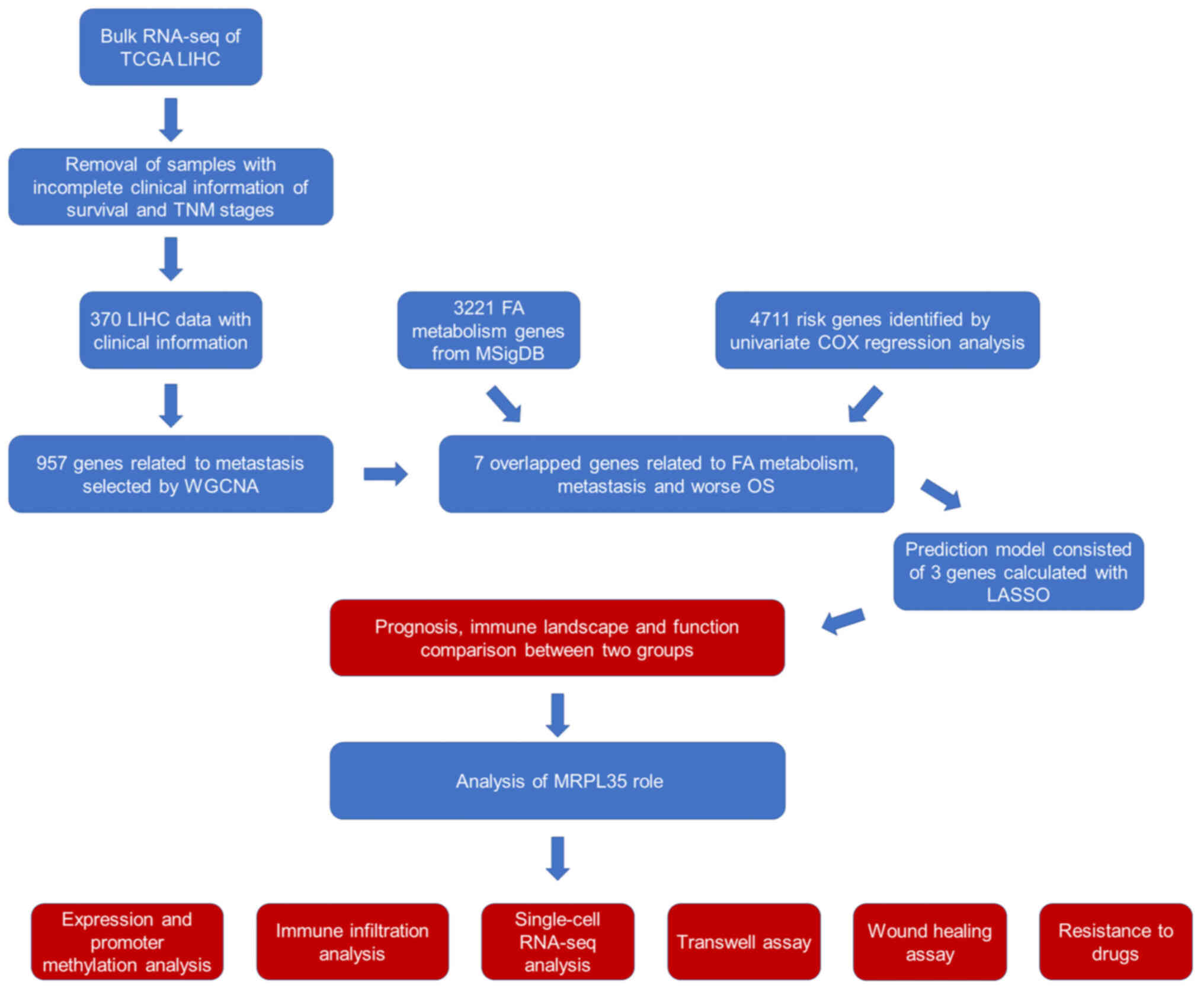
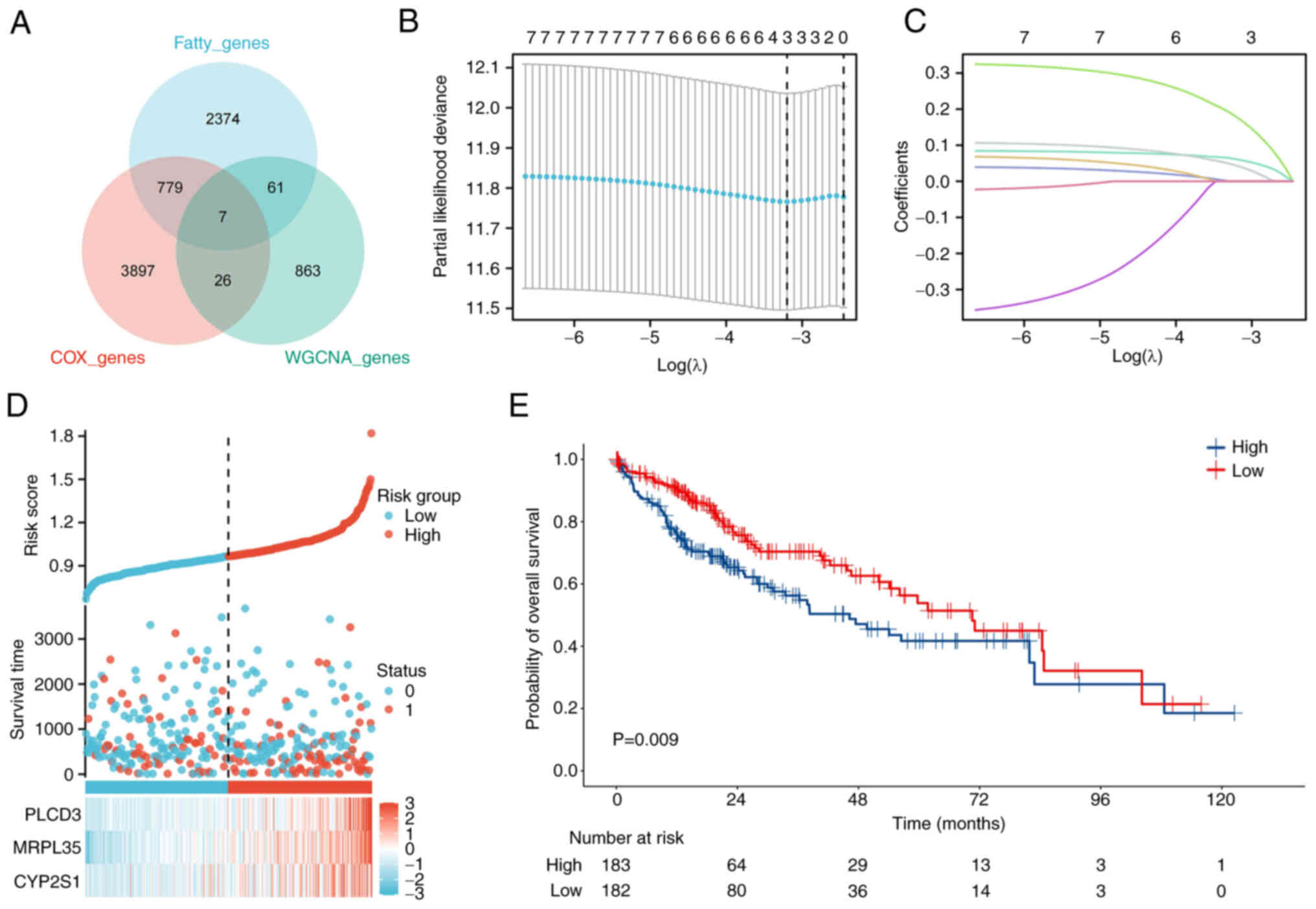
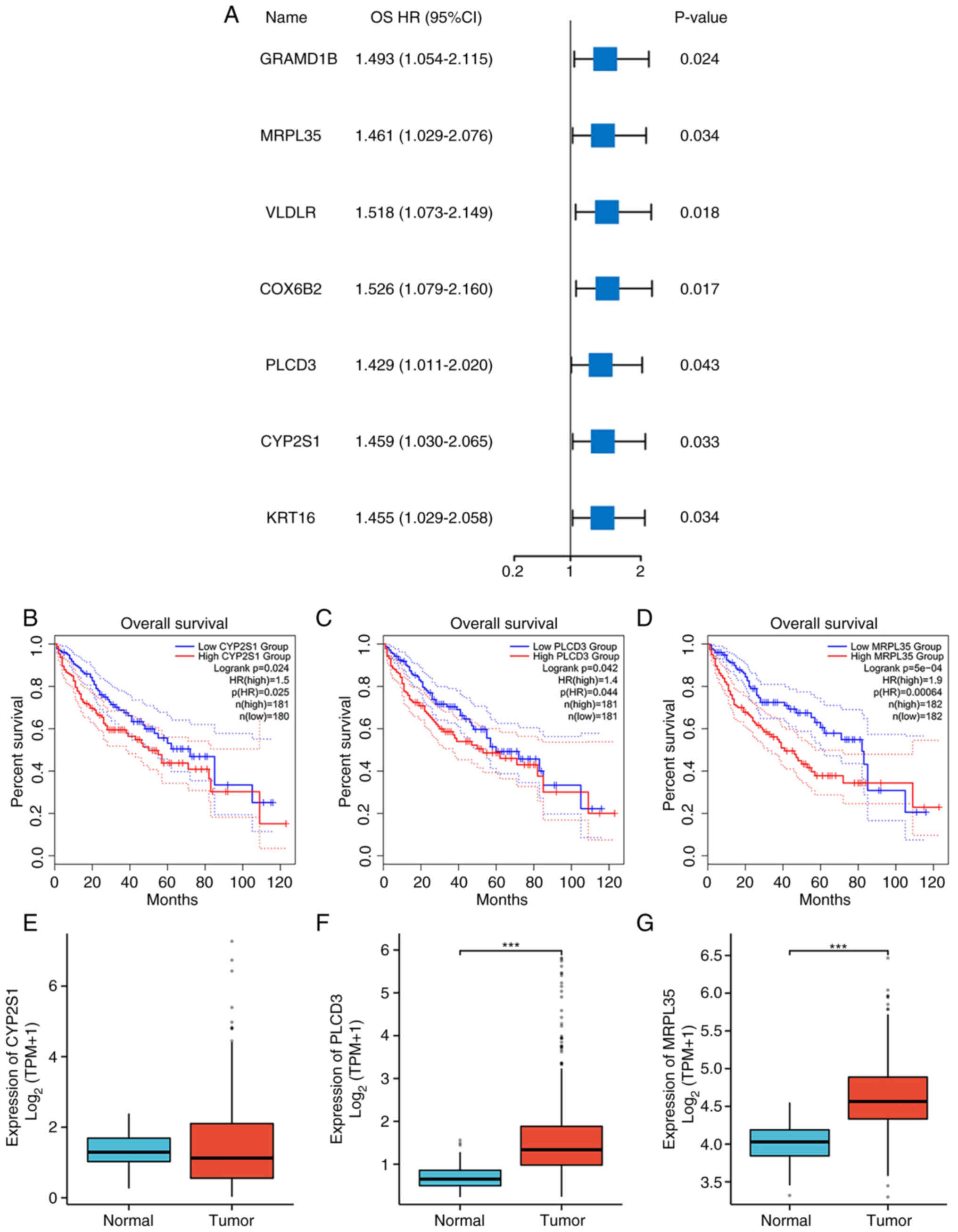
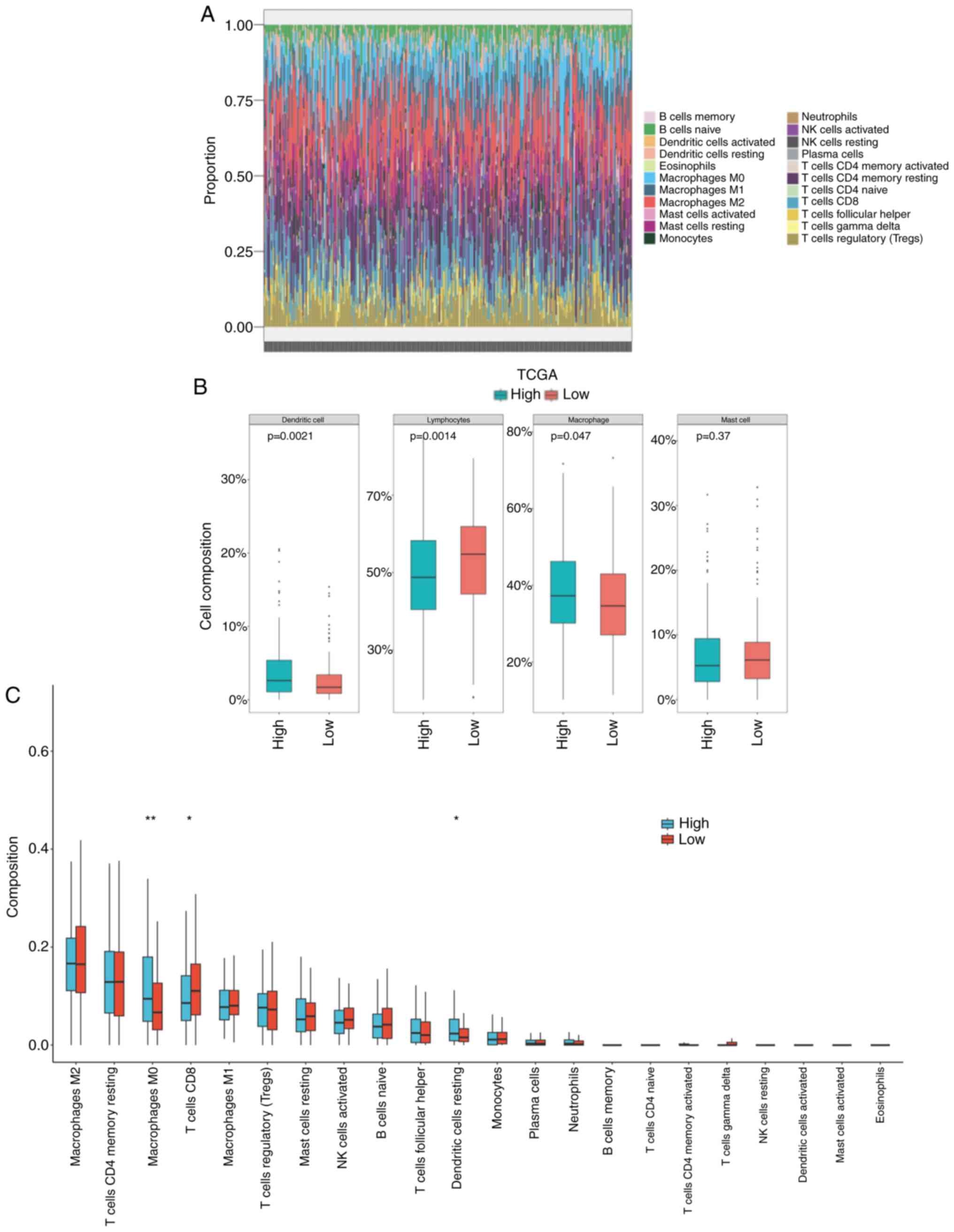
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000-14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reig M, Forner A, Rimola J, Ferrer-Fàbrega
J, Burrel M, Garcia-Criado Á, Kelley RK, Galle PR, Mazzaferro V,
Salem R, et al: BCLC strategy for prognosis prediction and
