Introduction
Normal hematopoietic cells differentiate into mature
granulocyte, monocyte-macrophage, erythroid, or megakaryocyte
lineages, strictly according to the inherent procedure (1). The occurrence of leukemia is mainly
due to the failure of the differentiation at a certain stage of
hematopoietic cells, which enables them to retain only their
hyperproliferative capacity and blocks their terminal
differentiation (1,2). If this disordered differentiation of
leukemia cells can be reversed using drugs, these drugs can then
perhaps be used in clinical treatment (3). In terms of the committed
differentiation of myeloid leukemia, in addition to the successful
induction of the terminal differentiation of leukemic cells into
granulocytes in vitro using certain drugs, such as all trans
retinoic acid (4), there have been
some reports on the committed differentiation of leukemic cells
into monocytes-macrophages. However, the majority of these
processes are limited to the induction of partial differentiation
to varying degrees, which limits their clinical application
(5). Therefore, an in-depth
investigation of the key mechanisms of the committed
differentiation of leukemic cells may help to identify new targets
which can be used to interfere with the directed differentiation of
leukemic cells.
Macrophages are a highly heterogeneous group of
cells, which can be polarized by two types of macrophages under
various tissue environments and physiological and pathological
conditions; for example, M1 and M2 are the two types of macrophages
(6–9). Different types of macrophages often
exhibit different phenotypes and functions, or even function via
opposite mechanisms. For example, M1 type macrophages mainly induce
Th1 type immune responses, producing potent antitumor effects
(10). M2 type macrophages are
divided into three subtypes, M2a, M2b and M2c (11,12).
M2a macrophages mainly induce Th2 type immune response and M2b and
M2c macrophages mainly exert immunomodulatory effects. The
cytokines, GM-CSF and M-CSF, are also involved in the polarization
process of macrophages (13);
monocytes exposed to granulocyte-macrophage colony-stimulating
factor (GM-CSF) exhibit characteristics of M1-type macrophages,
whereas those exposed to macrophage colony-stimulating factor
(M-CSF) exhibit characteristics of M2-type macrophages (14,15).
It may be possible to induce leukemic cells to differentiate into
mature macrophages, thus achieving directed differentiation
induction therapy; at the same time, the induction of mature M1
macrophages may also play a role in anti-leukemia immunity. As
regards the molecular mechanisms of the polar differentiation of
macrophages, some associated transcription factors and signal
transduction pathways have been found to be involved. Currently, it
is well recognized that the PU.1 and C/EBP families are key
transcription factors that regulate the committed differentiation
of monocyte/macrophage lineages (16,17).
Second, the Janus kinase (JAK)/STAT, PI3K/AKT/mTOR and Notch
signaling pathways have also been reported to play a role in the
polarization of macrophages (18–20).
Although the proto-oncogene c-myc has been shown to induce or
suppress the expression of hundreds of genes, including regulating
macrophage activation functions (21,22),
the specific molecular mechanisms of c-myc in the polarization and
differentiation of leukemic cells into M1 type macrophages have not
yet been fully reported.
The authors have previously confirmed that c-myc is
involved in the committed differentiation of THP-1 leukemic cells
into monocytes/macrophages (23).
Following validation in a preliminary experiment that no
significant apoptosis occurred during the differentiation induction
process, the present study aimed to further explore the changes and
roles in directed differentiation. In addition, non-coding RNAs
(ncRNAs) have been found to play a critical role in cell
proliferation and differentiation in recent years (24). Among these, microRNAs (miRNAs/miRs)
are often involved in the post-transcriptional regulation of
downstream target genes by functioning as oncogenes or tumor
suppressor genes (25); they are
also involved in some key pathological processes, such as the
abnormal proliferation, differentiation or apoptosis of leukemic
cells (26,27). Therefore, it was hypothesized that
some miRNAs may regulate the committed differentiation of leukemic
cells into monocytes/macrophages by targeting the c-myc gene. It
was predicted that miR-let-7 family members are negatively
associated with c-myc; by performing differential miRNA Chip
screening, Gene Expression Omnibus (GEO) database analysis and a
PubMed literature research, it was found that the possible upstream
miRNA of c-myc was miR-let-7c-5p. The present study aimed to
further investigate the role of the miR-let-7c-5p/c-myc signaling
axis in the differentiation of leukemic cells into
monocytes/macrophages.
Materials and methods
Materials
The human acute myeloid leukemia THP-1 cell line was
purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. Phorbol 12-myristate 13-acetate (PMA;
cat. no. P1585-1MG), lipopolysaccharide (LPS; cat. no. L2880-10MG)
and interferon (IFN)-γ (cat. no. C600039-0100) were purchased from
MilliporeSigma. Fetal bovine serum (FBS), the BCA protein assay kit
and the SDS-PAGE gel rapid preparation kit were purchased from
Shanghai Biyuntian Biotechnology Co., Ltd. RPMI-1640 medium was
purchased from Hyclone (Cytiva). Double antibody, RIPA protein
lysis solution and protein loading buffer were purchased from
Beijing Solarbio Science & Technology Co., Ltd. The inverted
microscope was purchased from Olympus Corporation. Cell Counting
Kit-8 (CCK-8) solution was purchased from Dojindo Laboratories,
Inc. The microplate reader was purchased from Tecan Group, Ltd.
Standard protein marker and the Lipofectamine 2000®
transfection kit were purchased from Invitrogen (Thermo Fisher
Scientific, Inc.). The ECL luminescence kit was purchased from
Shandong Sparkjade Scientific Instruments Co., Ltd. Anti-c-myc
(cat. no. sc-47694) antibody was purchased from Santa Cruz
Biotechnology Inc; anti-β-actin antibody (cat. no. 20536-1-AP) was
obtained from Cell Signaling Technology, Inc. HRP-labeled rabbit
secondary antibody (cat. no. ZB-2301) was purchased from OriGene
Technologies, Inc. Primer design was provided by Sangon Biotech
Co., Ltd. The reverse transcription PrimeScript RT reagent kit with
gDNA Eraser (Perfect Real-Time), the chimeric fluorescence
detection kit and TB Green Premix Ex Taq (Tli RNaseH Plus) kit were
purchased from Takara Biomedical Technology (Beijing) Co., Ltd. The
cycle kit was purchased from Jiangsu KGI Biotechnology Co., Ltd.
PE-CD11b (cat. no. 301306) and FITC-CD14 (cat. no. 301804)
fluorescent-conjugated antibodies were purchased from BioLegend,
Inc. TRIzol® reagent was purchased from Thermo Fisher
Scientific, Inc. Trichloromethane was purchased from Sinopharm
Chemical Reagent Co., Ltd.
The committed differentiation from
THP-1 cells into monocytes/macrophages induced by PMA + LPS +
IFN-γ
THP-1 cells were grown in RPMI-1640 complete medium
containing 10% FBS at 37°C and 5% CO2 in a six-well
plate. When the cells grew to the logarithmic phase, the
appropriate amount of THP-1 cell suspension and PMA + LPS + IFN-γ
solution were added to 96-well plates. The final concentration of
each well cell was 1×105/ml and the final concentration
of PMA + LPS + IFN-γ induction mixture was 10, 100 and 20 µg/l. The
cells were cultured at 37°C and 5% CO2 for 48 h. In
addition to observing the cell growth density directly under an
inverted microscope, 10 µl Cell Counting Kit-8 (CCK-8) solution was
added to each well followed by incubation at 37°C for 2 h. The
optical density (OD) values at 450 nm were measured using a
microplate reader.
Detection of CD11b and CD14
differentiation antigens using flow cytometry
THP-1 cells induced by PMA + LPS + IFN-γ for 48 h
were collected into flow tubes, suspended with PBS, washed twice by
centrifugation at 200 × g for 3 min at 4°C and the cell
concentration was adjusted to 1×106 cells/ml. Each tube
was supplemented with 100 µl PBS and 5 µl PE-labeled mouse
anti-human CD11b (cat. no. 301306, 1:1,000, BioLegend, Inc.) and
FITC-labeled mouse anti-human CD14 fluorescent antibody (cat. no.
301804, 1:1,000, BioLegend, Inc.) were added, respectively,
followed by incubation at 4°C for 30 min in the dark. The cells
were resuspended in 200 µl PBS and fixed in 1% paraformaldehyde at
4°C for 20 min. The expression levels of CD11b and CD14 in the
different treatment groups were analyzed on a FACSVerse (BD
Biosciences) flow cytometer. Isotypic rat IgG (BioLegend, Inc.) was
also used to examine non-specific binding. The experiment was
repeated three times. According to the different detection
antigens, they were divided into the CD11b PBS control group, CD11b
PMA + LPS + IFN-γ experimental group, CD14 PBS control group and
the CD14 PMA + LPS + IFN-γ experimental group.
