Introduction
Esophageal cancer ranks sixth in terms of cancer
deaths with an estimated 572,000 new cases and 509,000 deaths
yearly worldwide (1). Evidence has
shown that preoperative chemotherapy with cisplatin and
5-fluorouracil (CF) followed by surgical resection improves
survival of patients with esophageal squamous cell carcinoma (ESCC)
(2–4). However, some patients show resistance
to chemotherapy, the mechanism of which chemoresistance unclear,
thereby hindering successful outcomes.
CircRNAs are a class of endogenous noncoding RNAs
composed of single-stranded, covalently closed RNAs lacking 5′ and
3′ tails. It has been over 40 years since the existence of circRNAs
in viroid had been initially reported (5). CircRNAs are generated through
backsplicing events from linear primary transcripts, are resistant
to exonucleases, are typically nonpolyadenylated, and are highly
specific to cell type and developmental stage (6). Studies have revealed that circRNAs are
abundant, conserved, and stable with a tissue-specific expression
pattern (7–11). CircRNAs have been used as
potentially ideal biomarkers of disease owing to various
characteristics including universality, stability, conservatism,
and specificity. With the development of high-throughput
nonpolyadenylated and RNaseR-treated RNA sequencing (RNA-seq)
(12) and the mapping method using
bioinformatic tools to identify arrangements in an unbiased
assessment (13–16), it has become possible to establish
the profile of circRNAs expression, including novel transcript
isoforms.
Memczak demonstrated that circRNAs form a large
class of post-transcriptional regulators functioning as endogenous
miRNA sponges (6). Several
researches have revealed that the interaction between circRNAs and
miRNAs are associated with proliferation in cancer (17–19).
Furthermore, studies have shown that the interaction between
circRNA and miRNA plays an important role in the resistance of
gastric cancer to cisplatin (20).
In this study, we focused on the relationship
between circRNA and the resistance of ESCC to cisplatin in order to
provide new insights into circRNAs as potential biomarkers and
therapeutic targets for ESCC. Firstly, we analyzed the expression
profile of circRNAs in ESCC cells and cisplatin-resistant ESCC
cells using RNA-seq. Next, we examined the characteristic
expression of hsa_circ_0004365 in ESCC tissues and investigated its
function in vitro.
Materials and methods
Cell culture
Human ESCC cell lines TE8 (RBRC-RCB2098) and TE11
(RBRC-2100) (RIKEN BioResource Center, Ibaraki, Japan) were
cultured in RPMI-1640 medium supplemented with 10% fetal calf serum
(Thermo Fisher Scientific, Waltham, MA, USA) and 100 U/ml
penicillin and 100 µg/ml streptomycin (Nacalai Tesque, Kyoto,
Japan) in a humidified atmosphere containing 5% CO2 at
37°C. To generate cisplatin-resistant cell lines, TE8 and TE11
cells were cultured with increasing concentrations of cisplatin for
24 weeks. The established cisplatin-resistant cell lines, TE8R and
TE11R, were maintained under a constant cisplatin concentration of
6 µM. TE11 cells were also cultured with increasing concentrations
of 5-fluorouracil and docetaxel for 24 weeks. TE11 5-FU was
maintained at a 5-fluorouracil concentration of 4.0 µM and TE11 DTX
was maintained at a docetaxel concentration of 3.0 ng/ml.
Patients and tissues
In total, 82 samples of ESCC tissues were collected
from patients who were histologically diagnosed with primary
thoracic ESCC at Osaka University Hospital (Osaka, Japan) between
2017 April and 2020 September. The 47 samples were obtained via
biopsy before treatment, and the 35 samples were obtained by
surgical resection after neoadjuvant chemotherapy (NAC). All
samples were collected after obtaining written informed consent,
and this study was approved by the ethics committee of Osaka
University (approval no. 16305-4). Clinicopathologic findings were
classified according to the UICC-TNM Classification, seventh
edition (21). At our institution,
patients with ESCC primarily receive a DCF regimen as NAC, which
consisted of docetaxel 70 mg/m2 and cisplatin 70
mg/m2 on day 1 and 5-FU 700 mg/m2/day on days
1–5, every 3 weeks (22).
RNA preparation and quantitative
real-time polymerase chain reaction (qPCR)
Total RNA was extracted from ESCC cells or tissues
using TRIzol Reagent (Invitrogen, CA, USA) according to the
manufacturer's instructions. The quantity and quality of RNA
samples were measured using NanoDrop ND-1000 (Thermo Scientific,
DE, USA). The samples were reverse transcribed into cDNA using a
reverse transcription system (Promega, Madison, WI) according to
the manufacturer's protocol. Reverse transcription quantitative
real-time polymerase chain reaction (RT-qPCR) was performed on a
ViiA™ 7 real-time PCR system (Applied Biosystems, Foster City, CA,
USA) using SYBR Green (TOYOBO, Osaka, Japan) via the following
three-step protocol: 95°C for 2 min, (95°C for 15 sec, 65°C for 15
sec, and 72°C for 45 sec) ×40 cycles, melting curve analysis from
60 to 95°C with reads every 0.5°C. PCR was performed in triplicate,
with the expression values normalized to the mRNA expression of
β-actin calculated using the 2−ΔΔCt method (23). The genes and primers are specified
in Table SI.
Next-generation RNA sequencing and
analysis
Total RNAs were treated using the Epicenter
Ribo-Minus rRNA Removal Kit (Illumina, CA, USA) and RNAse R
(Epicenter, CA, USA) to remove ribosomal and linear RNA (6). Thereafter, RNA-seq libraries were
constructed using TruSeq Stranded mRNA Library Prep (Illumina, CA,
USA) following the manufacturer's protocol while skipping the
process of purifying the polyA-containing mRNA molecules with
oligo-dT attached magnetic beads. The BioAnalyzer 2100 system
(Agilent Technologies, Santa Clara, CA, USA) was used to confirm
the quality and quantity of the libraries. The RNA-seq library was
sequenced using an Illumina HiSeq 3000 instrument (Illumina, San
Diego, CA), and 150 bp paired-end reads were generated. Sequencing
reads were mapped independently using CIRCexplorer2 (13,14),
which is one of the circular RNA detection output tools based on
Python, a general-purpose programming language. Cirexplorer uses
TopHat (15) and TopHat-Fusion
(16) to detect backsplicing
junction reads. To compare differences in circRNA expression
profiles between TE11 and TE11R, we calculated the fold change
between the groups for each circRNA. A t-test was used to estimate
the statistical significance of the differences. A significant
difference in the expression of circRNA was defined by a
fold-change >3.0 or <0.5 with a P-value of <0.05.
CircRNA-microRNA-mRNA network
analysis
mRNAs involved in the pathways associated with
conferring cisplatin resistance to tumor cells were selected for
CircRNA-microRNA-mRNA network construction. The binding sites in
the 3′-UTR between differentially expressed miRNAs and the selected
mRNA were determined using Targetscan (http://www.targetscan.org/) to predict the mRNAs
targeted by miRNAs filtered using a cumulative weighted context
score of ≤0.8. We used Cytoscape software to visualize the data
(24) and the bioinformatic
database CircInteractome to predict the sequence of miRNA-binding
sites of circ_0004365 (25).
