Introduction
Colorectal cancer (CRC) is the third most common
malignancy worldwide and the second most common cause of
cancer-associated mortality (1).
After the locoregional cancer is treated, CRC often causes distant
metastasis, with the liver being the most common site (2). Due to anatomical reasons, patients
with rectal cancer (RC) have a higher risk of local recurrence
compared with patients with colon cancer, and locally recurrent RC
is associated with a poor prognosis (3–5).
Therefore, various neoadjuvant therapy approaches have been
attempted for stage II or stage III RC to prevent distant
metastasis and local recurrence, including neoadjuvant chemotherapy
(6–8), neoadjuvant chemoradiotherapy (9–11) and
total neoadjuvant therapy (12–14).
Previous studies have indicated that tumor
regression detected through imaging after preoperative treatment is
associated with long-term prognosis in patients with RC (15,16).
In a prospective cohort study, the tumor regression grade assessed
through imaging was significantly associated with overall survival
and disease-free survival (15).
Various efforts have been made to explore the usefulness of
serological or genetic biomarkers as predictors of the response to
preoperative treatment, although such biomarkers are costly
(17,18). The use of regular examinations to
predict the response of preoperative treatment is desirable in
terms of cost-effectiveness and objectivity. Colonoscopy is
typically performed on nearly all patients suspected of having
colorectal cancer before surgery to assess tumor size, depth and
circumference, followed by biopsy to confirm the diagnosis
(19,20). Patients undergoing preoperative
treatment are assessed for treatment response by comparing
