Introduction
Colorectal cancer (CRC) is a significant global
health concern, accounting for ~10% of all new cancer cases
worldwide and ranking as the second leading cause of cancer-related
mortality (1). In 2020, there were
an estimated 1.9 million new cases of CRC worldwide, leading to
~935,000 deaths globally (2). The
majority of CRCs arise from pre-existing colorectal adenomas (CRAs)
(3). CRC development often follows
the adenoma-carcinoma sequence, a process that can take several
years. Therefore, early detection and surgery are critical in
controlling this disease. However, the majority of patients present
with advanced disease and have a poor prognosis, and even with
surgery the recurrence rate remains high. Overall, <20% of
patients diagnosed with advanced CRC survive beyond 5 years
(4). CRC is thought to be triggered
by various factors, including genetic predisposition. For instance,
mutations in genes such as APC, TP53 and KRAS have been implicated
in increasing the risk of CRC (5).
In addition, certain environmental factors, such as smoking,
obesity, sedentary lifestyle and exposure to certain chemicals or
toxins, have also been associated with an increased risk of CRC
development. For instance, long-term alcohol abuse may cause damage
to the intestinal mucosa and increase the risk of colorectal cancer
(6). A previous study has shown
that the gut microbiota has a significant impact on human health
and disease (7). Alterations in the
intestinal microecosystem have been identified as a key factor
influencing the development of CRC (8). Such alterations may cause ecological
dysbiosis, an imbalance in microbial composition that disturbs the
host-microbiota interactions and may drive the development of
CRC.
In a previous study, the contribution of gut
microbiota to the development and progression of CRC was examined
using macro-genomic analysis (9).
The phyla Bacteroides, Firmicutes and Proteobacteria were found to
be abundant in patients with CRC compared with healthy individuals.
Certain microorganisms reach a relatively stable abundance or
composition when cancer progresses to an advanced stage, supporting
their role in cancer progression. Recent evidence also suggests the
role of the gut microenvironment in CRC development. For example,
alterations in the gut microbiota, such as Bacteroides
fragilis and Escherichia coli, can promote excessive
colon cell proliferation and drive cancer development (10). Fusobacterium nucleatum, a
common organism that is abundant in CRC, has been shown to induce
host cell epigenetic modifications and microsatellite instability
(11). While fecal coliforms are
normally not associated with disease, a previous study has shown a
higher abundance of fecal coliforms in stool samples from patients
with CRC compared with healthy controls (HC). A higher abundance of
fecal coliforms has also been observed in tumor tissue samples and
adjacent mucosa of patients with CRC compared with tissues of HC,
confirming the association between CRC and fecal coliforms
(12). In addition, intestinal
bacteria produce a series of metabolites during the reproduction
process that can directly or indirectly affect the metabolism of
the host (13). For instance,
Escherichia coli is another microorganism that is abundant
in CRC, which has been reported to induce CRC-associated DNA
methylation by producing trimethylamine in the intestine, thereby
enhancing carcinogenicity (14).
Methylphenidate sulfate is another genotoxic metabolite secreted by
bacteria that affects cell cycle kinetics and induces DNA damage in
colonic epithelial cells (15).
Furthermore, bacterial metabolites are of increasing
interest in CRC research. Yachida et al (16) compared the abundance of microbial
metabolic genes in the feces of HC and patients at different stages
of CRC development. In patients with precancerous polyps, an
increase in the abundance of genes involved in amino acid and
sulfur metabolism and a decrease in the abundance of genes involved
in methane metabolism was observed compared to the HC group.
Furthermore, patients with CRC also exhibited a higher abundance of
amino acid-related genes compared to the HC group. This is
consistent with the long-standing hypothesis that a gut microbial
environment that favors protein hydrolysis over glycolysis may
increase the risk of CRC (17). As
bacteria convert dietary intake into metabolic byproducts, diet
contributes significantly to the metabolites secreted by bacteria
(18). However, to date, the
involvement of fecal metabolites in CRC has still not been fully
elucidated. Furthermore, data on intestinal metabolites have not
been associated with potential clinical benefits in patients with
colorectal cancer, to the best of our knowledge. Therefore,
elucidating the macro-metabolome of feces may aid in the
understanding of CRC development.
The present study conducted a metabolomics analysis
on fecal samples using liquid chromatography-mass spectrometry
(LC-MS), with an aim to identify differential fecal metabolites
among patients in the CRC, CRA, and HC groups. Subsequently,
profiling of the gut metabolome of a group of patients with CRC was
performed, comparing long-term survivors (LTS) vs. short-term
survivors (STS). The objective of the current study was to
determine whether certain metabolites were associated with improved
survival by analyzing clinical data alongside gut metabolite
data.
Materials and methods
Human fecal sample collection
Fecal samples were collected prospectively from
patients who underwent colonoscopy and histopathological
examination at the Department of Gastroenterology, Tianyou
Hospital, Wuhan University of Science and Technology (Wuhan,
China), between January and December 2017. The subjects were
divided into three groups: i) Patients with colorectal
adenocarcinoma (n=35) were classified into the CRC group; ii)
patients with CRA (n=37) were classified into the adenoma group;
and iii) patients without colorectal pathology (n=30) were used as
the HC group. Participants who had taken antibiotics or
microecological drugs within 2 months prior to enrollment were
excluded, as were subjects with bowel infections, gastrointestinal
symptoms, hypertension, heart disease, diabetes or a history of
colonoscopy, adjuvant radiotherapy or surgical treatment prior to
sampling. The present study was approved by the Ethics Committee of
Tianyou Hospital, Wuhan University of Science and Technology
(Wuhan, China), and all subjects provided written informed consent.
Patients with CRC were followed up, and their survival data were
collected until August 2022 during the follow-up period.
Sample preparation
The fecal samples were stored at −80°C until
required and then thawed on ice prior to extraction. A total of 400
µl of methanol was added to 20 mg of each sample, and each sample
was homogenized in a blender for 3 min at 25°C. The homogenized
samples were then centrifuged at 7,900 × g for 10 min at 4°C. The
supernatant of each sample was collected and subjected to a second
centrifugation at 7,900 × g for 3 min at 4°C. The supernatant
obtained after the second centrifugation was used for ultra-high
performance liquid chromatography tandem mass spectrometry
(UPLC-MS/MS) analysis.
Metabolite analysis by UPLC-MS/MS
The analytical conditions were as follows: UPLC was
performed using a Waters ACQUITY UPLC™ HSS T3 C18 column (1.8 µm,
2.1×100.0 mm) by SCIEX Corp., with a solvent system consisting of
water (0.04% acetic acid) and acetonitrile (0.04% acetic acid) and
a gradient program of 100:0 v/v at 0 min, 5:95 v/v at 10.0 min,
5:95 v/v at 11.0 min, 95:5 v/v at 11.0 min and 95:5 v/v at 15.0
min. The flow rate was 0.00035 l/min, the temperature was 40°C, and
the injection volume was 5 µl. The effluents were then
alternatively connected to an electrospray ionization (ESI)-triple
quadrupole-linear ion trap (QTRAP)-MS/MS. Linear ion trap (LIT) and
triple quadrupole (QQQ) scans were obtained using an API 4500 QTRAP
LC/MS/MS system equipped with an ESI turbo ion-spray interface by
SCIEX Corp. The ionisation mode used was both negative and
positive, with the other parameters set the same. The ESI source
operating parameters were as follows: Ion source, turbojet; source
temperature, 550°C; ion injection voltage, 5,500 V; ion source gas
I, gas II and curtain gas were set to 55, 60 and 25.0 psi,
respectively; and collision gas was set to high. Instrument tuning
and mass calibration were performed in QQQ and LIT modes using 10
and 100 µmol/l polypropylene glycol solutions, respectively. QQQ
scans were obtained as multiple reaction monitoring (MRM)
experiments, and collision gas (nitrogen) was set to 5 psi. Further
optimization was performed for declustering potential and collision
energy of individual MRM conversions by determining the initial
settings. A specific set of MRM transitions was monitored based on
the metabolites eluted within each period.
Quality control (QC) of samples
The first and second order spectra detected by MS
were qualitatively analyzed using the Metware database (https://cloud.metware.cn/) and Human Metabolome
database (HMDB; http://hmdb.ca/). After obtaining MS
analysis data from different samples, the peak areas of all MS
peaks were integrated and the peaks of the same metabolites in
different samples were corrected using Analyst 1.6.3 software
(SCIEX Corp.). The peak area of each chromatographic peak
represents the relative content of the corresponding metabolite.