Silencing of circ_0004365
Small interfering RNA (siRNA) of circ_0004365 was
synthesized by Sigma-Aldrich (St. Louis, MO) targeting to sequence
of the fusion region of circ_0004365. TE11 and TE11R cells were
transfected with circ_0004365 siRNAs using the
Lipofectamine® RNAiMAX transfection reagent (Thermo
Fisher Scientific, Waltham, MA, USA) according to the
manufacturer's protocol. Briefly, cells were seeded at 2
×105 cells/well in 6-well plates added to a final
concentration of 50 nM using Lipofectamine® RNAiMAX
(Thermo Fisher Scientific, Waltham, MA, USA) and cultured for 24 h
at 37°C in 5% CO2. The sequence of circ_0004365 siRNA
was sense CUGUUUCUCGGAACAGGACdT dT and antisense
GUCCUGUUCCGAGAAACAGdTdT.
In vitro drug sensitivity assay
A total of 4.0×103 cells per well were
seeded in 96-well plates. Cell viability was then assessed using
the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide
(MTT) assay 48 h after the addition of cisplatin. Absorbance of the
converted dye was measured at a wavelength of 550 nm with
background subtraction at 665 nm using an iMark™ Microplate
Absorbance Reader (BIO-RAD, Tokyo, Japan). The concentration at
which the drug produced 50% growth inhibition (IC50) was estimated
using the relative survival curve.
Evaluation of histopathological
responses to chemotherapy
Histopathological tumor response was evaluated
according to the histological criteria of the Japanese Esophageal
Society (26) and was classified
into five categories according to the proportion of tumor
degeneration and necrosis: Grade 3 (markedly effective; no viable
cancer cells); grade 2 (moderately effective; viable cancer cells
accounting for less than 1/3 of the tumor tissue while other cancer
cells showed severe degeneration or necrosis); grade 1 (slightly
effective; apparently viable cancer cells accounting for 1/3 or
more of the tumor tissue, but some evidence of degenerating cancer
tissue or cells was present), and grade 0 (ineffective; denoting no
discernible therapeutic effect on cancer tissue or cells). Grade 1
lesions can also be subclassified into grade 1a (viable cancer
cells accounting for 2/3 or more of the tumor tissue) and grade 1b
(viable cancer cells accounting for 1/3 or more, but less than 2/3,
of the tumor tissue).
Statistical analysis
To test for statistically significant differences
between the two groups, continuous data were compared using the
unpaired Student's t-test. Differences were considered significant
at two-sided P-values of <0.05. All analyses were performed
using JMP version 14.0 (SAS Institute, Cary, NC).
Results
RNA-seq analysis revealed circRNA
expression profile in TE11 and cisplatin-resistant TE11
To identify circRNA expression patterns associated
with cisplatin resistance in ESCC cells, we performed RNA-seq of
three pairs of TE11 and cisplatin-resistant TE11 (TE11R). A total
of 10451 circRNAs were consistent with Circexplorer2 (16). Thereafter, we compared circRNAs
expression profiles between TE11 and TE11R to extract candidate
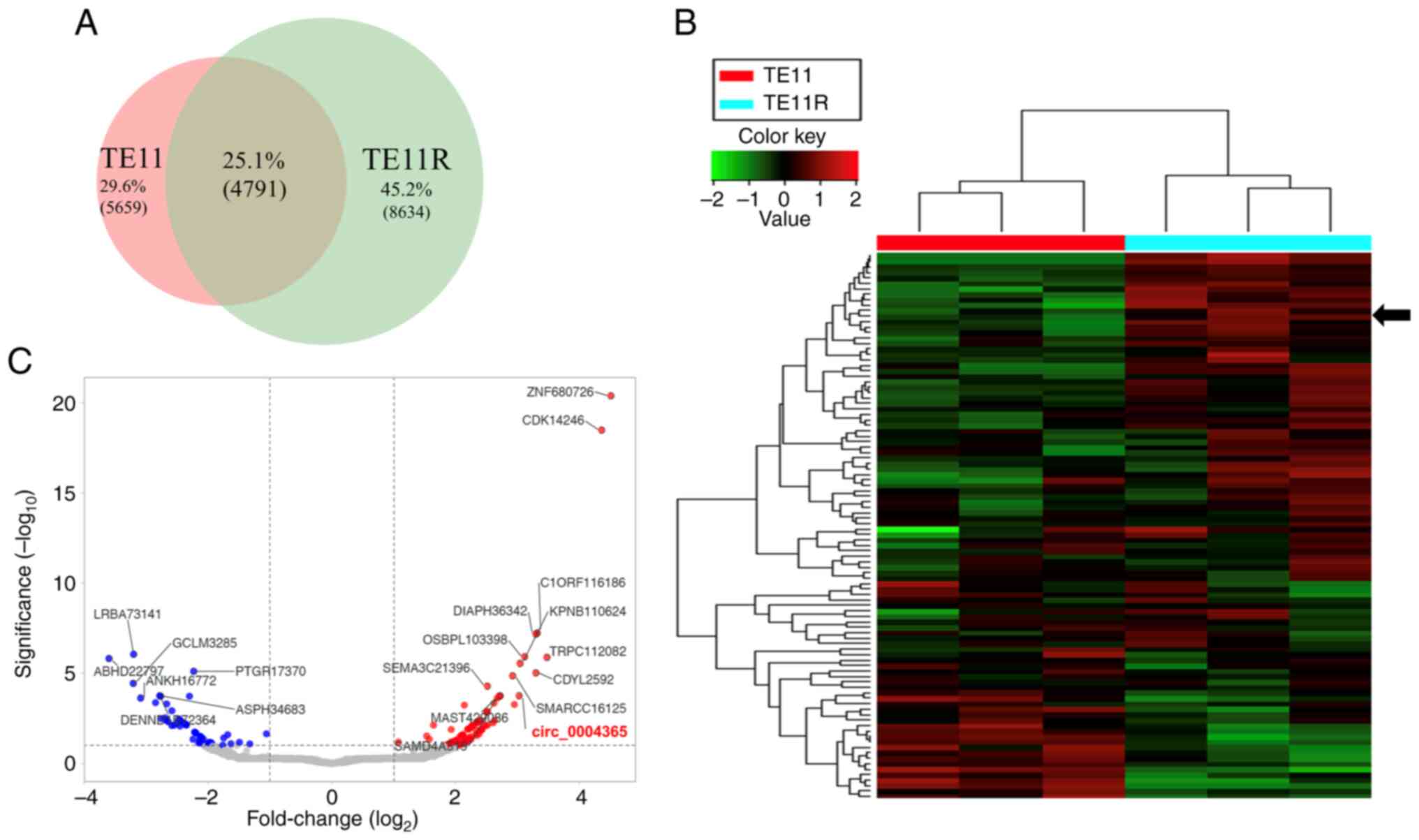
circRNA. Accordingly, 4791 circRNAs were expressed both in TE11 and
TE11R cells (Fig. 1A). Through
expression intensity sorting within TE11 and TE11R cells, the
mostly greater and lesser circRNAs in TE11R cells compared to those
in TE11 cells are shown via hierarchical clustering (Fig. 1B). Variations in circRNA expression
are demonstrated in the volcano plot (Fig. 1C). Among them, we selected 10
candidate circRNAs that were significantly upregulated and
downregulated in TE11R relative to TE11 (Tables I and II).
 | Table I.Top 10 upregulated circRNAs in TE11R
as compared with TE11. |
Table I.
