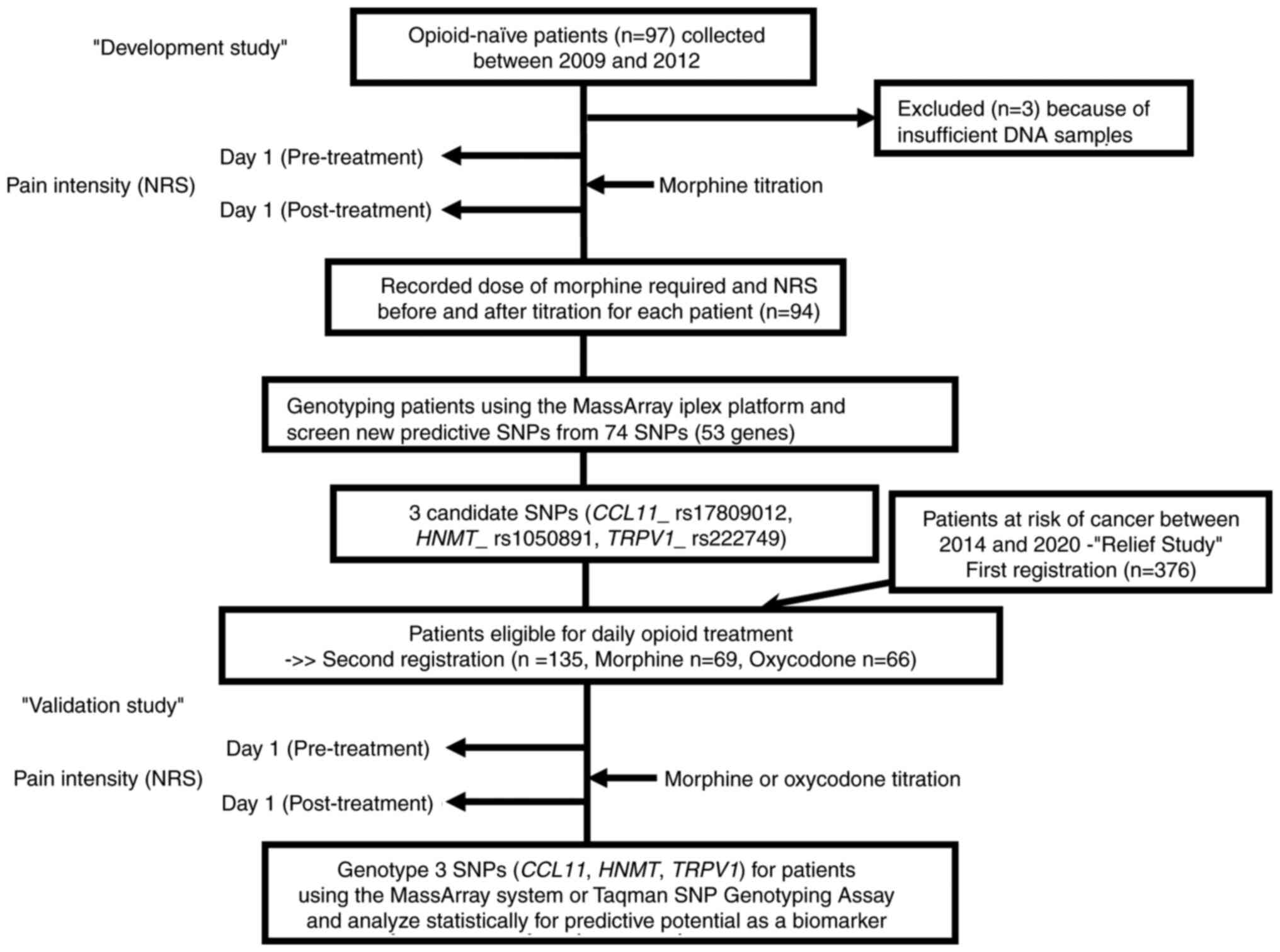
were enrolled in the current study (Fig. 1). Our cohorts do not include any
families affecting genotype independency. The development study to
screen for SNPs was conducted on the patients enrolled in our
prospective study performed from 2009 to 2012 at Kindai University
Faculty of Medicine and Sakai Hospital; these patients were treated
with morphine alone. The characteristics of the patients have been
described previously (14–16). Of the original 97 morphine-naïve
patients who met inclusion criteria (15), 94 were genotyped as mentioned below,
after excluding the remaining 3 patients because of inadequate DNA
samples.
The validation study was performed in the 135
patients who fulfilled the second registration due to the criteria
of suffering from cancer pain that necessitated daily treatment
with opioids, from among the subjects (n=378) who were originally
enrolled based on the first registration criteria in RELIEF study,
a randomized controlled trial conducted recently by us (Trial
registration number: UMIN000015579) (12). The second registration patients were
randomized (1:1) based on the COMT rs4680 SNP (GG or non-GG)
into a morphine group (Group M; n=70) or an oxycodone group (Group
O; n=70), in such a way that patients with the GG and non-GG (GA or
AA) genotypes were equally distributed in each group. The optimal
sample size was calculated as 140 based on our preliminary analysis
for the COMT rs4680 SNP (14), which has been described in detail in
a previous report (12). We finally
included 135 cases, after excluding 5 cases (because the data
description was incomplete in 1 case and the trial was not underway
in time for the genotyping in the remaining 4 cases), reducing the
number of subjects in Group M to 69 and that in Group O to 66. The
baseline characteristics of these 135 patients are presented in
Table I. The inclusion and
exclusion criteria for patients were as listed previously (12). CYP inducers such as rifampicin or
carbamazepine or inhibitors such as itraconazole or SSRI were not
administered to the participants.
 | Table I.Baseline characteristics of the
patients. |
Table I.
