Introduction
Patients with metastatic urothelial carcinoma (mUC)
have a poor prognosis, with a 5-year survival rate of ~15% using
conventional treatment regimens (1,2). The
current standard treatment for patients with mUC, cisplatin-based
chemotherapy, has a long history for mUC treatment but median
overall survival (OS) times for patients with mUC remain at 14–15
months (3). Patients demonstrating
mUC recurrence after first-line treatment, especially those with
progression during first-line treatment, have a particularly poor
prognosis. The introduction of immune checkpoint inhibitors (ICIs)
as a second-line treatment has resulted in longer durations of
treatment response and improved prognoses, but only for responders;
the proportion of those patients is limited to 15–30% (4–6).
Therefore, a non-invasive biomarker to evaluate response to ICIs is
urgently required.
Regarding tissue-based biomarkers for ICI responses,
several studies have reported the significance of tumor programmed
death-1 (PD-L1) expression, tumor mutation burden (TMB) and
microsatellite instability (MSI) (7–9).
However, tissue biopsies present several problems in terms of
invasiveness, accessibility and frequency of use in contrast to
blood-based biomarkers obtained by liquid biopsy (10). Of note, plasma circulating tumor DNA
(ctDNA) from liquid biopsies has shown particular promise in
screening patients who may benefit from ICIs (11). In addition, ctDNA is useful for
detecting minimal residual disease, monitoring disease status
during treatment and predicting response to therapeutic drugs with
a significant lead time to radiographic imaging (12–14).
However, the significance of plasma ctDNA as a biomarker to
evaluate pembrolizumab response in patients with mUC has not yet
been fully investigated.
Therefore, the present study investigated the
association between plasma ctDNA analysis and response to
pembrolizumab in patients with mUC.
Materials and methods
Patients and sample collection
A total of 25 plasma samples from 16 patients with
mUC were prospectively collected using PAX gene circulating free
DNA (cfDNA) tubes (PreAnalytiX GmbH) at the University of Tsukuba
Hospital (Tsukuba, Japan) between February 2018 and April 2021. Of
these 25 samples, 16 were collected prior to administration of
pembrolizumab (used as a second-line therapy) and nine were
collected after administration of pembrolizumab in conjunction with
computed tomography evaluation of clinical response. All patients
provided informed, written consent to participate in the present
study and the protocol was approved by the University of Tsukuba
Hospital Institutional Review Board (Tsukuba, Japan; approval no,
#H28-104). Clinical data were obtained from hospital charts.
Response to pembrolizumab was evaluated according to the Response
Evaluation Criteria in Solid Tumor (RECIST) guidelines (version
1.1) (15). In the present study,
response to pembrolizumab was classified as complete response (CR),
partial response (PR) and stable disease (SD).
Survival analysis
For analysis of OS, death from any cause was defined
as an event and patients without any events were censored at the
last follow-up visit. For progression-free survival (PFS) analysis,
progressive disease (PD) evaluated by RECIST was defined as an
event. The number of months from the date of administration of
pembrolizumab to the event or censored date was calculated for OS
and PFS.
Sample preparation and targeted
sequencing
The extracted plasma cfDNA was subjected to a ctDNA
analysis using the Avenio ctDNA Expanded Kit (Roche Diagnostics)
designed to detect 77 genetic alterations, including single
nucleotide variants (SNVs), indels and copy number variants (CNVs),
according to the manufacturer's instructions. DNA concentration was
assessed using a Qubit fluorometer (Thermo Fisher Scientific,
Inc.). Prepared libraries were sequenced using a NextSeq 500
(Illumina, Inc) and analyzed using Roche AVENIO ctDNA Analysis
Software (version 2.0.0; Roche Sequencing Solutions). Somatic
variants were called and filtered with human reference genome hg38
using the same software and default logical and operation filter
sets according to the manufacturer's instructions. Patients who
carried ≥2 alterations (SNV, indels or CNVs) were defined as
ctDNA-positive.
Statistical analysis
All statistical analyses were performed using R
(version 4.0.2; R Foundation) and GraphPad Prism (version 8;
GraphPad Software; Dotmatics). Kaplan-Meier survival curves were
generated and the differences between the curves were compared with
the log-rank test. For continuous variables, differences between
groups were compared using the Mann-Whitney U-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient characteristics
The characteristics of patients with mUC (n=16)
treated with second-line pembrolizumab are presented in Table I. The median age of the patient pool
was 70.0 years (range, 51–81 years) and the majority were male
(75.0%). The most common histology was UC (n=13), followed by UC
with small cell component (n=1) and UC with sarcomatoid variant
(n=1). In terms of tumor location, 10 (62.5%) patients had bladder
tumors and six (37.5%) had upper urinary tract tumors. Half of all
patients presented with visceral metastasis.
 | Table I.Patient characteristics (n=16). |
Table I.