Top 10 upregulated circRNAs in TE11R
as compared with TE11.
| Gene symbol | ID | Fold-change | P-value |
|---|
| ANKRD17 |
hsa_circ_0007883 | 3.12378 | 0.043183 |
| CREBBP |
hsa_circ_0007637 | 4.312371 | 0.045842 |
| FAM185A |
hsa_circ_0008271 | 3.093274 | 0.00134 |
| GRHL2 |
hsa_circ_0085173 | 3.102087 | 0.000498 |
| NFATC3 |
hsa_circ_0005615 | 4.819771 | 0.008801 |
| NPEPPS |
hsa_circ_0004622 | 3.09124 | 0.017307 |
| RACGAP1 |
hsa_circ_0009035 | 3.424382 | 0.039387 |
| SEMA3C |
hsa_circ_0004365 | 5.401536 | 0.001506 |
| TNFRSF21 |
hsa_circ_0001610 | 4.042647 | 0.004966 |
| ZRANB1 |
hsa_circ_0000268 | 3.42829 | 0.038204 |
 | Table II.Top 10 downregulated circRNAs in
TE11R as compared with TE11. |
Table II.
Top 10 downregulated circRNAs in
TE11R as compared with TE11.
| Gene symbol | ID | Fold-change | P-value |
|---|
| ASPH |
hsa_circ_0084615 | 0.442011529 | 0.049328 |
| PUM12831 |
hsa_circ_0000043 | 0.492310713 | 0.012358 |
| PTGR1 |
hsa_circ_0008043 | 0.364584794 | 0.039294 |
| PTGR1 |
hsa_circ_0003731 | 0.192529045 | 0.000263 |
| RAPGEF5 |
hsa_circ_0001681 | 0.321428355 | 0.010674 |
| PTGR1 |
hsa_circ_0088030 | 0.266651031 | 0.040861 |
| CELSR1 |
hsa_circ_0063809 | 0.383964597 | 0.030733 |
| VWA8 |
hsa_circ_0000605 | 0.410925792 | 0.00274 |
| IFAM53B |
hsa_circ_0000267 | 0.175883872 | 0.001147 |
| PSD3 |
hsa_circ_0004458 | 0.313919316 | 0.018126 |
Circ_0004365 is upregulated in
cisplatin-resistant TE11 cells
To validate the expression profile established by
RNA-seq and CIRCexplorer2 (16), we
designed divergent primers of each circRNA candidate to
specifically target the circular junction and performed RT-qPCR
using ESCC cells and tissues. Consistent with the RNA-seq results,
we found that the expression of circ_0004365, mapped on the SEMA3C
gene (907 bp) (Fig. 2A, B), was
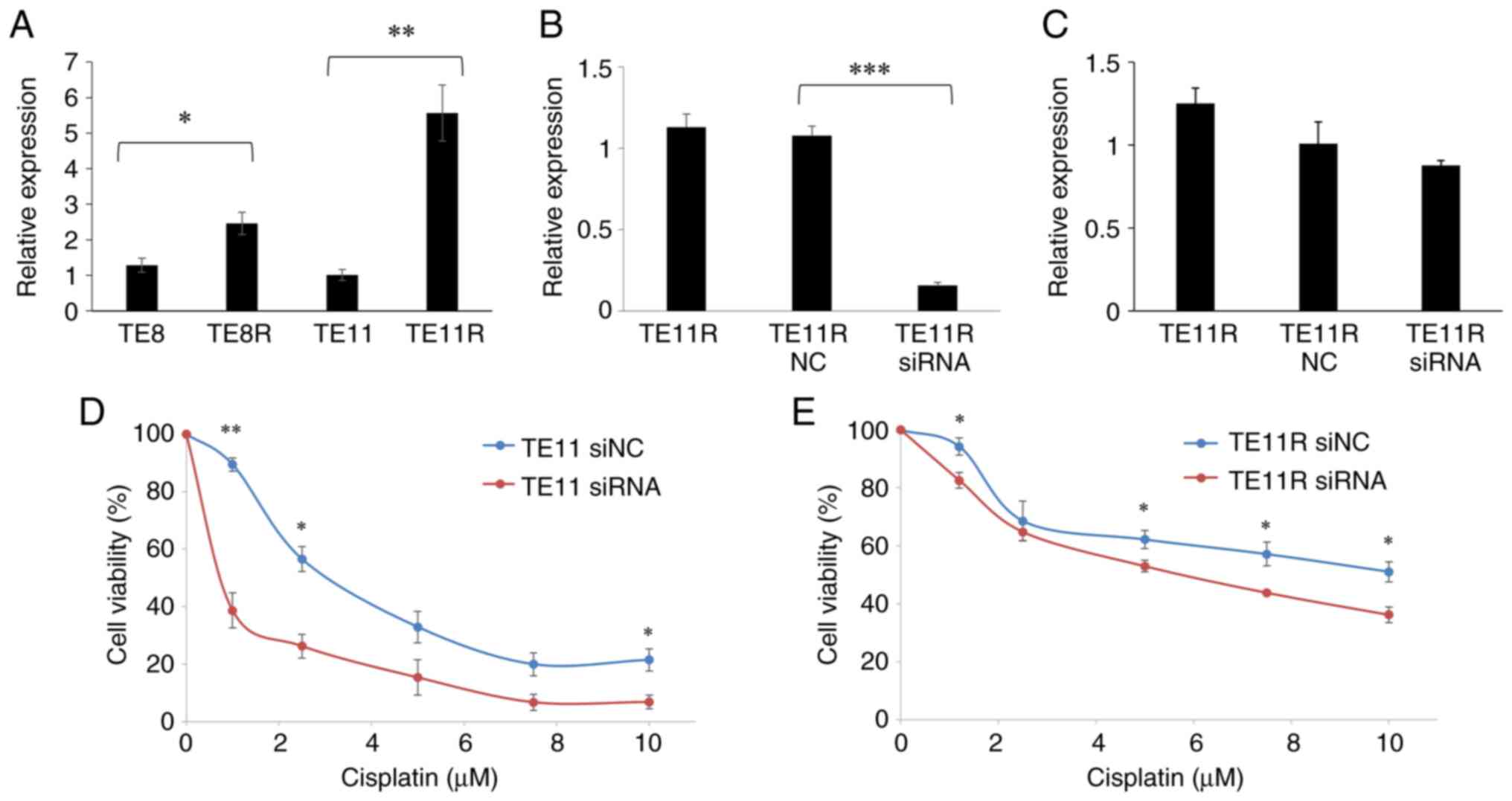
significantly greater in TE11R than in TE11. At the same time, the
upregulating expression pattern of circ_0004365 in resistant cells
was observed in TE8 and TE8R (Fig.