Baseline characteristics of the
patients.
|
|
| Morphine_group
(n=69) | Oxycodone_group
(n=66) |
|---|
|
|
|
|
|
|---|
| Item | Number of
patients | Low dose
(n=39) | High dose
(n=30) | P-value | Low dose
(n=51) | High dose
(n=15) | P-value |
|---|
| Age, n
(%)a |
|
|
| 0.028 |
|
| 1.000 |
| <70
years | 62 | 23 (59.0) | 9 (30.0) |
| 23 (45.1) | 7 (46.7) |
|
|
≥70 | 73 | 16 (41.0) | 21 (70.0) |
| 28 (54.9) | 8 (53.3) |
|
| Sex, n
(%)a |
|
|
| 0.217 |
|
| 0.256 |
|
Male | 77 | 21 (53.8) | 21 (70) |
| 25 (49.0) | 10 (66.7) |
|
|
Female | 58 | 18 (46.2) | 9 (30) |
| 26 (51.0) | 5 (33.3) |
|
| Performance status,
n (%)a,c |
|
|
| 0.805 |
|
| 1.000 |
| 0 | 10 | 3 (7.7) | 0 (0) |
| 3 (5.9) | 4 (26.7) |
|
| 1 | 79 | 22 (56.4) | 18 (60) |
| 31 (60.8) | 6 (40.0) |
|
| 2 | 33 | 10 (25.6) | 8 (26.7) |
| 13 (25.5) | 3 (20.0) |
|
| 3 | 10 | 3 (7.7) | 3 (10) |
| 2 (3.9) | 2 (13.3) |
|
| 4 | 4 | 1 (2.6) | 1 (3.3) |
| 2 (3.9) | 0 (0) |
|
| Pre-NRS, median
(IQR)b |
| 5 (3–6) | 6 (4–7) | 0.032 | 5 (4–6) | 7 (6–8) | 0.0007 |
| HADS, median
(IQR)b |
| 13 (11–21) | 14.5 (11–23) | 0.682 | 14 (9–21) | 15 (12–22) | 0.619 |
| SF-MPQ-2, median
(IQR)b |
| 30 (17.5–56) | 35 (21.5–56) | 0.566 | 42 (21–77) | 66 (58–82) | 0.015 |
| CCL11, n
(%)a |
|
|
| 0.03 |
|
| 0.78 |
| AA | 70 | 14 (35.9) | 19 (63.3) |
| 28 (54.9) | 9 (60.0) |
|
|
AG/GG | 65 | 25 (64.1) | 11 (36.7) |
| 23 (45.1) | 6 (40.0) |
|
| HNMT, n
(%)a |
|
|
| 0.81 |
|
| 0.016 |
| AA | 75 | 20 (51.3) | 17 (56.7) |
| 25 (49.0) | 13 (86.7) |
|
|
AG/GG | 60 | 19 (48.7) | 13 (43.3) |
| 26 (51.0) | 2 (13.3) |
|
| TRPV1, n
(%)a |
|
|
| 0.23 |
|
| 1 |
| CC | 71 | 24 (61.5) | 14 (46.7) |
| 25 (49.0) | 8 (53.3) |
|
|
CT/TT | 64 | 15 (38.5) | 16 (53.3) |
| 26 (51.0) | 7 (46.7) |
|
Titration and classification of the
patients
For opioid titration following cancer pain onset,
the opioid-naïve patients were administered intermediate release
(IR) opioids according to the guidelines for titration (NCCN
Guidelines™, Adult Cancer Pain) (2,17) by
specialized palliative care physicians. The 97 patients in the
development study were administered IR morphine, and the 135
patients in the validation study received either IR morphine or IR
oxycodone (groups M or O). We define the day of titration as day 1.
The administrations of the minimum standard dose of IR opioids,
that is, 5 mg/dose for morphine and 2.5 mg/dose for oxycodone (3.75
mg equivalent of IR morphine) were repeated for dose titration of
the patients until the pain decreased by ≥33% on the Numerical
Rating Scale (NRS, 0=no pain to 10=maximal pain), or the NRS score
decreased to ≤3 from pre- to post-titration on day 1. Patients
requiring 10 mg or more of IR morphine, or 7.5 mg or more of IR
oxycodone were classified into the high-dose group and patients
requiring 5 mg of IR morphine or 5 mg or less of IR oxycodone were
classified into the low-dose group. Patients in whom the NRS score
did not decrease to ≤3 or by ≥33% on the NRS after treatment on day
1 were categorized into the high-dose group, even if the dose was 5
mg of IR morphine or 5 mg of IR oxycodone (12). The post-titration NRS scores were
recorded one or two hours after the final titration in the patients
treated with morphine (Tmax=0·9 h) or oxycodone (Tmax=1·9 h),
respectively.
Genotyping
Genomic DNA was isolated from the blood samples, as
described previously (16).
Genotyping of the patients in the validation study was performed
for 74 SNPs of 54 genes (Table
SI), which were selected based on previous reports linking them
to pain sensitivity and/or opioid efficacy (18–26).
We focused on both chemokines and cytokines because they have come
to be more and more accepted as the major mediators that activate
glial cells to interact with neurons, which is emerging as a key
mechanism underlying chronic pain (21,22).
Several chemokines implicated could be missing from our list if
they have no appropriate SNPs within their genes.
Genes encoding transient receptor potential (TRP)
ion channels, which represent the major group of molecules involved
in nociception and development of pathological pain, were also
included as candidate genes (24).
In addition to several well-known molecules
modifying the mechanisms of opioid signaling or pain development
(20,21,26),
novel members likely to be associated with pain modulation such as
the oxytocin receptor (OXTR) (23),
molecules associated with increased morphine requirement
(serine/threonine-protein kinase TAOK3) (19) or with histamine degradation that is
thought to be important in nociception at the periphery [histamine
N-methyltransferase (HNMT) and amiloride-sensitive amine oxidase
(AOC1)] (18) were also added to
the list (Table SI). Analysis of
these genes were conducted in the 94 patients in the first study
(development study in Fig. 1),
using the MassARRAY iPLEX platform (27). The primer sequences used for the
MassARRAY genotyping will be offered on demand. The CYP450 enzymes
and the ATP-binding cassette transporters that have been thought to
be involved in the metabolism and efflux of opioids, respectively,
could also be candidates (26,28).
The SNPs of such genes are comprehensively covered in the DMET
Platform (29), which we are using
to genotype and analyze the same patients elsewhere (in
preparation). Before proceeding to the analysis, we performed a
quality control check on the data. We eliminated subjects with a
genotype call rate of <90%, deviation of the Hardy-Weinberg
equilibrium (P<1×10−5) (29), or if the minor allele frequency
(MAF) was less than 5% (Table
SI).
In the validation study, the 3 candidate SNPs
(CCL11 rs17809012, HNMT rs1050891, and TRPV1
rs222749) which were selected from among the 74 SNPs identified in
the development study were analyzed in second registration patients
selected from the RELIEF study (n=135) (12), using the MassARRAY iPLEX
platform.
Statistical analysis
In the development study, we screened 74 SNPs by
estimating the differences in the required dose of morphine at
titration (high or low) between the patients with the major
genotype (major-allele homozygotes) and the patients with the
non-major genotypes (heterozygotes plus minor-allele homozygotes)
using Fisher's exact test at a two-tailed significance level of 5%.