Patient characteristics (n=16).
| Characteristic | Value |
|---|
| Age, years | 70.0 (51–81) |
| Sex |
|
| Male | 12 (75.0) |
| Female | 4 (25.0) |
| Follow-up time,
months | 9.5 (1.0–24.7) |
| Histology |
|
| UC | 13 (81.3) |
| UC with
small cell component | 2 (12.5) |
| UC with
sarcomatoid variant | 1 (6.2) |
| Chemotherapy
regimen |
|
| GC | 9 (56.3) |
|
GCa | 4 (25.0) |
|
GC+GCa | 2 (12.5) |
| EP | 1 (6.2) |
| Performance
status |
|
| 0 | 7 (43.8) |
| 1 | 4 (25.0) |
| NA | 5 (31.2) |
| Clinical
response |
|
| PR | 4 (25.0) |
| SD | 5 (31.3) |
| PD | 7 (43.7) |
| Tumor location |
|
|
Bladder | 10 (62.5) |
| Upper
urinary tract | 6 (37.5) |
| Visceral
metastasis |
|
|
Present | 8 (50.0) |
|
Absent | 8 (50.0) |
As first-line chemotherapy before pembrolizumab
administration, nine (56.3%) patients received a combination
therapy of gemcitabine and cisplatin (GC), four (25.0%) patients
received a combination therapy of gemcitabine and carboplatin
(GCa), and one (6.2%) patient received a combination therapy of
etoposide and cisplatin (EP). The remaining two patients (12.5%)
received GC followed by GCa therapy as first- and second-line
chemotherapies. Most patients (68.8%) demonstrated a good initial
performance status of 0–1 at the time of pembrolizumab
administration and second-line therapy resulted in nine (four PR
and five SD; 56.3%) responders and seven non-responders (PD;
43.8%).
Genomic alteration profiles in plasma
ctDNA and clinical outcomes
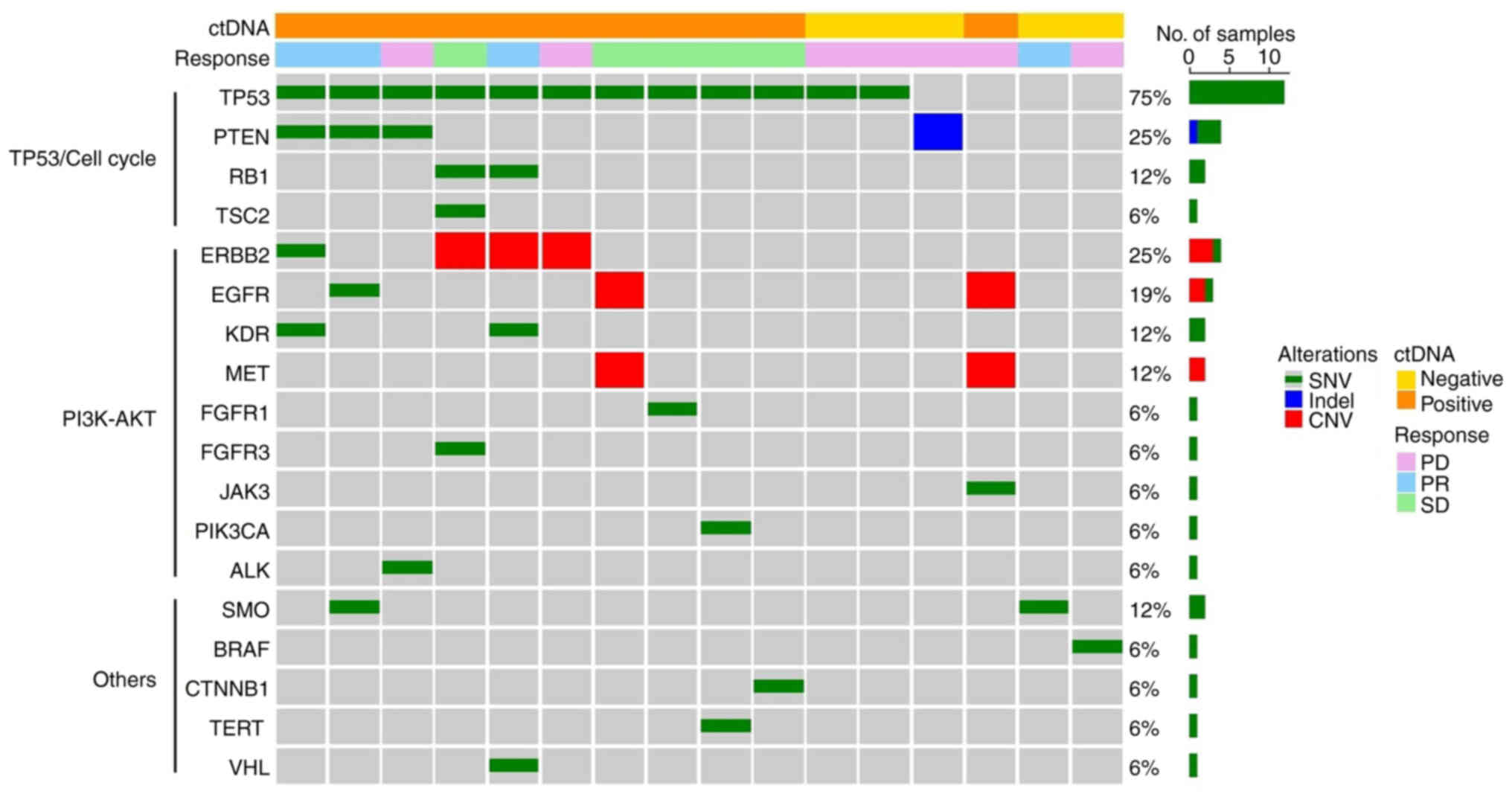
The genomic alteration profiles in plasma ctDNA
collected from the 16 patients at baseline were analyzed.