3A). In the meanwhile, there was no difference in circ_0004365
expression between TE11, TE11 5-FU, and TE11 DTX (Fig. S1). Furthermore, the expression of
circ_0004365 was broadly detected in human ESCC and nontumor
tissues both before and after NAC (Fig. S2A, B). Interestingly, the relative
expression of circ_0004365 was significantly greater in ESCC tumor
tissues after NAC than in matched tumor tissues before NAC
(Fig. S2C). Thus, the present
study focused on the relationship between cisplatin resistance and
the expression of circ_0004365 in ESCC.
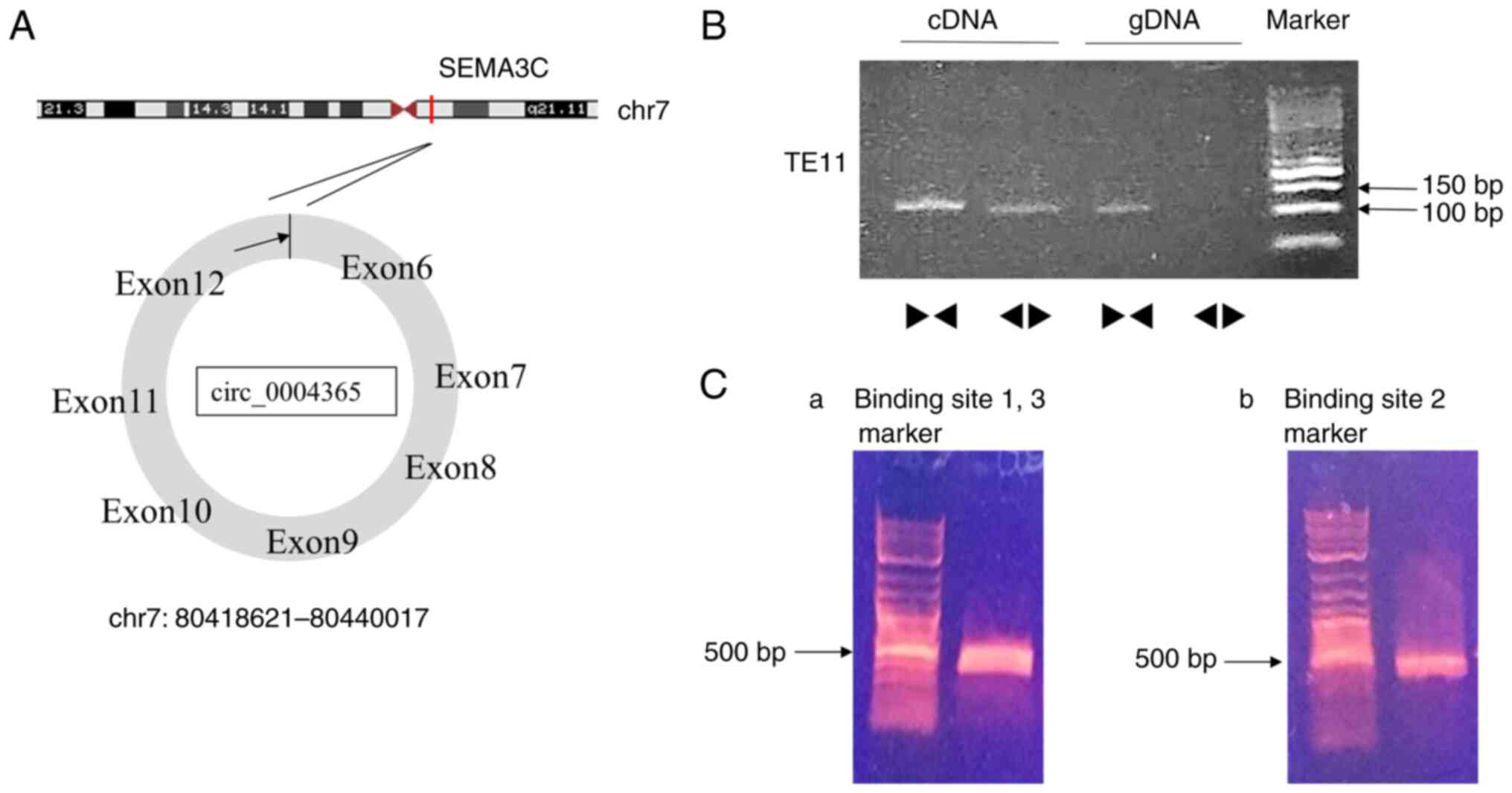 | Figure 2.(A) A schematic illustration showing
that circ_0004365 is derived from exons 6, 7, 8, 9, 10, 11, and 12
of the SEMA3C gene. (B) RT-qPCR products of TE11 with divergent
primers showing circularization of circ_0004365. (Ca) RT-qPCR
products with divergent primers showing binding sites 1, 3 of
circ_0004365. (Cb) RT-qPCR products with divergent primers showing
binding sites 2 of circ_0004365. circRNA, circular RNA; SEMA3C,
Semaphorin 3C; cDNA, complementary DNA; gDNA, genomic DNA; RT-qPCR,
reverse transcription-quantitative PCR. |
Circ_0004365 and cisplatin resistance
in vitro
Firstly, we designed siRNA oligonucleotides
targeting the specific junction of circ_0004365. RT-qPCR confirmed
that they successfully knocked down circ_0004365 expression
(Fig. 3B) without affecting the
levels of endogenous linear SEMA3C transcript (Fig. 3C). Thereafter, the MTT assay
revealed that TE11 with circ_0003465 knockdown had significantly
lower resistance to cisplatin compared to control-transfected cells
(Fig. 3D).
Similarly, TE11R cells transfected with circ_0003465
siRNA exhibited significantly lower resistance to cisplatin
compared to control-transfected cells (Fig. 3E). Consequently, our findings
suggested that circ_0004365 regulates the resistance of ESCC cells
to cisplatin.
Circ_004365 expression in ESCC
tissues
RT-qPCR revealed that circ_0004365 expression was
observed in ESCC tissues and nontumor esophageal tissues. Firstly,
no significant difference in the expression level of circ_0004365
was noted between ESCC and nontumor esophageal tissues (Fig. S2A, B). However, a comparison of the
15 pairs of ESCC tissues obtained before NAC and after NAC showed
that the expression of circ_0004365 was significantly upregulated
in tissues after NAC (P=0.03, Fig.