Detection of protein expression of
c-myc using western blot analysis
The cells in the control and experimental groups
were collected and washed twice with pre-cooled PBS. Total protein
was extracted and the protein concentration was determined by BCA
assay; 5X protein loading buffer was added and the cells were
denatured by boiling at 95°C for 10 min. SDS-PAGE (volume fraction
10%) electrophoresis was performed with 20 µg protein. The
separated proteins were transferred onto a PVDF membrane and
blocked with 5% skimmed milk for 90 min at room temperature. The
corresponding primary antibodies, c-myc (1:800) and β-actin
(1:2,000) were then added and incubated at 4°C overnight; this was
followed by the addition of goat anti-rabbit IgG antibody
(1:20,000) and incubation at room temperature for 90 min. The
membranes were then washed with 1X TBST including 0.2% Tween-20 for
30 min and finally washed for 30 min for imaging observation using
an ECL kit (Shandong Sparkjade Scientific Instruments Co., Ltd.).
ImageJ v1.51j8 was used for densitometry (National Institutes of
Health). The experiment was repeated three times.
Bioinformatics cross analysis of
upstream miRNAs targeting c-myc
On the basis of confirming the reduced expression of
c-myc in THP-1 cells following the induction of differentiation,
the potential miR-let-7 family spliceosomes with targeted binding
to c-myc were screened using TargetScan (Targetscan.org), StarBase
database (starbase.sysu.edu.cn/), a literature research (PubMed)
and differential microarray detection (Agilent miRNA Chip). For
differential microarray detection using the Agilent miRNA Chip
assay, the leukemic cells were collected before and 48 h following
induction, respectively and Affymetrix 3′ IVT expression profiling
microarray data were analyzed to obtain miRNAs with a higher
likelihood of binding to c-myc and a negative correlation with
c-myc. At the same time, to further demonstrate the association
between the expression of candidate miRNAs and c-myc, seven
different time points during the induction of differentiation of
THP-1 cells were randomly selected and the expression levels of
mRNAs were detected using reverse transcription-quantitative (RT-q)
PCR; correlations were analyzed using Pearson's correlation
analysis.
Detection of changes in gene
expression using RT-qPCR
Total RNA was extracted using TRIzol®
reagent (Thermo Fisher Scientific, Inc.). The concentration of
total RNA was measured using an ultramicro nucleic acid protein
analyzer (Thermo Fisher Scientific, Inc.). cDNA was synthesized
according to the instructions provided with the PrimeScript RT
reagent kit with gDNA Eraser (Perfect Real-Time). For reverse
transcription, samples were incubated in an Eppendorf PCR system at
42°C for 30 min, then at 90°C for 5 min and at 5°C for 5 min. cDNA
was used as a template for PCR amplification. The primers used were
as follows: miR-let-7c-5p sense, 5′-CGTCATCCTGAGGTAGTAGGTTGT-3′ and
antisense, 5′-TATGGTTTTGACGACTGTGTGAT-3′; U6 sense,
5′-AGAGAAGATTAGCATGGCCCCTG-3′ and antisense,
5′-AGTGCAGGGTCCGAGGTATT-3′. These two primers were designed by
stem-loop method. The downstream and reverse transcription primers
of stem-loop method were artificially added (Sangon Biotech Co.,
Ltd.). c-myc sense primer 5′-CCCCTACCCTCTCAACGACA-3, antisense
5′-CTTCTTGTTCCTCCTCAGAGTCG-3′ and GAPDH sense,
5′-ACAACTTTGGTATCGTGGAAGG-3′ and antisense,
5′-GCCATCACGCCAGTTTC-3′. Real-time fluorescence quantitative
amplification reaction was performed according to the instructions
provided with the TB Green Premix Ex Taq (Tli RNaseH Plus) kit. PCR
was conducted under the following conditions: 10 sec at 95°C; 40
cycles of 5 sec at 60°C and 10 sec at 72°C; 34 sec at 60°C.
Relative quantitative analysis was performed using the
2−ΔΔCq method (28).
Secondly, the transfection efficiency of miR-let-7c-5p mimics was
also detected and confirmed using RT-qPCR for miR-let-7c-5p
expression.
Dual-luciferase reporter gene
analysis
The bioinformatics prediction website was used to
predict the 3′UTR binding sequence of miR-let-7c-5p to c-myc. Dual
luciferase reporter genes were used to detect the targeted binding
of miR-let-7c-5p on the 3′UTR of c-myc. Briefly, the wild-type (WT)
plasmid, pmirGLO-c-myc-wt and the mutant plasmid, pmirGLO-c-myc-MUT
were constructed by Biosune Biotechnology (shanghai) Co.,Ltd. The
WT plasmid, the mutant (MUT) plasmid and the miR-let-7c-5p mimics
(UGAGGUAGUAGGUUGUAUGGUU) or NC mimics (UUCUCCGAACGUGUCACGUTT) were
co-transfected into the cells by transfection reagent kit
(jetPRIME; Polyplus). The activity of luciferase was determined
using the Dual-Luciferase Reporter Assay System (Envision;
PerkinElmer, Inc.) following 48 h of culture and Renilla
luciferase was used as an internal control.
Cell transfection
After using FAM fluorescence labeled si-NC as
negative parallel control to determine transfection efficiency,
THP-1 cells were transfected in accordance with the instructions
and the appropriate amount of si-c-myc and its corresponding
negative control were mixed with the transfection reagent
Lipofectamine 2000® transfection kit (Invitrogen; Thermo
Fisher Scientific, Inc.) to form the transfection complex, which
was added to the six-well plate to inoculate the cells for 48 h.
si-c-myc: sense 5′-CCACACAUCAGCACAACUATT-3′, antisense
5′-UAGUUGUGCUGAUGUGUGGTT-3′ was co-selected for synthesis. At the
same time, si-NC: sense 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense
5′-ACGUGACACGUUCGGAGAATT-3′ was taken as the negative control. The
two were synthesized by BioSune Biotechnology Co., Ltd.
Statistical analysis
GraphPad Prism 8 software (GraphPad Software, Inc.)
was used for data processing and the Shapiro-Wilk test was used to
assess normal distribution of the data. Each experiment was
repeated three times and the measurement data are expressed as the
mean ± standard deviation. Comparisons between two groups were
performed using an independent samples t-test and one-way ANOVA was
used for multiple-group comparisons. The Bonferroni test was used
as the post-hoc test following one-way ANOVA. Pearson's correlation
analysis was used to determine the correlation between
miR-let-7c-5p and c-myc. All data were analyzed using two-tailed
tests. P<0.05 was considered to indicate a statistically
significant difference.
Results
Proliferation of THP-1 cells is
inhibited by PMA + LPS + IFN-γ
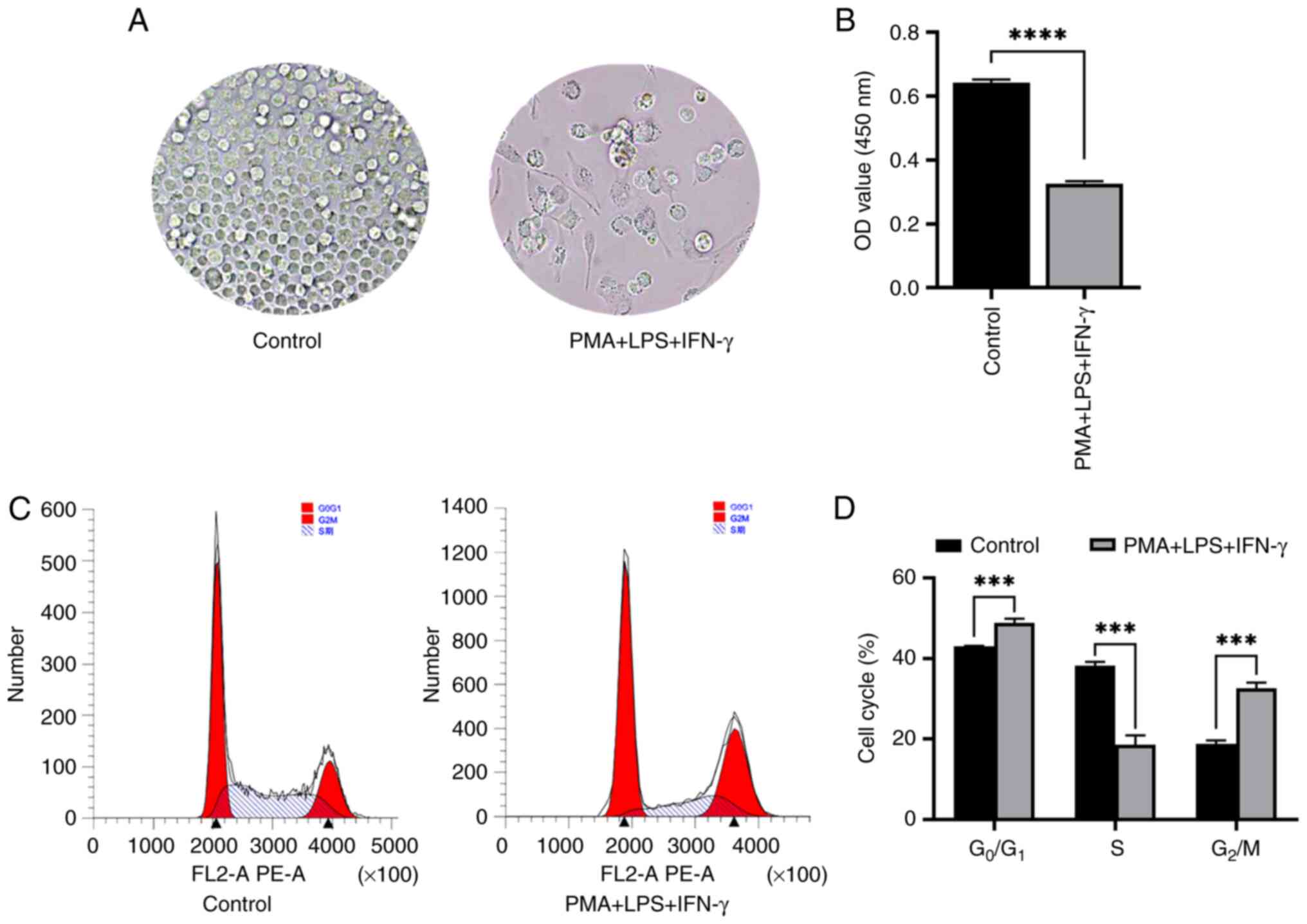
The changes in the growth density of THP-1 cells
induced by PMA + LPS + IFN-γ for 48 h were observed under an
inverted microscope. The results revealed that the cell density in
the PMA + LPS + IFN-γ experimental group was significantly lower
than that in the PBS control group and the cell growth gradually
changed from growth in suspension to adherent growth (Fig. 1A). The results of CCK-8 assay
revealed that the proliferative ability of the cells in the PMA +
LPS + IFN-γ experimental group was lower compared with those in the
PBS control group (Fig. 1B). Flow
cytometry for cell cycle distribution demonstrated that the number
of cells in the G0/G1 phase in the control
group was lower than that in the PMA + LPS + IFN-γ group. In
addition, the number of cells in the S phase in the PBS control
group was higher than that in the PMA + LPS + IFN-γ group and the
number of cells in the G2/M phase in the PBS control
group was also lower than that in the PMA + LPS + IFN-γ group
(Fig. 1C and D). These experimental
data demonstrated that the proliferation of the THP-1 cells was
inhibited by PMA + LPS + IFN-γ.