Unfiltered and filtered somatic variant data for each patient were
collected. In total, 43 somatic variants (35 SNVs, one indel and
seven CNVs) were identified with a median VAF of 2.0% (range,
0.08–55.8%) (Table SI). The
observed genomic alteration profiles of analyzed plasma ctDNA were
examined (Fig. 1). The median
number of genomic alterations was 2.5 (range, 1–6) and all patients
demonstrated ≥1 alterations. The most commonly altered gene was
TP53 (75%), followed by ERBB2 (25%), PTEN
(25%) and EGFR (19%). In terms of TP53 mutations, the
most frequent subtype was missense (67%), followed by nonsense
(25%) and splicing mutations (8%). Among ctDNA-positive patients,
defined as those who demonstrated ≥2 genomic alterations, 10/11
(91%) had TP53 SNVs. There were no significant differences
in cell-free DNA concentration after stratification by ctDNA
positivity (Fig. S1). The
characteristics of patients with mUC were also stratified by ctDNA
status (Table II). The
neutrophil-lymphocyte ratio of patients in the ctDNA-negative group
was significantly higher than that in the ctDNA-positive group
(7.28 vs. 2.65).
 | Table II.Patient characteristics stratified by
ctDNA status. |
Table II.
Patient characteristics stratified by
ctDNA status.
| Characteristic | cthNA-positive
(n=11) | cthNA-negative
(n=5) |
|---|
| Age, years | 71.0 (59–81) | 69.0 (51–74) |
| Sex |
|
|
|
Male | 8 (72.7) | 4 (80.0) |
|
Female | 3 (27.3) | 1 (20.0) |
| Follow-up time,
months | 10.4
(1.0–24.7) | 5.6 (2.4–11.9) |
| Chemotherapy
regimen |
|
|
| GC | 6 (54.5) | 3 (60.0) |
|
GCa | 2 (18.2) | 0 (0.0) |
|
GC+GCa | 2 (18.2) | 2 (40.0) |
| EP | 1 (9.1) | 0 (0.0) |
| Performance
status |
|
|
| 0 | 6 (54.5) | 1 (20.0) |
| 1 | 2 (18.2) | 2 (40.0) |
| NA | 3 (27.3) | 2 (40.0) |
| Tumor location |
|
|
|
Bladder | 8 (72.7) | 2 (40.0) |
| Upper
urinary tract | 3 (27.3) | 3 (60.0) |
| Visceral
metastasis |
|
|
|
Present | 5 (45.5) | 3 (60.0) |
|
Absent | 6 (54.5) | 2 (40.0) |
| WBC count | 6,000
(2,900–9,700) | 8,900
(3,600–22,400) |
| Neutrophil
count | 3,830
(1,302–6,383) | 6,602
(2,045–20,026) |
| Lymphocyte
count | 1,428