endoscopic images taken before and after preoperative treatment.
Colonoscopy images are thus among the most readily available tumor
images.
Conventional artificial intelligence (AI) techniques
and machine learning have major limitations in analyzing natural
data (21). However, deep learning,
which has emerged in previous years, excels in learning complex
structures with high-dimensional data and has enabled significant
advancements in various fields (21,22).
Deep learning is applicable to medicine, and AI tools developed in
collaboration with AI experts, specialized facilities, companies
and physicians are gradually being implemented in practical
settings (23–25).
The present research developed a deep learning
prediction model to assess the treatment response of RC to
neoadjuvant chemotherapy using endoscopic images taken during the
initial examination. The prediction accuracy of the model was
further investigated. To the best of our knowledge, no report has
verified whether deep learning of colonoscopy images from initial
examinations can predict the response of RC to neoadjuvant
chemotherapy.
Materials and methods
Patients and datasets
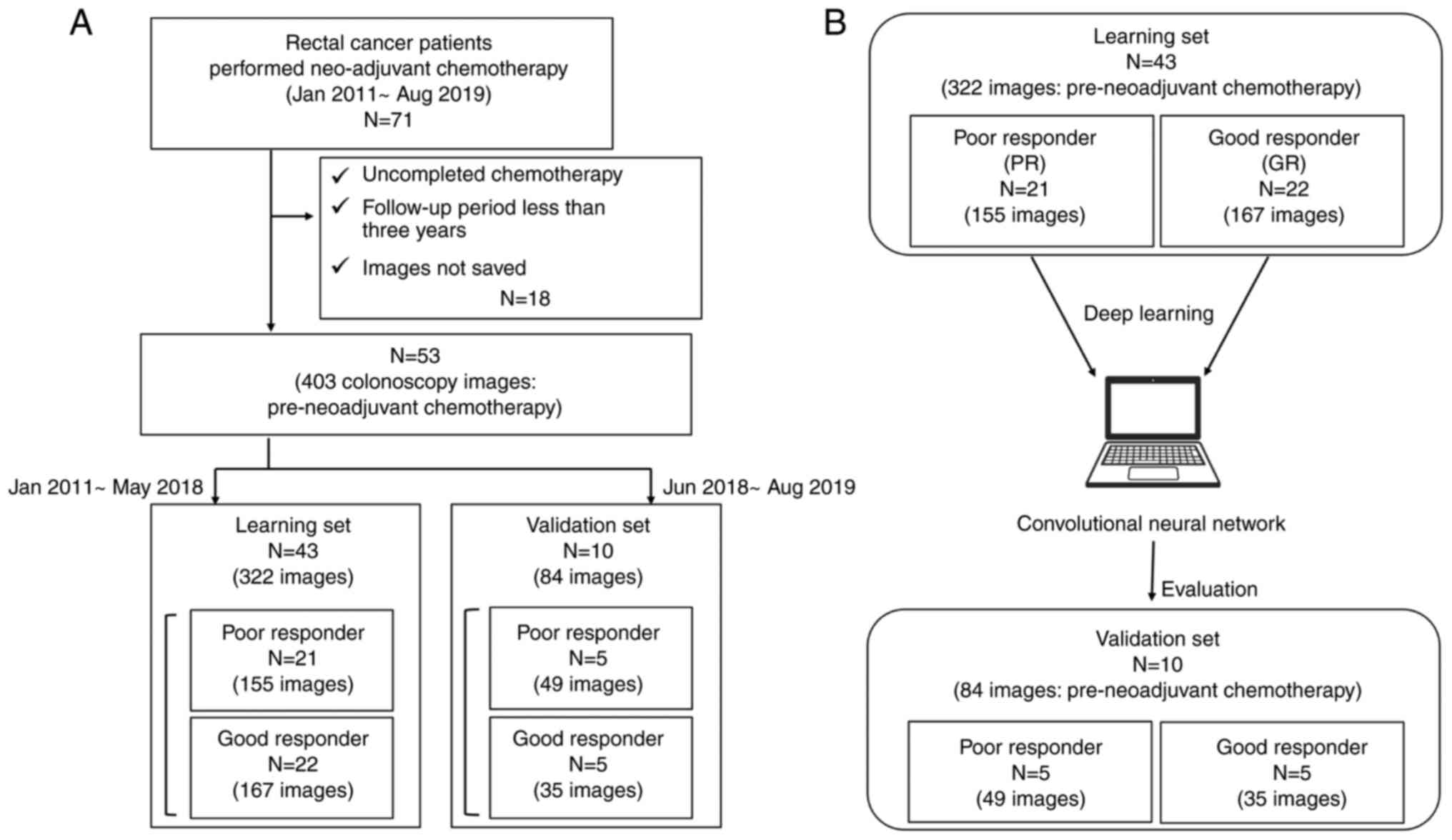
The present retrospective study included patients
who underwent radical resection for advanced RC after neoadjuvant
chemotherapy at Osaka University Hospital (Suita, Japan) between
January 2011 and August 2019. The exclusion criteria were
incomplete planned neoadjuvant chemotherapy, lack of preserved
endoscopic images or missing clinical data. The patient
characteristics are summarized in Tables I and SI. The mean age of the patients was 60.2
years (range, 19–77 years), with 33 men and 20 women.
 | Table I.Clinicopathological backgrounds. |
Table I.
Clinicopathological backgrounds.