PMA + LPS + IFN-γ induces the
committed differentiation of THP-1 cells into
monocytes/macrophages
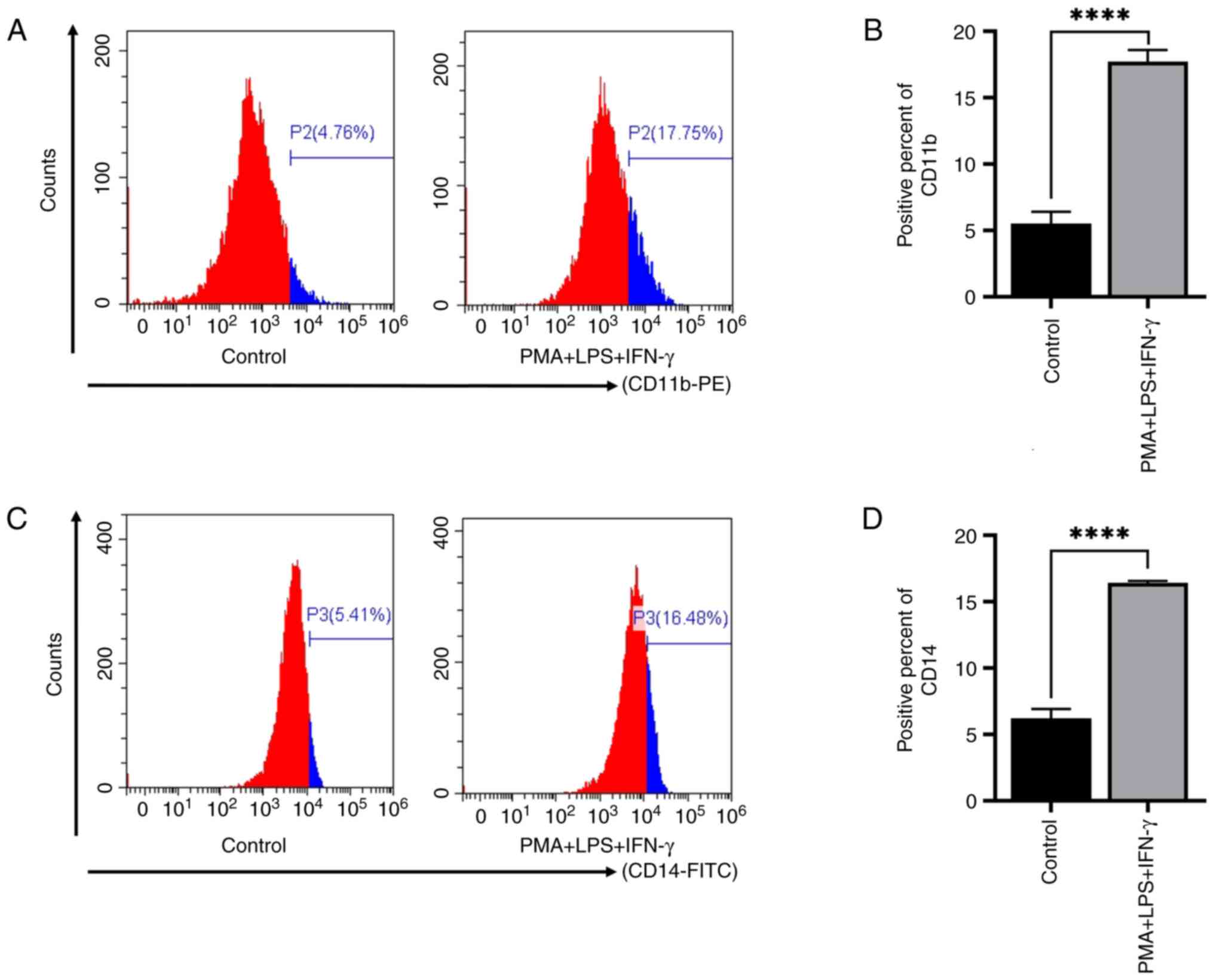
According to the data from the literature research
and the results of the preliminary experiments, PMA + LPS + IFN-γ
was used to induce the committed differentiation of THP-1 cells
into monocytes/macrophages. The results of flow cytometry revealed
that the average positive rates of CD11b in the PMA + LPS + IFN-γ
group were markedly increased compared with those in the PBS
control group (Fig. 2A and B). The
expression rates of CD14 in the PMA + LPS + IFN-γ group were also
increased compared with those in the PBS control group (Fig. 2C and D). These results indicated
that PMA + LPS + IFN-γ induced the differentiation of THP-1 cells
into monocytes/macrophages.