(378–2,408) | 907
(417–1,958) |
| NLR | 2.7 (1.3–12.1) | 7.3
(1.7–40.0)a |
Association between mutation frequency
data from target sequencing and response to pembrolizumab
The association between mutation profiles and
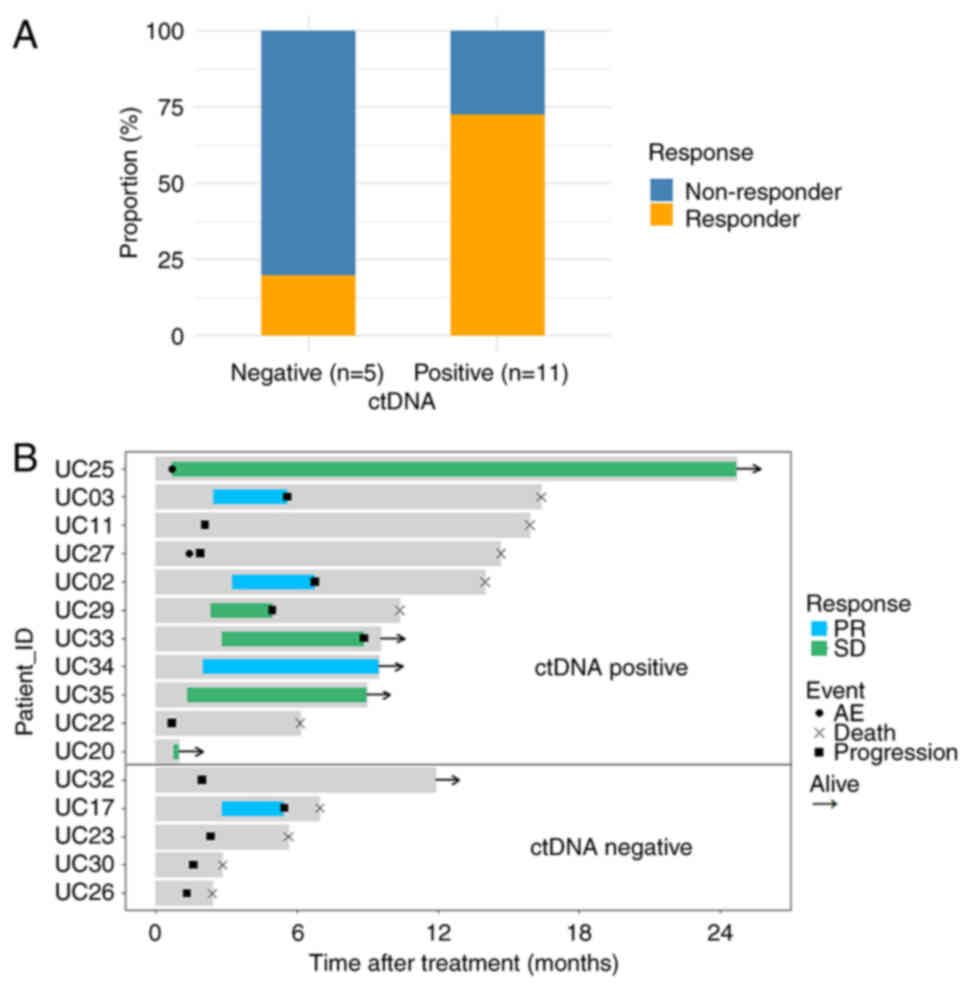
pembrolizumab response was analyzed. The proportion of responsive
patients in the ctDNA-positive group was higher than that in the
ctDNA-negative group (72.7 vs. 20.0%) (Fig. 2A). After stratification by ctDNA
positivity, median response times in the ctDNA-positive group were
significantly longer compared with those in the ctDNA-negative
group (3.2 vs. 0 months) (Fig. 2B).
Furthermore, 4/11 (36.4%) patients in the ctDNA-positive group
demonstrated a sustained response at data cutoff, while none in the
ctDNA-negative group had such a response.
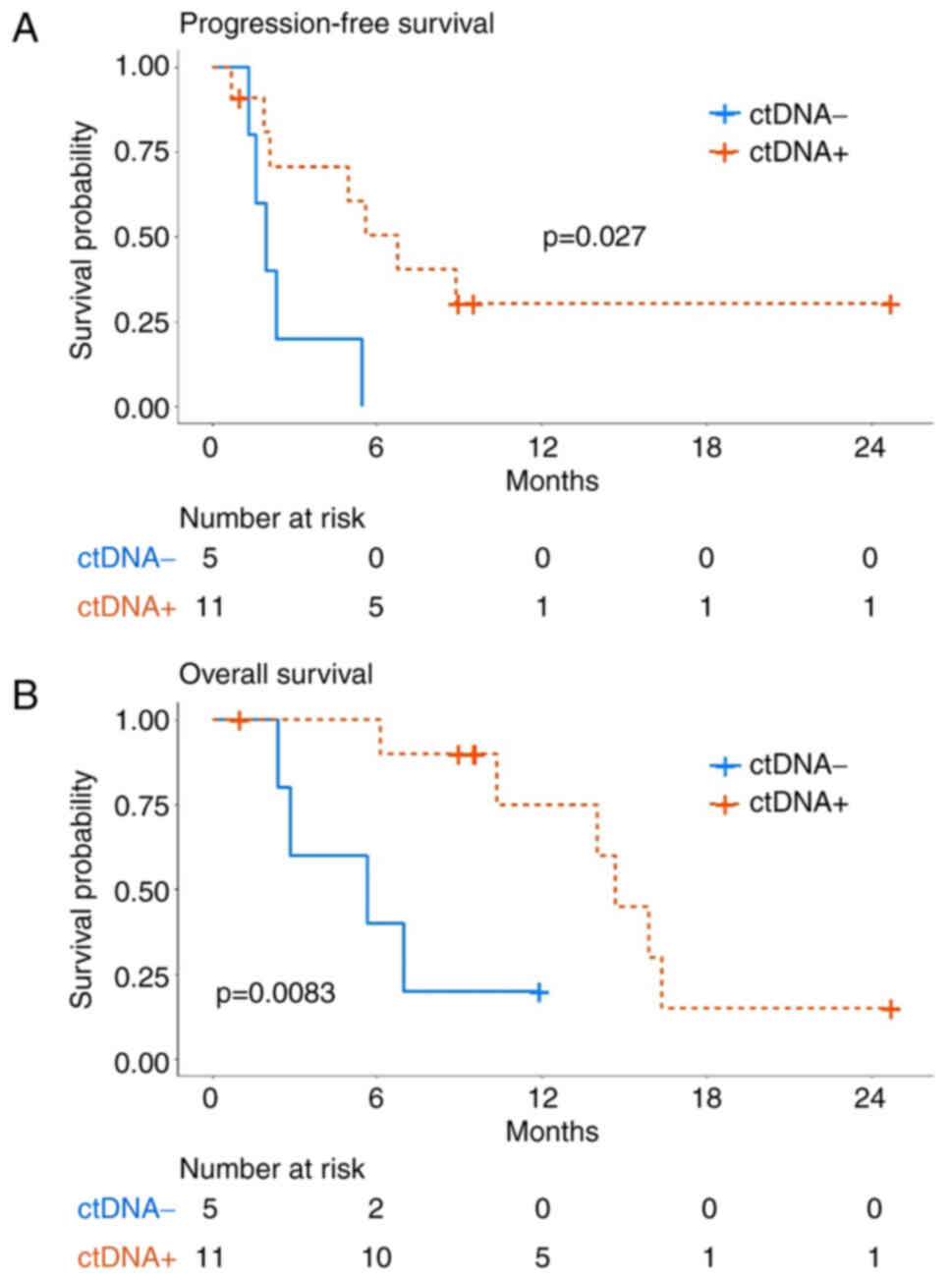
The association between ctDNA positivity and patient
prognoses, as well as response to pembrolizumab, was analyzed. The
median PFS of patients in the ctDNA-positive group was
significantly longer compared with that in the ctDNA-negative group
(3.9 vs. 2.0 months) (Fig. 3A) and
the median OS demonstrated a similar trend (14.7 vs. 5.6 months)
(Fig. 3B). These results suggested
that the mutation frequency in plasma ctDNA was associated with
clinical response to pembrolizumab and disease outcomes.
Association between changes in the VAF
of each mutation in plasma ctDNA and pembrolizumab response
The association between mutation frequency and
clinical response during pembrolizumab treatment using samples from
eight (50.0%) of 16 patients collected during pembrolizumab
treatment was examined. There was no association between changes in
the mutation frequency and individual clinical responses (Fig. S2). Next, the association between
changes in the VAF of each mutation and individual clinical
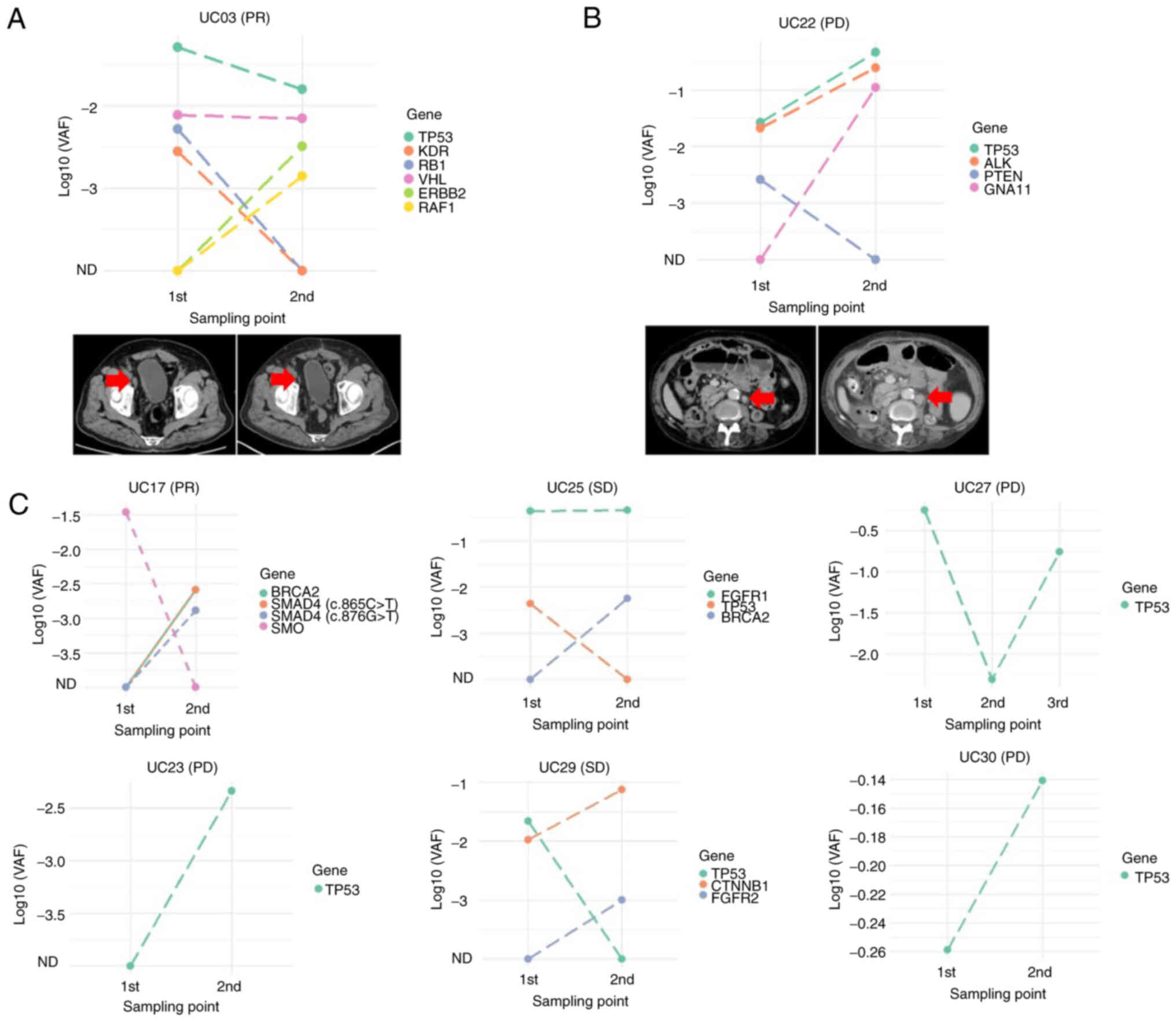
responses was examined. The UC03 patient who demonstrated PR with
shrinking lymph node metastasis had decreased VAF in TP53,
RB1 and KDR, whereas the VAF in ERBB2 and
RAF1 was increased (Fig.
4A). Another patient (UC22), who demonstrated PD with