The odds ratio (OR) and 95% confidence interval (CI) were obtained
for each SNP.
For the analysis in the validation study, we
characterized the 3 candidate SNPs (CCL11 rs17809012,
HNMT rs1050891, and TRPV1 rs222749) by performing
simple regression analyses separately for Group M and Group O. The
primary outcome was pain relief. The objective variables examined
were the ΔNRS. The ΔNRS was defined as the difference in the NRS
score before and after the titration. Therefore, the greater the
ΔNRS, the greater the pain relief.
In addition to genotype (major-allele homozygotes or
heterozygotes plus minor-allele homozygotes) of the 3 SNPs,
independent variables considered were age (<70 or ≥70 years),
sex, performance status (ps;1/≥2), pre-NRS (1–10),
total scores on the HADS (Hospital Anxiety and Depression Scale)
(15), SF-MPQ-2 (Short-Form McGill
Pain Questionnaire-2) (30), and
the required dose (high or low), of which pre-NRS, HADS, and
SF-MPQ-2 were ordinal variables. The differences in the required
dose (high or low) of each opioid were estimated using Fisher's
exact test for categorical variables or using Mann-Whitney U test
for ordinal data.
We also analyzed the three SNPs for the overall
subject population, in which the variable of ‘dose’ was omitted
because of a different conversion ratio of 3:2 between oral
morphine and oral oxycodone formulations due to their incompatible
dosage forms, as has been mentioned (12). Instead, ‘treatment’ (morphine or
oxycodone) is added as an independent variable. For the analysis of
the overall subject population, a simple regression analysis,
together with multiple regression analyses to adjust for
confounding variables, were performed. We performed two ways of
multiple regression analyses. Either all of the above-mentioned
variables or selected variables (pre_NRS, treatment, HADS and
SF-MPQ-2) was entered into each of the analyses.
The variance inflation factor (VIF) was used to
diagnose multicollinearity problems. A P<0.05 was considered as
denoting statistical significance. The analyses were performed
using the JMP statistical software (v14.2, SAS Institute, Cary, NC,
USA).
Results
First screening of SNPs
In our development study conducted on 94 patients,
33% (n=31) of the patients were classified as requiring high-dose
morphine while 67% (n=63) were classified into the low-dose
morphine group. Correlations of the required dose of morphine with
the gene polymorphisms were analyzed using 74 SNPs, without
corrections for multiple comparisons. A total of 7 SNPs that did
not meet the quality control criterion [deviation from the
Hardy-Weinberg equilibrium (n=2), call rate <90% (n=2), and
minor allele frequency <5% (n=3)] were not considered in the
subsequent analyses (Table
SI).
The patients were genotyped for the 74 SNPs to
compare the high- and low-dose groups for the numbers of patients
who were homozygous for the major allele and the numbers of
patients with non-major (heterozygous plus homozygous for minor
allele) alleles. This analysis showed that 3 SNPs (TRPV1
rs222749, CCL11 rs17809012, HNMT rs1050891) out of
the 74 SNPs were associated with the morphine dose requirement
(P≤0.05), suggesting that these SNPs could be biomarkers to predict
the morphine controllability in cancer pain for individual patients
(Table II).
 | Table II.Correlation between genotypes and
morphine dose requirementa. |
Table II.
Correlation between genotypes and
morphine dose requirementa.
| Gene | SNP | Genotypes | OR (95%
CI)b |
P-valuec |
|---|
| CCL11 | rs17809012 | AA // AG + GG | 0.32
(0.11–0.85) | 0.014 |
| TRPV1 | rs222749 | CC // CT + TT | 3.13
(1.27–7.74) | 0.020 |
| HNMT | rs1050891 | AA // AG + GG | 0.40
(0.16–0.99) | 0.050 |
| TRPV1 | rs224534 | AA // AG + GG | 0.44
(0.17–1.07) | 0.079 |
| ADRB2 | rs1042713 | AA // AG + GG | 3.22
(0.86–12.1) | 0.101 |
| KCNS1 | rs734784 | AA // AG + GG | 2.62
(0.95–7.26) | 0.102 |
| IL-1RN | rs2234677 | GG // GA + AA | 2.80
(0.72–11.3) | 0.110 |
| COMT | rs4680 | GG // GA + AA | 0.47
(0.20–1.14) | 0.125 |
| TRPM8 | rs17868387 | AA // AG + GG | 2.39
(0.80–7.09) | 0.145 |
| GCH1 | rs3783641 | TT // TA + AA | 0.47
(0.15–1.37) | 0.165 |
| AIF1 | rs2844475 | TT // TC + CC | 0.48
(0.19–1.22) | 0.165 |
| CCL8 | rs1133763 | AA // AC + CC | 1.96
(0.76–5.17) | 0.187 |
Validation study
The 3 candidate SNPs thus extracted in the first
screening were then examined in the total of 135 patients enrolled
in our validation study. We first determined if these SNPs were
strongly linked to the analgesic effect of morphine, as the ability
of morphine to relieve the pain was the first selection criterion
in the development study. A simple regression analysis for Group M
showed that the three SNPs were correlated with the difference in
the ΔNRS between the genotype groups. The patients homozygous for
the major allele of CCL11 rs17809012 (AA) or HNMT
rs1050891 (AA) showed a significantly reduced ΔNRS by 0.63 or 0.48,
on average, as compared with patients carrying rs17809012 (AG/GG)
or rs1050891 (AG/GG), with P-values of 0.007 and 0.041,
respectively (Table III). For the
patients who were homozygous for the major allele of TRPV1
rs222749 (CC), the ΔNRS was higher by 0.43 as compared with that in
patients with rs222749 (CT/TT); the P-value was 0.071. Much lower
ΔNRS differences were observed between genotype groups for these
SNPs in Group O (Table III),
confirming that these SNPs are specific biomarkers for
morphine-induced analgesia. The difference in the analgesic effect
between the CCL11 genotype groups was also shown to be
particularly pronounced for morphine, by plotting the
post-titration NRS against the pre-titration NRS (Fig. S1A). Regardless of the opioids that
were used in the overall subject population, these SNPs appeared to
affect the ΔNRS, although the differences were statistically
insignificant in the simple regression model (Table SII). However, multiple linear
regression model to adjust for age, sex, ps,pre-NRS, treatment,
genotypes, total scores on the HADS and SF-MPQ-2 showed that
significant ΔNRS differences were observed between genotype groups
of CCL11 rs17809012 and HNMT rs1050891 (0.26 and
0.30, with P-values of 0.038 and 0.021, respectively), and there
seemed no strong confounding variables, with all the VIF values
evenly low (<1.5) (Table SIII,
multiple regression model 1). We also chose independent variables
that were likely to directly influence on pain sensitivity (and
thus the ΔNRS) such as pre NRS, treatment, HADS and SF-MPQ-2
(15,30). Multiple linear regression model
using these variables with the three genotypes showed that the ΔNRS
differences between genotypes for the two SNPs remained significant
(0.27 and 0.29, with P-values of 0.034 and 0.022 for CCL11
rs17809012 and HNMT rs1050891, respectively; (Table SIII, multiple regression model
2).
 | Table III.Simple regression analyses for
determinants of the ΔNRS on day 1 in the Morphine and Oxycodone
groups. |
Table III.