treatment recommendation: The 2022 update. J Hepatol. 76:681–693.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Swinnen JV, Brusselmans K and Verhoeven G:
Increased lipogenesis in cancer cells: New players, novel targets.
Curr Opin Clin Nutr Metab Care. 9:358–365. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Santos CR and Schulze A: Lipid metabolism
in cancer. FEBS J. 279:2610–2623. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Medes G, Thomas A and Weinhouse S:
Metabolism of neoplastic tissue. IV. A study of lipid synthesis in
neoplastic tissue slices in vitro. Cancer Res. 13:27–29.
1953.PubMed/NCBI
|
|
7
|
Alannan M, Fayyad-Kazan H, Trézéguet V and
Merched A: Targeting lipid metabolism in liver cancer.
Biochemistry. 59:3951–3964. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Budhu A, Roessler S, Zhao X, Yu Z, Forgues
M, Ji J, Karoly E, Qin LX, Ye QH, Jia HL, et al: Integrated
metabolite and gene expression profiles identify lipid biomarkers
associated with progression of hepatocellular carcinoma and patient
outcomes. Gastroenterology. 144:1066–1075.e1. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sangineto M, Villani R, Cavallone F,
Romano A, Loizzi D and Serviddio G: Lipid metabolism in development
and progression of hepatocellular carcinoma. Cancers (Basel).