enlargement of lymph node metastasis, had increased VAF in TP53,
AKL and GNA11 and decreased VAF in PTEN (Fig. 4B). Among these mutated genes, only
the VAF change in TP53 was associated with clinical
response. In terms of the VAF changes for each mutated gene in the
other six patients, only TP53 was associated with clinical
response during pembrolizumab treatment (Fig. 4C). These results suggested that the
VAF changes in TP53 during pembrolizumab treatment
accurately reflected the therapeutic response.
Discussion
The present study, using genomic alteration
profiling of plasma ctDNA collected from 16 patients with mUC at
baseline, demonstrated that ctDNA-positive status was associated
with patient response to pembrolizumab treatment. Furthermore,
analysis of consecutive samples from eight patients during
treatment demonstrated that decreases in TP53 VAF accurately
reflected therapeutic response. These results suggested that
profiling of plasma ctDNA was useful in predicting and monitoring
the therapeutic pembrolizumab response in patients with mUC.
Blood-based biomarkers, which may be detected by
liquid biopsy in a non-invasive manner, are useful in screening
responsive patients (11); however,
there are no currently established and approved blood-based
biomarkers for predicting responses to ICI. While TMB is recognized
as a tissue-based biomarker for ICI response (8,16),
previous studies suggest that elevated TMB in blood, a liquid
biopsy-style marker as detected by ctDNA assays, is also associated
with responses and prognoses in patients treated with ICI (17–19).
Si et al (18) previously
reported that blood TMB 320 mutations/megabase was
useful in predicting clinical benefit from ICI treatment compared
with chemotherapy. In addition, Kim et al (19) reported that ctDNA mutational load
scores correlate with clinical pembrolizumab responses in
metastatic gastric cancer. Although the present study did not
examine the blood TMB, an association between ctDNA-positive status
and ICI response was demonstrated. On the other hand, Laukhtina
et al (20) reported an
association between high baseline ctDNA levels and worse