Simple regression analyses for
determinants of the ΔNRS on day 1 in the Morphine and Oxycodone
groups.
|
| Morphine
(n=69) | Oxycodone
(n=66) |
|---|
|
|
|
|
|---|
| Variable | β | t-value | Partial regression
coefficient (95% CI) | P-value | β | t-value | Partial regression
coefficient (95% CI) | P-value |
|---|
| Age | 0.07 | 0.55 | 0.13 (−0.34 to
0.60) | 0.585 | 0.03 | 0.26 | 0.05 (−0.34 to
0.45) | 0.796 |
| Sex | 0.01 | 0.08 | 0.02 (−0.47 to
0.50) | 0.939 | −0.11 | −0.91 | −0.18 (−0.57 to
0.21) | 0.366 |
| Performance
status | −0.10 | −0.81 | −0.20 (−0.68 to
0.29) | 0.421 | 0.22 | 1.81 | 0.37 (−0.04 to
0.78) | 0.075 |
| Pre-NRS | 0.55 | 5.34 | 0.53 (0.33 to
0.73) | <0.0001 | 0.60 | 5.97 | 0.47 (0.31 to
0.63) | <0.0001 |
| HADS score | 0.09 | 0.73 | 0.02 (−0.04 to
0.08) | 0.469 | 0.16 | 1.27 | 0.03 (−0.02 to
0.08) | 0.209 |
| SF-MPQ-2 total
score | 0.23 | 1.92 | 0.02 (−0.00 to
0.03) | 0.059 | 0.28 | 2.32 | 0.01 (0.002 to
0.02) | 0.024 |
| Dose | −0.21 | −1.73 | −0.41 (−0.88 to
0.06) | 0.09 | −0.15 | −1.24 | −0.29 (−0.75 to
0.18) | 0.221 |
| Genotype |
|
|
|
|
|
|
|
|
|
CCL11 | −0.32 | −2.79 | −0.63 (−1.07 to
−0.17) | 0.007 | 0.09 | 0.70 | 0.14 (−0.26 to
0.53) | 0.486 |
|
HNMT | −0.25 | −2.09 | −0.48 (−0.94 to
−0.02) | 0.041 | −0.03 | −0.2 | −0.04 (−0.44 to
0.36) | 0.840 |
|
TRPV1 | 0.22 | 1.84 | 0.43 (−0.04 to
0.89) | 0.071 | 0.05 | 0.42 | 0.08 (−0.31 to
0.48) | 0.673 |
Predictive factors for opioid
selection
Next, we examined the genotype-treatment
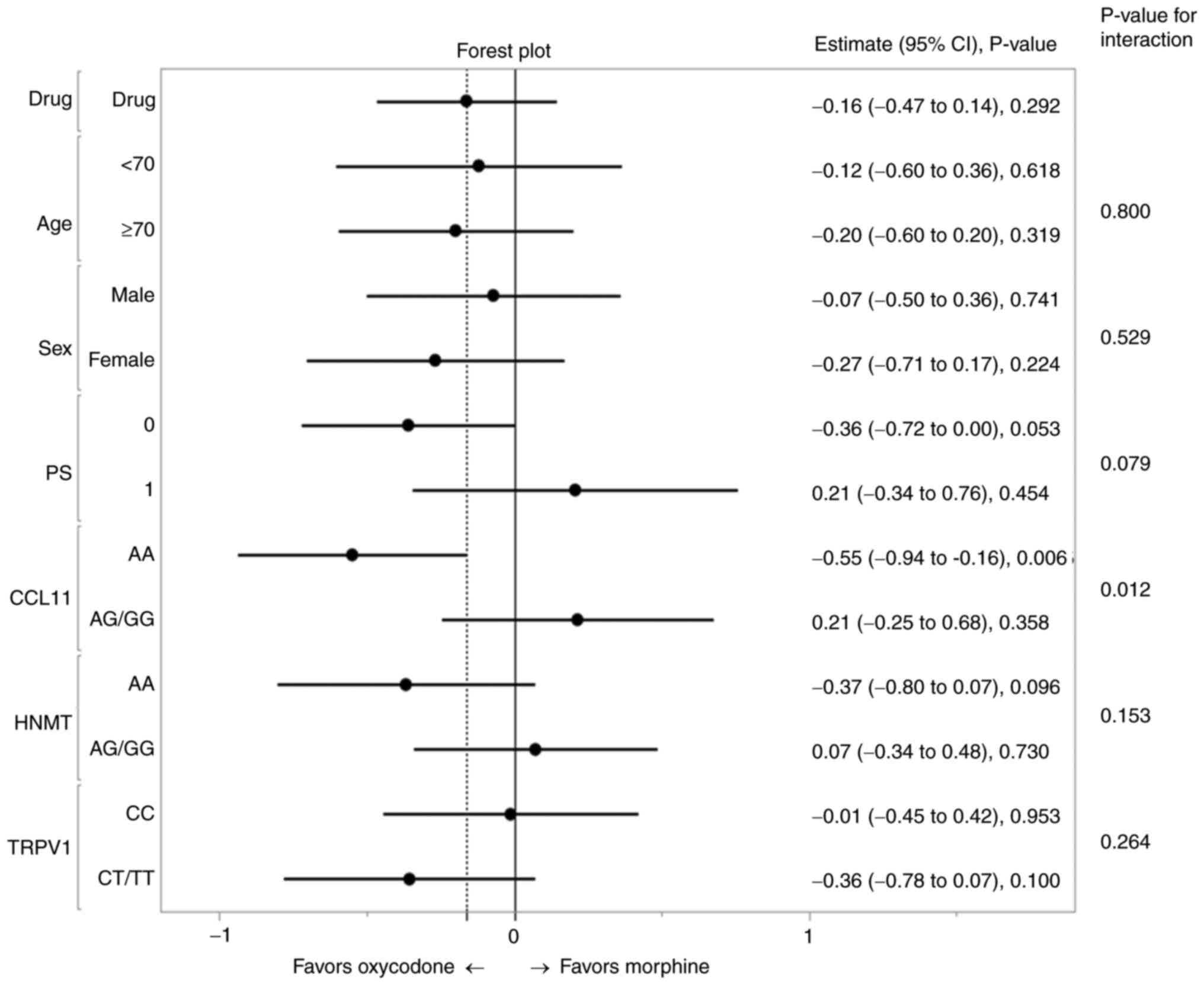
interactions for ΔNRS. A forest plot was drawn based on the
estimate (relative risk) with its 95% CI in 2 categorical groups
for each variable (Fig. 2). Better
efficacy was observed in Group O than in Group M in patients
homozygous for the major alleles of CCL11 or HNMT,
and in patients heterozygous or homozygous for minor alleles of the
TRPV1 genotype. A significant P-value was detected for the
interaction (0.012) between the CCL11 genotype and treatment
(Table SIV and Fig. 2). The Group M patients with
CCL11(AA), HNMT(AA), and TRPV1(CT/TT) showed
reduced least square means (LSM) of the ΔNRS as compared to
patients with other genotype and treatment combinations (Table IV). The superiority of one opioid
over another in terms of the analgesic effect was also shown to be
reversed between the two CCL11 genotype groups by plotting
the post-titration NRS against the pre-titration NRS, oxycodone
being the better treatment for patients with CCL11_AA and
morphine for patients with AG/GG (Fig.