c-myc expression is downregulated
during the monocyte/macrophage differentiation of THP-1 cells
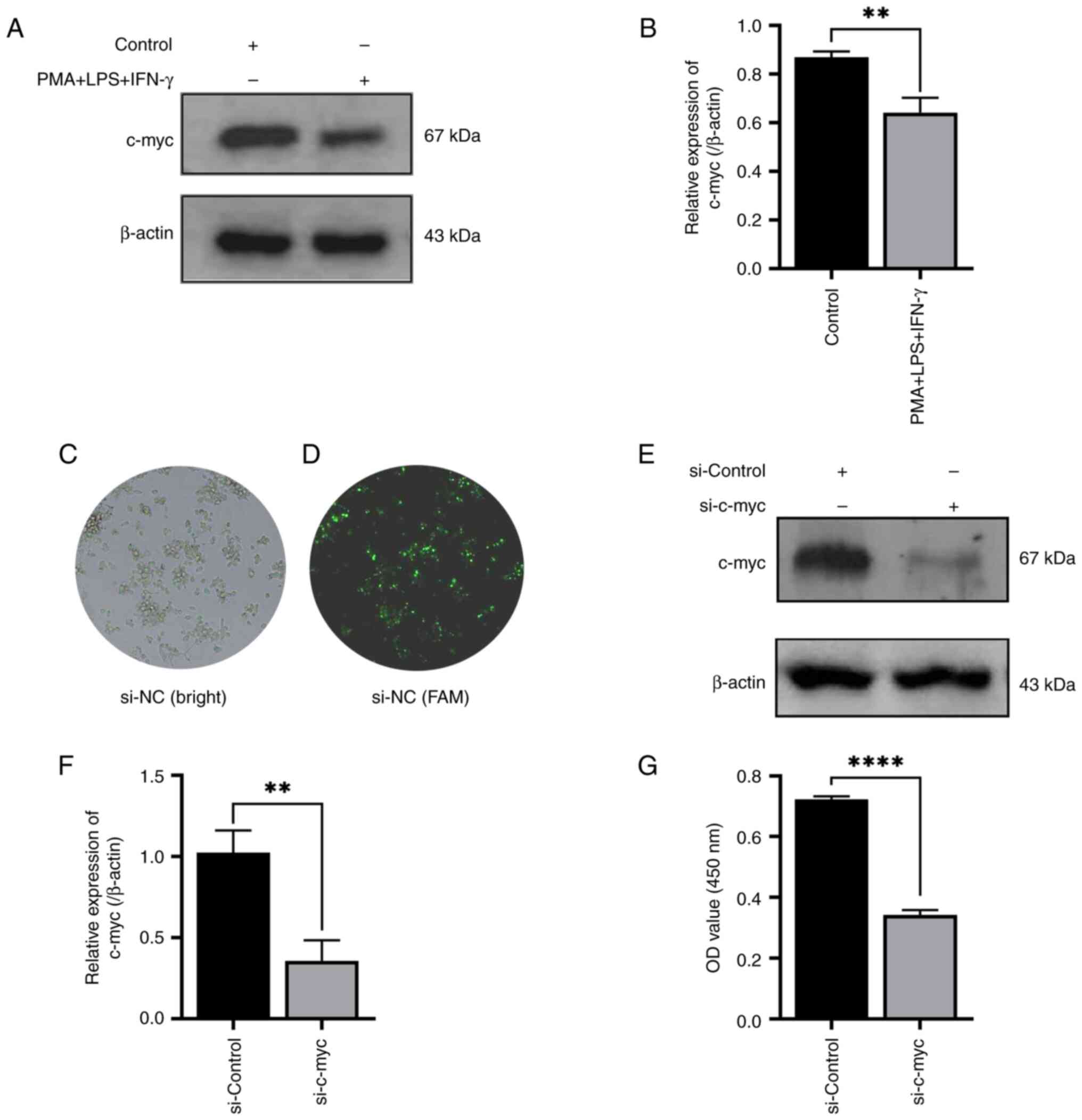
As c-myc protein was involved in the differentiation
of leukemic cells into granulocytes, the present study aimed to
further investigate whether c-myc gene was also involved in the
macrophage differentiation of leukemic cells and whether this type
of change affects the proliferation and differentiation process of
leukemic cells. The committed differentiation into
monocytes/macrophages was obtained by stimulating the THP-1 cells
with PMA + LPS + IFN-γ for 48 h. The results revealed that the
relative expression levels of c-myc were downregulated following
exposure to PMA + LPS + IFN-γ, as compared with the PBS control
group (Fig. 3A and B). Furthermore,
to elucidate the role of c-myc in the aforementioned committed
differentiation of leukemic cells, the expression of c-myc was
knocked down using siRNA (si-c-myc); FAM fluorescence labeled si-NC
as a negative parallel control to observe the transfection
efficiency through fluorescence observation and confirm the
successful transfection (Fig 3C and
D). The results indicated that si-c-myc significantly inhibited
the protein expression of c-myc in THP-1 cells as compared with the
si-control group (Fig. 3E and F).
The results of CCK-8 assay also indicated that the proliferation of
THP-1 cells was significantly inhibited following transfection with
si-c-myc (Fig. 3G).
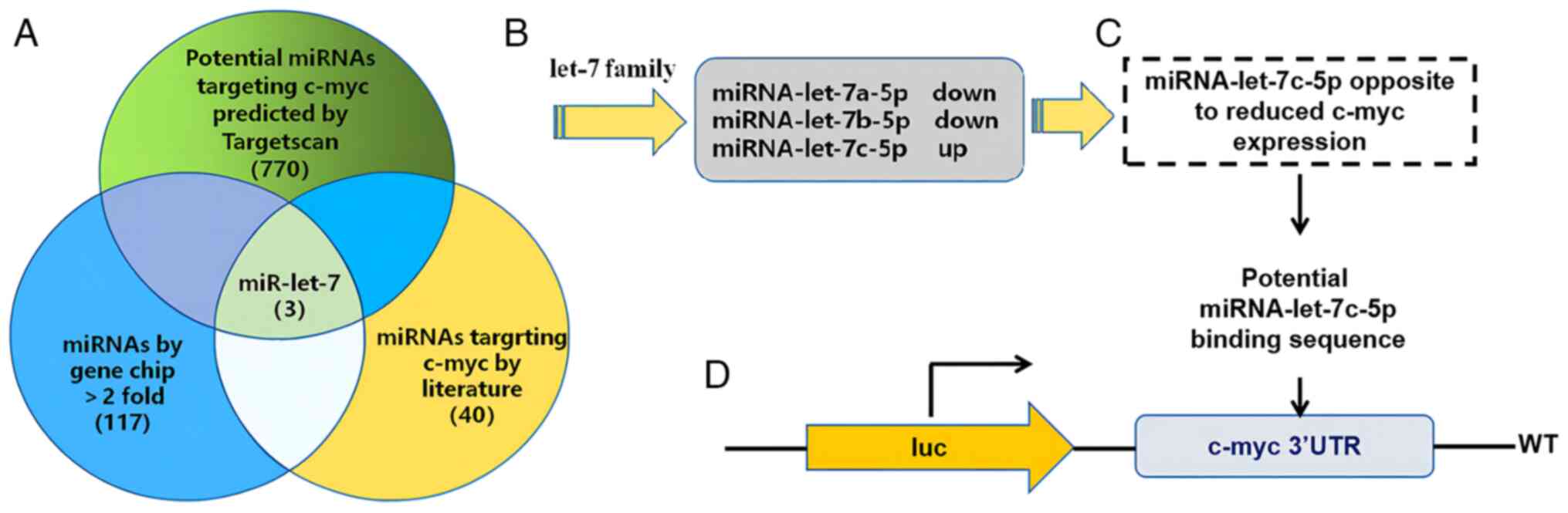
c-myc upstream miRNA bioinformatics
cross analysis
As it was confirmed that the expression of c-myc was
reduced in THP-1 cells following the induction of differentiation,
the potential miR-let-7 family spliceosomes with targeted binding
to c-myc were screened using TargetScan, a PubMed literature review
and differential microarray detection. Following the
cross-sectional analysis of the aforementioned three data sources
(Fig. 4A), it was found that
miR-let-7a-5p and miR-let-7b-5p expression was decreased, whereas
miR-let-7c-5p expression was elevated (Fig. 4B). miR-let-7c-5p was identified as a
possible candidate miRNA for the targeted regulation of c-myc,
based on the fact that miRNAs target downstream genes to suppress
their expression. A total of seven different time points at which
PMA + LPS + IFN-γ induced the differentiation of THP-1 cells were
randomly selected and the expression levels of miR-let-7c-5p and
c-myc were detected using RT-qPCR. Pearson's correlation analysis
was performed to confirm the negative correlation between
miR-let-7c-5p and c-myc (Fig. 4C);
the correlation was statistically significant. Following target
prediction using TargetScan, the c-myc gene 3′UTR region was found
to have a sequence basis for miR-let-7c-5p targeted binding
(Fig. 4D).