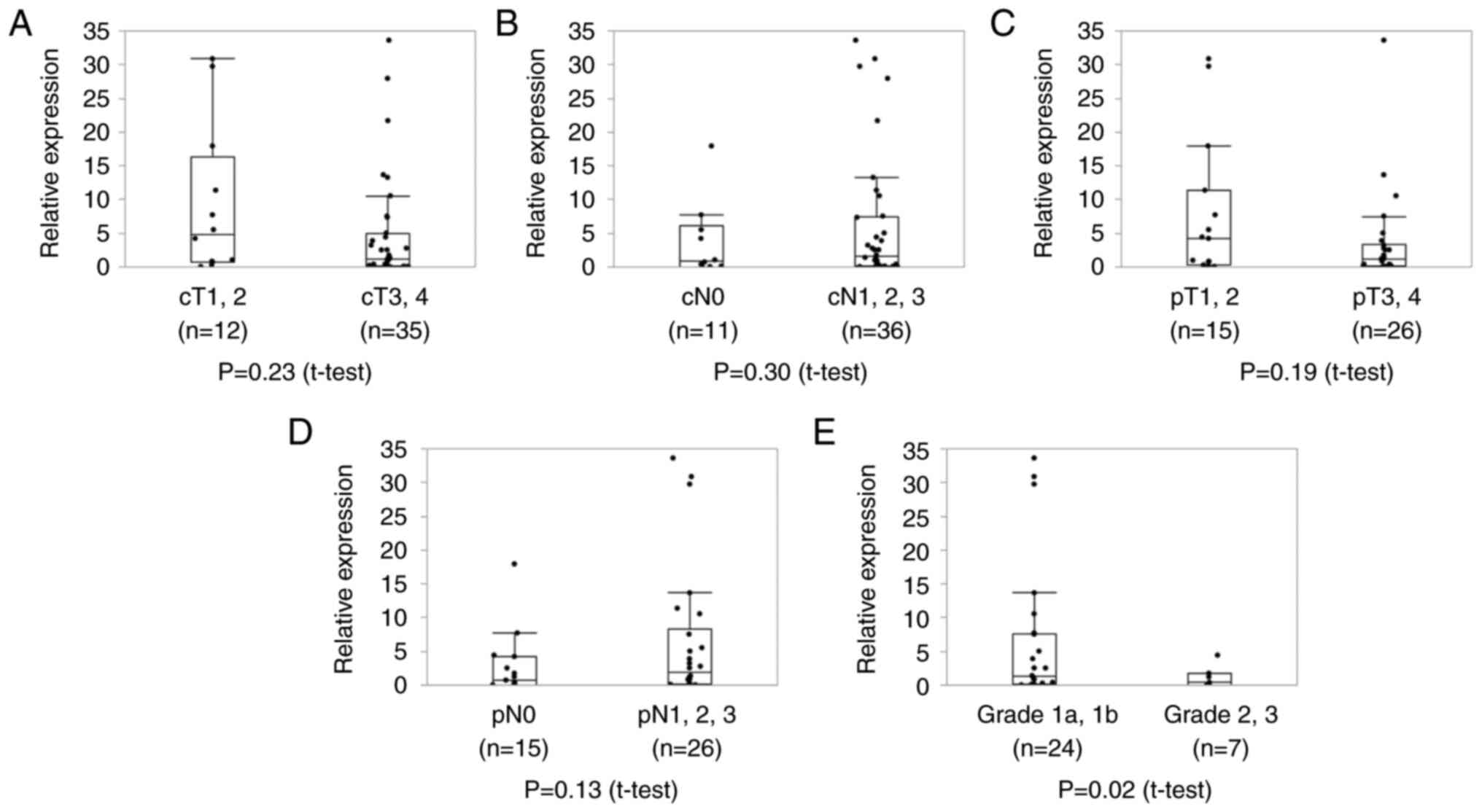
S2C). Next, we analyzed whether circ_0004365 expression was
correlated with the characteristics of ESCC patients (Table III). Accordingly, no significant
correlation was observed between circ_0004365 expression level
before NAC and cStage and pStage (Fig.
4A-D). Interestingly, the expression of circ_0004365 before NAC
was significantly upregulated in patients with poor pathological
response (P=0.02, Fig. 4E).
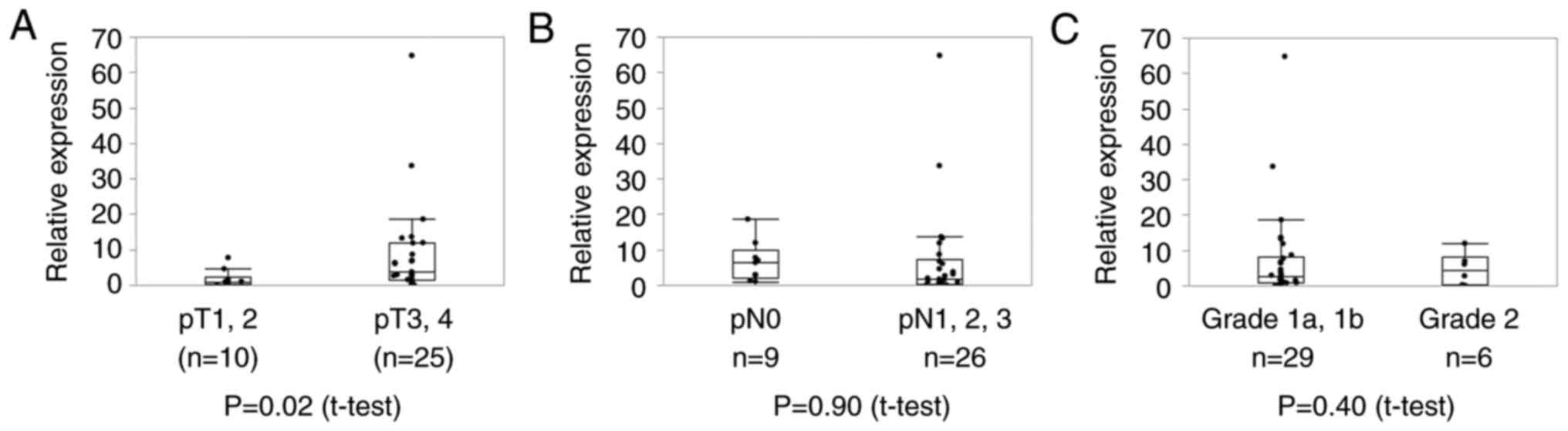
Furthermore, patients with advanced pT Stage showed an upregulation
of circ_0004365 expression after NAC (P=0.02, Fig. 5A). No significant difference was
noted between the expression level of circ_0004365 after NAC and pN
and pathological response (Fig. 5B,
C). Together, these data suggested that increased circ_0004365
expression in ESCC patients was correlated with resistance to
chemotherapy, including cisplatin.
 | Table III.The clinical characteristics of
patients with esophageal squamous cell carcinoma. |
Table III.