S1A). Such interaction between opioid and genotype was not
observed for the HNMT SNP (Fig.
S1B).
 | Table IV.LSMs of ΔNRS for patients in terms of
their treatment and genotype interactions. |
Table IV.
LSMs of ΔNRS for patients in terms of
their treatment and genotype interactions.
|
Variablea | Groupb | LSM | Standard error | 95% confidence
interval |
|---|
|
CCL11*treatment | AA+morphine | 2.33 | 0.30 | 1.74–2.93 |
|
| AG/GG+morphine | 3.58 | 0.29 | 3.01–4.16 |
|
| AA+oxycodone | 3.43 | 0.29 | 2.87–4.00 |
|
|
AG/GG+oxycodone | 3.16 | 0.32 | 2.52–3.79 |
| HNMT*treatment | AA+morphine | 2.54 | 0.29 | 1.97–3.11 |
|
| AG/GG+morphine | 3.50 | 0.31 | 2.88–4.11 |
|
| AA+oxycodone | 3.28 | 0.29 | 2.71–3.84 |
|
|
AG/GG+oxycodone | 3.36 | 0.33 | 2.70–4.02 |
|
TRPV1*treatment | CC+morphine | 3.37 | 0.29 | 2.80–3.94 |
|
| CT/TT+morphine | 2.52 | 0.32 | 1.89–3.14 |
|
| CC+oxycodone | 3.39 | 0.31 | 2.78–4.00 |
|
|
CT/TT+oxycodone | 3.23 | 0.31 | 2.62–3.84 |
Discussion
In the current study, we explored predictive
biomarkers for selecting the most suitable opioid for treatment of
cancer pain. From our development study that included 94 patients,
three (TRPV1 rs222749, CCL11 rs17809012, HNMT
rs1050891) out of 74 SNPs were selected as new biomarker candidates
for predicting analgesic response to morphine. The subsequent
validation study confirmed these SNPs as being involved in the
analgesic effect of morphine in Group M, but not in that of
oxycodone. Such discrepancy in the effects of the SNPs between
these drugs could also be anticipated from the notion that morphine
and oxycodone exert their antinociceptive effects through distinct
opioid receptor populations (31,32)
despite the structural and functional similarities between the two
opioids (33).
Multiple regression analysis of data from the 135
patients in the validation study suggested the CCL11 and
HNMT genotypic variants as the major determinants of the
choice of opioid. Further analysis of these SNPs revealed a
significant interaction with the treatment effect for the
CCL11 genotype. The partial regression coefficient for the
interaction term (CCL11*treatment) was −0.38, which was a
significantly large value compared with that of the CCL11
(−0.24) or the treatment (−0.17) alone (Table SIV). The least square means (LSM)
of the ΔNRS calculated for patients with AA+morphine (patients with
AA treated with morphine) and patients with AG/GG+morphine were
2.33 and 3.59, while those with AA+oxycodone and AG/GG+oxycodone
were 3.43 and 3.16, respectively (Table IV). The patients with AA treated
with morphine showed a considerably reduced ΔNRS, suggesting that
oxycodone should be administered to patients with the AA genotype
of CCL11 to ease pain with an additional ~1.0 reduction in
the post-titration NRS. This procedure may be more critical for
patients with high pre-NRS scores who require immediate
analgesia.
Existence of relationships has been reported between
CCL11 rs17809012 (also known as eotaxin1) with
inflammatory-related diseases, such as asthma (34), fibromyalgia (25), and even ischemic stroke (35) and schizophrenia (36). Zhang et al demonstrated that
expression of this chemokine caused by inflammation in various
sites throughout the body amplifies and prolongs the inflammatory
condition, leading to fibromyalgia, a chronic pain syndrome
(25). The SNP rs17809012 is
located in the CCL11 promoter region, and a significantly
higher mRNA expression level was observed for the A allele than the
G allele (25,34). These data suggest that subjects with
rs17809012 AA are more responsive to pathological inflammation than
those with the AG/GG genotype. Moreover, contribution of the C-C
chemokine receptors (CCRs) to the pathogenesis of neuropathic pain
has recently been reported (37,38).
Upregulation of CCL11, one of the endogenous ligands of CCR3, binds
and activates CCR3 in the neurons or microglia to produce
neuropathic pain (38). Cancer pain
can be a mixture of nociceptive and less morphine-responsive
neuropathic pain (39). Oxycodone
has been reported to provide clinically meaningful relief in
patients with neuropathic pain (5–7), and
this may be more applicable to those with the rs17809012 AA
genotype for CCL11, who tend to have pains with more
neuropathic properties that are refractory to morphine
treatment.
Our study had some limitations. First, we conducted
genotypic analysis for 74 SNPs that were carefully selected from
among pain- and/or opioid-related genes. However, the heterogeneous
causes of cancer pain cannot be expected to be covered by these
SNPs. Some other related SNPs identified more recently could be
also candidates. Such additional examples include SNPs of CCL2,
CCL4, CCL7, CCL24, CCL26, CXCL2, CXCL10, CXCR3, CXCR4, IL-2,
IL-4, and IL-8 (37,40,41),
which remain to be investigated in the future.