For fecal samples, aliquots of each individual sample were combined
and extractions were performed as aforementioned. A mixed QC sample
was included every 10 test samples during instrumental analysis to
monitor the reproducibility of the analysis. The stability of the
samples was evaluated by measuring the total ion current (TIC) as
part of the QC procedure. The mixed QC samples showed a stable TIC
within a specific range, indicating that no decomposition or other
adverse reactions occurred during the analysis. The high overlap of
the curves for the detection of metabolites’ TIC met the QC
criteria. The percentage of substances with a coefficient of
variation (CV)< the reference value was analyzed using the
empirical cumulative distribution function (ECDF). The proportion
of substances with a coefficient of CV less than 0.5 was >85%,
indicating that the experimental data were relatively stable.
Statistical analysis
Unsupervised multivariate principal component
analysis (PCA) was performed on the fecal samples to obtain a
preliminary understanding of the overall metabolic differences
between the three groups (CRC, CRA and HC) (19). Orthogonal partial least squares
discriminant analysis (OPLS-DA) was used to confirm the
differentiation of metabolite profiles between the groups (20). The OPLS-DA model was validated using
a random permutation test. This involved randomly permuting the
sample labels 200 times and recalculating the model's evaluation
metrics (21). R2Y, Q2 and R2X are
important evaluation metrics used in the validation of the OPLS-DA
model. A variable importance in projection (VIP) value ≥1 was used
to identify metabolites for further analysis. Univariate analysis
of metabolite differences was conducted for each metabolite using
Cox proportional hazards models and fold change (FC) values were
calculated to determine significance. Metabolites with VIP ≥1 and
|log2FC| >1 were considered significantly different.
Venn analysis was performed to determine the overlapping
differentially abundant metabolites. Participants were stratified
into subgroups based on smoking status, alcohol consumption, and
red meat intake for stratified analysis. The identified metabolites
were annotated using the Kyoto Encyclopedia of Genes and Genomes
(KEGG) database (https://www.kegg.jp/) and Metabolite
Set Enrichment Analysis (MSEA; http://www.msea.com.cn/). The diagnostic value was
assessed using receiver operating characteristic (ROC) curves.
Overall survival (OS) was defined as the time from the first
diagnosis of CRC to the date of death from any cause, and was used
as the clinical endpoint. Patients with no events/death were
reviewed at the last follow-up date (August 2022). We stratified
CRC patients into two subgroups based on their OS: Those with an OS
≤5 years and those with an OS >5 years. This allowed us to
perform a stratified analysis based on the duration of survival in
patients with CRC. Univariate Cox regression and Kaplan-Meier
analysis were used to predict the prognostic ability of patients
with CRC. Patients were categorized into high and low groups based
on the median relative abundance of metabolites for Cox regression
and Kaplan-Meier analysis. The P-values were obtained using the
Wald test for Cox regression and the log-rank test for Kaplan-Meier
analysis. The relative abundance data of metabolites were
normalized using unit variance scaling. PCA and OP-LSDA analyses
were performed using R 3.5.1. Statistical analysis was performed
using SPSS 26 (IBM Corp.) and P-values were two-tailed. The
differences in baseline data, such as age, gender, area, among the
three groups (CRC, CRA and HC), were analyzed using the chi-square
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Clinical characteristics
The present study enrolled a total of 35 patients
with CRC, 37 patients with CRA and 30 HC. Among the patients with
CRC, there were 19 males (54.3%) and 16 females (45.7%), with a
median age of 57 years (range, 37–81 years). In the CRA group,
there were 23 males (62.2%) and 14 females (37.8%), with a median
age of 52 years (range, 30–75 years). In the HC group, there were
12 males (40%) and 18 females (60%), with a median age of 45 years
(range, 23–67 years). There were no significant differences in
baseline characteristics such as age, gender and area among the
three groups. The baseline data are presented in Table I. For the 35 patients with CRC,
baseline characteristics and survival information were collected
during the follow-up period. Among the patients followed up, 17
individuals had a total survival period of >5 years, accounting
for 48.6%. The characteristics of the study cohort and their
survival information are summarized in Table II.
 | Table I.Baseline characteristics. |
Table I.
Baseline characteristics.
|
| Number of
cases |
|
|
|---|
|
|
|
|
|
|---|
| Variable | Colorectal
cancer | Colorectal
adenoma | Healthy
control |
χ2-value | P-value |
|---|
| Age, years |
|
|
|
|
|
|
<60 | 21 | 28 | 24 | 3.657 | 0.161 |
|
≥60 | 14 | 9 | 6 |
|
|
| Sex |
|
|
| 3.305 | 0.192 |
|
Male | 19 | 23 | 12 |
|
|
|
Female | 16 | 14 | 18 |
|
|
| Body mass index,
kg/m2 |
|
|
| 2.302 | 0.316 |
|
<18.5 | 6 | 10 | 10 |
|
|
|
≥18.5 | 29 | 27 | 20 |
|
|
| Area |
|
|
| 0.970 | 0.616 |
|
Urban | 15 | 19 | 12 |
|
|
|
Rural | 20 | 18 | 18 |
|
|
| Marital status |
|
|
| 0.208 | 0.901 |
| No | 7 | 9 | 7 |
|
|
|
Yes | 28 | 28 | 23 |
|
|
| Smoking |
|
|
| 0.113 | 0.945 |
|
Heavy | 15 | 16 | 14 |
|
|
|
Light | 20 | 21 | 16 |
|
|
| Alcohol
consumption |
|
|
| 0.148 | 0.929 |
|
Heavy | 16 | 17 | 15 |
|
|
|
Light | 19 | 20 | 15 |
|
|
| Vegetable
intake |
|
|
| 5.547 | 0.062 |
|
High | 22 | 13 | 15 |
|
|
|
Low | 13 | 24 | 15 |
|
|
| Red meat
intake |
|
|
| 0.805 | 0.669 |
|
High | 14 | 18 | 15 |
|
|
|
Low | 21 | 19 | 15 |
|
|
| White meat
intake |
|
|
| 1.848 | 0.397 |
|
High | 18 | 15 | 17 |
|
|
|
Low | 17 | 22 | 13 |
|
|
| Processed meat
intake |
|
|
| 1.769 | 0.413 |
|
High | 20 | 16 | 13 |
|
|
|
Low | 15 | 21 | 17 |
|
|
| Exercise |
|
|
| 1.390 | 0.499 |
|
Regular | 20 | 16 | 15 |
|
|
|
Irregular | 15 | 21 | 15 |
|
|
 | Table II.Characteristics of patients with
colorectal cancer. |
Table II.
Characteristics of patients with
colorectal cancer.