c-myc is a direct target gene of
miR-let-7c-5p
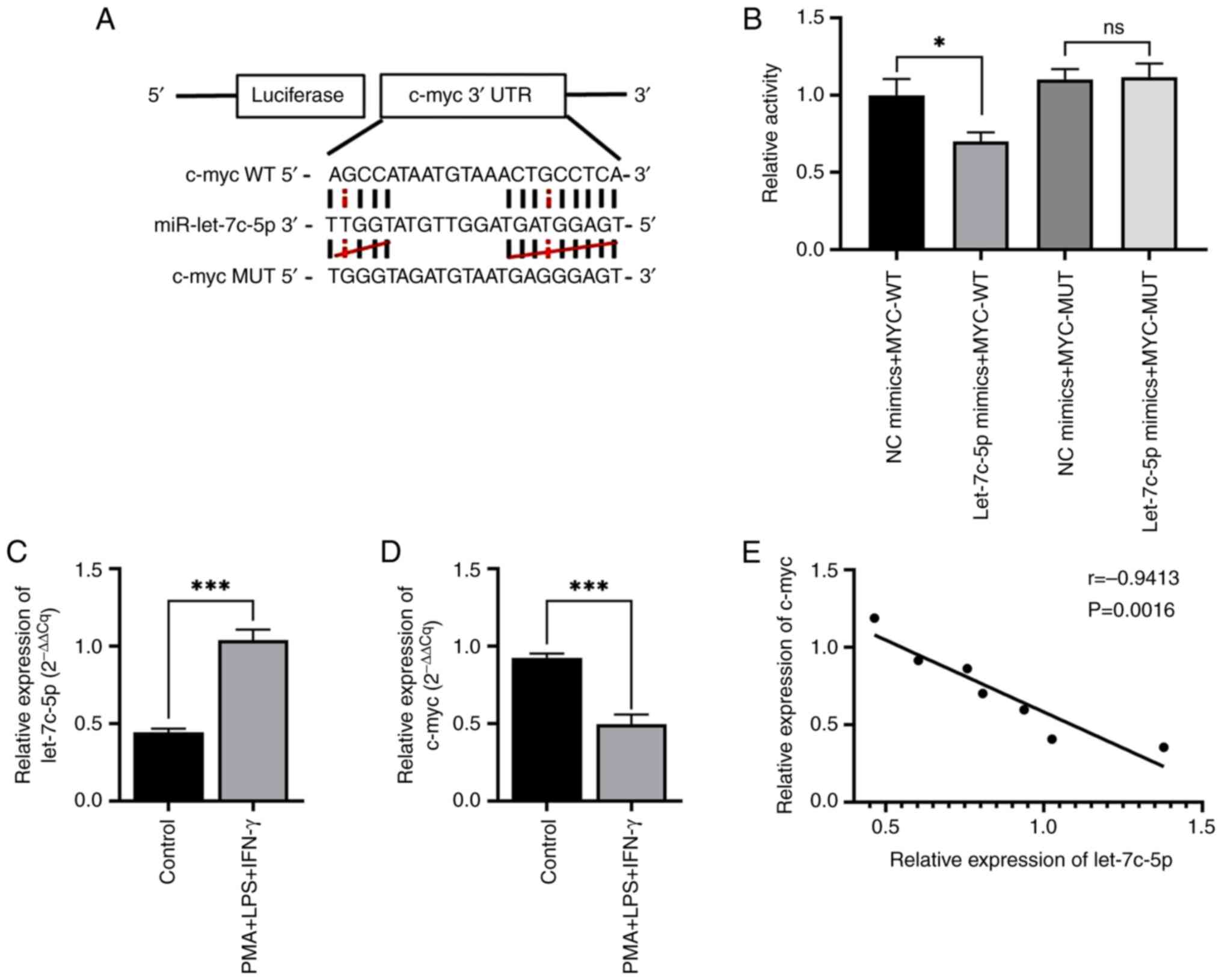
A binding region between miR-let-7c-5p and c-myc was
identified using the StarBase database; the paired sites were then
predicted using the TargetScan database (Fig. 5A). To examine the direct binding of
miR-let-7c-5p with c-myc, plasmids with WT or MUT 3′UTR of c-myc
were constructed, which were co-transfected with miR-let-7c-5p
mimics or NC for dual-luciferase reporter assay. The results of
dual luciferase reporter gene assay revealed that the luciferase
activity of the miR-let-7c-5p mimics + c-myc-WT group was
significantly lower than that of the mimics NC + c-myc-WT group.
There was no significant difference in the luciferase activity of
the miR-let-7c-5p mimics + c-myc-MUT group and the mimics NC +
c-myc-MUT group. It was found that miR-let-7c-5p mimics
significantly decreased the luciferase reporter activity of the WT
3′UTR plasmid of c-myc, although it did not affect that of the MUT
3′UTR plasmid. These data indicated that c-myc was a direct target
gene of miR-let-7c-5p (Fig. 5B). At
48 h following the induction of the differentiation of THP-1 cells
in vitro, the relative expression levels of miR-let-7c-5p
were examined using RT-qPCR. It was found that the expression of
miR-let-7c-5p in the PMA + LPS + IFN-γ group was upregulated
compared with that in the control group (Fig. 5C). However, the relative expression
levels of the c-myc gene in the PMA + LPS + IFN-γ group were lower
than those in the control group (Fig.
5D). Following the random selection of seven time points for
miR-let-7c-5p vs. c-myc gene expression, Pearson's correlation
analysis indicated that the two presented a negative correlation
(Fig. 5E).
miR-let-7c-5p is involved in the
induction of the differentiation of THP-1 cells and in regulating
the expression of c-myc
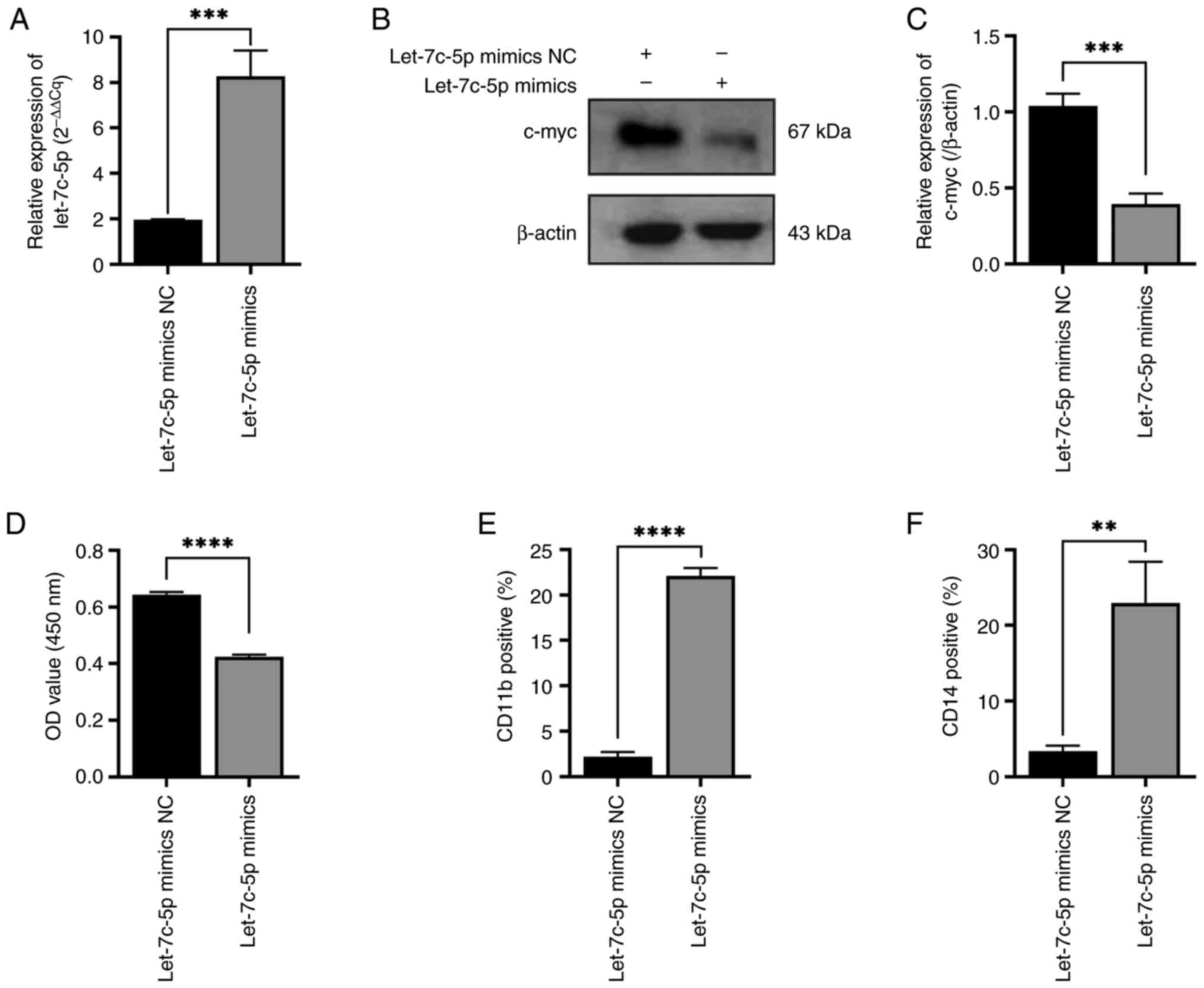
To demonstrate whether miR-let-7c-5p is involved in
the committed differentiation of THP-1 cells into
monocytes/macrophages and in regulating the expression of c-myc,
miR-let-7c-5p mimics were transfected into the THP-1 cells and the
transfection efficiency of miR-let-7c-5p mimics was confirmed using
RT-qPCR assay as indicated in Fig.
6A. The expression level of c-myc was found to be decreased in
the miR-let-7c-5p mimics group compared with the miR-let-7c-5p
mimics NC group (Fig. 6B and C).
The proliferation of the THP-1 cells transfected with miR-let-7c-5p
mimics was decreased compared with the cells transfected with
miR-let-7c-5p mimics NC (Fig. 6D).
The positive rates of CD11b expression in the miR-let-7c-5p mimics
groups were higher than those in the miR-let-7c-5p mimics NC group
(Fig. 6E). The positive rates of
CD14 expression in the miR-let-7c-5p mimics group were also higher
than those in the miR-let-7c-5p mimics NC group (Fig. 6F).