The clinical characteristics of
patients with esophageal squamous cell carcinoma.
| Characteristic | Cases, n (%)
Tissues before NAC | Tissues after
NAC |
|---|
| Sex |
|
|
|
Male | 38 (81) | 28 (72) |
|
Female | 9 (19) | 11 (18) |
| Ethnicity |
|
|
|
Asian | 47 | 39 |
|
Others | 0 | 0 |
| Tumor location |
|
|
| Ut | 5 (11) | 3 (8) |
| Mt | 21 (45) | 16 (41) |
| Lt | 21 (45) | 20 (51) |
| Histology |
|
|
|
Poor | 0 | 7 (18) |
|
Well/moderate | 42 (89) | 29 (74) |
| Not
evaluable | 5 (11) | 3 (8) |
| cTa |
|
|
| 1 | 4 (9) | 1 (3) |
| 2 | 8 (17) | 5 (13) |
| 3 | 28 (60) | 27 (69) |
| 4 | 7 (15) | 6 (14) |
| cNa |
|
|
| 0 | 11 (23) | 7 (18) |
| 1 | 24 (51) | 18 (46) |
| 2 | 11 (23) | 14 (36) |
| 3 | 1 (2) | 0 |
| cMa |
|
|
| 0 | 40 (85) | 31 (79) |
| 1 | 7 (15) | 8 (21) |
| cStagea |
|
|
| I | 7 (15) | 4 (10) |
| II | 7 (15) | 5 (13) |
|
III | 26 (55) | 22 (56) |
| IV | 7 (15) | 8 (21) |
| Preoperative
chemotherapy |
|
|
|
Present | 34 (72) | 39 |
|
Absent | 13 (28) | 0 |
| Operation |
|
|
|
Absent | 5 (11) | 3 (8) |
|
Present | 42 (89) | 36 (92) |
Analysis of the regulatory network of
circRNAs, miRNAs, and mRNAs
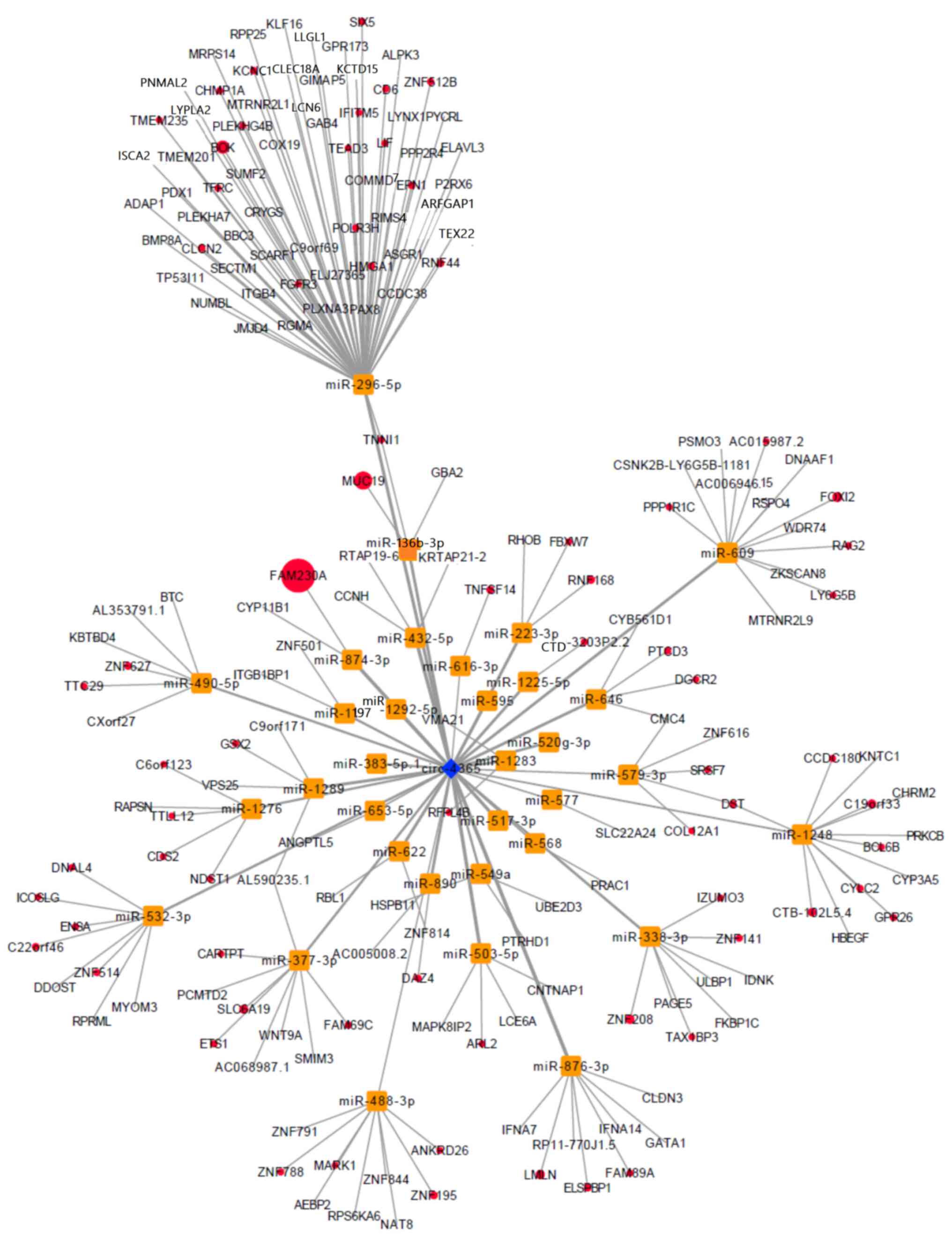
Biological prediction and analysis predicted that
circ_0004365 binds 33 miRNAs to regulate the expression of 187
mRNAs (Table SII), and the ceRNA
network, circRNAs, miRNAs, and mRNAs network was visualized
(Fig. 6). CircInteractome (26) predicted that miR-503 had the most
binding sites for circ_0004365 (Fig.
S3A). The sequence of 3 miRNA-binding sites of circ_0004365 and
miR-503 is shown in Fig. S3B. We
designed 2 pairs of divergent primers to specifically target the
binding sites (Table SI). The one
targets binding site 1 and 3 continuously and the other targets
binding site 2. RT-PCR products with divergent primers showed
binding 3 sites of circ_0004365 (Fig.
2C a, b). Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analysis revealed that hsa04390: Hippo signaling pathway,
hsa04650: Natural killer cell mediated cytotoxicity, hsa04623:
Cytosolic DNA-sensing pathway, and hsa04514: Cell adhesion
molecules were enriched (Table
SIII).
Discussion
Cisplatin is among the most predominant drugs for
chemotherapy (27). Several studies
have shown that preoperative chemotherapy including cisplatin
followed by surgical resection improves survival from ESCC
(4). However, the prognosis of ESCC
still remains poor, with it ranking sixth in terms of cancer deaths
worldwide (1). As such, elucidating
the mechanisms causing resistance to cisplatin in ESCC could
improve the prognosis of ESCC patients. The present study initially
clarified the role of circ_0004365 in ESCC. In line with this, we
performed RNA-seq analysis, with CIRCexproler showing that several
circRNAs were significantly upregulated in cisplatin-resistant TE11
cells than in parental TE11. Among those validated by RT-qPCR,
circ_0004365 was greater in cisplatin-resistant TE11 cells than in
parental TE11 cells. This increased expression pattern in
cisplatin-resistant cells was also observed in cisplatin-resistant
TE8 cells. Knockdown of circ_0004365 by siRNA enhanced cisplatin
sensitivity in TE11 cells and cisplatin-resistant TE11 cells,
suggesting that circ_0004365 may play an important role in the
development of ESCC resistance to cisplatin.