| Variables | Number of cases
(%) |
|---|
| Sex |
|
|
Male | 19 (54.3) |
|
Female | 16 (45.7) |
| Age, years |
|
|
<60 | 12 (34.3) |
|
≥60 | 23 (65.7) |
| CA 19-9, U/ml |
|
|
≥27 | 21 (60.0) |
|
<27 | 14 (40.0) |
| CEA, ng/ml |
|
| ≥5 | 20 (57.1) |
|
<5 | 15 (42.9) |
| Degree of
differentiation |
|
|
Low | 17 (51.4) |
|
High | 18 (48.6) |
| Lymph node
metastasis |
|
|
Yes | 15 (42.9) |
| No | 20 (57.1) |
| Distant
metastasis |
|
|
Yes | 11 (31.4) |
| No | 24 (68.6) |
| Type |
|
|
Colon | 23 (65.7) |
|
Rectal | 12 (34.3) |
| Overall survival,
years |
|
| ≤5 | 18 (51.4) |
|
>5 | 17 (48.6) |
| Treatment
options |
|
| Surgery
and chemotherapy | 10 (28.6) |
| Surgery
only | 8 (22.9) |
|
Chemotherapy only | 12 (34.3) |
|
Palliative care | 5 (14.3) |
QC
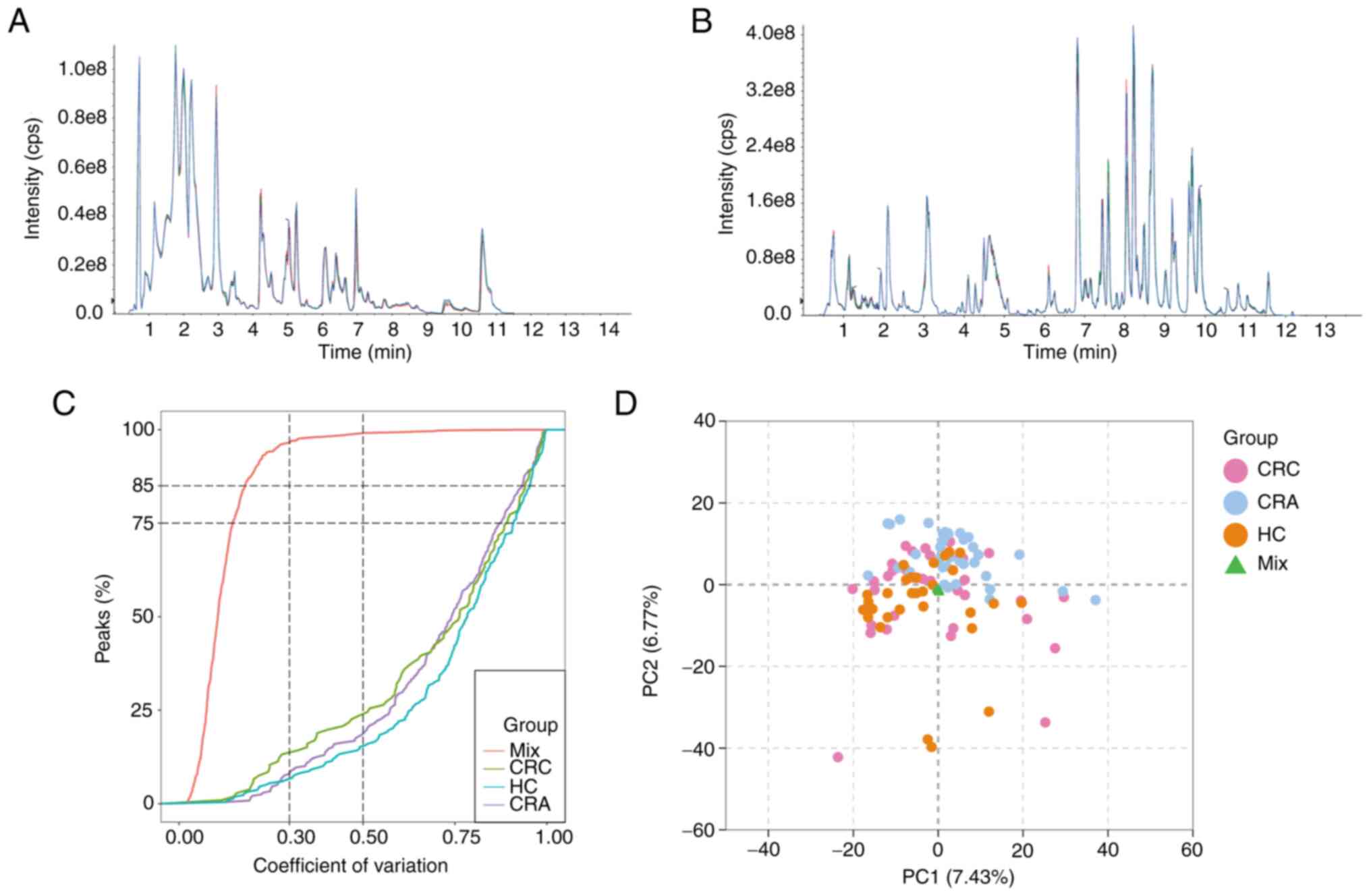
To evaluate the repeatability of metabolite
extraction and detection, overlapping display analysis of TIC plots
was performed on MS/MS data of different QC samples (Fig. 1A and B). TIC measures the total
intensity of all ions detected at each retention time, and the TICs
of all QC samples were compared to ensure their consistency. ECDF
analysis revealed that the percentage of substances with CV values
<0.5 in QC samples was >85% (Fig.
1C). These results confirmed the reproducibility and stability
of the proposed method, indicating that the significant differences
observed between the two groups using multivariate statistical
analysis were more likely to be caused by genuine metabolite
changes rather than technical errors.
A comprehensive targeted metabolomics analysis of
the collected stool samples was conducted using a targeted
UPLC-MS/MS method, resulting in the detection of 1,641 metabolites.
Unsupervised multivariate PCA was utilized to assess trends among
all groups in the initial cohort and potential outliers in the
data. PCA results demonstrated no clear trend of separation among
the CRC group and the other two groups (Fig. 1D). However, the mixed samples used
for QC were gathered at one point, further confirming the
reliability of the assay.
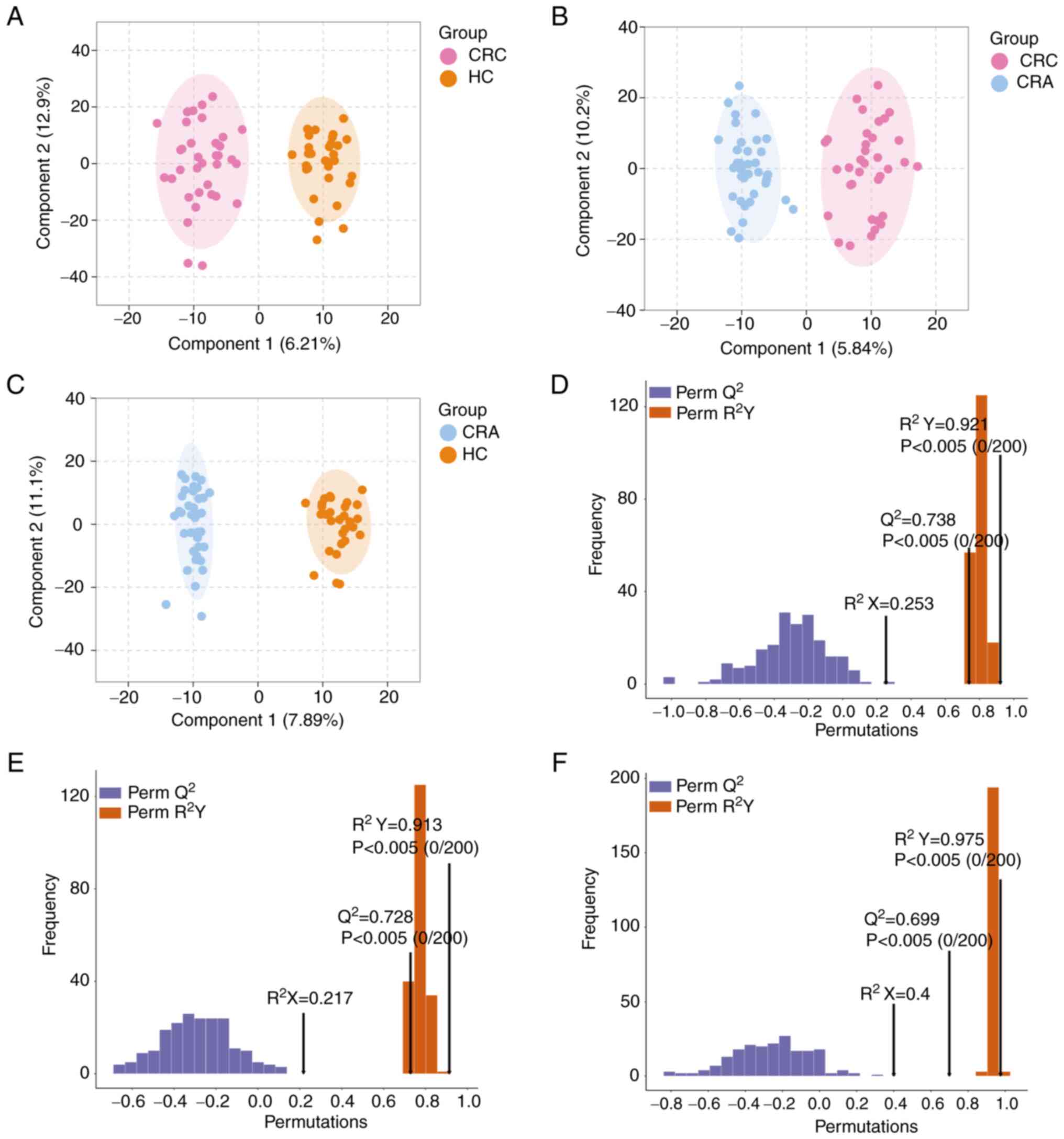
OPLS-DA model and validation
To identify metabolites that may significantly
contribute to CRC development, two-by-two comparisons were
performed among the three groups. Metabolomics analysis was carried
out using the OPLS-DA model, which maximizes the separation between
samples. OPLS-DA successfully differentiated the CRC vs. HC group,
CRC vs. CRA group and CRA vs. HC group (Fig. 2A-C). The discriminatory ability of
the model was confirmed by constructing cross-validated OPLS-DA
models. The 200-time permutation test revealed Q2 values
of 0.738, 0.728 and 0.699, respectively, indicating that these
models were not overfitting (Fig.
