Introduction
Amongst all cancers diagnosed in women, breast
cancer accounts for 11.7% of cases and is the leading cause of
cancer-associated death in women worldwide (1). Breast cancer can be classified into
four subtypes, depending on the receptor status: Luminal A, luminal
B, HER2 positive, and triple-negative (2). The incidence of triple-negative breast
cancer (TNBC) is estimated to be 15–20% of all breast cancer cases
(3), and is characterized by a high
rate of early recurrence and distant metastases compared with other
types of breast cancer (4). Owing
to the lack of expression of estrogen receptor (ER), progesterone
receptor (PgR), and ERBB2 (HER2), the current
treatment methods for TNBC primarily include surgery, radiotherapy,
and chemotherapy (5). Although
significant progress has been achieved in the treatment modalities
and management strategies, the survival rates of patients have not
significantly improved. Thus, further investigation of the
underlying molecular mechanisms and biological behaviors of TNBC,
which will provide a solid foundation for developing new
therapeutic strategies, is vital.
Nuclear factor erythroid-derived 2-like 3 (Nrf3),
also known as NFE2L3, first identified in 1999, is a member of the
Cap‘n'Collar (CNC) family, which is comprised mainly of Nrf1, Nrf2,
BACH1, and BACH2 (6). The CNC
family is similar to leucine zippers in basic regions, indicating
their function may be similar (7).
Recent research on Nrf1 and Nrf2 has attracted notable attention,
showing that the ability of Nrf2 (8,9) to
mediate metabolic reprogramming and increase antioxidant capacity
underlies the malignant behaviors of NRF2-activated cancer cells,
and that Nrf1 (10) protects cancer
cells from proteotoxicity induced by anticancer proteasome
inhibitors. However, there are few reports on Nrf3, partly due to a
lack of evident abnormalities, such as in gross anatomy and
multiple blood parameters in Nrf3-deficient mice (11). Nonetheless, the role of Nrf3 differs
from that of other CNC family members, and is similar to the
definitive role of Nrf2 in oxidative stress (12). Nrf1 maintains protein stability by
maintaining proteasome gene expression (13). Nrf3 is implicated in multiple
cellular processes, such as tumorigenesis (14), inflammation, wound healing (15), and stem cell differentiation
(16). Consequently, Nrf3 has been
identified as a critical component of multiple types of cancer,
including colon cancer (17) and
pancreatic cancer (18). Nrf3 may
be involved in tumor growth and invasion via the β-catenin/TCF4
signaling pathway, which degrades the tumor suppressor p53 and Rb
in a ubiquitin-independent manner (19). Nrf3 also affects the cell cycle via
the NF-κB-DUX4-CDK1 signaling pathway (17). Additionally, Nrf3 knockdown in
hepatocellular carcinoma cells resulted in increased migration and
epithelial-mesenchymal transition (EMT) (20). These studies suggest a potential
involvement of Nrf3 in tumorigenesis. However, there is
insufficient evidence to explain how Nrf3 plays a role in TNBC
development.
As a group of plasma membrane-associated lipid
kinases, three classes of PI3K exist: Class I, class II, and class
III, each characterized by their unique structure and specific
substrate. The class I PI3Ks are classified as class A and class B.
A class IA PI3K has two subunits: A regulatory subunit (P85) and a
catalytic subunit (P110). The catalytic subunits in class IA PI3K
are divided into three types: p110α, p110β, and p110δ, which
correspond to the genes PIK3CA, PIK3CB, and PIK3CD, respectively
(21). Among these PI3K genes,
breast cancer is associated with p110 abnormal activation. The
PI3K/AKT/mTOR cascade, one of the primary signaling pathways of
tyrosine kinases, controls several cellular functions, such as
proliferation and protein synthesis. Additionally, the
overactivation of the PI3k/AKT/mTOR signaling pathway results in
various biological changes to breast cancer cells (22), and P110α enhances the proliferation
and migratory ability of breast cancer cells by activating the EMT
signaling pathway (23,24).
In the present study, TNBC specimens showed higher
Nrf3 expression compared with other types of breast cancer tissues
and adjacent noncancerous tissues, and its expression was
negatively associated with survival. Furthermore, Nrf3 knockdown
reduced cell proliferation and migration in vitro and in
vivo. In comparison, overexpression of Nrf3 had the opposite
effect. Moreover, the results showed that Nrf3 increased P110α,
p-AKT, and p-mTOR expression and regulated the expression of
EMT-related proteins. The findings suggest that Nrf3 may enhance
the proliferation and migration of TNBC by modulating PI3K/AKT/mTOR
signaling, thereby highlighting a novel therapeutic target for the
management of TNBC.
Materials and methods
Clinical samples and bioinformatics
analysis
Breast cancer and adjacent cancer tissues (n=105)
were harvested at the Affiliated Hospital of North Sichuan Medical
College between October 2018 and October 2021. It was confirmed
that all cancer cases were breast cancer, and the subtypes were
also confirmed. There were 35 TNBC cases, 35 Her2+
cases, and 35 Lumina cases. The Cancer Genome Atlas (TCGA) and GTEX
were used to examine Nrf3 expression in breast cancer and normal
tissues. Preoperative radiotherapy and chemotherapy were not
administered to any patients. Related clinical data were collected.
All patients provided informed consent for the use of clinical
research materials.
Immunohistochemistry
Tissues were first fixed at room temperature for 48
h in 4% formaldehyde solution, dehydrated using an increasing
series of alcohol solutions, deparaffinized using a hyaline agent,
embedded in paraffin and finally sectioned into 4-µM thick sections
and stored at 4°C. After sectioning, antigen retrieval was
performed using sodium citrate for 25 min at 100°C in a water bath,
followed by drying, dewaxing and hydration using a decreasing
series of alcohol solutions. The sections were immersed in 3%
hydrogen peroxide for 15 min to quench endogenous peroxidase
activity and immediately incubated at 4°C overnight with primary
antibodies (rabbit anti-Nrf3 antibody; cat. no. PA5-99083; 1:50;
Thermo Fisher Scientific, Inc.). The following day, the samples
were incubated with the horseradish peroxidase-conjugated secondary
antibodies (cat. no. AB0101; Abways Technology) for 1 h at 37°C.
The slides were then stained with DAB (DAB Plus Kit from MXB
Biotechnologies) solution for 2 min at room temperature,
counterstained with hematoxylin for 30 sec and dried for blocking.
Two experienced pathologists evaluated the results according to the
percentage of positively stained tumor cells and staining
intensity.
Positive staining degree was classified as 0,
<10%; 1, 10–20%; 2, 21–50%; and 3, >50%. The staining
intensity was graded as follows: 0, no staining; 1, weak staining;
2, moderate staining; and 3, strong staining. To calculate the
final scores, the score for the percentage stained was added to the
score for the staining intensity. The final scores were stratified
as ≥6 and <6, indicating high and low expression,
respectively.
Cell culture and lentivirus
transfection
MDA-MB-231 and MDA-MB-468 (triple-negative breast
cancer cell lines) were provided by the National Collection of
Authenticated Cell Cultures. The PI3K pathway inhibitor, LY294002
(Selleck Chemicals), was purchased in powder form, dissolved to 100
mM using DMSO, and then stored at −80°C. The Nrf3 lentiviral vector
(sc-404543-LAC) and negative control were obtained from Santa Cruz
Biotechnology, Inc. Two shRNA target sequences for Nrf3 were
designed and inserted into the pLKO.1 vector. shRNA plasmids were
transfected with lentiviruses to generate a recombinant lentivirus.
The Nrf3 shRNA interference target sequences were based on our
previous study (25).
A total of 2×105 MDA-MB-231 or MDA-MB-468
cells were seeded in each well, and cultured to 40% confluency. In
addition to the complete medium, lentiviral particles were added at
a final concentration of polybrene (5 µg/ml). After mixing, the
original medium was replaced, and infection was allowed for 24 h.
After 24 h, the medium was replaced and puromycin hydrochloride was
added to allow for screening for 48 h. The effect of overexpression
and knockdown of Nrf3 was determined using western blotting.
Western blotting
Cells were lysed using western & IP lysis buffer
(cat. no. P0013; Beyotime Instiute of Biotechnology). The proteins
were loaded on a 10% SDS gel, resolved using SDS-PAGE, and then
transferred to PVDF membranes. Membranes were blocked in TBST with
5% skimmed milk. Next, membranes were blocked with primary
antibodies at 4°C overnight, including anti-Nrf3 (cat. no.
PA5-102015; 1:1,000; Thermo Fisher Scientific, Inc.), anti-PI3K
p110α (cat. no. 4249; 1:1,000; Cell Signaling Technology, Inc.),
anti-phospho-Akt (S473) (cat. no. 4060; 1:1,000; Cell Signaling
Technology, Inc.), rabbit anti-phospho-Akt (Thr308) (cat. no.