| Variables | Learning set
(n=43) | Validation set
(n=10) | P-value |
|---|
| Age, years |
|
|
|
| Median
(IQR) | 64 (56–71) | 59 (37–65) | 0.069a |
| Sex, n (%) |
|
|
|
|
Male | 28 (65.1) | 5 (50.0) | 0.475b |
|
Female | 15 (34.9) | 5 (50.0) |
|
| Body mass
index |
|
|
|
| Median
(IQR) | 22 (20.4–24.5) | 24.1
(16.3–25.5) | 0.139a |
| Carcinoembryonic
antigen, ng/ml |
|
|
|
| Median
(IQR) | 5 (2–9) | 4.5 (3–17.8) | 0.731a |
| Carbohydrate
antigen 19-9, U/ml |
|
|
|
| Median
(IQR) | 18 (6–39) | 27.5
(8.6–52.9) | 0.413a |
| Tumor location, n
(%) |
|
|
|
| Ra | 4 (9.3) | 1 (10.0) |
>0.999b |
| Rb | 39 (90.7) | 9 (90.0) |
|
| Histological grade,
n (%) |
|
|
|
| Tub, no
evidence of cancer | 38 (88.4) | 9 (90.0) |
>0.999b |
| Muc,
Por | 5 (11.6) | 1 (10.0) |
|
| Lymphatic invasion,
n (%) |
|
|
|
|
Present | 19 (44.2) | 5 (50.0) |
>0.999b |
|
Absent | 24 (55.8%) | 5 (50.0) |
|
| Vascular invasion,
n (%) |
|
|
|
|
Present | 13 (30.2) | 3 (30.0) |
>0.999b |
|
absent | 30 (69.8) | 7 (70.0) |
|
| Pathological tumor
invasion, n (%) |
|
|
|
| T0,
Tis, T1, T2 | 22 (51.2) | 5 (50.0) |
>0.999b |
| T3,
T4 | 21 (48.8) | 5 (50.0) |
|
| Pathological lymph
node metastasis, n (%) |
|
|
|
|
Present | 15 (34.9) | 2 (20.0) | 0.471b |
|
Absent | 28 (65.1) | 8 (80.0) |
|
| Pathological tumor
regression grade, n (%) |
|
|
|
| Grade
0, 1, 2 | 6 (14.0) | 3 (30.0) | 0.346b |
| Grade
3 | 37 (86.0) | 7 (70.0) |
|
| Clinical response
to NAC, n (%) |
|
|
|
| Poor
response | 22 (51.2) | 5 (50.0) |
>0.999b |
| Good
response | 21 (48.8) | 5 (50.0) |
|
| Regimens of NAC, n
(%) |
|
|
|
|
Capecitabine, oxaliplatin
containing | 42 (97.7) | 10 (100.0) |
>0.999b |
|
Capecitabin and
Bevacizumab | 1 (2.3) | 0 (0.0) |
|
The information and images of the patients were
obtained from electronic medical records. Clinical and pathological
factors were determined according to the 8th edition of the Union
for International Cancer Control Tumor-Node-Metastasis
classification (26). Multidetector
row computed tomography (MDCT) and magnetic resonance imaging (MRI)
were used to preoperatively diagnose progression, including tumor
invasion and lymph node metastasis. Lymph nodes with a short axis
diameter of ≥7 mm on MDCT were considered positive (27). The MDCT parameters were rotation
speed of 0.6 s/r, helical pitch of 17.5 mm/r and slice thickness of
0.625 mm. The reconstruction intervals were set to 0.5 mm. MRI was
performed using a thin, 3-mm section turbo spin-echo T2-weighted
technique with a surface pelvic phased-array coil and a small field
of view. Bowel preparation, air insufflation or intravenous
antispasmodic agents were not routinely used.
The location and histological grade in response to
neoadjuvant chemotherapy were determined according to the Japanese
Classification of Colorectal, Appendiceal and Anal Carcinoma
(28). The classification of
pathological tumor regression grade was based on the guidelines
provided by the National Comprehensive Cancer Network (29). R0 resection was defined as no
evidence of tumor within 1-mm of the distal, proximal or radial
margins, as assessed by the review of the pathologists who were
independent from the present study.
Evaluation of clinical response to
neoadjuvant chemotherapy
Clinical response to neoadjuvant chemotherapy was
assessed by MRI conducted before and after chemotherapy according
to the Response Evaluation Criteria in Solid Tumors version 1.1
(30). To simplify the end points,
progressive disease and stable disease were defined as poor
responder RC (PR-RC), and partial response and complete response as
good responder RC (GR-RC).