2D-F).
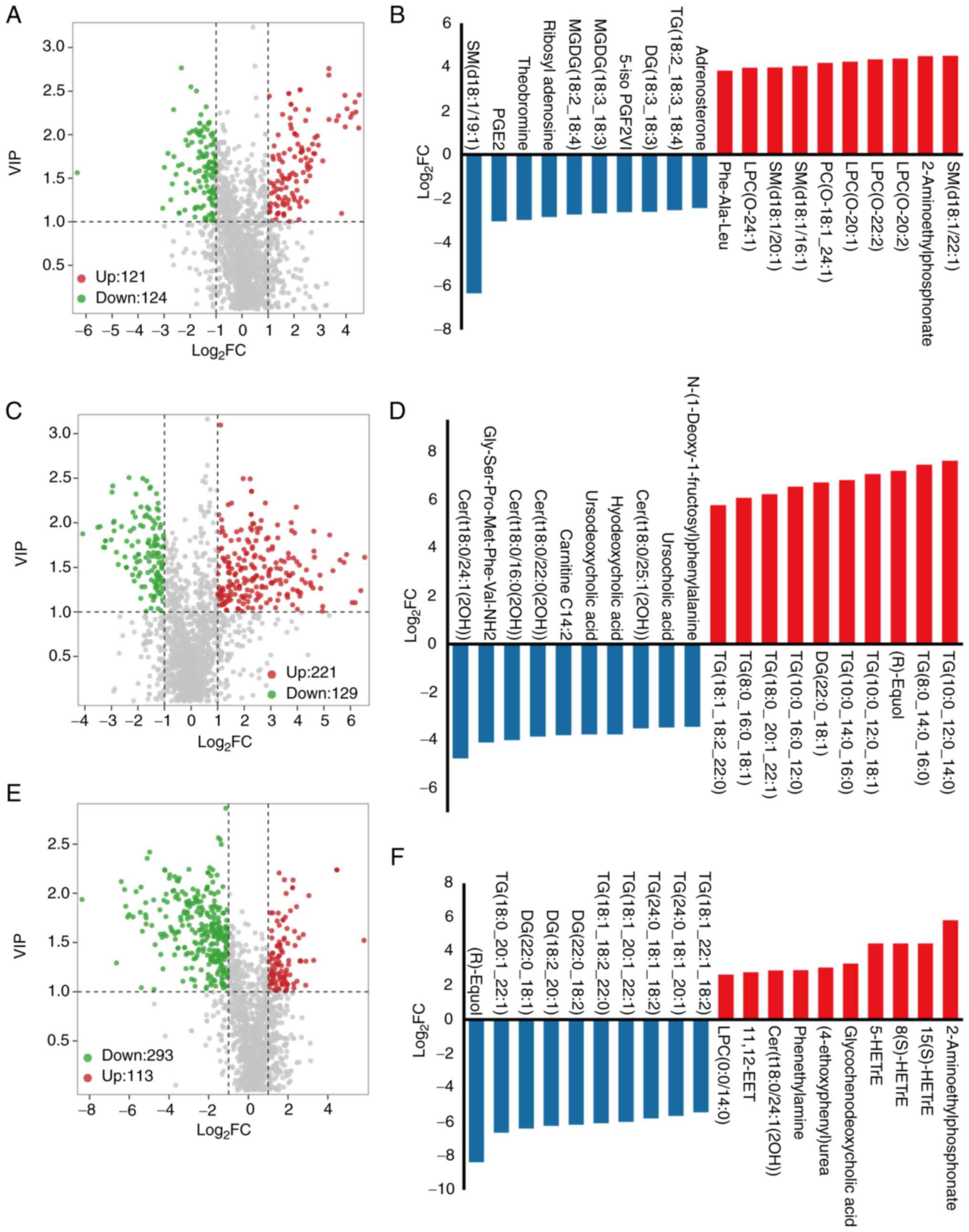
Metabolite profiling
The aim of the present study was to identify
metabolites that may significantly affect CRC development. To
achieve this, metabolomics analysis was carried out and two-by-two
comparisons were performed using the OPLS-DA model, the results of
which confirmed the maximum separation between specimens.
Differential metabolites were discovered using a VIP threshold ≥1
and |log2FC| >1.
The results of the current study showed that when
comparing HC with CRC (Table SI),
245 differential metabolites were identified, including 121
upregulated and 124 downregulated metabolites (Fig. 3A). Sphingomyelin (SM; d18:1/22:1)
and 2-aminoethylphosphonate were the top two most increased
metabolites in patients with CRC compared with the HC group. By
contrast, SM (d18:1/19:1), prostaglandin E2, theobromine and
ribosyl adenosine were among the metabolites significantly
downregulated in patients with CRC.
When comparing CRA with CRC (Table SII), 350 metabolites were found to
be differentially produced, while certain metabolites, such as
ceramide, carnitine, amino acids and their metabolites and small
peptides, were decreased in patients with CRC compared with those
with CRA. Certain glycerophospholipids, such as triglyceride
(10:0_12:0_14:0), were found to be increased in patients with CRC
compared to those with CRA (Fig.
3B). A total of 406 metabolites were found to be significantly
altered in patients with CRA compared with those in the HC group
(Table SIII; Fig. 3C). Patients with CRA showed an
increase in 2-aminoethylphosphonate, 5-hydroxyeicosatetraenoic acid
glycochenodeoxycholic acid, phenethylamine and 4-ethoxyphenyl. By
contrast, (R)-equol was the most decreased of the identified
metabolites in patients with CRA compared with the HC group.
Fig. 3D-F illustrates the top 10
upregulated and top 10 downregulated metabolites between each of
the two groups compared.
Upon further analysis of the significantly altered
metabolites using Venn analysis, the same differential metabolites
from these three comparisons were found (Fig. 4A). Details of the 20 metabolites
that were significantly changed in all three comparisons are
provided in Table SIV. These
metabolites may potentially contribute to the progression of colon
tumorigenesis, characterizing CRC tumors from a metabolic
perspective.