13038; 1:1,000; Cell Signaling Technology, Inc.), anti-Akt (cat.
no. 9272; 1:1,000; Cell Signaling Technology, Inc.),
anti-phospho-mTOR (Ser2448) (cat. no. 5536; 1:1,000; Cell Signaling
Technology, Inc.), anti-mTOR (cat. no. 2972; 1:1,000; Cell
Signaling Technology, Inc.), anti-E-cadherin (cat. no. 20874-1-AP;
1:1,000; ProteinTech Group, Inc.), anti-N-cadherin (cat. no.
22018-1-AP; 1:1,000; ProteinTech Group, Inc.), anti-Vimentin (cat.
no. 10366-1-AP; 1:1,000; ProteinTech Group, Inc.), anti-MMP-3 (cat.
no. 17873-1-AP; 1:1,000; ProteinTech Group, Inc.), anti-MMP-9 (cat.
no. 10375-2-AP; 1:1,000; ProteinTech Group, Inc.) and anti-GAPDH
(cat. no. AB0036; 1:5,000; Abways Technology) antibody. The
following day, the membranes were incubated with the relevant
secondary antibody. Between and after incubation with the
antibodies, membranes were washed with TBST. Signals were
visualized using Immobilon ECL Ultra Western HRP (cat. no.
WBULS0100, MilliporeSigma). The results were analysed using a
ChemiDoc XRS + Gel Imaging System (Bio-Rad Laboratories, Inc).
Transcriptome sequencing and
analysis
MDA-MB-231 and MDA-MB-231/Nrf3 cells were collected
and prepared for three paired biological replications. The samples
were send to oebiotech company (https://www.oebiotech.com/) for sequencing. Total RNA
was extracted using the mirVana miRNA Isolation Kit (cat. no. 1561;
Ambion; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocols. RNA integrity was evaluated using the
Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). The samples
with RNA Integrity Number (RIN) ≥7 were subjected to the subsequent
analysis. The libraries were constructed using TruSeq Stranded mRNA
LTSample Prep Kit (15 cycles; cat. no. RS-122-2103; Illumina, Inc.)
according to the manufacturer's instructions. The purified library
products were evaluated using the Agilent 2200 Tape Station and
Qubit®2.0 (Life Technologies; Thermo Fisher Scientific,
Inc.) and then diluted to 10 pM for cluster generation in situ on
the pair-end flow cell followed by HiSeq2500 sequencing (2×150 bp).
Raw data (raw reads) of fastq format were firstly processed using
Trimmomatic (version 0.36) (26)
and the low quality reads were removed to obtain the clean reads.
The clean reads were mapped to the human genome (GRCh38) using
HISAT2.2.1 (27). The fragments per
kilobase of transcript per million mapped reads of each gene were
calculated using Cufflinks 2.2.1 (28) and the read counts of each gene were
obtained by HTSeq-count version 2.0.4 (29) Differential expression analysis was
performed using the DESeq (2012) R package (30). P<0.05 and fold-change >2 and
<0.5 were set as the thresholds for significant differential
expression. Hierarchical cluster analysis of differentially
expressed genes was performed to demonstrate the expression pattern
of genes in different groups. Gene set variation analysis (GSVA
1.24.2) (R package) (31) was
utilized to perform GSVA. The gene set ‘hall.v7.0.symbols.gmt’ was
selected as the reference gene set. P<0.05 was considered to
indicate a statistically significant difference. The transcriptome
raw data has been deposited in the Sequence Read Archive database
(https://www.ncbi.nlm.nih.gov/sra;
submission no. PRJNA965920).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from MDA-MB-231 cells using
TRIzol®. The RNA was converted to cDNA using a
RevertAid First Strand cDNA Synthesis Kit according to the
manufacturer's protocol. qPCR was performed on a CFX Connect
Real-Time PCR Detection System and SuperRealPreMix Plus SYBR Green.
Based on the 2ΔΔCq method (32), the relative RNA expression was
calculated. Table SI displays the
detailed primer sequences included in this study. The thermocycling
conditions were as follows: Initial denaturation at 94°C for 2 min,
followed by 40 cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C
for 30 sec, and then a final extension at 72°C for 10 min.
Cell proliferation and migration
assay
CCK-8 experiments (assays obtained from Dojindo
Molecular Technologies, Inc.) were used to determine cell
proliferation. Approximately 5×103 cells per well were
plated into a 96-well plate with complete medium and cultured in an
incubator. The spectrometric absorbance was determined using a
microplate reader at 450 nm. Each experiment was repeated three
times.
For migration assays, wound healing, and Transwell
assays were used to investigate cell migration. Transwell assays
were performed as follows: 5×103 cells in 100 µl
serum-free medium was added to the upper chamber of a Transwell
insert without Matrigel. Supplemented medium was added to the lower
chamber as a chemoattractant. A cotton swab was used to remove the
cells on the upper side after 24 h of incubation. A mixture of 4%
formaldehyde and 0.1% crystal violet was used to fix and stain
migrated cells at room temperature for 20 min. Finally, the number
of cells in three independent fields was counted under a
brightfield microscope (IX73; Olympus Corporation) (magnification,
×100).
For the wound healing assay, 5×103 cells
per well were added to a six-well plate. Once a confluent monolayer
of cells had formed, the monolayer was scratched and imaged after
becoming adherent, and then cultured in serum-free medium for a
further 24 h. The cells were imaged again in the same location
after washing with PBS.
Luciferase reporter assay
Nrf3, a transcript factor, is known to bind
antioxidant response element (ARE) sites, with the core sequence of
an ARE being 5′-TGACNNNGC-3′. The p110α promoter sequence was
loaded to find multiple ARE sites. Thus, Nrf3 may active P110α by
binding ARE sites. siRNA knockdown of Nrf3 expression and pCMV-Nrf3
overexpression vector were purchased from Cyagen Biosciences. The
p110α promoter was cloned into pEZX (−1,000 bp from the
transcription initiation site). The resulting full-length reporter
plasmid was termed pEZX-1000, which may contain multiple
Nrf3-binding sites. The deletion mutation reporters (pEZX-714 and
pEZX-418) were generated using this plasmid. Plasmids and siRNAs
were transfected into MDA-MB-231 and MDA-MB-468 cells using
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific,
Inc.).
The MDA-MB-231 and MDA-MB-468 cells were seeded in
six-well plates and transfected with a p110α full-length reporter
plasmid or deletion mutant promoter plasmid and Nrf3-overexpressing
vector, siNrf3, and the negative control vector together with the
PGL 4.74 TK-Luc reporter. After transfection (48 h later), the cell
lysates were prepared. As directed by the manufacturer,
Firefly/Renilla luciferase values were determined using a
Dual-Luciferase Reporter Assay System (cat. no. E1910; Promega
Corporation). A GloMax-96 plate reader (Promega Corporation) was
used to measure the luciferase activity.
ChIP
Pierce magnetic ChIP assays were performed using the
Pierce Magnetic ChIP Kit (cat. no. 88848; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions.
Immunoprecipitation was performed using chromatin with anti-Nrf3
and a normal rabbit-IgG antibody. ChIP-enriched DNA was amplified
and quantified using qPCR. The primer sets at the Nrf3-binding
sites for the p110α promoter were: Primer 1 forward,
5′-AGCAAAAGGTCTCCACGAAGTGAG-3′ and reverse,
5′-CGCCGCTGTCAGTGGCTAAC-3′; and primer 2 forward,
5′-TCTCTACCCCAGCTCGCCTG-3′ and reverse,
5′-GATGCGCAAAGAAGCGGAAG-3′.
Animal experiments
The MDA-231, MDA-231/Nrf3, MDA-231/ShNrf3#1, and
MDA-231/shNrf3#2 tumor models were established to investigate the
role of Nrf3 in the proliferation and metastasis of TNBC. A total
of 20 nude female mice were obtained from the Beijing HFK
Biotechnology Co., Ltd. and maintained in micro isolator cages to
analyze tumor growth. For subcutaneous inoculation,
5×106 cells were resuspended in PBS and injected
subcutaneously into 5-week-old mice (100 µl per mouse); four groups
were formed as follows: MDA-231, MDA-231/Nrf3, MDA-231/ShNrf3#1,
and MDA-231/shNrf3#2 (5 mice per group). The tumors were measured
every 3 days after their initial appearance. The mice were
sacrificed 40 days after the inoculation. Recovery experiments were
performed as described previously (25). When tumors appeared, the mice were
further divided into three groups of mice: MDA-231(DMSO),
MDA-231/Nrf3 (DMSO), MDA-231/Nrf3 (LY294002, 20 mg/kg/day) (5 mice
per group). Treatment was performed for 14 days, and the mice were
sacrificed at the end of treatment by injection of sodium
pentobarbital (150 mg/kg).