Colonoscopy
Colonoscopy was performed before and after
neoadjuvant chemotherapy. The images were obtained using the EVIS
LUCERA video system (Olympus Corporation) and the following
colonoscopes: CF-HQ290I, CF-Q260AI, PCF-H290I, PCF-H290I,
PCF-Q260AI, CF-H290I and CF-H260AI (Olympus Corporation). All these
endoscopes were equipped with high-definition-compatible
charge-coupled devices that enabled high-quality imaging. Although
the endoscopes had different scope diameters, viewing angles and
focal lengths, they yielded images of similar quality. All images
had at least one RC lesion, and multiple images of the same lesion
were produced to illustrate the differences in angle, distance and
extension of the mucosa. Images that include at least part of the
tumor area within the field of view were selected. The present
study did not limit images of the target by the distance from the
tumor surface. All images were captured under white light after
adjusting white balance. Poor-quality images due to halos, blurred
focus or mucus were excluded from the current study.
Deep learning
A model was constructed using deep learning of
colonoscopy images to predict PR-RC or GR-RC. Overall, two models
were constructed, one based on pre-treatment images and the other
based on post-treatment images (Figs.
1B and S1A). AlexNet (31) in Matlab 2022b (MathWorks) was used
as the network for building the models (32). Deep Learning Toolbox (MathWorks;
http://jp.mathworks.com/products/deep-learning.html)
and Image Processing Toolbox (MathWorks; http://jp.mathworks.com/products/image.html) were
used as toolboxes for constructing the deep learning models. To
eliminate unnecessary background information and focus on the
tumor, each image was cropped to a square shape and resized to a
predetermined size prior to the analysis. Occlusion was used to
assess the impact input images and determine their influence on the
classification results.
Statistical analyses
The differences in clinicopathological factors
between the two groups were analyzed using Fisher's exact test.
Continuous variables that had a non-parametric distribution were
analyzed using the Mann-Whitney U test. The Shapiro-Wilk test was
used to test for normality. The continuous variables are presented
as the median ± interquartile range. P<0.05 was considered to
indicate a statistically significant difference for all analyses.
All statistical analyses were performed using the JMP Pro version
16 (SAS Institute, Inc.).
Results
A total of 53 of the 71 patients were included in
the present study. A total of 322 pre-treatment images from 43
patients who underwent neoadjuvant chemotherapy for advanced RC
between January 2011 and March 2018 were included in the learning
set (Table I; Fig. 1). A validation set was created that
included 84 images obtained during the pre-treatment examination of
10 patients who underwent neoadjuvant chemotherapy between June
2018 and August 2019. All patients underwent total mesorectal
excision with R0 resection 3 to 6 weeks after completing
neoadjuvant chemotherapy. In the learning set, patients with PR-RC
had more advanced pathological tumor invasion and pathological
lymph node metastasis compared with that observed among patients
GR-RC, reflecting the clinical efficacy of neoadjuvant chemotherapy
(Table II).
 | Table II.Clinicopathological features of the
learning set. |
Table II.
Clinicopathological features of the
learning set.