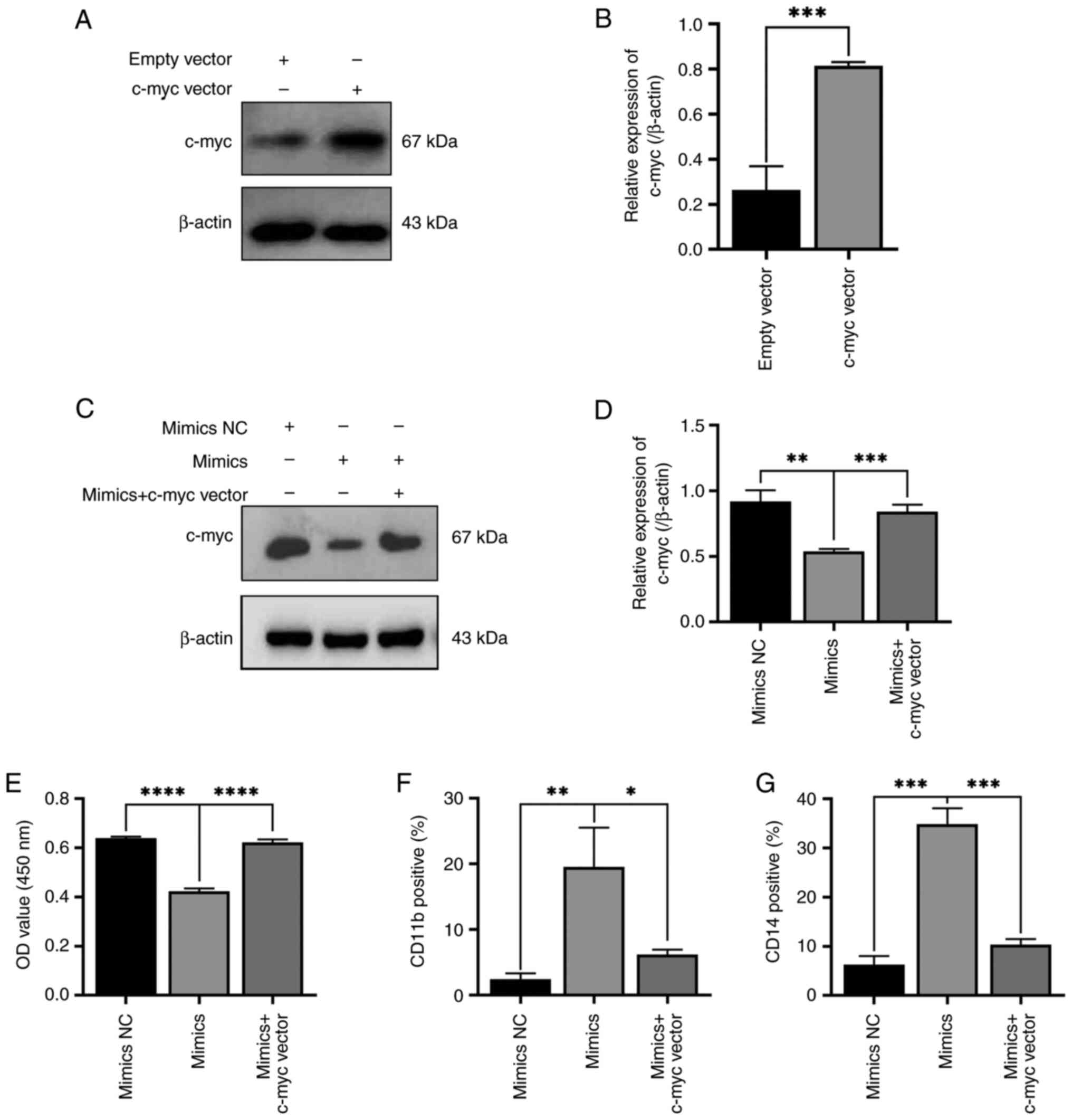
The rescue experiment of miR-let-7c-5p
on differentiation of THP-1 cells and reversed by transfection of
c-myc vector
As it was demonstrated that miR-let-7c-5p mimics
play a role in promoting the differentiation of THP-1 cells by
reducing c-myc expression, the present study aimed to further
demonstrate whether miR-let-7c-5p promotes the committed
differentiation leukemic cells into monocytes/macrophages through
the c-myc pathway. Thus, a rescue experiment was performed.
Firstly, parallel transfection was performed using c-myc vector and
its empty vector and western blotting was used to detect the
expression level of c-myc protein after transfection, verifying the
success and efficiency of transfection (Fig. 7A and B). miR-let-7c-5p mimics and
c-myc overexpression plasmid were co-transfected into THP-1 cells.
It was found that the expression of c-myc in the mimics NC group
was significantly higher than that in the miR-let-7c-5p mimics
group and its expression in the miR-let-7c-5p mimics + c-myc vector
group was significantly higher than that in the miR-let-7c-5p
mimics group. No significant difference in c-myc expression was
found between the miR-let-7c-5p mimics NC group and the
miR-let-7c-5p mimics + c-myc vector group (Fig. 7C and D). As regards the
proliferation of THP-1 cells, cell proliferation in the
miR-let-7c-5p mimics NC group was higher compared with
miR-let-7c-5p mimics group and that in the miR-let-7c-5p mimics
group was lower compared with the miR-let-7c-5p mimics + c-myc
vector group. There was no significant difference in cell
proliferation between the miR-let-7c-5p mimics NC group and the
miR-let-7c-5p mimics + c-myc vector group (Fig. 7E). The positive rates of CD11b
expression in the miR-let-7c-5p mimics NC group were lower than
those in the miR-let-7c-5p mimics group and those in the
miR-let-7c-5p mimics group were higher than those in the
miR-let-7c-5p mimics + c-myc vector group; in addition, the
positive rates of CD11b expression in the miR-let-7c-5p mimics NC
group were slightly lower than those in the miR-let-7c-5p mimics +
c-myc vector group (Fig. 7F). The
positive rates of CD14 expression in the miR-let-7c-5p mimics NC
group were lower than those in the miR-let-7c-5p mimics group and
those in the miR-let-7c-5p mimics group were higher than those in
the miR-let-7c-5p mimics + c-myc vector group; in addition, the
positive rates of CD14 expression in the miR-let-7c-5p mimics NC
group were slightly lower than those in the miR-let-7c-5p mimics +
c-myc vector group (Fig. 7G). These
results demonstrated that the effects of miR-let-7c-5p mimics on
the proliferation and differentiation of THP-1 cells were markedly
reversed by the overexpression of c-myc. These data indicated that
miR-let-7c-5p promoted the differentiation of monocytes/macrophages
by suppressing c-myc.
Discussion
As regards the development of normal hematopoietic
cells, they can undergo committed differentiation and eventually
differentiate into mature granulocytic, monocytic, erythroid, or
megakaryocytic cells in strict accordance with the inherent
procedure (1). It has been proved
that a number of transcription factors are involved in this process
of differentiation and determine the genetic program of each mature
phenotype (29). Among them,
increasing evidence suggests that c-myc is one of the main
transcription factors involved. c-myc is a necessary factor for
maintaining a balance between self-renewal and differentiation of
hematopoietic stem cells. It is widely hypothesized that
overexpression of c-myc is mainly beneficial for promoting cell
proliferation and has an inhibitory effect on the directed
differentiation of hematopoietic cells (30). In various malignant tumor cells, the
expression level of myc gene often shows an overexpression state,
which is one of the important causes of the occurrence and
development of malignant tumors (31). In acute lymphocytic leukemia and
myeloid leukemia and other hematological tumors, c-myc also
exhibits overexpression and is closely related to disease
progression. The results of a number of in vitro studies
also confirm that c-myc mainly manifests as an oncogene,
participating in the proliferation, differentiation and apoptosis
processes of leukemia cells (32,33).
Therefore, controlling the expression of c-myc gene can interfere
with the proliferation and differentiation process of leukemia
cells. Among them, macrophage is an important immune effector cell,
a group of highly heterogeneous cells with two types of
macrophages, such as M1 and M2 macrophages (6–10). For
patients with tumor and leukemia, it is more important to induce
macrophages to differentiate into M1 macrophages, which are
associated with anti-tumor activity (34,35).
As aforementioned, c-myc plays an important role in modulating the
differentiation of leukemia cells, therefore it was hypothesized
that c-myc may also be involved in the maturation and
differentiation of leukemia cells into monocytes and macrophages.
However, the specific molecular mechanism of c-myc this committed
differentiation remains to be elucidated. In the present study, to
demonstrate the role of c-myc in macrophage polarization, THP-1
cells were induced to differentiate into more matured macrophages
and found the expression of c-myc in THP-1 leukemia cells was
downregulated after being induced to differentiate into
macrophages. These results suggested that the downregulation of
c-myc contributed to the inhibition of proliferation and induction
of differentiation of THP-1 cells.
In recent years, ncRNAs have been found to be
closely associated with development of leukemia. Among them, miRNAs
are often involved in regulation of target genes by acting as
oncogenes or tumor suppressor genes (25), including pathological processes such
as abnormal proliferation, differentiation, or apoptosis of
leukemic cells (26,27). For example, about 50% of miRNAs are
located near or within cancer translocation genes and can inhibit
or promote tumorigenesis by downregulating the expression of
oncogenes or tumor suppressor gene (27). miRNAs also play an important role in
the pathogenesis of leukemia and it has been demonstrated that
miRNA expression profiling can be used as a biomarker for the
diagnosis, prognosis and efficacy of leukemia patients (27,36–38).