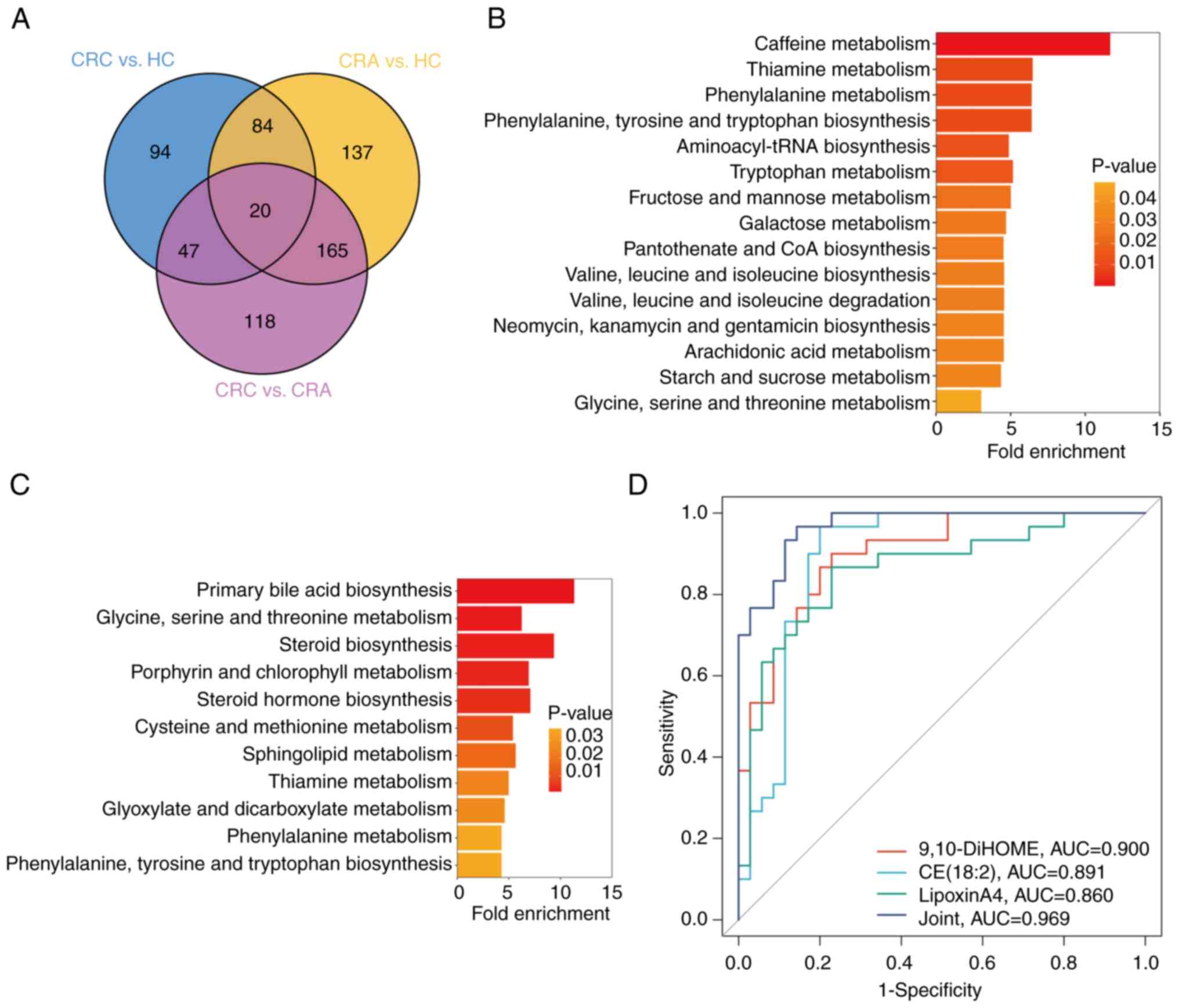 | Figure 4.Kyoto Encyclopedia of Genes and
Genomes pathway and ROC analysis of differential metabolites. (A)
Venn diagram of significantly differentially expressed metabolites
in the three groups. Metabolite Set Enrichment Analysis in (B) CRC
vs. HC group and (C) CRC vs. CRA group. (D) ROC curves for the
diagnosis of gut metabolites between CRC and HC cases. CRC,
colorectal cancer; CRA, colorectal adenoma; HC, healthy control;
ROC, receiver operating characteristic curve; AUC, area under the
ROC curve; 9,10-DiHOME, 9,10-dihydroxy-12-octadecenoic acid, CE,
cholesterol ester. |
In addition, stratified analysis based on smoking
status, alcohol consumption and red meat intake was performed to
better understand the association between intestinal metabolites
and colon cancer. SM (d18:1/19:1) was found significantly enriched
in the heavy smoking subgroup among patients with CRC compared with
the HC group. Furthermore, 3-sulfocatechol was significantly
elevated in the heavy alcohol consumption subgroup among patients
with CRC compared with the HC group. In the high red-meat intake
subgroup, significant elevations in multiple saturated and
unsaturated fatty acids such as free fatty acid (34:6) and
α-carboxy-α-methylbutyrylhydroxamic acid were observed among
patients with CRC compared with the HC group (Table SV).
Enrichment analysis for
metabolites
To gain insight into the functional roles of
significantly altered metabolites in each group, MSEA was
performed. Significant differences were observed in several
pathways related to metabolism among the three groups analyzed.
Specifically, the differential metabolites between the CRC and HC
groups were found to be enriched in pathways related to ‘caffeine
metabolism’, ‘thiamine metabolism’, ‘phenylalanine metabolism’ and
‘phenylalanine, tyrosine and tryptophan biosynthesis’ (Fig. 4B). The significantly enriched
metabolites between the CRC and CRA groups were related to ‘primary
bile acid biosynthesis’, ‘glycine, serine and threonine
metabolism’, ‘steroid biosynthesis’ and ‘porphyrin and chlorophyll
metabolism’ (Fig. 4C). These
results suggested that significant changes in metabolic pathway
patterns were observed during the ongoing tumor progression.
Diagnostic value of metabolites
To assess the potential diagnostic value of
gut-associated metabolites in CRC, ROC analysis was conducted to
evaluate the sensitivity, specificity and the area under the curve
(AUC), thereby providing a quantitative measurement of the
diagnostic performance of differential metabolites. Differential
metabolites with high AUC values can effectively differentiate
between the CRC and HC groups. Notably, the top three metabolites
9,10-dihydroxy-12-octadecenoic acid, cholesterol ester (18:2) and
lipoxinA4 had AUC values of 0.900, 0.891 and 0.860, respectively.
Furthermore, the combined quantification of these three metabolites
showed an AUC of 0.969 for CRC diagnosis (Fig. 4D). These findings demonstrated the
potential of the combined diagnostic group as a non-invasive
approach for the early detection of CRC.
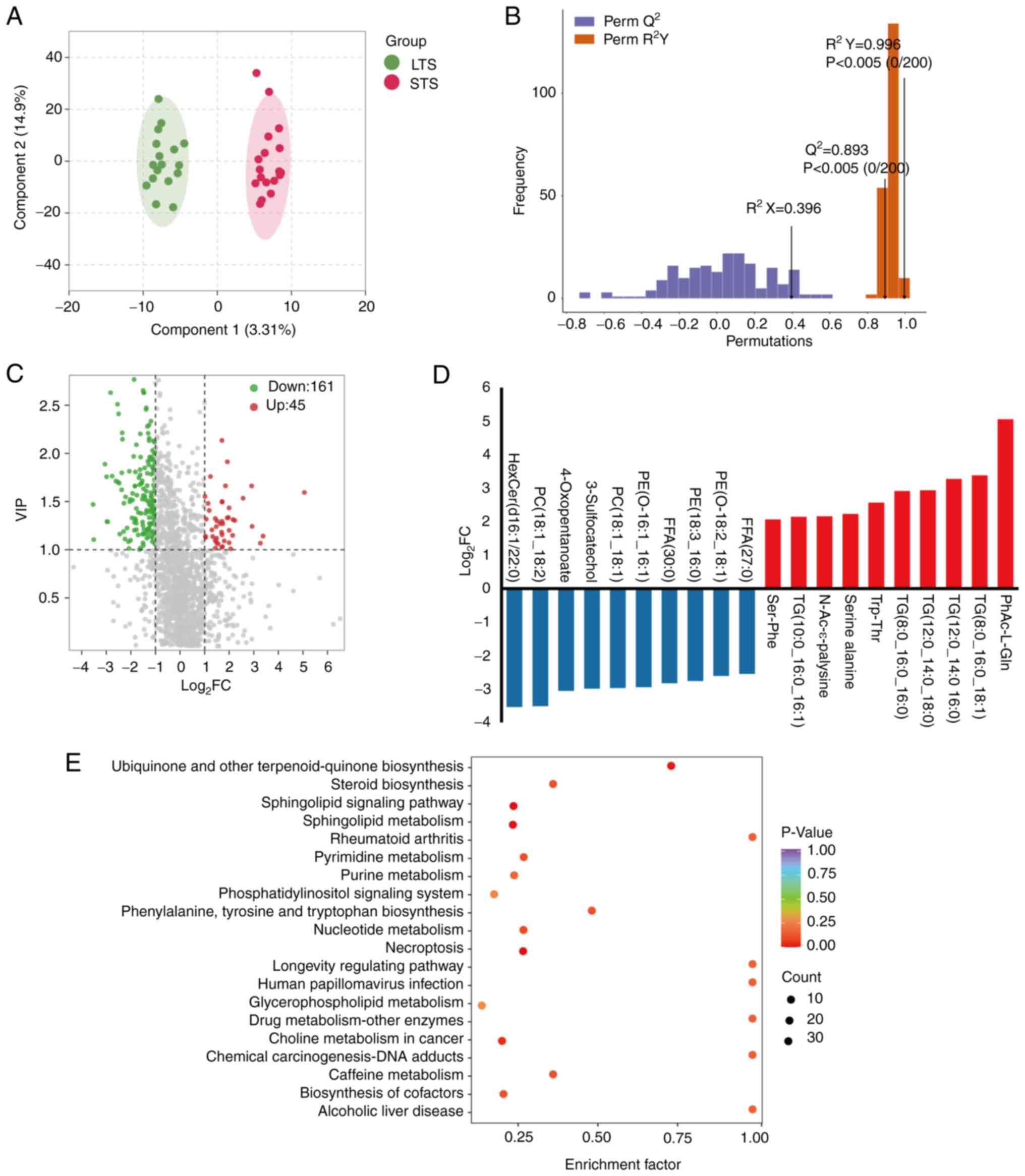
Gut metabolite composition for
patients stratified by survival
To investigate the role of gut metabolites in
modulating clinical outcomes (particularly OS) in patients with
CRC, a cohort of 18 STS who survived ≤5 years was compared with 17
LTS who survived >5 years. The differences in gut metabolites
were evaluated and significant alterations in metabolites between
the two groups were identified. Unsupervised OPLS-DA was conducted
to further differentiate the metabolite profiles and screen for key
metabolites. As shown in the score chart and validation plot
(Fig. 5A and B), the LTS group was
clearly separated from the STS group, indicating that metabolic
changes were STS- or LTS-specific. Volcano plots and bar graphs
showed 45 significantly upregulated and 161 significantly
downregulated metabolites in the STS compared with the LTS group
and revealed a potential metabolic discrimination of top
metabolites between these two groups (Fig. 5C and D). These differential
metabolites characterized LTS and STS from a metabolic perspective,
particularly the dysregulation of hexosylceramide (d16:1/22:0)
(Table SVI).