disease-free survival and OS. This may be due to different
profiling of the mutated genes, including the percentage of
TP53 mutations, between the previous studies and the present
study. A number of studies previously reported that TP53
mutations are associated with superior clinical benefits to ICI
treatment (21–23). In line with these studies, the
present study demonstrated that TP53 mutations were
associated with an improved patient response to ICI. Sun et
al (22) reported that
TP53 missense mutations were associated with better clinical
benefit in their local clinical cohort, but nonsense mutations were
not. Similar trends were also reported in the MSK study cohort,
while the Checkmate-012 study cohort reported an opposite trend
(22). In the present study,
TP53 missense mutations were the most common subtype of
TP53 mutations.
A recent study reported that ctDNA clearance during
ICI treatment in advanced urothelial carcinoma is associated with
better clinical outcomes (24),
which led the present study to investigate VAF changes for each
mutation during second-line pembrolizumab treatment. The tumor
suppressor TP53 is the most frequently mutated
cancer-associated gene in urothelial bladder cancer (25), a result also demonstrated by the
present study. Thorsson et al (26) suggested that driver mutations, such
as TP53, by inducing genomic instability, may alter the
immune environment through the production of neoantigens and
paradoxically produce better response to ICIs. However, among the
genomic alterations detected in the present study, decreases in
TP53 VAF were associated with improved clinical response
during pembrolizumab treatment. Similarly, Parkinson et al
(27) reported that, in high-grade
serous ovarian carcinoma, TP53 VAF changes during
chemotherapy correlated with clinical response. The findings of the
present study, which suggested that decreases in TP53 VAF
during pembrolizumab treatment were associated with therapeutic
response, open up new possibilities for frequent liquid biopsies to
screen first-line non-responders for second-line feasibility.