Second, we screened the SNP candidates for our
development study only from among patients who were treated with
morphine. It would be desirable to also identify SNPs specifically
associated with oxycodone sensitivity, which, in combination with
the present results, may help in optimization of the pain
treatment. Further analysis is needed with a larger number of
patients treated with oxycodone.
Third, analyses of SNPs normally use models assuming
dominant inheritance (major-allele homozygotes vs. heterozygotes
plus minor-allele homozygotes) and recessive inheritance
(major-allele homozygotes plus heterozygotes vs. minor-allele
homozygotes), but we only used the former model. Our sample size
was too small to include a sufficient number of minor-allele
homozygotes for every SNP. We may have missed some latent candidate
SNPs because of the small sample size.
Fourth, we omitted the variable of ‘dose’ when
analyzing the total subject population due to the incompatible
dosage forms between oral morphine and oral oxycodone formulations.
This precluded us from comparing the opioids for their quantitative
effects on pain relief, which might not be unreasonable given that
the opioid for each patient is selected before the dose.
In conclusion, this is an extension of the RELIEF
study to analyze the correlations between some SNPs and the
efficacy (or suitability) of opioid drugs in patients with cancer
pain. We identified three SNPs as biomarkers, and found that the
CCL11 rs17809012 SNP, in particular, was highly correlated
with the pain controllability in patients treated with morphine or
oxycodone. Further studies using larger sample sizes are needed to
analyze and confirm the individual as well as synergistic effects
of these SNPs. Measurements of the serum concentrations of the
candidate proteins (i.e. CCL11 or HNMT) in patients and/or
biochemical analyses of these molecules in cultured cells or
animals may be expected to pave the way for the development of
personalized pain management in cancer patients.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Mrs. Mami Kitano,
Mrs. Haruka Sakamoto, Mrs. Yume Shinkai (all from Department of
Medical Oncology, Kindai University Faculty of Medicine) and Dr
Masato Terashima (Department of Genome Biology, Kindai University
Faculty of Medicine) for technical support and Dr Marco A. De
Velasco (Department of Genome Biology, Kindai University Faculty of
Medicine) for critically reading the manuscript.
Funding
This study was financially supported by the Health Labor
Sciences Research Grant (Grant for Innovative Clinical Cancer
Research: H26-Innovative Cancer-General-056; grant no.
16ck0106059h0003) and the Japan Agency for Medical Research and
Development (Innovative Clinical Cancer Research: grant no.
17ck0106328h0001).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YF, HM, YC, JTs, AK, KNi and KNa designed the study.
YF performed the experiments and collected the data. HM, JTs, TY,
KS, MN, RS, CM, YO, KT, HH, MT, TO, NT, KH, TT and JTa collected
the clinical data. YF, HM, YC, JTs and TY analyzed and interpreted
the data. YF and HM drafted the manuscript. YF, HM, YC, JTs, AK,
KNi and KNa revised the manuscript critically. YF, HM and JTs
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The study was conducted according to the guidelines
of The Declaration of Helsinki and the Japanese ethical guidelines
for clinical research, and was approved by the Ethical Committee of
Kindai University Faculty of Medicine (approval no. 26-130).
Written informed consent was obtained from all participants
involved in the study.
Patient consent for publication
The publication of data was approved in writing by
all patients.
Competing interests
Dr Tsurutani reports research fundings from Daiichi
Sankyo, Eisai, Taiho, Chugai, Nihonkayaku, Eli Lilly, Pfizer and
MSD outside the submitted work. Dr Hayashi reports honoraria from
Amgen K.K., AstraZeneca K.K., Boehringer Ingelheim Japan Inc.,
Bristol-Myers Squibb Co. Ltd., Chugai Pharmaceutical Co. Ltd.,
Daiichi Sankyo Co. Ltd., Eli Lilly Japan K.K., Janssen
Pharmaceutical K.K., Kyorin Pharmaceutical Co. Ltd., Merck
Biopharma Co. Ltd., MSD K.K., Novartis Pharmaceuticals K.K., Ono
Pharmaceutical Co. Ltd., Taiho Pharmaceutical Co. Ltd. and Takeda
Pharmaceutical Co. Ltd., and research funding from AstraZeneca
K.K., Astellas Pharma Inc., MSD K.K., Ono Pharmaceutical Co. Ltd.,
Nippon Boehringer Ingelheim Co. Ltd., Novartis Pharma K.K., Pfizer
Japan Inc., Bristol-Myers Squibb Co. Ltd., Eli Lilly Japan K.K.,
Chugai Pharmaceutical Co. Ltd., Daiichi Sankyo Co. Ltd., Merck
Serono Co. Ltd., Merck Biopharma Co. Ltd., Takeda Pharmaceutical
Co. Ltd., Taiho Pharmaceutical Co. Ltd., SymBio Pharmaceuticals
Ltd., AbbVie Inc., inVentiv Health Japan, ICON Japan K.K.,
Gritstone Oncology Inc., Parexel International Corp., Kissei
Pharmaceutical Co. Ltd., EPS Corp., Syneos Health, Pfizer R&D
Japan G.K., A2 Healthcare Corp., Quintiles Inc./IQVIA Services
Japan K.K., EP-CRSU Co. Ltd., Linical Co. Ltd., Eisai Co. Ltd.,
CMIC Shift Zero K.K., Kyowa Hakko Kirin Co. Ltd., Bayer Yakuhin
Ltd., EPS International Co. Ltd. and Otsuka Pharmaceutical Co. Ltd.