Based on the differential metabolites between the
LTS group and STS group, KEGG pathway enrichment analysis was
performed (Fig. 5E), highlighting
several pathways, including ‘ubiquinone and other terpenoid -
quinone biosynthesis’, ‘steroid biosynthesis’, ‘sphingolipid
signaling pathway’, ‘rheumatoid arthritis’, ‘pyrimidine metabolism’
and ‘purine metabolism’. These pathways may be involved in the
survival of patients with CRC. The results suggested that the gut
metabolic composition determines the differential enrichment of
metabolic functional pathways between the LTS and STS groups, which
may ultimately affect patient survival.
Prognostic ability of metabolites
Subsequently, the association between metabolites
and OS in the CRC cohort was investigated by dividing patients into
two groups based on their relative metabolite content. As expected,
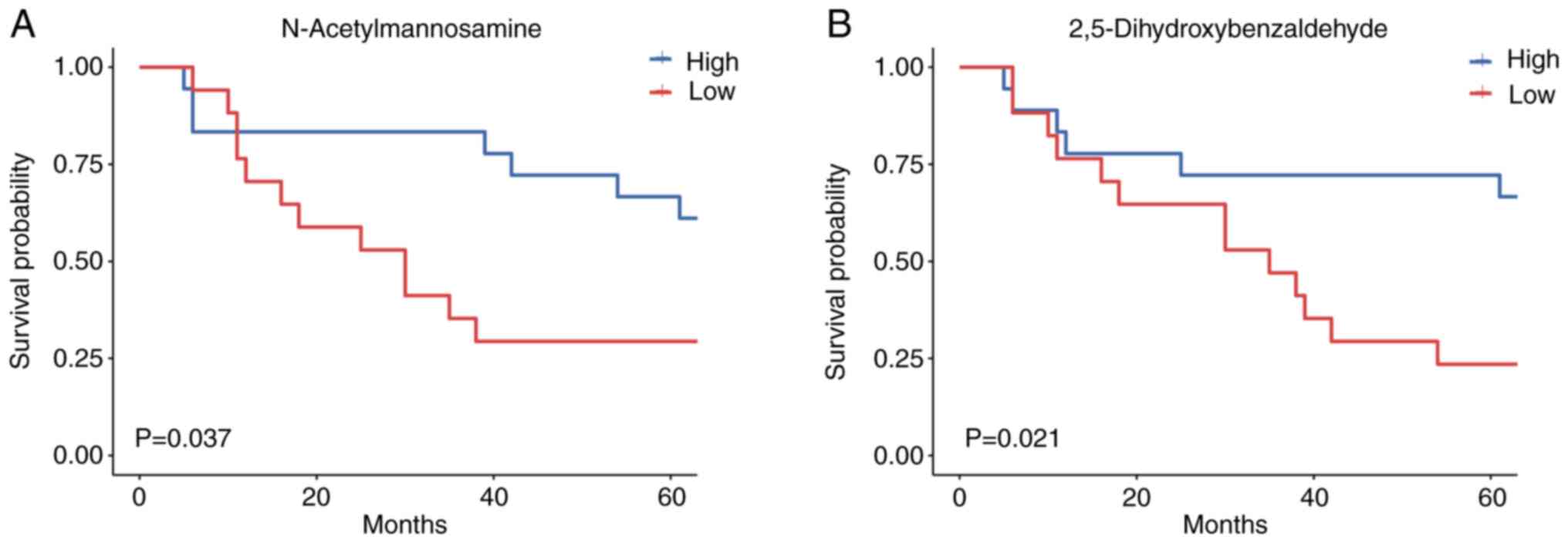
N-acetylmannosamine [hazard ratio (HR), 0.381; 95% confidence
interval (CI), 0.147–0.985] and 2,5-dihydroxybenzaldehyde (HR,
0.336; 95% CI, 0.126–0.895) had significant clinical prognostic
value, according to univariate Cox proportional hazards models
(Table III). Potential
confounders, including gender, age, carbohydrate antigen 19-9
(CA19-9), carcinoembryonic antigen (CEA), differentiated degree,
lymph node metastasis, distant metastasis and type, were evaluated,
but significant associations with CRC were not found (Table III). Importantly, an increased
abundance of N-acetylmannosamine and 2,5-dihydroxybenzaldehyde was
associated with better OS (Fig. 6A and
B). The findings from the present study suggested that gut
metabolites may serve as predictors of survival outcomes in
patients with CRC, indicating the potential relevance of the
metabolic composition in mediating CRC progression.
 | Table III.Cox regression analysis of overall
survival. |
Table III.
Cox regression analysis of overall
survival.
| Univariate
analysis | Hazard ratio (95%
confidence interval) | P-value |
|---|
| Sex (female vs.
male) | 0.643
(0.170–2.438) | 0.516 |
| Age (≥60 vs. <60
years) | 1.118
(0.355–3.521) | 0.849 |
| CA 19-9 (≥27
vs.< 27 U/ml) | 0.725
(0.231–2.275) | 0.581 |
| CEA (≥5 vs. <5
ng/ml) | 2.790
(0.599–13.002) | 0.191 |
| Differentiated
degree (high vs. low) | 1.864
(0.462–7.516) | 0.382 |
| Lymph node
metastasis (yes vs. no) | 0.887
(0.193–4.083) | 0.877 |
| Distant metastasis
(yes vs. no) | 2.500
(0.636–9.828) | 0.189 |
| Type (colon vs.
rectal) | 0.357
(0.105–1.207) | 0.097 |
| N-Acetylmannosamine
(high vs. low) | 0.381
(0.147–0.985) | 0.046a |
|
2,5-Dihydroxybenzaldehyde (high vs.
low) | 0.336
(0.126–0.895) | 0.029a |
Discussion
The high prevalence of CRC is a growing challenge in
China, and specific biomarkers are needed, especially in the early
stages of the disease, to enable early detection and improve
patient outcomes (22). Gut
microecology dysregulation has been associated with the development
of CRC (23). Proximal feces,
located in the colorectal mucosa, can represent structural and
metabolic alterations associated with disease progression (24). Elucidating the association between
the intestinal metabolome and the molecular characteristics of
tumors in individuals with CRC is necessary to understand the role
of the metabolome in colorectal carcinogenesis. The aim of the
present study was to identify potential biomarkers by analyzing
fecal metabolites from patients with CRC and comparing them with
those of pre-cancerous patients with CRA and healthy subjects, and
to explore the impact of metabolites on the ongoing progression of
CRC.