The present study has several limitations. First,
the number of patients studied was small, so larger patient cohorts
are necessary for future studies. Second, technological
restrictions limited the assessable genes to 77; therefore, the
blood TMB and MSI status of samples could not be identified. Third,
as the study protocol did not include any tissue analysis, the TMB
and PD-L1 expression could not be evaluated, both of which have
been previously reported to correlate with ICI response (7–9).
Studies have reported that sequential ctDNA analysis can identify
responders to ICI therapy faster when compared with radiographic
imaging (12,13). As the present study protocol did not
determine the timings of liquid biopsies and radiographic
evaluation, it was difficult to assess the time saved for ctDNA
analysis over radiographic imaging. Finally, variants associated
with clonal hematopoiesis of indeterminate potential could be
mistaken for tumor-specific variants.
In conclusion, plasma ctDNA analysis in patients
with mUC treated with pembrolizumab demonstrated an association of
TP53 mutation frequency with clinical response of patients.
Furthermore, decreases in TP53 VAF during pembrolizumab
treatment was associated with clinical response. These results
suggest that plasma ctDNA analysis of patients with mUC could
potentially be used for predicting both pembrolizumab response and
monitoring disease status.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Mr. Noriyo Ishibashi
(Department of Inspection, Tsukuba i-Laboratory LLP), Mrs. Noriko
Kunita (Technical Assistant, Department of Urology, University of
Tsukuba) and Mrs. Naoko Ueki (Technical Assistant, Department of
Urology, University of Tsukuba) for their technical support in
sample processing and library preparation.
Funding
This work was supported by a grant from the Japan Society for
the Promotion of Science KAKENHI (grant no. 21K09365).
Availability of data and materials
Sequence data generated during the current study are
available in the Japanese Genotype-phenotype Archive (https://ddbj.nig.ac.jp/resource/jga-study/JGAS000622).
The generated somatic variant data for each patient is presented in
Table SI.
Authors' contributions
YN, TK and BJM contributed to the study design and
technical and material support. KH, YN, SK, KT, MS, AH, HNe and HNi
performed sample preparation. KH, YN and TK analyzed the acquired
data. KH and YN drafted the manuscript. KH, YN, SK, BJM, HNe and
HNi revised the manuscript critically. KH and YN confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The study protocol and data processing were approved
by the University of Tsukuba Hospital Institutional Review Board
(approval no. H28-104). Written informed consent was obtained from
all patients involved in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
von der Maase H, Sengelov L, Roberts JT,
Ricci S, Dogliotti L, Oliver T, Moore MJ, Zimmermann A and Arning
M: Long-term survival results of a randomized trial comparing
gemcitabine plus cisplatin, with methotrexate, vinblastine,
doxorubicin, plus cisplatin in patients with bladder cancer. J Clin
Oncol. 23:4602–4608. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Babaian RJ, Johnson DE, Llamas L and Ayala
AG: Metastases from transitional cell carcinoma of urinary bladder.
Urology. 16:142–144. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Clark PE, Spiess PE, Agarwal N, Bangs R,
Boorjian SA, Buyyounouski MK, Efstathiou JA, Flaig TW, Friedlander
T, Greenberg RE, et al: NCCN guidelines® insights
bladder cancer, version 2.2016 featured updates to the NCCN
guidelines. J Natl Compr Canc Netw. 14:1213–1224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Balar AV, Galsky MD, Rosenberg JE, Powles
T, Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J,
Perez-Gracia JL, et al: Atezolizumab as first-line treatment in
cisplatin-ineligible patients with locally advanced and metastatic
urothelial carcinoma: A single-arm, multicentre, phase 2 trial.
Lancet. 389:67–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bellmunt J, de Wit R, Vaughn DJ, Fradet Y,
Lee JL, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK,
et al: Pembrolizumab as second-line therapy for advanced urothelial
carcinoma. N Engl J Med. 376:1015–1026. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sharma P, Retz M, Siefker-Radtke A, Baron
A, Necchi A, Bedke J, Plimack ER, Vaena D, Grimm MO, Bracarda S, et
al: Nivolumab in metastatic urothelial carcinoma after platinum
therapy (CheckMate 275): A multicentre, single-arm, phase 2 trial.