| Variables | Poor responder
(n=21) | Good responder
(n=22) | P-value |
|---|
| Age, years |
|
|
|
| Median
(IQR) | 62 (56–71) | 64.5 (54–70) | 0.913a |
| Sex, n (%) |
|
|
|
|
Male | 16 (76.2) | 12 (54.5) | 0.203b |
|
Female | 5 (23.8) | 10 (45.5) |
|
| Body mass
index |
|
|
|
| Median
(IQR) | 22.8
(20.4–24.8) | 21.9
(20.1–23.6) | 0.671a |
| Carcinoembryonic
Antigen, ng/ml |
|
|
|
| Median
(IQR) | 6 (3–10) | 3 (2–9.75) | 0.413a |
| Carbohydrate
antigen 19-9, U/ml |
|
|
|
| Median
(IQR) | 18 (5.75–46.5) | 12 (5–40) | 0.913a |
| Tumor location, n
(%) |
|
|
|
| Ra | 2 (9.5) | 0 (0.0) | 0.233b |
| Rb | 19 (90.5) | 22 (100.0) |
|
| Histological grade,
n (%) |
|
|
|
| Tubular
adenocarcinoma | 17 (81.0) | 21 (95.5) | 0.185b |
| Muc or
por | 4 (19.0) | 1 (4.5) |
|
| Lymphatic invasion,
n (%) |
|
|
|
|
Present | 11 (52.4) | 8 (36.4) | 0.364b |
|
Absent | 10 (47.6) | 14 (63.6) |
|
| Vascular invasion,
n (%) |
|
|
|
|
Present | 8 (38.1) | 5 (22.7) | 0.332b |
|
Absent | 13 (61.9) | 17 (77.3) |
|
| Pathological tumor
invasion, n (%) |
|
|
|
| T1,
T2 | 6 (28.6) | 16 (72.7) | 0.006b,c |
| T3,
T4 | 15 (71.4) | 6 (7.3) |
|
| Pathological lymph
node metastasis, n (%) |
|
|
|
|
Present | 11 (52.4) | 4 (18.2) | 0.027b,c |
|
Absent | 10 (47.6) | 18 (81.8) |
|
| Pathological tumor
regression grade, n (%) |
|
|
|
| Grade
0, 1, 2 | 1 (4.8) | 5 (22.7) | 0.185b |
| Grade
3 | 20 (95.2) | 17 (77.3) |
|