Dr Takeda reports honoraria from Ono Pharmaceutical Co., Boehringer
Ingelheim Japan Inc. and Novartis Pharma K.K. outside the submitted
work. Dr Haratani reports grants from AS ONE Corporation outside
the submitted work. Dr Takahama reports research funding from
Takeda and Pfizer outside the submitted work. Dr. Nishio reports
grants from Eli Lilly Japan K.K., Otsuka Pharmaceutical Co., Ltd.,
Nippon Boehringer Ingelheim Co., Ltd., Eli Lilly Japan K.K., Ignyta
Inc., Astellas Pharma Inc., Thoracic Oncology Research Group, and
North East Japan Study Group outside the submitted work. Dr
Nakagawa reports grants from Takeda Pharmaceutical Co., Ltd., Taiho
Pharmaceutical Co., Ltd., SymBio Pharmaceuticals Ltd., AbbVie Inc.,
inVentiv Health Japan, ICON Japan K.K., Gritstone Oncology Inc.,
Parexel International Co., Kissei Pharmaceutical Co., Ltd., EPS
Co., Syneos Health, Pfizer R&D Japan G.K., A2 Healthcare Co.,
Quintiles Inc./IQVIA Services JAPAN K.K., EP-CRSU Co., Ltd.,
Linical Co., Ltd., Eisai Co., Ltd., CMIC Shift Zero K.K., Kyowa
Hakko Kirin Co., Ltd., Bayer Yakuhin, Ltd., EPS International Co.,
Ltd., Otsuka Pharmaceutical Co., Ltd., AstraZeneca K.K., Astellas
Pharma Inc., MSD K.K., Ono Pharmaceutical Co., Ltd., Nippon
Boehringer Ingelheim Co., Ltd., Novartis Pharma K.K., Pfizer Japan
Inc., Bristol Myers Squibb Co., Eli Lilly Japan K.K., Chugai
Pharmaceutical Co., Ltd., Daiichi Sankyo Co., Ltd. and Merck Serono
Co., Ltd./Merck Biopharma Co., Ltd. outside the submitted work. Drs
Fujita, Matsuoka, Chiba, Sakai, Yoshida, Nakura, Sakamoto,
Makimura, Ohtake, Tanaka, Okuno, Takegawa, Tanizaki and Koyama have
nothing to disclose.
Glossary
Abbreviations
Abbreviations:
|
SNP
|
single nucleotide polymorphism
|
|
COMT
|
catechol-O-methyltransferase
|
|
IR
|
immediate-release
|
|
NRS
|
Numerical Rating Scale
|
|
CCL11
|
C-C motif chemokine ligand 11
|
|
HNMT
|
histamine N-methyltransferase
|
|
TRPV1
|
transient receptor potential V1
|
|
HADS
|
hospital anxiety and depression
scale
|
|
SF-MPQ-2
|
Short-Form McGill Pain
Questionnaire-2
|
References
|
1
|
Fallon M, Giusti R, Aielli F, Hoskin P,
Rolke R, Sharma M and Ripamonti C; ESMO Guidelines Committee, :
Management of cancer pain in adult patients: ESMO clinical practice
guidelines. Ann Oncol. 29 (Suppl 4):iv166–iv191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Swarm RA and Dans M: NCCN frameworks for
resource stratification of nccn guidelines: Adult cancer pain and
palliative care. J Natl Compr Canc Netw. 16:628–631. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Caraceni A, Hanks G, Kaasa S, Bennett MI,
Brunelli C, Cherny N, Dale O, De Conno F, Fallon M, Hanna M, et al:
Use of opioid analgesics in the treatment of cancer pain:
Evidence-based recommendations from the EAPC. Lancet Oncol.
13:e58–e68. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Owusu Obeng A, Hamadeh I and Smith M:
Review of opioid pharmacogenetics and considerations for pain
management. Pharmacotherapy. 37:1105–1121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Coluzzi F, Caputi FF, Billeci D, Pastore
AL, Candeletti S, Rocco M and Romualdi P: Safe use of opioids in
chronic kidney disease and hemodialysis patients: Tips and tricks
for non-pain specialists. Ther Clin Risk Manag. 16:821–837. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kalso E and Vainio A: Morphine and
oxycodone hydrochloride in the management of cancer pain. Clin
Pharmacol Ther. 47:639–646. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Watson CPN, Moulin D, Watt-Watson J,
Gordon A and Eisenhoffer J: Controlled-release oxycodone relieves
neuropathic pain: A randomized controlled trial in painful diabetic
neuropathy. Pain. 105:71–78. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bell GC, Donovan KA and McLeod HL:
Clinical implications of opioid pharmacogenomics in patients with
cancer. Cancer Control. 22:426–432. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rakvåg TT, Klepstad P, Baar C, Kvam TM,
Dale O, Kaasa S, Krokan HE and Skorpen F: The Val158Met
polymorphism of the human catechol-O-methyltransferase (COMT) gene
may influence morphine requirements in cancer pain patients. Pain.
116:73–78. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rakvåg TT, Ross JR, Sato H, Skorpen F,
Kaasa S and Klepstad P: Genetic variation in the
catechol-O-methyltransferase (COMT) gene and morphine requirements
in cancer patients with pain. Mol Pain. 4:642008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reyes-Gibby CC, Shete S, Rakvåg T, Bhat
SV, Skorpen F, Bruera E, Kaasa S and Klepstad P: Exploring joint
effects of genes and the clinical efficacy of morphine for cancer
pain: OPRM1 and COMT gene. Pain. 130:25–30. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matsuoka H, Tsurutani J, Chiba Y, Fujita
Y, Sakai K, Yoshida T, Nakura M, Sakamoto R, Makimura C, Ohtake Y,
et al: Morphine versus oxycodone for cancer pain using a
catechol-O-methyltransferase genotype biomarker: A multicenter,
randomized, open-label, phase III clinical trial (RELIEF Study).
Oncologist. 28:278–e166. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Crews KR, Monte AA, Huddart R, Caudle KE,
Kharasch ED, Gaedigk A, Dunnenberger HM, Leeder JS, Callaghan JT,
Samer CF, et al: Clinical pharmacogenetics implementation
consortium guideline for CYP2D6, OPRM1, and COMT genotypes and
select opioid therapy. Clin Pharmacol Ther. 110:888–896. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matsuoka H, Arao T, Makimura C, Takeda M,
Kiyota H, Tsurutani J, Fujita Y, Matsumoto K, Kimura H, Otsuka M,
et al: Expression changes in arrestin β 1 and genetic variation in
catechol-O-methyltransferase are biomarkers for the response to
morphine treatment in cancer patients. Oncol Rep. 27:1393–1399.
2012.PubMed/NCBI
|
|
15
|
Matsuoka H, Yoshiuchi K, Koyama A,
Makimura C, Fujita Y, Tsurutani J, Sakai K, Sakamoto R, Nishio K
and Nakagawa K: Expectation of a decrease in pain affects the
prognosis of pain in cancer patients: A prospective cohort study of
response to morphine. Int J Behav Med. 24:535–541. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matsuoka H, Makimura C, Koyama A, Fujita
Y, Tsurutani J, Sakai K, Sakamoto R, Nishio K and Nakagawa K:
Prospective replication study implicates the
catechol-O-methyltransferase Val158Met polymorphism as a
biomarker for the response to morphine in patients with cancer.