12:14192020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumagai S, Togashi Y, Kamada T, Sugiyama
E, Nishinakamura H, Takeuchi Y, Vitaly K, Itahashi K, Maeda Y,
Matsui S, et al: The PD-1 expression balance between effector and
regulatory T cells predicts the clinical efficacy of PD-1 blockade
therapies. Nat Immunol. 21:1346–1358. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou L, Xia S, Liu Y, Ji Q, Li L, Gao X,
Guo X, Yi X and Chen F: A lipid metabolism-based prognostic risk
model for HBV-related hepatocellular carcinoma. Lipids Health Dis.
22:462023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feng T, Wu T, Zhang Y, Zhou L, Liu S, Li
L, Li M, Hu E, Wang Q, Fu X, et al: Stemness analysis uncovers that
the peroxisome proliferator-activated receptor signaling pathway
can mediate fatty acid homeostasis in sorafenib-resistant
hepatocellular carcinoma cells. Front Oncol. 12:9126942022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bensaad K, Favaro E, Lewis CA, Peck B,
Lord S, Collins JM, Pinnick KE, Wigfield S, Buffa FM, Li JL, et al:
Fatty acid uptake and lipid storage induced by HIF-1α contribute to
cell growth and survival after hypoxia-reoxygenation. Cell Rep.
9:349–365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murai H, Kodama T, Maesaka K, Tange S,
Motooka D, Suzuki Y, Shigematsu Y, Inamura K, Mise Y, Saiura A, et
al: Multiomics identifies the link between intratumor steatosis and
the exhausted tumor immune microenvironment in hepatocellular
carcinoma. Hepatology. 77:77–91. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gualdoni GA, Mayer KA, Göschl L, Boucheron
N, Ellmeier W and Zlabinger GJ: The AMP analog AICAR modulates the
Treg/Th17 axis through enhancement of fatty acid oxidation. FASEB
J. 30:3800–3809. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen L, Zhou Q, Liu J and Zhang W: CTNNB1
alternation is a potential biomarker for immunotherapy prognosis in
patients with hepatocellular carcinoma. Front Immunol.
12:7595652021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen S, Khatiwada S, Behary J, Kim R and
Zekry A: Modulation of the gut microbiome to improve clinical
outcomes in hepatocellular carcinoma. Cancers (Basel). 14:20992022.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bartha Á and Győrffy B: TNMplot.com: A web
tool for the comparison of gene expression in normal, tumor and
metastatic tissues. Int J Mol Sci. 22:26222021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ranstam J and Cook JA: Statistical nugget
LASSO regression. Br J Surg. 105:13482018. View Article : Google Scholar
|
|
24
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46(D1): D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen W, Song Z, Zhong X, Huang M, Shen D,
Gao P, Qian X, Wang M, He X, Wang T, et al: Sangerbox: A
comprehensive, interaction-friendly clinical bioinformatics
analysis platform. iMeta. 1:e362022. View Article : Google Scholar
|
|
26
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48((W1)): W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun D, Wang J, Han Y, Dong X, Ge J, Zheng
R, Shi X, Wang B, Li Z, Ren P, et al: TISCH: A comprehensive web
resource enabling interactive single-cell transcriptome
visualization of tumor microenvironment. Nucleic Acids Res. 49(D1):
D1420–D1430. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Reinhold WC, Sunshine M, Liu H, Varma S,
Kohn KW, Morris J, Doroshow J and Pommier Y: CellMiner: A web-based
suite of genomic and pharmacologic tools to explore transcript and
drug patterns in the NCI-60 cell line set. Cancer Res.
72:3499–3511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jeon YG, Kim YY, Lee G and Kim JB:
Physiological and pathological roles of lipogenesis. Nat Metab.