For metastasis assays, a total of 20 female nude
mice provided by Beijing HFK Biotechnology Co., Ltd. PBS was to
resuspend 1×107 cells per ml, of which 100 µl was
injected into the tail veins of each mouse. Mice were divided into
four groups as follows: MDA-231, MDA-231/Nrf3, MDA-231/ShNrf3#1,
and MDA-231/shNrf3#2 (5 mice per group). After 60 days, the mice
were sacrificed, lungs were excised and imaged. Subsequently, the
lung tissues were fixed in 4% formalin and embedded in paraffin,
and then hematoxylin and eosin staining was performed. Mice were
maintained in pathogen-free conditions with a controlled
temperature (23±2°C), relative humidity (45–65%) and light/dark
cycle (12/12 h) with free access to food and water at all times.
All animal procedures were performed in accordance with the North
Sichuan Medical College's Animal Care Committee and the guidelines
of the Animal Protection Law of the People's Republic of China
(2009). All animal experiments were approved by the Affiliated
Hospital of North Sichuan Medical College (approval no.
2022033).
Statistical analysis
Data are presented as the mean ± standard deviation.
A one-way ANOVA followed by a post-hoc Tukey's test or a Student's
t-test was used to compare differences between ≥3 or 2 groups,
respectively. The χ2 test was performed to analyze the
relationship between clinical characteristics and Nrf3 expression.
P<0.05 was considered to indicate a statistically significant
difference. Statistical analysis was performed using IBM SPSS
version 23 (IBM Corp.).
Results
Nrf3 expression is increased in TNBC,
and its expression is negatively associated with survival
Bioinformatics was used to analyze Nrf3 mRNA
expression in different types of breast cancer. The findings
revealed that Nrf3 expression in patients with TNBC was upregulated
compared to its expression in other types of breast cancer and
adjacent tissues, based on data obtained from TCGA and GTEX
(Fig. 1A-C). According to the
Kaplan-Meier survival curves, patients with TNBC expressing high
levels of Nrf3 had shorter survival times (Fig. 1D). The correlation between Nrf3 and
EMT-related genes was also analyzed; the results are shown in
Fig. S1A-D.
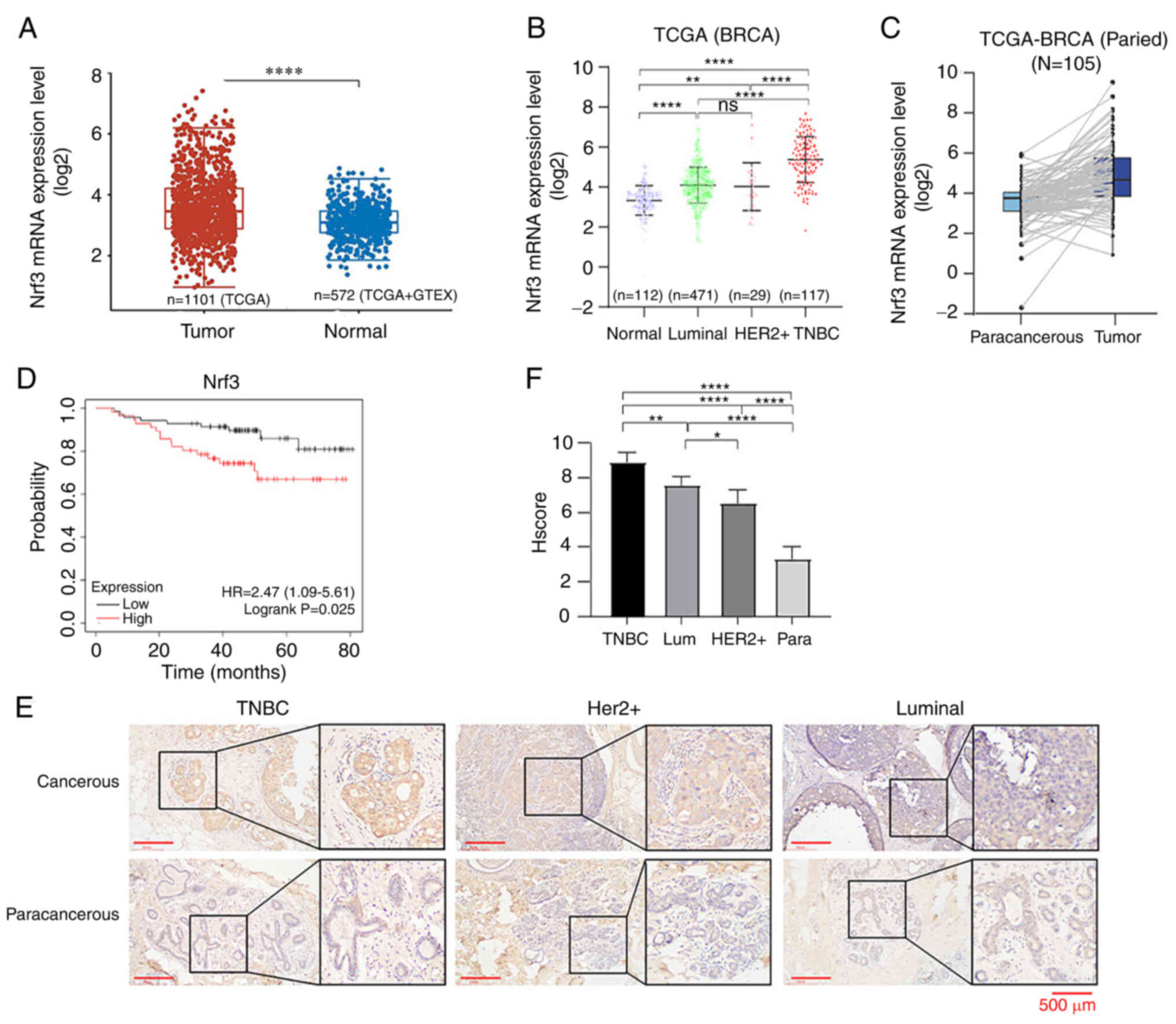 | Figure 1.Nrf3 is upregulated in
triple-negative breast cancer, and its expression is negatively
associated with survival prognosis. (A) Differential expression of
Nrf3 mRNA levels in breast cancer and the paracancerous tissues.
Normal tissues, n=112; tumor tissues, n=1,066. (B) Nrf3 mRNA
expression levels in normal breast tissue and different
pathological types of breast cancer tissues. Normal tissues, n=112;
tumor tissues, n=467; luminal, n=471; Her2+, n=29; TNBC,
n=117. (C) Differential expression of Nrf3 mRNA levels in breast
cancer and the paracancerous tissues. (D) Kaplan-Meier survival
analysis between low and high Nrf3 expression breast cancer
patients. Nrf3-low, n=54; Nrf3-high, n=72. P=0.025. (E)
Representative images of immunohistochemical staining and the (F)
immunohistochemical score in 105 breast cancer patients with
different pathological types. TNBC, n=35; Her2+,0 n=35;
luminal, n=35. *P<0.05, **P<0.01 and ****P<0.0001. Nrf3,
nuclear factor erythroid 2-related factor 3; TNBC, triple-negative
breast cancer; TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue
Expression; BRCA, breast cancer. |
A total of 105 patients with breast cancer of
different pathological types were examined for expression of Nrf3
in their pathological tissues using immunohistochemistry. Higher
expression of Nrf3 was detected in TNBC tissues compared with other
types of breast cancer and adjacent tissues (Fig. 1E and F). Analysis of the correlation
between clinical characteristics and Nrf3 expression was conducted
using a χ2 test. The score for Nrf3 expression in breast
cancer was divided into high expression (scores ≥6) and low
expression (scores <6). There was no association between Nrf3
expression and age, lymph node metastases, tumor sizes, or TNM
stage (Table I).
 | Table I.Association between Nrf3 expression
and clinicopathological factors. |
Table I.
Association between Nrf3 expression
and clinicopathological factors.
|
| Nrf3 expression,
n |
|
|---|
|
|
|
|
|---|
| Factors | Higha | Lowb | P-value |
|---|
| Age, years |
|
| 0.618 |
|
<50 | 31 | 14 |
|
|
≥50 | 44 | 16 |
|
| Pathological
grade |
|
| 0.096 |
| 1 | 4 | 0 |
|
| 2 | 62 | 22 |
|
| 3 | 9 | 8 |
|
| Tumor size, cm |
|
| 0.993 |
|
<2 | 36 | 14 |
|
| ≥2 | 39 | 16 |
|
| Lymph node
metastasis |
|
| 0.429 |
|
Present | 26 | 8 |
|
|
Absent | 49 | 22 |
|
| TNM stage |
|
| 0.225 |
|
I/II | 67 | 29 |
|
|
III/IV | 8 | 1 |
|
Overexpression of Nrf3 contributes to
the proliferation and migration of TNBC
TNBC cell lines stably overexpressing Nrf3 or
with Nrf3 expression knocked down were constructed to determine
whether Nrf3 played a role in the proliferation and migration of
breast cancer cells. Overexpression and knockdown of Nrf3 were
confirmed using western blotting (Fig.