The present study provided evidence that intestinal
metabolites are closely associated with CRC progression. As
colorectal tumors progress along the adenoma-carcinoma sequence,
gut metabolite homeostasis is disrupted, and key metabolic pathways
are altered in CRC pathogenesis. Disruption of intestinal metabolic
pathways plays an important role in colorectal tumorigenesis
(25). Cholesterol metabolites and
sphingolipids were the most highly upregulated metabolites and
significantly associated with CRC. Furthermore, fat-rich diets and
increased amounts of cholesterol are associated with the
development of CRC (26). It has
been shown that reprogramming of amino acid and lipid metabolism in
the tumor microenvironment and metabolic crosstalk between
pathogens and host cells contribute to the rapid growth of cancer
cells and tumor formation (27).
Enhanced lipid synthesis or uptake leads to uncontrolled tumor
development (28).
The metabolomic analysis of the present study
revealed that several metabolites, such as 2-aminoethylphosphonate,
were enriched in the CRA and CRC groups compared with the HC group.
2-Aminoethylphosphonate is a potentially nutritionally active
phosphatidyl sphingolipid that significantly increases the content
of hydroxy ceramides, which play an important role in the
expression of genes related to its biosynthesis-related genes in
the skin (29). Amino acids play an
important role in several steps of protein biosynthesis, where they
maintain redox homeostasis as both electron donors and acceptors
and act as energy sources (30).
The abundance of amino acid derivatives is essential to promote
cancer cell proliferation. They provide crucial building blocks for
protein synthesis and contribute to energy generation and
nucleotide synthesis (31). By
contrast, L-carnitine was reported to ameliorate symptoms of
cachexia in tumor patients, which may be a point of ambiguity in
the function of the complex network of intestinal metabolites
(32).
The present study suggested that fecal metabolites
have the potential to aid in the non-invasive diagnosis of CRC, as
indicated from the set of metabolomic markers that showed the
potential to accurately differentiate between patients with CRC and
HC. Microbiome alterations associated with CRC have been considered
a promising source of diagnostic biomarkers, with several studies
focusing on this aspect. For instance, Zeller et al
(33) provided evidence for the
potential of such biomarkers by developing highly predictive and
accurate models using up to 22 CRC-associated microbial taxa and
validating their findings in multiple independent cohorts from
different countries. Combining metabolomics and microbiome data can
help determine the metabolic alterations that contribute to CRC
progression. Previous studies have used such combined data to infer
potential metabolic alterations in CRC (34). For example, Yachida et al
(16) reported an increase in
methane metabolism in the gut of CRC patients compared to HC.
We conducted a prospective analysis of metabolomics
data in patients with CRC explore the association between gut
ecology and survival in patients with cancer. This sets our study
apart from others as most studies have been cross-sectional in
nature (35,36). Several investigators have analyzed
cancer progression and the distribution of bacterial abundance,
demonstrating an association between the increase in specific flora
and favorable clinical outcomes. For instance, in 43 patients with
advanced melanoma, an increase in Clostridium perfringens
was associated with a favorable response to anti-PD-1 therapy
(37). Similarly, the association
between Agrobacterium tumefaciens and the clinical response
to anti-PD-1 therapy was validated in a Japanese cohort of patients
with advanced non-small lung cancer (NSCLC), indicating a lack of
beneficial effect (38).
The present study utilized comprehensive
macro-metabolomic analysis to demonstrate higher levels of
phenylacetyl-L-glutamine, triglyceride (8:0_16:0_18:1),
tryptophan-threonine and serine-alanine in STS. By contrast,
hexosylceramide (d16:1/22:0), phosphatidylcholine (18:1_18:2),
4-oxopentanoate and 3-sulfocatechol were more abundant in the feces
of LTS. Sphingolipids are being investigated for their role in
carcinogenesis and cancer therapy, and have emerged as a topic of
interest for anticancer therapies (39,40).
Gut microbial metabolites are believed to associate the gut
microbiome with systemic activity. For example, Agrobacterium
tumefaciens and Barnesiella intestinihominis have been
shown to promote therapeutic immune modulation induced by
cyclophosphamide (41). A crucial
metabolic function of the gut microbiome involves the conversion of
dietary fiber and mucopolysaccharides ingested by the host into
short-chain fatty acids (42).
Short-chain fatty acids have been shown to exhibit protective
effects in animal models of colitis and colitis-induced CRC, as
well as exerting anti-proliferative effects on cancer cells
(43).
A recent study examining plasma tryptophan
metabolites in patients with NSCLC treated with immunotherapy found
that low levels of 3-hydroxyaminobenzoic acid were significantly
associated with longer median progression-free survival (44). Furthermore, patients with pancreatic
ductal adenocarcinoma and a rare LTS phenotype had significantly
higher tumor bacterial diversity than patients with more typical
shorter survival. The diversity of the microbiome was also found to
have a significant impact on the survival of tumor patients,
supporting a causal role for the gut microbiome in shaping tumor
progression (45). However, certain
metabolites, including 2-pentanone and tridecane, have been
reported to adversely affect therapeutic efficacy in CRC patients
(46). The current study identified
N-acetylmannosamine and 2,5-dihydroxybenzaldehyde as potential
prognostic markers for patients with CRC. While the association
between these metabolites and CRC progression remains unclear, the
findings of the current study suggest that they may have value in
predicting CRC prognosis.
It is important to note that the current study was
limited by its small sample size and single-center design, and
larger trials are needed to confirm the present results.
In summary, metabolomic data were utilized to track
the dynamic changes in metabolites during the progression of CRC.
The findings of the present suggest that gut metabolites may play a
critical role in CRC development and clinical outcomes; however,
more extensive studies are required to validate these observations.
These outcomes could have significant implications for the
development of future diagnostic and treatment strategies for
CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural Science
Foundation of China (grant no. 81573239).
Availability of data and materials
The raw metabolomics data generated and/or analyzed
during the current study are available in the MetaboLights
repository under accession no. MTBLS7833 (https://www.ebi.ac.uk/metabolights/MTBLS7833).
All other datasets used and/or analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
HLo, QWu and ZX designed the study and enrolled
patients. RZ, XH, FY, SJ and QH collected patient information and
analyzed the data. DW, HLi, QWa and ZX confirmed the authenticity
of the raw data. ZX, RZ, DW, HLi and QWa wrote the manuscript and
evaluated the results. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
All patients signed written informed consents. The
collection and use of samples were approved by the Ethics Committee
of Tianyou Hospital, Wuhan University of Science and Technology
(Wuhan, China). All aspects of the study complied with The
Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li N, Lu B, Luo C, Cai J, Lu M, Zhang Y,
Chen H and Dai M: Incidence, mortality, survival, risk factor and
screening of colorectal cancer: A comparison among China, Europe,
and northern America. Cancer Lett. 522:255–268. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lynch JP and Hoops TC: The genetic
pathogenesis of colorectal cancer. Hematol Oncol Clin North Am.
16:775–810. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Biller LH and Schrag D: Diagnosis and
treatment of metastatic colorectal cancer: A review. JAMA.
325:669–685. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Keum N and Giovannucci E: Global burden of
colorectal cancer: Emerging trends, risk factors and prevention
strategies. Nat Rev Gastroenterol Hepatol. 16:713–732. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Patel SG, Karlitz JJ, Yen T, Lieu CH and
Boland CR: The rising tide of early-onset colorectal cancer: A
comprehensive review of epidemiology, clinical features, biology,
risk factors, prevention, and early detection. Lancet Gastroenterol
Hepatol. 7:262–274. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jakobsson HE, Rodríguez-Piñeiro AM,
Schütte A, Ermund A, Boysen P, Bemark M, Sommer F, Bäckhed F,
Hansson GC and Johansson ME: The composition of the gut microbiota
shapes the colon mucus barrier. EMBO Rep. 16:164–177. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang J, Wei H, Zhou Y, Szeto CH, Li C, Lin
Y, Coker OO, Lau HCH, Chan AWH, Sung JJY and Yu J: High-fat diet
promotes colorectal tumorigenesis through modulating gut microbiota
and metabolites. Gastroenterology. 162:135–149.e2. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu W, Zhang R, Shu R, Yu J, Li H, Long H,
Jin S, Li S, Hu Q, Yao F, et al: Study of the relationship between
microbiome and colorectal cancer susceptibility using 16SrRNA
sequencing. Biomed Res Int. 2020:78283922020.PubMed/NCBI
|
|
10
|
Vacante M, Ciuni R, Basile F and Biondi A:
Gut microbiota and colorectal cancer development: A closer look to
the adenoma-carcinoma sequence. Biomedicines. 8:4892020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mima K, Nishihara R, Qian ZR, Cao Y,
Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al:
Fusobacterium nucleatum in colorectal carcinoma tissue and patient
prognosis. Gut. 65:1973–1980. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lucas C, Barnich N and Nguyen HTT:
Microbiota, inflammation and colorectal cancer. Int J Mol Sci.