In recent years, the regulatory relationship between ncRNA and
c-myc has attracted more attention: 25 miRNAs of 20 different
families so far have been found to regulate c-myc, as well as 33
miRNAs regulated by c-myc and most of the above miRNAs tend to bind
the 3′UTR of the c-myc gene in a traditional manner and exert their
regulatory effects (39,40). Of which, miR-let-7 family members
are also involved in the proliferation and differentiation of
leukemic cells (41,42). Some studies have shown that ncRNAs
play an important role in the transcription and translation
regulation of this gene, with some ncRNAs directly interacting with
c-myc (43), for example, as to the
miRNA-let-7 family, Wong et al (44) demonstrated that miRNA let-7
suppresses nasopharyngeal carcinoma cells proliferation through
downregulating c-myc expression. Sampson et al (45) found that miRNA let-7a downregulates
myc and reverts myc-induced growth in Burkitt lymphoma cells.
Buechner et al (46) showed
that tumor-suppressor miRNA let-7 could target the proto-oncogene
myc and inhibit cell proliferation in MYCN-amplified neuroblastoma.
Lan et al (47) indicated
that Hsa-let-7g inhibits proliferation of hepatocellular carcinoma
cells by downregulation of c-myc and upregulation of p16. He et
al (48) demonstrated that
let-7a miRNA protects against the growth of lung carcinoma by
suppression of k-Ras and c-myc in nude mice. Zhang et al
(49) found miRNA let-7g inhibited
hypoxia-induced proliferation of PASMCs via
G0/G1 cell cycle arrest by targeting c-myc.
However, there are few reports on the role of c-myc in the directed
differentiation of leukemia cells into macrophages.
On the basis of the aforementioned results on the
downregulation of c-myc in committed differentiation of THP-1 cells
towards macrophages and the role of different members of let-7
family in targeting modulation of c-myc according to the results
reported in the literature, it was hypothesized that the let-7
family mediates the directed differentiation of leukemia cells into
macrophages through targeted regulation of c-myc. The present study
detected and predicted the upstream miR-let-7 family spliceosomes
with the potential for targeting to c-myc by crosstalk analysis by
TargetScan, PubMed literature research and differential microarray
detection. After intersection analysis of the above three data
sources, it was found that miR-let-7a-5p and miR-let-7b-5p
expression was decreased, while miR-let-7c-5p expression was
increased following induction of differentiation. The miR-let-7c-5p
spliceosome was identified as a possible candidate miRNA for
targeted regulation of c-myc, based on the fact that miRNAs often
target to suppress the expression of downstream genes. At the same
time, the relative expression levels of miR-let-7c-5p and c-myc
gene in THP-1 cells induced to differentiate into macrophages and
the dual luciferase reporter gene experiment confirmed that
miR-let-7c-5p could bind 3′UTR of c-myc and regulate its promoter
activity, which was involved in the committed macrophage
differentiation of THP-1 cells.
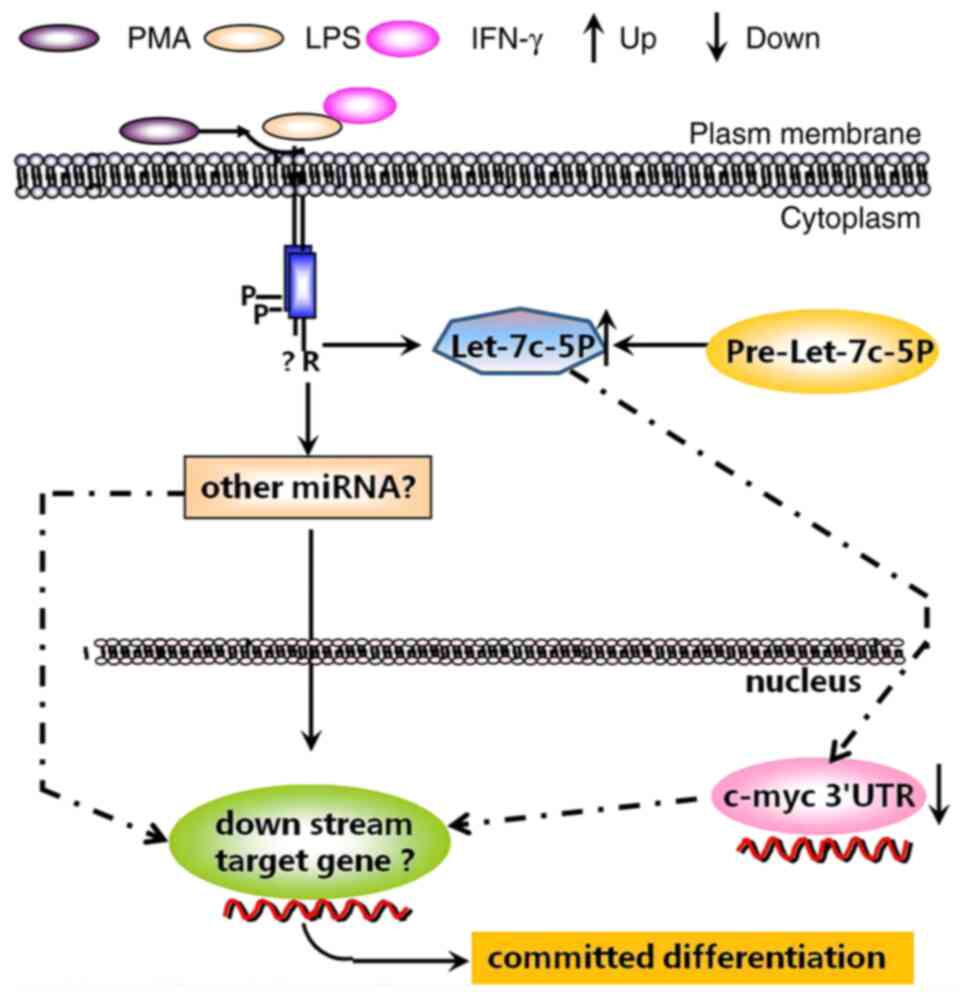
In summary, the present study revealed for the first
time that expression of miR-let-7c-5p was upregulated in THP-1
cells induced to differentiate into macrophages, which could
downregulate the expression of c-myc by targeting c-myc 3′UTR
(Fig. 8). The miR-let-7c-5p/c-myc
axis may be a potential target in macrophage polarization and
highlights an improved understanding for potential mechanism of
pathogenesis and progression of leukemia. As the in vitro
experimental results have preliminarily confirmed the important
role of miR-let-7c-5p in targeted monocyte/macrophage
differentiation of leukemia cells, the focus of the next clinical
research on miR-let-7c-5p will to detect the potential role of
miR-let-7c-5p in promising biomarkers for prognosis of leukemia
patients, or to verify whether it can serve as a biological
indicator for independent risk assessment. More importantly, will
be to further explore its potential as a therapeutic target for
leukemia through in vitro and in vivo experiments.
Meanwhile, it should be noted that since the regulation of
committed differentiation of leukemia cells is complicated and the
role of miR-let-7c-5p/c-myc axis in modulation of differentiation
of leukemia cells still requires further in-depth research. In
addition, in-depth research on downstream target genes of
miR-let-7c-5p/c-myc axis not only enriches the action pattern of
this signal axis, but also has a guiding role in increasing
effective targets for targeted intervention therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the ‘Twelfth Five-Year’
National Science and Technology Support Program (grant no.
2013BAI07B02), Natural Science Foundation of China (grant no.
81573467), Natural Science Foundation of Shandong (grant nos.
ZR2020QH160 and ZR2021MH080), The Foundation for Jinan's Clinical
Science and Technology Innovation (grant no. 202134001).
Cultivation Fund of the first affiliated hospital of Shandong First
Medical University (Shandong Qianfoshan Hospital; grant no.
QIPY2020NSFC0819).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GSJ, KHB, QL, RJS and YFW made substantial
contributions to the conception and design and also critically
reviewed the study. RJS, YFW, CZW, YHW and PCD performed the
experiments. RJS, YFW, CZW and XLS analyzed the data and wrote the
manuscript. GSJ, KHB and QL confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Carroll D and St Clair DK: Hematopoietic
stem cells: Normal versus malignant. Antioxid Redox Signal.
29:1612–1632. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khwaja A, Bjorkholm M, Gale RE, Levine RL,
Jordan CT, Ehninger G, Bloomfield CD, Estey E, Burnett A,
Cornelissen JJ, et al: Acute myeloid leukaemia. Nat Rev Dis
Primers. 2:160102016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bruserud O and Gjertsen BT: New strategies
for the treatment of acute myelogenous leukemia: Differentiation
induction-present use and future possibilities. Stem Cells.
18:157–165. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stubbins RJ and Karsan A: Differentiation
therapy for myeloid malignancies: Beyond cytotoxicity. Blood Cancer
J. 11:1932021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gocek E and Marcinkowska E:
Differentiation therapy of acute myeloid leukemia. Cancers (Basel).
3:2402–2420. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sica A and Mantovani A: Macrophage
plasticity and polarization: In vivo veritas. J Clin Invest.
122:787–795. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sica A, Erreni M, Allavena P and Porta C:
Macrophage polarization in pathology. Cell Mol Life Sci.