S2). Cells overexpressing Nrf3 were termed MDA-MB-231/Nrf3 and
MDA-MB-468/Nrf3. The cells in which Nrf3 expression was knocked
down were termed MDA-MB-231shNrf3#1, MDA-MB-231shNrf3#2,
MDA-MB-468shNrf3#1, and MDA-MB-468shNrf3#2.
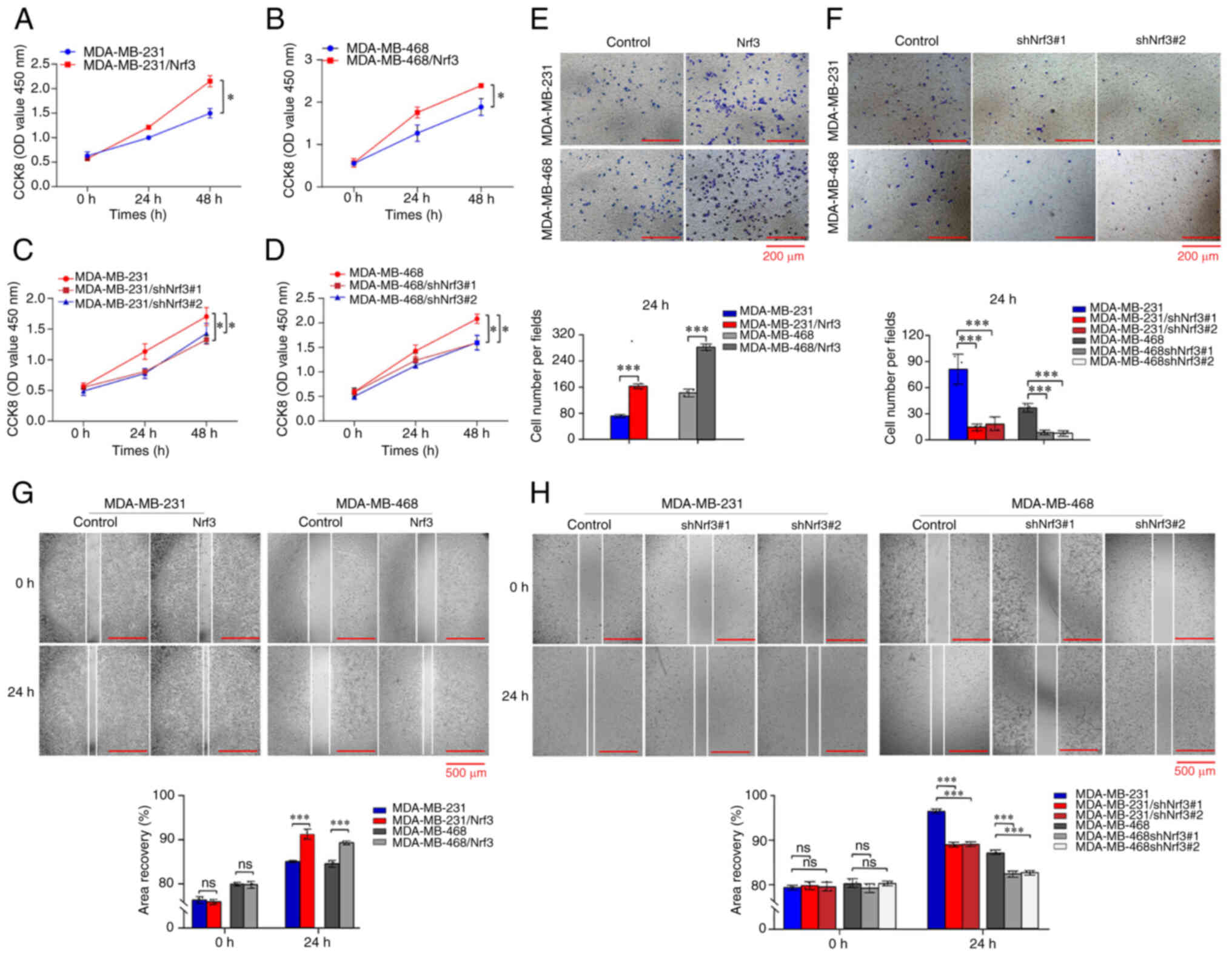
CCK-8 assays showed that overexpression of Nf3
significantly promoted the proliferation of TNBC cells, whereas
knockdown of Nrf3 reduced cell proliferation (Fig. 2). The role of Nrf3 in migration of
TNBC cells was examined using wound healing and Transwell migration
assays. The wound healing assays showed that the migratory rate of
MDA-MB-231/Nrf3 cells increased 7.3±0.2% compared to that of
MDA-MB-231, whereas the migration rate in MDA-MB-231/shNrf3 showed
a decrease of 8.2±0.15% (Fig. 2G).
Additionally, the migratory rate of MDA-MB-468/shNrf3 cells
decreased by 4.15±0.85% compared to that of MDA-MB-468 cells,
whereas the migratory rate of MDA-MB-468/Nrf3 cells increased by
5.4±0.2% (Fig. 2H). For
overexpression of Nrf3, the number of cells passing through the
Transwell membranes in the different groups was 62±11 per field
(MDA-MB-231), 161±17 per field (MDA-MB-231/Nrf3), 142±8.7 per field
(MDA-MB-468), and 259±3.6 per field (MDA-MB-468/Nrf3) (Fig. 2E). For Nrf3 knockdown, the number of
cells passing through the Transwell membranes was 82±29 per field
(MDA-MB-231), 19±8.8 per field (MDA-MB-231/shRNA#1), 14±5 per field
(MDA-MB-231/shRNA#2), 42±8.7 per field (MDA-MB-468), 12±2.6 per
field (MB-468/shRNA#1), and 14±3 per field (MDA-MB-468/shRNA#2)
(Fig. 2F). These results
demonstrated that overexpression of Nrf3 increased the invasive
ability of TNBC cells, whereas knockdown of Nrf3 decreased the
invasive ability of the cells.
Tumor growth and metastasis in
vivo
To confirm the role of Nrf3 in promoting the
proliferation and migration of TNBC in vivo,
Nrf3-overexpression, Nrf3-knockdown MDA-MB-231, and MDA-MB-231
cells were injected. MDA-MB-231shNrf3#1 and MDA-MB-231shNrf3#2
cells failed to form subcutaneous tumors; whereas only 4 out of 5
mice developed tumors in the MDA-MB-231 group, whereas five out of
five mice developed tumors in the MDA-MB-231/Nrf3 group. The
average tumor weights in the different groups at the endpoint were
0.091±0.070 g (MDA-MB-231/Nrf3) and 0.017±0.007 g (MDA-MB-231)
(Fig. 3A and B).
Immunohistochemistry was used to detect the expression of p110α,
Nrf3, E-cadherin, N-cadherin, and vimentin. The results revealed
that the expression of p110α, Nrf3, N-cadherin, and vimentin in the
MDA-MB-231/Nrf3 group was substantially higher compared with the
control group, whereas the expression of E-cadherin exhibited the
opposite trend (Fig. 3C and D).
Fig. 3E-G shows that the number of
lung metastases in the different groups was as follows: 3.2±0.4
(MDA-MB-231), 4.6±0.6 (MDA-MB-231/Nrf3), 0.4±0.5
(MDA-MB-231shNrf3#1), and 0.6±0.5 (MDA-MB-231shNrf3#2). The images
of the entire metastatic tumour experiment are shown in Fig. S3. These data indicated that Nrf3
facilitated the growth and migration of TNBC cells in
vivo.