According to our information, the present report's
initial goal was to analyze the association between circ_0004365
and cancer drugs. Exons 6, 7, 8, 9, 10, 11, and 12 of the
SEMA3C gene were used to create circ_0004365. SEMA3C
is reportedly an oncogene, and it has been shown in certain studies
that high levels of SEMA3C expression are associated with cancer
development and poor prognosis in several cancers, such as breast,
prostate, gastric, liver, pancreatic, and lung cancers (28). It was shown that SEMA3C is
overexpressed in 85% of glioblastomas and helps to maintain glioma
cancer stem-like cell self-renewal and drive tumor progression by
promoting Wnt signaling (29,30).
In the present study, the expression of SEMA3C in TE11R was not
affected by knocking down circ_0004365 expression. There may be no
association between SEMA3C expression and circ_0004365 expression
so that we didn't quantify SEMA3C expression in ESCC tissues.
A novel class of noncoding RNAs, circRNAs are
expected to be biomarkers of disease owing to their universality,
stability, conservatism, and specificity (7–11).
Studies have already reported that some circRNAs are potential
diagnostic and prognostic markers in patients with ESCC. Xu et
al demonstrated that that hsa-circ_0000654 expression in ESCC
tissues was significantly higher compared to adjacent nontumor
tissues and that high circ_0000654 expression was notably
correlated with the higher T stage and local lymph node metastasis
(31). Shi Y reported that
hsa-circ_0006168 expression level was notably greater in ESCC
tissues than in matched normal tissues and that high
hsa_circ_0006168 expression was markedly correlated with the TNM
stage and lymph node metastasis of ESCC patients (32). Furthermore, more and more in
vitro studies have reported that some circRNAs can regulate the
proliferation, migration, invasion, apoptosis, cell cycle, and
epithelial-mesenchymal transition (EMT) (31–35).
Evidence accumulated to date have shown that the expression of some
circRNAs in ESCC are certainly associated with the progression of
ESCC; however, only a handful reports have clinically demonstrated
the value of circRNAs as biomarkers. For instance, Hu et al
reported (36) that the plasma
levels of circGSK3β were reduced after surgery and that circGSK3β
levels were much higher in patients with recurrence/metastasis 10
months after surgery than in those without recurrence/metastasis.
This suggests that plasma circGSK3β level may be a valuable
clinical predictor of ESCC. The present study also demonstrated the
possible utility of circRNA as a clinical predictor of cisplatin
sensitivity in ESCC. Evidence has shown that circ_0004365
expression before NAC was significantly upregulated in patients
with poor pathological response. This suggests that the
upregulation of circ_0004365 may predict the sensitivity to
cisplatin in ESCC. Furthermore, a significantly upregulation of
circ_0004365 expression after NAC was observed in patients with
advanced pT Stage. This suggests that cisplatin-resistant patients
might still have numerous tumor cells remaining given that the
expression of circ_0004365 was still upregulated after NAC. Taken
together, these results suggest that increased circ_0004365
expression in ESCC patients may be correlated with resistance to
cisplatin and that circ_0004365 could potentially be used as a
clinical biomarker or a therapeutic target.
Over the past 50 years, cisplatin has become a
common chemotherapeutic drug for numerous tumors, including ESCC
and testicular, lung, bladder, ovarian, and liver cancers, among
others (31,37). However, resistance to cisplatin
often develops with continuous application. In line with this, the
mechanism of cisplatin resistance has been widely studied to
improve the prognosis of the patients with various cancers. There
also have been several reports indicating the correlation between
circRNAs and cisplatin resistance. Zou FW reported that circ_001275
was upregulated in cisplatin-resistant esophageal cancer tissues
and cell lines and that circRNA_001275 promoted the proliferation
of cisplatin-resistant cells (38).
Chang et al found that circ_0007142 was increased in
cisplatin-resistant ESCC and that silencing of circ_0007142
enhanced cisplatin sensitivity in ESCC (39).
With the development of high-throughput sequencing
technology and broad use of bioinformatics analysis, more and more
circular RNAs and their regulatory downstream have been predicted
and validated. In the present study, we constructed a
circRNA-miR-mRNA network in ESCC using bioinformatics analysis. The
bioinformatic database CircInteractome (26) predicted that miR-503 had the most
binding sites for circ_0004365, which RT-PCR products with
divergent primers showed. Visualization of the circRNA-miR-mRNA
network also revealed the interaction between circ_0004365 and
miR-503. Interestingly, Qiu T reported that miR-503 regulated the
resistance of non-small cell lung cancer cells to cisplatin and
cell apoptosis, at least in part, by targeting Bcl-2 (40). Evidence has suggested that
circ_0004365 could possibly influence cisplatin resistance through
downstream processing as may be corroborated by the aforementioned
results, which showed that circ_0004365 has binding sites with
miR-503 for sponging that may affect the cisplatin resistance. KEGG
analysis demonstrated that BBC3, TEAD3, WNT9A, LLGL1, and BMP8A
genes were enriched in the Hippo signaling pathway. It has been
reported that Hippo signaling pathway is frequently mutated in
ESCC. Song et al previously reported that the Hippo
signaling pathway plays an important regulatory role in the
development of esophageal cancer (41). Furthermore, Zhou et al
demonstrated that Hippo signaling pathway transcription factor YAP
as a molecular target for arsenic induced synthetic effects with
cisplatin treatment in ESCC (42).
More and more studies have revealed the biological
mechanism of circRNAs as competitive endogenous noncoding RNAs, not
only for sponging miR but also for regulating parental gene
expression, transcriptional translation, and protein modification
(7,9,12,14,43–45).
Future studies should analyze the mechanisms by which circ_0004365
influences cisplatin resistance. Furthermore, there is a need to
evaluate a larger number of samples and long-term prognosis such as
survival and recurrence.
In conclusion, we identified a novel circ_0004365
derived from SEMA3C via RNA-seq. Our findings suggested that
circ_0004365 may play an important role in the cisplatin resistance
of ESCC, which may provide new insights into its use as a potential
biomarker and therapeutic target for patients with
cisplatin-resistant ESCC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by JSPS KAKENHI (grant no.
JP18K16310).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request. Nucleotide sequence data reported are available in the
DDBJ Sequenced Read Archive under the accession number
PRJDB16229.
Authors' contributions
MoY and KT conceived and designed the present study.
MoY conducted the majority of the molecular and cellular
experiments. MoY, KT, MaY, KoY, TM, TS, KaY, TT, YK, KN, HE and YD
contributed to data acquisition and analysis. KeY, HM and YO
performed bioinformatics, statistical analysis and interpretation
of the data. MoY and KT were major contributors in writing the
manuscript and confirm the authenticity of all the raw data. All
authors provided supervision of the manuscript, and read and
approved the final manuscript.