The present study constructed a prediction model
based on deep learning of pre-treatment images and clinical
responses in the learning set. The accuracy of pre-chemotherapy
deep learning model in predicting the response to chemotherapy in
the validation set is shown in Table
III. In the validation dataset, 49 images were taken from five
patients who were clinically diagnosed as PR-RC, and 35 images were
taken from five patients who were clinically diagnosed as GR-RC.
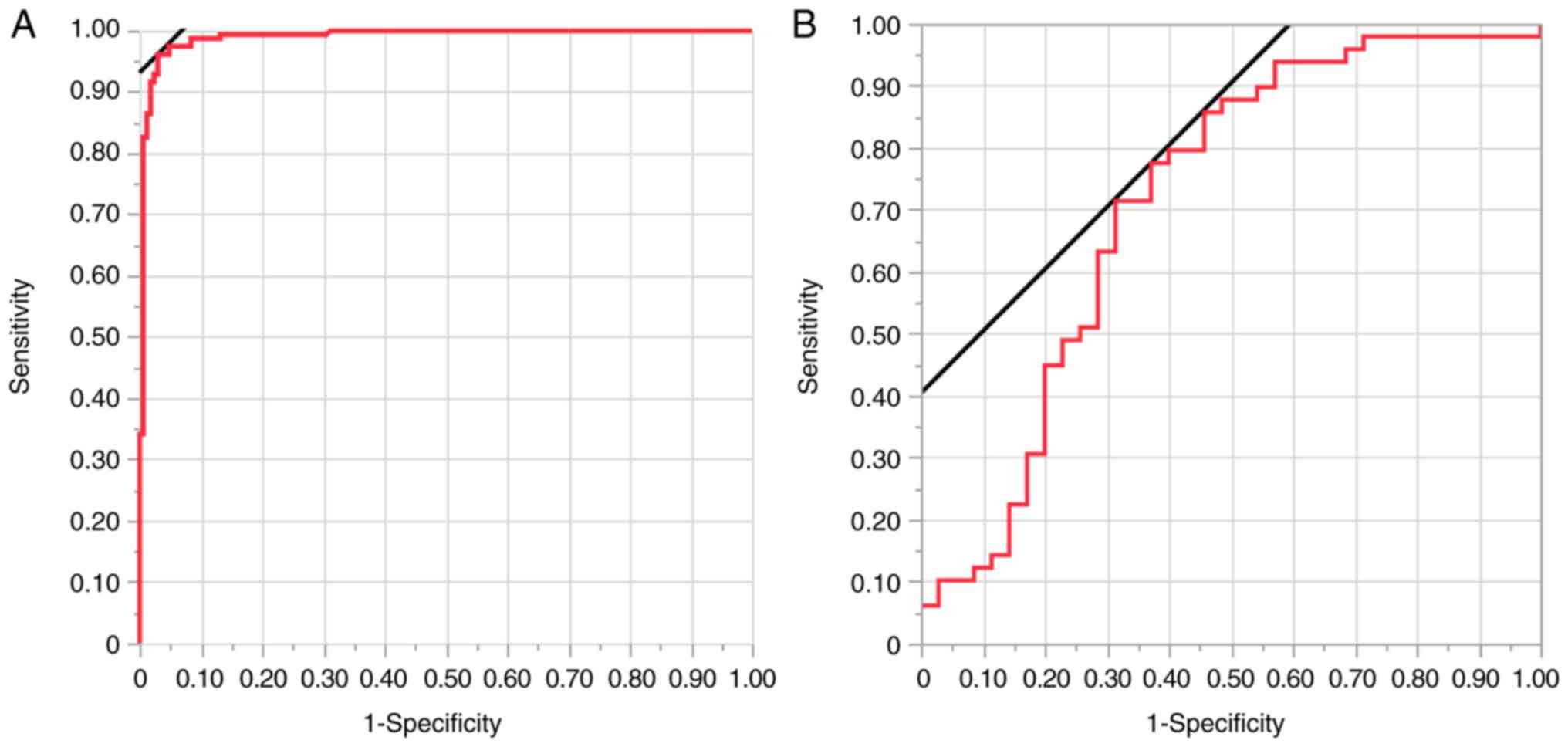
When using the validation set, the sensitivity, specificity,
positive predictive value, accuracy and area under the curve (AUC)
of the model in predicting PR-RC status were 77.6% (38/49), 62.9%
(22/35), 74.5% (38/51), 71.4% (60/84) and 0.713, respectively
(Table II; Fig. 2).
 | Table III.Prediction accuracy in the validation
set of AI prediction models constructed by deep learning of
colonoscopy images before neoadjuvant chemotherapy. |
Table III.
Prediction accuracy in the validation
set of AI prediction models constructed by deep learning of
colonoscopy images before neoadjuvant chemotherapy.
|
| AI prediction |
|
|---|
|
|
|
|
|---|
| Clinical
response | Poor responder | Good responder | Total |
|---|
| Poor responder | 38 images | 11 images | 49 images |
| Good responder | 13 images | 22 images | 35 images |
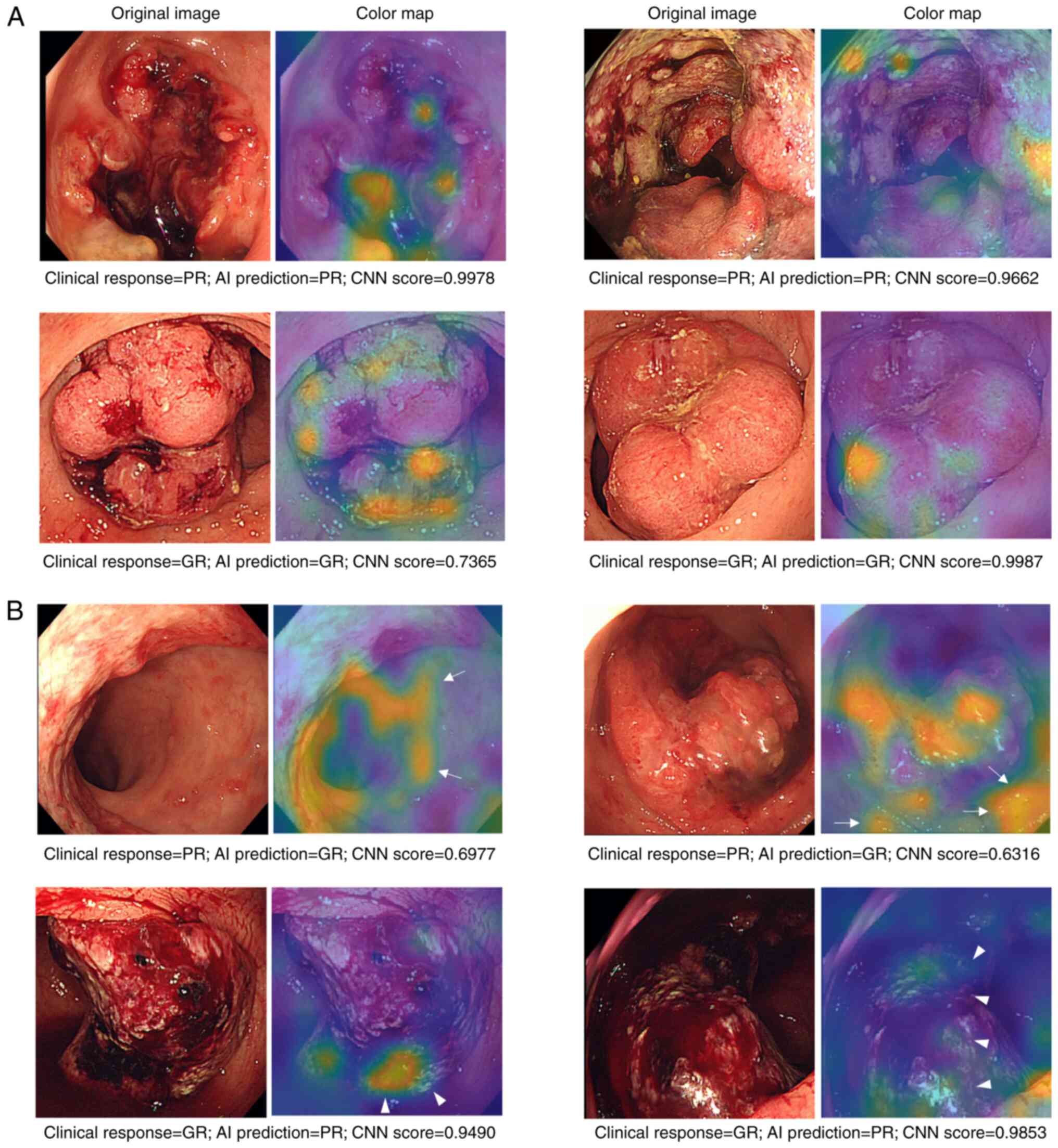