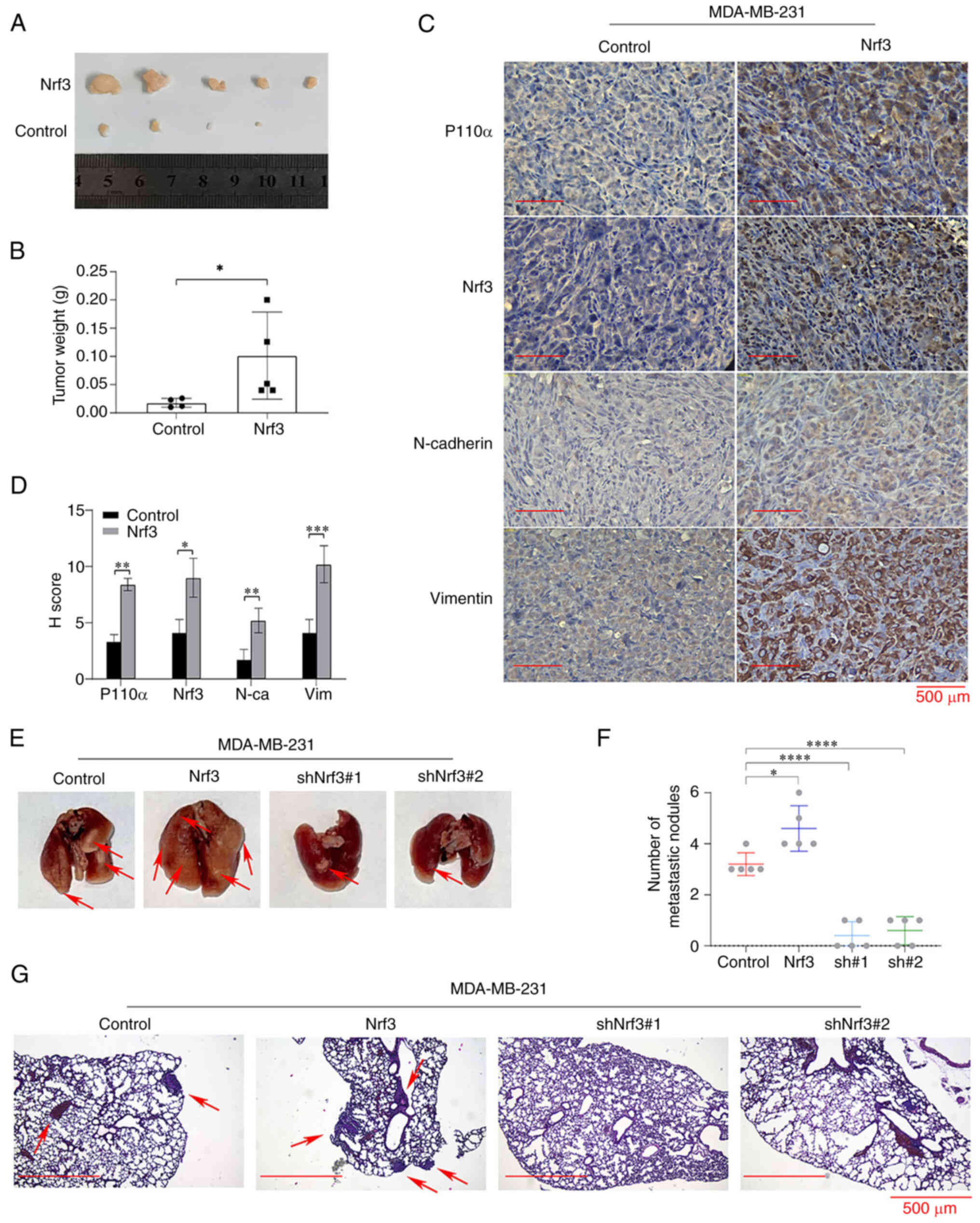 | Figure 3.Nrf3 promotes TNBC cell proliferation
and metastasis in vivo. (A) Representative images of tumors
in the different groups. (B) Mean tumor weight in each group. (C)
p110α, Nrf3, N-cadherin, and vimentin expression were detected by
immunostaining in the xenografted tumors. Magnification, ×400. (D)
Mean staining intensity of p110α, Nrf3, N-cadherin, and vimentin
(E) Representative images of the lungs excised from the mice in the
different groups. (F) The number of metastatic lung sites
(indicated by arrows) was counted. Data are presented as the mean ±
SEM of 5 repeats. (G) Representative images of H&E-stained
sections in the dissected lungs 60 days after inoculation.
Magnification, ×100. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. Nrf3, nuclear factor erythroid 2-related factor 3;
TNBC, triple-negative breast cancer; sh, short hairpin; N-ca,
N-Cadherin; Vim, Vimentin. |
Nrf3 regulates the PI3K/AKT/mTOR
pathway and expression of EMT-related proteins
To explore the underlying mechanisms of Nrf3 in
enhancing the growth and invasion of breast cancer cells,
transcriptome sequencing was performed using three paired
biological replications of MDA-MB-231 and MDA-MB-231/Nrf3 cells.
Based on the sequencing data, 215 significantly differentially
expressed genes, including 159 upregulated and 66 downregulated
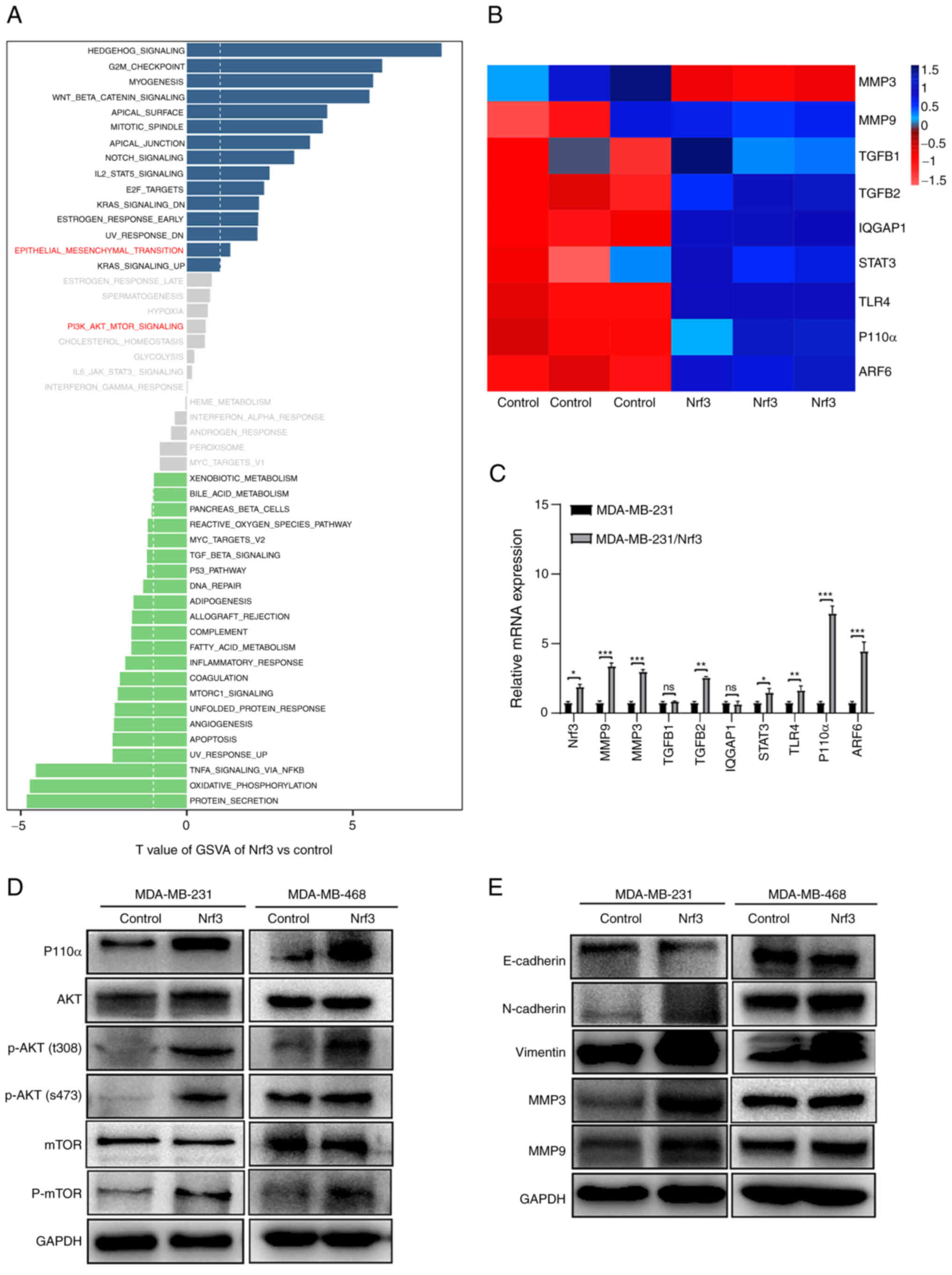
genes (Fig. S4). GSVA showed that
EMT-related genes were enriched, which is a critical mechanism for
cancer metastasis (Fig. 4A). The
genes related to proliferation and migration were clustered and are
displayed using thermography (Fig.
4B). To validate this result, RT-qPCR was used to examine the
mRNA levels of the above-related genes (p110α, MMP3, MMP9, TGF β1,
TGF β2, IQGAP1, STAT3, TLR4, and ARF6). The most significant change
among these candidate molecules was in p110α in the MDA-MB-231/Nrf3
cell line compared with MDA-MB-231 cells (Fig. 4C), which was consistent with
transcriptome sequencing. Additionally, the expression of the p110α
protein and its downstream molecules were further determined by
western blotting. Comparing the Nrf3 overexpression group with the
control group revealed that p-AKT and p-mTOR expression was
increased. However, the expression of AKT and mTOR were not
significantly altered (Fig. 4D).
Furthermore, owing to Nrf3 overexpression, N-cadherin, vimentin,
MMP3, and MMP9 expression increased, whereas E-cadherin expression
decreased (Fig. 4E).