Lancet Oncol. 18:312–322. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Petrelli F, Ghidini M, Ghidini A and
Tomasello G: Outcomes following immune checkpoint inhibitor
treatment of patients with microsatellite instability-high cancers:
A systematic review and meta-analysis. JAMA Oncol. 6:1068–1071.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Samstein RM, Lee CH, Shoushtari AN,
Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ,
Omuro A, et al: Tumor mutational load predicts survival after
immunotherapy across multiple cancer types. Nat Genet. 51:202–206.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reck M, Rodríguez-Abreu D, Robinson AG,
Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al: Pembrolizumab versus chemotherapy for PD-L1-positive
non-small-cell lung cancer. N Engl J Med. 375:1823–1833. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Snyder A, Morrissey MP and Hellmann MD:
Use of circulating tumor DNA for cancer immunotherapy. Clin Cancer
Res. 25:6909–6915. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bardelli A and Pantel K: Liquid biopsies,
what we do not know (yet). Cancer Cell. 31:172–179. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Goldberg SB, Narayan A, Kole AJ, Decker
RH, Teysir J, Carriero NJ, Lee A, Nemati R, Nath SK, Mane SM, et
al: Early assessment of lung cancer immunotherapy response via
circulating tumor DNA. Clin Cancer Res. 24:1872–1880. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Christensen E, Birkenkamp-Demtröder K,
Sethi H, Shchegrova S, Salari R, Nordentoft I, Wu HT, Knudsen M,
Lamy P, Lindskrog SV, et al: Early detection of metastatic relapse
and monitoring of therapeutic efficacy by ultra-deep sequencing of
plasma cell-free DNA in patients with urothelial bladder carcinoma.
J Clin Oncol. 37:1547–1557. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jardim DL, Goodman A, de Melo Gagliato D
and Kurzrock R: The challenges of tumor mutational burden as an
immunotherapy biomarker. Cancer Cell. 39:154–173. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rizvi NA, Cho BC, Reinmuth N, Lee KH, Luft
A, Ahn MJ, van den Heuvel MM, Cobo M, Vicente D, Smolin A, et al:
Durvalumab with or without tremelimumab vs standard chemotherapy in
first-line treatment of metastatic non-small cell lung cancer: The
MYSTIC phase 3 randomized clinical trial. JAMA Oncol. 6:661–674.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Si H, Kuziora M, Quinn KJ, Helman E, Ye J,
Liu F, Scheuring U, Peters S, Rizvi NA, Brohawn PZ, et al: A
blood-based assay for assessment of tumor mutational burden in
first-line metastatic NSCLC treatment: Results from the MYSTIC
study. Clin Cancer Res. 27:1631–1640. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim ST, Cristescu R, Bass AJ, Kim KM,
Odegaard JI, Kim K, Liu XQ, Sher X, Jung H, Lee M, et al:
Comprehensive molecular characterization of clinical responses to
PD-1 inhibition in metastatic gastric cancer. Nat Med.
24:1449–1458. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Laukhtina E, Hassler MR, Pradere B,
Yanagisawa T, Quhal F, Rajwa P, Sari Motlagh R, König F, Pallauf M,
Kawada T, et al: Circulating tumour DNA is a strong predictor of
outcomes in patients treated with systemic therapy for urothelial
carcinoma. Eur Urol Focus. 8:1683–1686. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hellmann MD, Nathanson T, Rizvi H, Creelan
BC, Sanchez-Vega F, Ahuja A, Ni A, Novik JB, Mangarin LMB,
Abu-Akeel M, et al: Genomic features of response to combination
immunotherapy in patients with advanced non-small-cell lung cancer.
Cancer Cell. 33:843–852.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun H, Liu SY, Zhou JY, Xu JT, Zhang HK,
Yan HH, Huan JJ, Dai PP, Xu CR, Su J, et al: Specific TP53 subtype
as biomarker for immune checkpoint inhibitors in lung
adenocarcinoma. EBioMedicine. 60:1029902020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu S, Geng S, Shi N, Zhang L, Xue W, Li Y
and Jiang K: Survival prediction of patients treated with immune
checkpoint inhibitors via KRAS/TP53/EGFR-single gene mutation.
Front Pharmacol. 13:8785402022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ravi P, Ravi A, Riaz IB, Freeman D, Curran
C, Mantia C, McGregor BA, Kilbridge KL, Pan CX, Pek M, et al:
Longitudinal evaluation of circulating tumor DNA using sensitive
amplicon-based next-generation sequencing to identify resistance
mechanisms to immune checkpoint inhibitors for advanced urothelial
carcinoma. Oncologist. 27:e406–e409. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lamy A, Gobet F, Laurent M, Blanchard F,
Varin C, Moulin C, Andreou A, Frebourg T and Pfister C: Molecular
profiling of bladder tumors based on the detection of FGFR3 and
TP53 mutations. J Urol. 176:2686–2689. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Thorsson V, Gibbs DL, Brown SD, Wolf D,
Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy
JA, et al: The immune landscape of cancer. Immunity.
48:812–830.e14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Parkinson CA, Gale D, Piskorz AM, Biggs H,
Hodgkin C, Addley H, Freeman S, Moyle P, Sala E, Sayal K, et al:
Exploratory analysis of TP53 mutations in circulating tumour DNA as
biomarkers of treatment response for patients with relapsed
high-grade serous ovarian carcinoma: A retrospective study. PLoS
Med. 13:e10021982016. View Article : Google Scholar : PubMed/NCBI
|