Ethics approval and consent to
participate
The current study was approved by the Human Ethics
Review Committee of the Osaka University School of Medicine
(approval no. 16305-4) and written informed consent was obtained
from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Medical Research Council Oesophageal
Cancer Working Group, : Surgical resection with or without
preoperative chemotherapy in oesophageal cancer: A randomised
controlled trial. Lancet. 359:1727–1733. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Allum WH, Stenning SP, Bancewicz J, Clark
PI and Langley RE: Long-term results of a randomized trial of
surgery with or without preoperative chemotherapy in esophageal
cancer. J Clin Oncol. 27:5062–5067. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ando N, Kato H, Igaki H, Shinoda M, Ozawa
S, Shimizu H, Nakamura T, Yabusaki H, Aoyama N, Kurita A, et al: A
randomized trial comparing postoperative adjuvant chemotherapy with
cisplatin and 5-fluorouracil versus preoperative chemotherapy for
localized advanced squamous cell carcinoma of the thoracic
esophagus (JCOG9907). Ann Surg Oncol. 19:68–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLOS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suzuki H, Zuo Y, Wang J, Zhang MQ,
Malhotra A and Mayeda A: Characterization of RNase R-digested
cellular RNA source that consists of lariat and circular RNAs from
pre-mRNA splicing. Nucleic Acids Res. 34:e632006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim D and Salzberg SL: TopHat-Fusion: An
algorithm for discovery of novel fusion transcripts. Genome Biol.
12:R722011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan W, Zhou R, Wang J, Han J, Yang X, Hao
Y, Lu H, Zhang X, Li P, Tao J, et al: Circular RNA Cdr1as
sensitizes bladder cancer to cisplatin by upregulating APAF1
expression through miR-1270 inhibition. Oncol. 13:1559–1576.
2019.
|
|
19
|
Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie
JW, Wang JB, Lin JX, Chen QY, Cao LL, et al: Circular RNA
circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to
facilitate gastric cancer invasion and metastasis. Cancer Lett.
471:38–48. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang X, Li Z, Zhang Q, Wang W, Li B, Wang
L, Xu Z, Zeng A, Zhang X, Zhang X, et al: Circular RNA AKT3
upregulates PIK3R1 to enhance cisplatin resistance in gastric
cancer via miR-198 suppression. Mol Cancer. 18:712019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sobin LH, Gospodarowicz MK and Wittekind
C: UICC international union against cancer. TNM classification of
malignant tumors. pp. 7Wiley-Blackwell; New York: 2009
|
|
22
|
Yamasaki M, Yasuda T, Yano M, Hirao M,
Kobayashi K, Fujitani K, Tamura S, Kimura Y, Miyata H, Motoori M,
et al: Multicenter Randomized phase II Study of cisplatin and
fluorouracil plus docetaxel (DCF) Compared with cisplatin and
fluorouracil plus Adriamycin (ACF) as preoperative Chemotherapy for
Resectable Esophageal squamous cell carcinoma (OGSG1003). Ann
Oncol. 28:116–120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Japan Esophageal Society, . Japanese
classification of esophageal cancer, 11th Edition: Part II and III.
Esophagus. 14:37–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dasari S and Tchounwou PB: Cisplatin in
cancer therapy: Molecular mechanisms of action. Eur J Pharmacol.
740:364–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hao J and Yu JS: Semaphorin 3C and its
receptors in cancer and cancer stem-like cells. Biomedicines.
6:422018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Man J, Shoemake J, Zhou W, Fang X, Wu Q,
Rizzo A, Prayson R, Bao S, Rich JN and Yu JS: Sema3C promotes the
survival and tumorigenicity of glioma stem cells through Rac1
activation. Cell Rep. 9:1812–1826. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hao J, Han X, Huang H, Yu X, Fang J, Zhao
J, Prayson RA, Bao S and Yu JS: Sema3C signaling is an alternative
activator of the canonical WNT pathway in glioblastoma. Nat Commun.
14:22622023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu Z, Tie X, Li N, Yi Z, Shen F and Zhang
Y: Circular RNA hsa_circ_0000654 promotes esophageal squamous cell
carcinoma progression by regulating the miR-149-5p/IL-6/STAT3
pathway. IUBMB Life. 72:426–439. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi Y, Guo Z, Fang N, Jiang W, Fan Y, He
Y, Ma Z and Chen Y: Hsa_circ_0006168 sponges miR-100 and regulates
mTOR to promote the proliferation, migration and invasion of
esophageal squamous cell carcinoma. Biomed Pharmacother.
117:1091512019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Z, Lin W, Gao L, Chen K, Yang C,
Zhuang L, Peng S, Kang M and Lin J: Hsa_circ_0004370 promotes
esophageal cancer progression through miR-1294/LASP pathway. Biosci
Rep. 39:BSR201823772019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lan X, Liu X, Sun J, Yuan Q and Li J:
CircRAD23B facilitates proliferation and invasion of esophageal
cancer cells by sponging miR-5095. Biochem Biophys Res Commun.
516:357–364. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xing Y, Zha WJ, Li XM, Li H, Gao F, Ye T,
Du WQ and Liu YC: Circular RNA circ-Foxo3 inhibits esophageal
squamous cell cancer progression via the miR-23a/PTEN axis. J Cell
Biochem. 121:2595–2605. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hu X, Wu D, He X, Zhao H, He Z, Lin J,
Wang K, Wang W, Pan Z, Lin H, et al: CircGSK3β promotes metastasis
in esophageal squamous cell carcinoma by augmenting β-catenin
signaling. Mol Cancer. 18:1602019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kartalou M and Essigmann JM: Mechanisms of
resistance to cisplatin. Mutat Res. 478:23–43. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zou FW, Yang SZ, Li WY, Liu CY, Liu XH, Hu
CH, Liu ZH and Xu S: circRNA_001275 upregulates Wnt7a expression by
competitively sponging miR-370-3p to promote cisplatin resistance
in esophageal cancer. Int J Oncol. 57:151–160. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chang N, Ge N, Zhao Y, Yang L, Qin W and
Cui Y: Hsa_circ_0007142 contributes to cisplatin resistance in
esophageal squamous cell carcinoma via miR-494-3p/LASP1 Axis. J
Clin Lab Anal. 36:e243042022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Qiu T, Zhou L, Wang T, Xu J, Wang J, Chen
W, Zhou X, Huang Z, Zhu W, Shu Y and Liu P: miR-503 regulates the
resistance of non-small cell lung cancer cells to cisplatin by
targeting Bcl-2. Int J Mol Med. 32:593–598. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Song S, Ajani JA, Honjo S, Maru DM, Chen
Q, Scott AW, Heallen TR, Xiao L, Hofstetter WL, Weston B, et al:
Hippo coactivator YAP1 upregulates SOX9 and endows esophageal
cancer cells with stem-like properties. Cancer Res. 74:4170–4182.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou W, Liu M, Li X, Zhang P, Li J, Zhao
Y, Sun G and Mao W: Arsenic nano complex induced degradation of YAP
sensitized ESCC cancer cells to radiation and chemotherapy. Cell
Biosci. 10:1462020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: CircRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen CY and Sarnow P: Initiation of
protein synthesis by the eukaryotic translational apparatus on
circular RNAs. Science. 268:415–417. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar : PubMed/NCBI
|