Nrf3 directly binds to the p110α
promoter and increases its expression
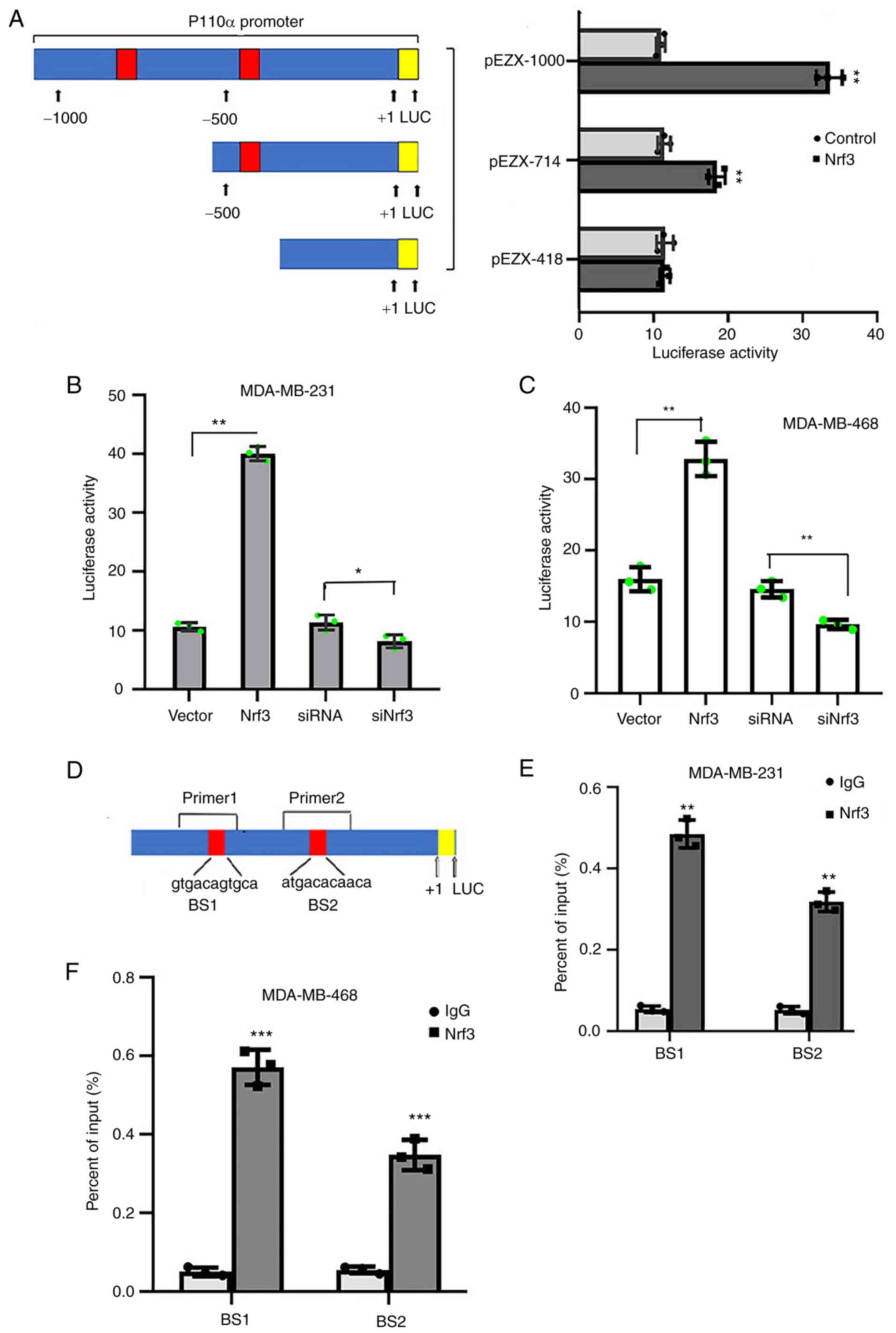
A p110α promoter was subcloned into the pEZX vector
to determine whether Nrf3 regulated p110α expression in TNBC cells.
The full-length reporter plasmid was designated pEZX-1000, which
contained two predicted Nrf3 binding sites (−725 to −715 and −429
to −419). Additionally, two deletion mutation reporters were
constructed using this plasmid (pEZX-714 and pEZX-418). pEZX-1000,
pEZX-714, and pEZX-418 plasmids were co-transfected with the Nrf3
expression vector into the MDA-MB-231 cells. The results showed
that Nrf3 enhanced the activity of the full-length reporter
(pEZX-1000) and the deletion mutant reporter (pEZX-714), whereas
the other deletion mutant reporter (pEZX-418) did not exhibit the
same activity (Fig. 5A). Further,
MDA-MB-231 and MDA-MB-468 cells were co-transfected with pEZX-1000
and an Nrf3 expression vector or siRNA Nrf3 (Fig. 5B and C) showed that overexpression
of Nrf3 enhanced p110α promoter activity. Conversely, Nrf3
knockdown attenuated the p110α promoter activity.
A ChIP assay was performed in MDA-MB-231 and
MDA-MB-468 cells to determine the mutual interactions between Nrf3
and potential Nrf3-binding sites in the p110α promoter region.
Primers were located at binding sites 1 (BS1) and 2 (BS2) of the
p110α promoter (Fig. 5D). An
anti-Nrf3 antibody immunoprecipitated significantly more chromatin
in MDA-MB-231/Nrf3 and MDA-MB-468/Nrf3 cells compared with the
control (Fig. 5E and F). These
results suggest that Nrf3 directly binds to the p110α promoter
region and affects the transcriptional activity of p110α in
MDA-MB-231 and MDA-MB-468 cells.
PI3K inhibitor partly reverses the effects of Nrf3
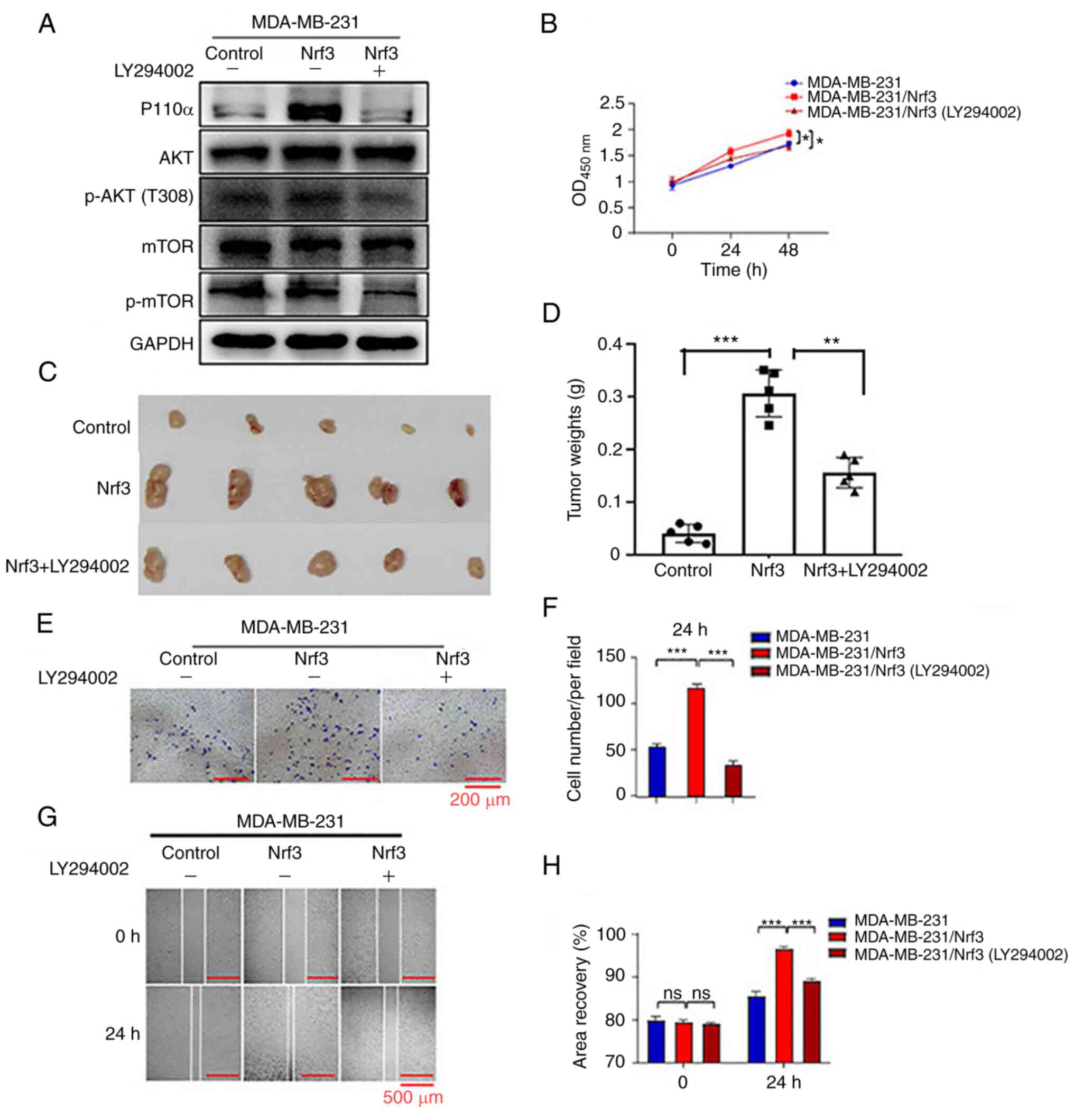
overexpression. MDA-MB-231/Nrf3 cells were used for the recovery
experiments. Western blot analysis was performed after cells were
treated with the PI3K inhibitor, LY294002, (Fig. 6A). The CCK-8 assays and animal
experiments showed that the PI3K inhibitor partly reversed the
proliferation of MDA-MB-231/Nrf3 cells in vitro and in
vivo (Fig. 6B-D). As measured
by wound healing and Transwell assays, the PI3K inhibitor
attenuated the effect of Nrf3, resulting in the migration of
MDA-MB-231 cells (Fig. 6E–H). Based
on these findings, PI3K may be involved in regulating Nrf3-mediated
proliferation and migration in breast cancer cells.