18:13102017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hanus M, Parada-Venegas D, Landskron G,
Wielandt AM, Hurtado C, Alvarez K, Hermoso MA, López-Köstner F and
De la Fuente M: Immune system, microbiota, and microbial
metabolites: The unresolved triad in colorectal cancer
microenvironment. Front Immunol. 12:6128262021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dalal N, Jalandra R, Bayal N, Yadav AK,
Harshulika, Sharma M, Makharia GK, Kumar P, Singh R, Solanki PR and
Kumar A: Gut microbiota-derived metabolites in CRC progression and
causation. J Cancer Res Clin Oncol. 147:3141–3155. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alhinai EA, Walton GE and Commane DM: The
role of the gut microbiota in colorectal cancer causation. Int J
Mol Sci. 20:52952019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yachida S, Mizutani S, Shiroma H, Shiba S,
Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M,
et al: Metagenomic and metabolomic analyses reveal distinct
stage-specific phenotypes of the gut microbiota in colorectal
cancer. Nat Med. 25:968–976. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu S, Han L, Hu X, Sun T, Xu D, Li Y, Chen
Q, Yao W, He M, Wang Z, et al: N6-methyladenosine reader IMP2
stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect:
Implication in colorectal cancer. J Hematol Oncol. 14:1882021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jonsson AL and Bäckhed F: Role of gut
microbiota in atherosclerosis. Nat Rev Cardiol. 14:79–87. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shuwen H, Yinhang W, Xingming Z, Jing Z,
Jinxin L, Wei W and Kefeng D: Using whole-genome sequencing (WGS)
to plot colorectal cancer-related gut microbiota in a population
with varied geography. Gut Pathog. 14:502022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang C, Zhou S, Chang H, Zhuang F, Shi Y,
Chang L, Ai W, Du J, Liu W, Liu H, et al: Metabolomic profiling
identified serum metabolite biomarkers and related metabolic
pathways of colorectal cancer. Dis Markers. 2021:68588092021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gu J, Xiao Y, Shu D, Liang X, Hu X, Xie Y,
Lin D and Li H: Metabolomics analysis in serum from patients with
colorectal polyp and colorectal cancer by 1H-NMR
spectrometry. Dis Markers. 2019:34918522019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luo XJ, Zhao Q, Liu J, Zheng JB, Qiu MZ,
Ju HQ and Xu RH: Novel genetic and epigenetic biomarkers of
prognostic and predictive significance in stage II/III colorectal
cancer. Mol Ther. 29:587–596. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rebersek M: Gut microbiome and its role in
colorectal cancer. BMC Cancer. 21:13252021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vymetalkova V, Vodicka P, Vodenkova S,
Alonso S and Schneider-Stock R: DNA methylation and chromatin
modifiers in colorectal cancer. Mol Aspects Med. 69:73–92. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang C, Wang XY, Zhang P, He TC, Han JH,
Zhang R, Lin J, Fan J, Lu L, Zhu WW, et al: Cancer-derived exosomal
HSPC111 promotes colorectal cancer liver metastasis by
reprogramming lipid metabolism in cancer-associated fibroblasts.
Cell Death Dis. 13:572022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vernia F, Longo S, Stefanelli G, Viscido A
and Latella G: Dietary factors modulating colorectal
carcinogenesis. Nutrients. 13:1432021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bian X, Liu R, Meng Y, Xing D, Xu D and Lu
Z: Lipid metabolism and cancer. J Exp Med. 218:e202016062021.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Munir R, Lisec J, Swinnen JV and Zaidi N:
Lipid metabolism in cancer cells under metabolic stress. Br J
Cancer. 120:1090–1098. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tomonaga N, Manabe Y, Aida K and Sugawara
T: Dietary ceramide 2-aminoethylphosphonate, a marine
sphingophosphonolipid, improves skin barrier function in hairless
mice. Sci Rep. 10:138912020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vučetić M, Cormerais Y, Parks SK and
Pouysségur J: The central role of amino acids in cancer redox
homeostasis: Vulnerability points of the cancer redox code. Front
Oncol. 7:3192017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vettore L, Westbrook RL and Tennant DA:
New aspects of amino acid metabolism in cancer. Br J Cancer.
122:150–156. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jiang F, Zhang Z, Zhang Y, Pan X, Yu L and
Liu S: L-carnitine ameliorates cancer cachexia in mice partly via
the carnitine palmitoyltransferase-associated PPAR-γ signaling
pathway. Oncol Res Treat. 38:511–516. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zeller G, Tap J, Voigt AY, Sunagawa S,
Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N, et
al: Potential of fecal microbiota for early-stage detection of
colorectal cancer. Mol Syst Biol. 10:7662014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Weir TL, Manter DK, Sheflin AM, Barnett
BA, Heuberger AL and Ryan EP: Stool microbiome and metabolome
differences between colorectal cancer patients and healthy adults.
PLoS One. 8:e708032013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Clos-Garcia M, Garcia K, Alonso C,
Iruarrizaga-Lejarreta M, D'Amato M, Crespo A, Iglesias A, Cubiella
J, Bujanda L and Falcón-Pérez JM: Integrative analysis of fecal
metagenomics and metabolomics in colorectal cancer. Cancers
(Basel). 12:11422020.2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen F, Dai X, Zhou CC, Li KX, Zhang YJ,
Lou XY, Zhu YM, Sun YL, Peng BX and Cui W: Integrated analysis of
the faecal metagenome and serum metabolome reveals the role of gut
microbiome-associated metabolites in the detection of colorectal
cancer and adenoma. Gut. 71:1315–1325. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tanoue T, Morita S, Plichta DR, Skelly AN,
Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, et
al: A defined commensal consortium elicits CD8 T cells and
anti-cancer immunity. Nature. 565:600–605. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao S, Gao G, Li W, Li X, Zhao C, Jiang
T, Jia Y, He Y, Li A, Su C, et al: Antibiotics are associated with
attenuated efficacy of anti-PD-1/PD-L1 therapies in Chinese
patients with advanced non-small cell lung cancer. Lung Cancer.
130:10–17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ogretmen B: Sphingolipid metabolism in
cancer signalling and therapy. Nat Rev Cancer. 18:33–50. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Govindarajah N, Clifford R, Bowden D,
Sutton PA, Parsons JL and Vimalachandran D: Sphingolipids and acid
ceramidase as therapeutic targets in cancer therapy. Crit Rev Oncol
Hematol. 138:104–111. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hayase E and Jenq RR: Role of the
intestinal microbiome and microbial-derived metabolites in immune
checkpoint blockade immunotherapy of cancer. Genome Med.
13:1072021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim S, Covington A and Pamer EG: The
intestinal microbiota: Antibiotics, colonization resistance, and
enteric pathogens. Immunol Rev. 279:90–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lavelle A and Sokol H: Gut
microbiota-derived metabolites as key actors in inflammatory bowel
disease. Nat Rev Gastroenterol Hepatol. 17:223–237. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Azuma K, Xiang H, Tagami T, Kasajima R,
Kato Y, Karakawa S, Kikuchi S, Imaizumi A, Matsuo N, Ishii H, et
al: Clinical significance of plasma-free amino acids and tryptophan
metabolites in patients with non-small cell lung cancer receiving
PD-1 inhibitor: A pilot cohort study for developing a prognostic
multivariate model. J Immunother Cancer. 10:e0044202022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Riquelme E, Zhang Y, Zhang L, Montiel M,
Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, San Lucas A, et
al: Tumor microbiome diversity and composition influence pancreatic
cancer outcomes. Cell. 178:795–806.e12. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Botticelli A, Vernocchi P, Marini F,
Quagliariello A, Cerbelli B, Reddel S, Del Chierico F, Di Pietro F,
Giusti R, Tomassini A, et al: Gut metabolomics profiling of
non-small cell lung cancer (NSCLC) patients under immunotherapy
treatment. J Transl Med. 18:492020. View Article : Google Scholar : PubMed/NCBI
|