Biomed Rep. 7:380–384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Swarm R, Abernethy AP, Anghelescu DL,
Benedetti C, Blinderman CD, Boston B, Cleeland C, Coyle N,
Deleon–Casasola OA, Eilers JG, et al: Adult cancer pain. J Natl
Compr Canc Netw. 8:1046–1086. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Agúndez JAG, Luengo A, Herráez O, Martínez
C, Alonso-Navarro H, Jiménez-Jiménez FJ and García-Martín E:
Nonsynonymous polymorphisms of histamine-metabolising enzymes in
patients with Parkinson's disease. Neuromolecular Med. 10:10–16.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cook-Sather SD, Li J, Goebel TK, Sussman
EM, Rehman MA and Hakonarson H: TAOK3, a novel genome-wide
association study locus associated with morphine requirement and
postoperative pain in a retrospective pediatric day surgery
population. Pain. 155:1773–1783. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Foulkes T and Wood JN: Pain genes. PLoS
Genet. 4:e10000862008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ji RR, Berta T and Nedergaard M: Glia and
pain: Is chronic pain a gliopathy? Pain. 154 (Suppl 1):S10–S28.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ramesh G, MacLean AG and Philipp MT:
Cytokines and chemokines at the crossroads of neuroinflammation,
neurodegeneration, and neuropathic pain. Mediators Inflamm.
2013:4807392013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Russo R, D'Agostino G, Raso GM, Avagliano
C, Cristiano C, Meli R and Calignano A: Central administration of
oxytocin reduces hyperalgesia in mice: Implication for cannabinoid
and opioid systems. Peptides. 38:81–88. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sousa-Valente J, Andreou AP, Urban L and
Nagy I: Transient receptor potential ion channels in primary
sensory neurons as targets for novel analgesics. Br J Pharmacol.
171:2508–2527. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Z, Feng J, Mao A, Le K, La Placa D,
Wu X, Longmate J, Marek C, St Amand RP, Neuhausen SL and Shively
JE: SNPs in inflammatory genes CCL11, CCL4 and MEFV in a
fibromyalgia family study. PLoS One. 13:e01986252018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Světlík S, Hronová K, Bakhouche H,
Matoušková O and Slanař O: Pharmacogenetics of chronic pain and its
treatment. Mediators Inflamm. 2013:8643192013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gabriel S, Ziaugra L and Tabbaa D: SNP
genotyping using the sequenom MassARRAY iPLEX platform. Curr Protoc
Hum Genet. Chapter 2: Unit 2.12. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matic M, de Wildt SN, Tibboel D and van
Schaik RHN: Analgesia and opioids: A pharmacogenetics shortlist for
implementation in clinical practice. Clin Chem. 63:1204–1213. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shiotani A, Murao T, Fujita Y, Fujimura Y,
Sakakibara T, Nishio K and Haruma K: Novel single nucleotide
polymorphism markers for low dose aspirin-associated small bowel
bleeding. PLoS One. 8:e842442013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Matsuoka H, Tsurutani J, Chiba Y, Fujita
Y, Terashima M, Yoshida T, Sakai K, Otake Y, Koyama A, Nishio K and
Nakagawa K: Selection of opioids for cancer-related pain using a
biomarker: A randomized, multi-institutional, open-label trial
(RELIEF study). BMC Cancer. 17:6742017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nakamura A, Hasegawa M, Minami K, Kanbara
T, Tomii T, Nishiyori A, Narita M, Suzuki T and Kato A:
Differential activation of the µ-opioid receptor by oxycodone and
morphine in pain-related brain regions in a bone cancer pain model.
Br J Pharmacol. 168:375–388. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nielsen CK, Ross FB, Lotfipour S, Saini
KS, Edwards SR and Smith MT: Oxycodone and morphine have distinctly
different pharmacological profiles: Radioligand binding and
behavioural studies in two rat models of neuropathic pain. Pain.
132:289–300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Klepstad P, Fladvad T, Skorpen F, Bjordal
K, Caraceni A, Dale O, Davies A, Kloke M, Lundström S, Maltoni M,
et al: Influence from genetic variability on opioid use for cancer
pain: A European genetic association study of 2294 cancer pain
patients. Pain. 152:1139–1145. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chang HS, Kim JS, Lee JH, Cho JI, Rhim TY,
Uh ST, Park BL, Chung IY, Park CS and Shin HD: A single nucleotide
polymorphism on the promoter of eotaxin1 associates with its mRNA
expression and asthma phenotypes. J Immunol. 174:1525–1531. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liang C, Ni G, Ma J, Liu H, Mao Z, Sun H
and Zhang X: Impact of tag single nucleotide polymorphisms (SNPs)
in CCL11 gene on risk of subtypes of ischemic stroke in xinjiang
han populations. Med Sci Monit. 23:4291–4298. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kang WS, Kim YJ, Park HJ, Kim SK, Paik JW
and Kim JW: Association of CCL11 promoter polymorphisms with
schizophrenia in a Korean population. Gene. 656:80–85. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pawlik K, Piotrowska A, Kwiatkowski K,
Ciapała K, Popiolek-Barczyk K, Makuch W and Mika J: The blockade of
CC chemokine receptor type 1 influences the level of nociceptive
factors and enhances opioid analgesic potency in a rat model of
neuropathic pain. Immunology. 159:413–428. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pawlik K, Ciechanowska A, Ciapała K,
Rojewska E, Makuch W and Mika J: Blockade of CC chemokine receptor
type 3 diminishes pain and enhances opioid analgesic potency in a
model of neuropathic pain. Front Immunol. 12:7813102021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lovo EE, Moreira A, Cruz C, Carvajal G,
Barahona KC, Caceros V, Blanco A, Mejias R, Alho E and Soto T:
Radiomodulation in mixed, complex cancer pain by triple target
irradiation in the brain: A preliminary experience. Cureus.
14:e254302022.PubMed/NCBI
|
|
40
|
Richards GC, Lluka LJ, Smith MT, Haslam C,
Moore B, O'Callaghan J and Strong J: Effects of long-term opioid
analgesics on cognitive performance and plasma cytokine
concentrations in patients with chronic low back pain: A
cross-sectional pilot study. Pain Rep. 3:e6692018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou TQ, Gao HY, Guan XH, Yuan X, Fang GG,
Chen Y and Ye DW: Chemokines and their receptors: Potential
therapeutic targets for bone cancer pain. Curr Pharm Des.
21:5029–5033. 2015. View Article : Google Scholar : PubMed/NCBI
|