Discussion
In the present study, Nrf3 was shown to play a
critical role in the proliferation and invasion of TNBC cells via
the PI3K/AKT/mTOR signaling pathway. First, Nrf3 expression was
evidently upregulated in TNBC tissues compared with the
noncancerous tissues, and its expression was associated with worse
survival times. Additionally, overexpression of Nrf3 was shown to
significantly enhance cell proliferation and migration in
vitro and in vivo, while Nrf3 knockdown produced the
opposite results. Moreover, Nrf3 activated the PI3K/AKT signaling
pathway and regulated EMT-related protein expression. Luciferase
and ChIP assays showed that Nrf3 positively regulated PI3K
expression. By inhibiting PI3K, the effect of Nrf3 on cell growth
and invasion was partially reversed.
Nrf3 is associated with the development and
occurrence of several types of cancer (33–35),
and its function remains contested. Chevillard et al
(36) suggested that Nrf3 has a
protective effect in lymphoma; however, further validation at the
molecular level is lacking. Other reports have shown that Nrf3 may
act as an oncogene to promote carcinogenesis, such as in colon
cancer (17), bladder cancer
(18), and hepatocellular carcinoma
(37). Therefore, Nrf3 can function
as both an oncogene or tumor suppressor gene dependent on the type
of cancer. The results of the present study showed that Nrf3, as an
oncogene, increased the proliferation and migration of TNBC cells
in vitro and in vivo. Although Nrf3 expression was
previously reported to be reduced in breast cancer (38), the results of the present study
demonstrated that Nrf3 expression in TNBC was higher than in
adjacent tissues and other cancer types. These differences in
research findings may be attributed to differences in sample sizes
and tumor types.
The underlying mechanism of Nrf3 in promoting
proliferation and migration may involve the PI3K/AKT/mTOR signaling
pathway. The genes related to proliferation and migration were
determined using transcriptome sequencing and qPCR. The results of
the present study showed that p110α expression was most notably
altered among the candidate molecules. p110α is the second most
frequently mutated gene in TNBC (39) and plays an important role in
regulating receptor tyrosine kinase (RTK) signaling, cell growth,
the cell cycle, and cell survival in breast cancer (40). Expression of p110α and its
downstream molecules was also detected. The PI3K/AKT/mTOR signaling
pathway was activated by Nrf3, which may be essential for the
multiple cellular activities observed (41), including cell metabolism, cell
growth and migration, and apoptosis. Hyperactivation of
PI3K/AKT/mTOR promotes cell growth and migration of TNBC cells
(42). Moreover, using the
luciferase reporter and ChIP assays, it was found that Nrf3
directly bound to the p110α promoter sequence. To the best of our
knowledge, this is the first study to show that Nrf3 is responsible
for the activation of p110α transcription.
During cancer metastasis, EMT also plays an
important role. EMT is characterized by the loss of epithelial
markers, an increase in mesenchymal markers, and increased cell
migration (43,44). Activation of EMT has been shown to
promote the phenotypic association of proliferative migration,
tumor induction, stem cell activity of tumor cells, and tumor drug
resistance in breast cancer (45).
Nrf3 knockdown reduces the migration of hepatocellular carcinoma
cells by inhibiting EMT (46). The
present study showed that the overexpression of Nrf3 upregulated
N-cadherin and vimentin expression whilst reducing E-cadherin
expression. In contrast, p110α also triggered EMT by inducing the
loss of E-cadherin (42). Thus, it
is hypothesized that Nrf3 activated EMT via PI3K/AKT/mTOR
signaling, although the precise mechanism still requires further
investigation.
The present study first revealed that Nrf3 promoted
the growth and migration of TNBC cells in vitro and in
vivo. Additionally, it was found that Nrf3 increased p110α,
p-AKT, and p-mTOR expression and regulated EMT expression-related
proteins. Luciferase reporter and ChIP assays showed that Nrf3
increased p110α promoter activity by directly binding to the
promoter region. A link between Nrf3 and p110α indicated that
Nrf3-mediated proliferation and migration of TNBC may be related to
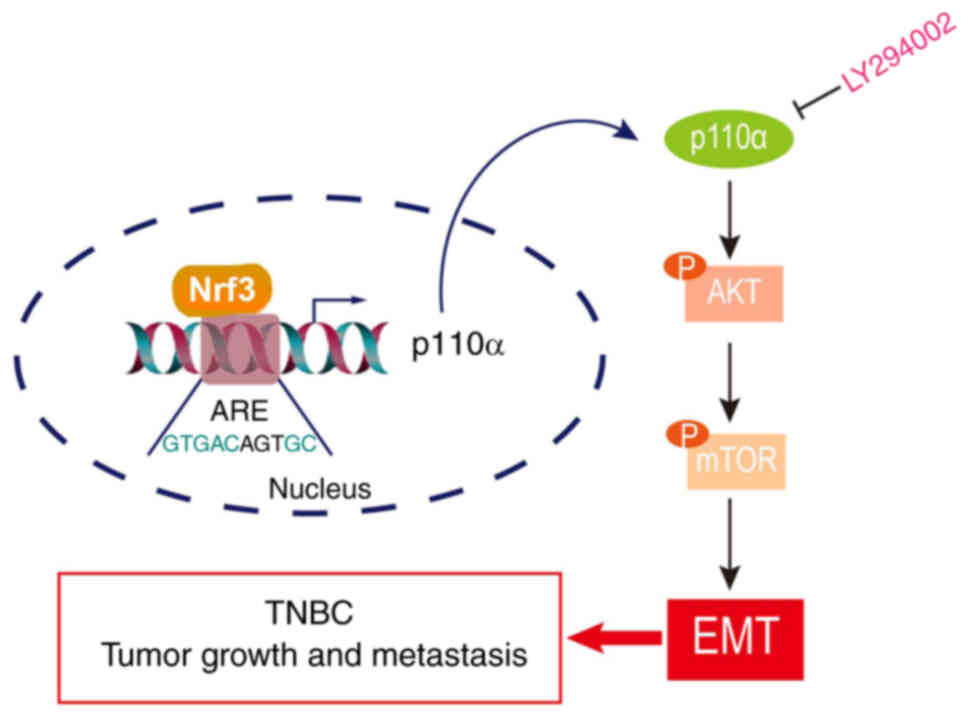
PI3K/AKT/mTOR signaling (Fig.
7).
In conclusion, the present study results
demonstrated that Nrf3 facilitates the proliferation and migration
of TNBC by activating PI3K/AKT/mTOR signaling and EMT, thereby
providing a potential novel therapeutic target for TNBC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Strategic Cooperation Research
Project of Nan Chong (grant no. 20SXCXTD004) and the Major Emphasis
Project of North of Sichuan Medical College (grant no. CBY22-ZDA02,
CBY22-ZD04).