5:735–759. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin L, Wen J, Lin B, Chen H, Bhandari A,
Qi Y, Zheng D and Wang O: Phospholipase C Delta 3 inhibits
apoptosis and promotes proliferation, migration, and invasion of
thyroid cancer cells via Hippo pathway. Acta Biochim Biophys Sin
(Shanghai). 53:481–491. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu H, Yin Y, Jiang B, Feng Z, Cai T and Wu
S: Cuproptosis signature and PLCD3 predicts immune infiltration and
drug responses in osteosarcoma. Front Oncol. 13:11564552023.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yuan L, Li JX, Yang Y, Chen Y, Ma TT,
Liang S, Bu Y, Yu L and Nan Y: Depletion of MRPL35 inhibits gastric
carcinoma cell proliferation by regulating downstream signaling
proteins. World J Gastroenterol. 27:1785–1804. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang L, Lu P, Yan L, Yang L, Wang Y, Chen
J, Dai J, Li Y, Kang Z, Bai T, et al: MRPL35 is up-regulated in
colorectal cancer and regulates colorectal cancer cell growth and
apoptosis. Am J Pathol. 189:1105–1120. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Aiyappa-Maudsley R, Storr SJ, Rakha EA,
Green AR, Ellis IO and Martin SG: CYP2S1 and CYP2W1 expression is
associated with patient survival in breast cancer. J Pathol Clin
Res. 8:550–566. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo H, Zeng B, Wang L, Ge C, Zuo X, Li Y,
Ding W, Deng L, Zhang J, Qian X, et al: Knockdown CYP2S1 inhibits
lung cancer cells proliferation and migration. Cancer Biomark.
32:531–539. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang C, Yue C, Herrmann A, Song J,
Egelston C, Wang T, Zhang Z, Li W, Lee H, Aftabizadeh M, et al:
STAT3 activation-induced fatty acid oxidation in CD8+ T
effector cells is critical for obesity-promoted breast tumor
growth. Cell Metab. 31:148–161.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Datta J, Dai X, Bianchi A, De Castro Silva
I, Mehra S, Garrido VT, Lamichhane P, Singh SP, Zhou Z, Dosch AR,
et al: Combined MEK and STAT3 inhibition uncovers stromal
plasticity by enriching for cancer-associated fibroblasts with
mesenchymal stem cell-like features to overcome immunotherapy
resistance in pancreatic cancer. Gastroenterology. 163:1593–1612.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xie F, Zhou X, Su P, Li H, Tu Y, Du J, Pan
C, Wei X, Zheng M, Jin K, et al: Breast cancer cell-derived
extracellular vesicles promote CD8+ T cell exhaustion
via TGF-β type II receptor signaling. Nat Commun. 13:44612022.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bourayou E and Golub R: Signaling pathways
tuning innate lymphoid cell response to hepatocellular carcinoma.
Front Immunol. 13:8469232022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Johnson DE, O'Keefe RA and Grandis JR:
Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev
Clin Oncol. 15:234–248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fang Y, Zhang Q, Lv C, Guo Y, He Y, Guo P,
Wei Z, Xia Y and Dai Y: Mitochondrial fusion induced by
transforming growth factor-β1 serves as a switch that governs the
metabolic reprogramming during differentiation of regulatory T
cells. Redox Biol. 62:1027092023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Povero D, Chen Y, Johnson SM, McMahon CE,
Pan M, Bao H, Petterson XT, Blake E, Lauer KP, O'Brien DR, et al:
HILPDA promotes NASH-driven HCC development by restraining
intracellular fatty acid flux in hypoxia. J Hepatol. 79:378–393.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Piao GH, Piao WH, He Y, Zhang HH, Wang GQ
and Piao Z: Hyper-methylation of RIZ1 tumor suppressor gene is
involved in the early tumorigenesis of hepatocellular carcinoma.
Histol Histopathol. 23:1171–1175. 2008.PubMed/NCBI
|
|
46
|
Chen G, Fan X, Li Y, He L, Wang S, Dai Y,
Bin C, Zhou D and Lin H: Promoter aberrant methylation status of
ADRA1A is associated with hepatocellular carcinoma. Epigenetics.
15:684–701. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Alshabi AM, Vastrad B, Shaikh IA and
Vastrad C: Identification of crucial candidate genes and pathways
in glioblastoma multiform by bioinformatics analysis. Biomolecules.
9:2012019. View Article : Google Scholar : PubMed/NCBI
|