72:4111–4126. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Walajtys-Rode E and Dzik JM:
Monocyte/macrophage: NK cell cooperation-old tools for new
functions. Results Probl Cell Differ. 62:73–145. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gurski CJ and Dittel BN: Myeloperoxidase
as a marker to differentiate mouse monocyte/macrophage subsets. Int
J Mol Sci. 23:82462022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao X, Di Q, Liu H, Quan J, Ling J, Zhao
Z, Xiao Y, Wu H, Wu Z, Song W, et al: MEF2C promotes M1 macrophage
polarization and Th1 responses. Cell Mol Immunol. 19:540–553. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang LX, Zhang SX, Wu HJ, Rong XL and Guo
J: M2b macrophage polarization and its roles in diseases. J Leukoc
Biol. 106:345–358. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yunna C, Mengru H, Lei W and Weidong C:
Macrophage M1/M2 polarization. Eur J Pharmacol. 877:1730902020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang X, Li Y, Fu M and Xin HB: Polarizing
macrophages in vitro. Methods Mol Biol. 1784:119–126. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Draijer C, Penke L and Peters-Golden M:
Distinctive effects of GM-CSF and M-CSF on proliferation and
polarization of two major pulmonary macrophage populations. J
Immunol. 202:2700–2709. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jaguin M, Houlbert N, Fardel O and
Lecureur V: Polarization profiles of human M-CSF-generated
macrophages and comparison of M1-markers in classically activated
macrophages from GM-CSF and M-CSF origin. Cell Immunol. 281:51–61.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qian F, Deng J, Lee YG, Zhu J, Karpurapu
M, Chung S, Zheng JN, Xiao L, Park GY and Christman JW: The
transcription factor PU.1 promotes alternative macrophage
polarization and asthmatic airway inflammation. J Mol Cell Biol.
7:557–567. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu L, Peng S, Duan M, Liu C, Li L, Zhang
X, Ren B and Tian H: The role of C/EBP homologous protein (CHOP) in
regulating macrophage polarization in RAW264.7 cells. Microbiol
Immunol. 65:531–541. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie J, Wu X, Zheng S, Lin K and Su J:
Aligned electrospun poly(L-lactide) nanofibers facilitate wound
healing by inhibiting macrophage M1 polarization via the JAK-STAT
and NF-κB pathways. J Nanobiotechnology. 20:3422022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vergadi E, Ieronymaki E, Lyroni K,
Vaporidi K and Tsatsanis C: Akt signaling pathway in macrophage
activation and M1/M2 polarization. J Immunol. 198:1006–1014. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Juhas U, Ryba-Stanislawowska M, Szargiej P
and Mysliwska J: Different pathways of macrophage activation and
polarization. Postepy Hig Med Dosw (Online). 69:496–502. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pello OM, De Pizzol M, Mirolo M, Soucek L,
Zammataro L, Amabile A, Doni A, Nebuloni M, Swigart LB, Evan GI, et
al: Role of c-MYC in alternative activation of human macrophages
and tumor-associated macrophage biology. Blood. 119:411–421. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pello OM: Macrophages and c-MYC cross
paths. Oncoimmunology. 5:e11519912016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang G, Albihn A, Tang T, Tian Z and
Henriksson M: Role of Myc in differentiation and apoptosis in HL60
cells after exposure to arsenic trioxide or all-trans retinoic
acid. Leuk Res. 32:297–307. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang P, Wu W, Chen Q and Chen M:
Non-coding RNAs and their integrated networks. J Integr Bioinform.
16:201900272019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hill M and Tran N: MiRNA interplay:
Mechanisms and consequences in cancer. Dis Model Mech.
14:dmm0476622021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Y, Cheng Z, Pang Y, Cui L, Qian T,
Quan L, Zhao H, Shi J, Ke X and Fu L: Role of microRNAs, circRNAs
and long noncoding RNAs in acute myeloid leukemia. J Hematol Oncol.
12:512019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Anelli L, Zagaria A, Specchia G, Musto P
and Albano F: Dysregulation of miRNA in Leukemia: Exploiting miRNA
expression profiles as biomarkers. Int J Mol Sci. 22:71562021.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Edginton-White B and Bonifer C: The
transcriptional regulation of normal and malignant blood cell
development. FEBS J. 289:1240–1255. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dhanasekaran R, Deutzmann A,
Mahauad-Fernandez WD, Hansen AS, Gouw AM and Felsher DW: The MYC
oncogene-the grand orchestrator of cancer growth and immune
evasion. Nat Rev Clin Oncol. 19:23–36. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dang CV: MYC on the path to cancer. Cell.
149:22–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pippa R and Odero MD: The role of MYC and
PP2A in the initiation and progression of myeloid leukemias. Cells.
9:5442020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song G, Li Y, Zhang Z, Ren X, Li H, Zhang
W, Wei R, Pan S, Shi L, Bi K and Jiang G: c-myc but not
Hif-1α-dependent downregulation of VEGF influences the
proliferation and differentiation of HL-60 cells induced by ATRA.
Oncol Rep. 29:2378–2384. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu YC, Zou XB, Chai YF and Yao YM:
Macrophage polarization in inflammatory diseases. Int J Biol Sci.
10:520–529. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Martinez FO and Gordon S: The M1 and M2
paradigm of macrophage activation: Time for reassessment.
F1000Prime Rep. 6:132014. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Murray PJ: Macrophage Polarization. Annu
Rev Physiol. 79:541–566. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mardani R, Abadi MHJ, Motieian M,
Taghizadeh-Boroujeni S, Bayat A, Farsinezhad A, Hayat SM, Motieian
M and Pourghadamyari H: MicroRNA in leukemia: Tumor suppressors and
oncogenes with prognostic potential. J Cell Physiol. 234:8465–8486.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Curtale G, Renzi TA, Mirolo M, Drufuca L,
Albanese M, De Luca M, Rossato M, Bazzoni F and Locati M:
Multi-step regulation of the TLR4 pathway by the
miR-125a~99b~let-7e cluster. Front Immunol. 9:20372018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kagawa T, Watanabe M, Inoue N, Otsu H,
Saeki M, Katsumata Y, Takuse Y and Iwatani Y: Increases of microRNA
let-7e in peripheral blood mononuclear cells in Hashimoto's
disease. Endocr J. 63:375–380. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Swier L, Dzikiewicz-Krawczyk A, Winkle M,
van den Berg A and Kluiver J: Intricate crosstalk between MYC and
non-coding RNAs regulates hallmarks of cancer. Mol Oncol. 13:26–45.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tao J, Zhao X and Tao J: c-myc-miRNA
circuitry: A central regulator of aggressive B-cell malignancies.
Cell Cycle. 13:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pelosi A, Careccia S, Lulli V, Romania P,
Marziali G, Testa U, Lavorgna S, Lo-Coco F, Petti MC, Calabretta B,
et al: miRNA let-7c promotes granulocytic differentiation in acute
myeloid leukemia. Oncogene. 32:3648–3654. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qi F, Wang X, Zhao S, Wang C, Sun R, Wang
H, Du P, Wang J, Wang X and Jiang G: miR-let-7c-3p targeting on
Egr-1 contributes to the committed differentiation of leukemia
cells into monocyte/macrophages. Oncol Lett. 24:2732022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wong TS, Man OY, Tsang CM, Tsao SW, Tsang
RK, Chan JY, Ho WK, Wei WI and To VS: MicroRNA let-7 suppresses
nasopharyngeal carcinoma cells proliferation through downregulating
c-Myc expression. J Cancer Res Clin Oncol. 137:415–422. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sampson VB, Rong NH, Han J, Yang Q, Aris
V, Soteropoulos P, Petrelli NJ, Dunn SP and Krueger LJ:
MicroRNA–let–7a down–regulates MYC and reverts MYC–induced growth
in Burkitt lymphoma cells. Cancer Res. 67:9762–9770. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Buechner J, Tømte E, Haug BH, Henriksen
JR, Løkke C, Flægstad T and Einvik C: Tumour–suppressor microRNAs
let–7 and mir–101 target the proto–oncogene MYCN and inhibit cell
proliferation in MYCN–amplified neuroblastoma. Br J Cancer.
105:296–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lan FF, Wang H, Chen YC, Chan CY, Ng SS,
Li K, Xie D, He ML, Lin MC and Kung HF: Hsa–let–7g inhibits
proliferation of hepatocellular carcinoma cells by downregulation
of c–myc and upregulation of p16(INK4A). Int J Cancer. 128:319–331.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
He XY, Chen JX, Zhang Z, Li CL, Peng QL
and Peng HM: The let–7a microRNA protects from growth of lung
carcinoma by suppression of k–Ras and c–Myc in nude mice. J Cancer
Res Clin Oncol. 136:1023–1028. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang WF, Xiong YW, Zhu TT, Xiong AZ, Bao
HH and Cheng XS: MicroRNA let-7g inhibited hypoxia-induced
proliferation of PASMCs via G0/G1 cell cycle arrest by targeting
c-myc. Life Sci. 170:9–15. 2017. View Article : Google Scholar : PubMed/NCBI
|