Availability of data and materials
Transcriptome sequencing datasets generated and/or
analyzed during the current study are available in the Sequence
Read Archive database (https://www.ncbi.nlm.nih.gov/sra) as submission number
PRJNA965920. The other datasets used and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
JCT, QYH, WMC and WH designed the study, performed
the experiments and wrote the manuscript. YHC and MWC performed the
statistical analysis. JCT, QYH and WMC confirm the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
In accordance with the guidelines of the Animal
Protection Law of the People's Republic of China-2009, procedures
on animals were approved by the Affiliated Hospital of North
Sichuan Medical College (approval no. 2022033).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brown M, Tsodikov A, Bauer KR, Parise CA
and Caggiano V: The role of human epidermal growth factor receptor
2 in the survival of women with estrogen and progesterone
receptor-negative, invasive breast cancer: The California Cancer
Registry, 1999-2004. Cancer. 112:737–747. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Duffy MJ, McGowan PM and Crown J: Targeted
therapy for triple-negative breast cancer: Where are we? Int J
Cancer. 131:2471–2477. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gradishar WJ, Anderson BO, Abraham J, Aft
R, Agnese D, Allison KH, Blair SL, Burstein HJ, Dang C, Elias AD,
et al: Breast cancer, version 3.2020, NCCN clinical practice
guidelines in oncology. J Natl Compr Canc Netw. 18:452–478. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kobayashi A, Ito E, Toki T, Kogame K,
Takahashi S, Igarashi K, Hayashi N and Yamamoto M: Molecular
cloning and functional characterization of a new Cap'n’ collar
family transcription factor Nrf3. J Biol Chem. 274:6443–6452. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Motohashi H, O'Connor T, Katsuoka F, Engel
JD and Yamamoto M: Integration and diversity of the regulatory
network composed of Maf and CNC families of transcription factors.
Gene. 294:1–12. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sekine H and Motohashi H: Roles of CNC
transcription factors NRF1 and NRF2 in cancer. Cancers (Basel).
13:5412021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yuan J, Zhang S and Zhang Y: Nrf1 is paved
as a new strategic avenue to prevent and treat cancer,
neurodegenerative and other diseases. Toxicol Appl Pharmacol.
360:273–283. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rojo de la Vega M, Chapman E and Zhang DD:
NRF2 and the hallmarks of cancer. Cancer Cell. 34:21–43. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Derjuga A, Gourley TS, Holm TM, Heng HH,
Shivdasani RA, Ahmed R, Andrews NC and Blank V: Complexity of CNC
transcription factors as revealed by gene targeting of the Nrf3
locus. Mol Cell Biol. 24:3286–3294. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He F, Ru X and Wen T: NRF2, a
transcription factor for stress response and beyond. Int J Mol Sci.
21:47772020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Radhakrishnan SK, Lee CS, Young P, Beskow
A, Chan JY and Deshaies RJ: Transcription factor Nrf1 mediates the
proteasome recovery pathway after proteasome inhibition in
mammalian cells. Mol Cell. 38:17–28. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kobayashi A: Roles of NRF3 in the
hallmarks of cancer: Proteasomal inactivation of tumor suppressors.
Cancers (Basel). 12:26812020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Braun S, Hanselmann C, Gassmann MG, Auf
Dem Keller U, Born-Berclaz C, Chan K, Kan YW and Werner S: Nrf2
transcription factor, a novel target of keratinocyte growth factor
action which regulates gene expression and inflammation in the
healing skin wound. Mol Cell Biol. 22:5492–5505. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pepe AE, Xiao Q, Zampetaki A, Zhang Z,
Kobayashi A, Hu Y and Xu Q: Crucial role of nrf3 in smooth muscle
cell differentiation from stem cells. Circ Res. 106:870–979. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bury M, Le Calvé B, Lessard F, Dal Maso T,
Saliba J, Michiels C, Ferbeyre G and Blank V: NFE2L3 controls colon
cancer cell growth through regulation of DUX4, a CDK1 inhibitor.
Cell Rep. 29:1469–1481.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qian J, Huang C, Zhu Z, He Y, Wang Y, Feng
N, He S, Li X, Zhou L, Zhang C and Gong Y: NFE2L3 promotes tumor
progression and predicts a poor prognosis of bladder cancer.
Carcinogenesis. 43:457–468. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Waku T, Nakamura N, Koji M, Watanabe H,
Katoh H, Tatsumi C, Tamura N, Hatanaka A, Hirose S, Katayama H, et
al: NRF3-POMP-20S proteasome assembly axis promotes cancer
development via ubiquitin-independent proteolysis of p53 and
retinoblastoma protein. Mol Cell Biol. 40:e00597–19. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yin L, Duan JJ, Bian XW and Yu SC:
Triple-negative breast cancer molecular subtyping and treatment
progress. Breast Cancer Res. 22:612020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Katso R, Okkenhaug K, Ahmadi K, White S,
Timms J and Waterfield MD: Cellular function of phosphoinositide
3-kinases: Iimplications for development, homeostasis, and cancer.
Annu Rev Cell Dev Biol. 17:615–675. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gjelaj E and Hamel PA: Distinct
epithelial-to-mesenchymal transitions induced by PIK3CA (H1047R)
and PIK3CB. J Cell Sci. 134:jcs2482942021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grille SJ, Bellacosa A, Upson J,
Klein-Szanto AJ, van Roy F, Lee-Kwon W, Donowitz M, Tsichlis PN and
Larue L: The protein kinase Akt induces epithelial mesenchymal
transition and promotes enhanced motility and invasiveness of
squamous cell carcinoma lines. Cancer Res. 63:2172–2178.
2003.PubMed/NCBI
|
|
25
|
Cai BQ, Chen WW, Zhao JA, Hou W and Tang
JC: Nrf3 Promotes 5-FU resistance in colorectal cancer cells via
the NF-κB/BCL-2 signaling pathway in vitro and in vivo. J Oncol.
2021:93555552021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Trapnell C, William BA, Pertea G,
AMortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and
Pachter L: Transcript assembly and quantification by RNA-Seq
reveals unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Anders S, Pyl PT and Huber W: HTSeq-a
Python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Anders S and Huber W: Differential
expression of RNA-Seq data at the gene level-the DESeq package.
EMBL; 2013
|
|
31
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chowdhury A, Katoh H, Hatanaka A, Iwanari
H, Nakamura N, Hamakubo T, Natsume T, Waku T and Kobayashi A:
Multiple regulatory mechanisms of the biological function of NRF3
(NFE2L3) control cancer cell proliferation. Sci Rep. 7:124942017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Immonen A, Haapasaari KM, Skarp S,
Karihtala P and Teppo HR: NRF3 decreases during melanoma
carcinogenesis and is an independent prognostic marker in melanoma.
Oxid Med Cell Longev. 2022:22402232022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Waku T, Katayama H, Hiraoka M, Hatanaka A,
Nakamura N, Tanaka Y, Tamura N, Watanabe A and Kobayashi A: NFE2L1
and NFE2L3 complementarily maintain basal proteasome activity in
cancer cells through CPEB3-mediated translational repression. Mol
Cell Biol. 40:e00010–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chevillard G, Paquet M and Blank V: Nfe2l3
(Nrf3) deficiency predisposes mice to T-cell lymphoblastic
lymphoma. Blood. 117:2005–2008. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yu MM, Feng YH, Zheng L, Zhang J and Luo
GH: Short hairpin RNA-mediated knockdown of nuclear factor
erythroid 2-like 3 exhibits tumor-suppressing effects in
hepatocellular carcinoma cells. World J Gastroenterol.
25:1210–1223. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun J, Zheng Z, Chen Q, Pan Y, Lu H, Zhang
H, Yu Y and Dai Y: NRF3 suppresses breast cancer cell metastasis
and cell proliferation and is a favorable predictor of survival in
breast cancer. Onco Targets Ther. 12:3019–3030. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xie G, Ji A, Yuan Q, Jin Z, Yuan Y, Ren C,
Guo Z, Yao Q, Yang K, Lin X and Chen L: Tumour-initiating capacity
is independent of epithelial-mesenchymal transition status in
breast cancer cell lines. Br J Cancer. 110:2514–2523. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tang Y and Wang S, Li Y, Yuan C, Zhang J,
Xu Z, Hu Y, Shi H and Wang S: Simultaneous glutamine metabolism and
PD-L1 inhibition to enhance suppression of triple-negative breast
cancer. J Nanobiotechnology. 20:2162022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Miricescu D, Totan A, Stanescu-Spinu II,
Badoiu SC, Stefani C and Greabu M: PI3K/AKT/mTOR signaling pathway
in breast cancer: From molecular landscape to clinical aspects. Int
J Mol Sci. 22:1732020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Costa RLB, Han HS and Gradishar WJ:
Targeting the PI3K/AKT/mTOR pathway in triple-negative breast
cancer: A review. Breast Cancer Res Treat. 169:397–406. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Aiello NM and Kang Y: Context-dependent
EMT programs in cancer metastasis. J Exp Med. 216:1016–1026. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chaffer CL, San Juan BP, Lim E and
Weinberg RA: EMT, cell plasticity and metastasis. Cancer Metastasis
Rev. 35:645–654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bobal P, Lastovickova M and Bobalova J:
The Role of ATRA, natural ligand of retinoic acid receptors, on
EMT-related proteins in breast cancer: Minireview. Int J Mol Sci.
22:133452021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ren YG, Wang YJ, Hao S, Yang YH, Xiong WD
and Qiu L: NFE2L3 promotes malignant behavior and EMT of human
hepatocellular carcinoma (HepG2) cells via Wnt/β-catenin pathway. J
Cancer. 11:6939–6949. 2020. View Article : Google Scholar : PubMed/NCBI
|