Prediction errors where the model was unable to
predict response occurred when the AI focused on areas other than
the tumor (Fig. 3). The percentages
of incorrect predictions where the AI focused on tumor area were
36.4% for PR-RC and 15.4% for GR-RC. The percentage of incorrect
predictions where the AI focused on the normal mucosa was 54.5% for
PR-RC and 46.1% for GR-RC (Table
IV). The percentages of incorrect predictions focused on
bleeding were 9.1% for PR-RC and 38.5% for GR-RC. AI focus on
non-tumor areas was considered to be partly responsible for the
incorrect AI predictions.
 | Table IV.Focused area of incorrect AI
prediction in the validation set. |
Table IV.
Focused area of incorrect AI
prediction in the validation set.
|
| Focused area of
incorrect AI prediction |
|
|---|
|
|
|
|
|---|
|
|
| Other area |
|
|---|
|
|
|
|
|
|---|
| Clinical
response | Tumor area | Normal mucosa | Bleeding | Total |
|---|
| Poor responder | 4 images
(36.4%) | 6 images
(54.5%) | 1 image (9.1%) | 11 images |
| Good responder | 2 images
(15.4%) | 6 images
(46.1%) | 5 images
(38.5%) | 13 images |
Finally, the present study constructed another
prediction model based on deep learning of clinical responses and
colonoscopy images of the same patient dataset after preoperative
chemotherapy, and its accuracy was verified. The same learning and
validation sets were used to test the model based on colonoscopy
images after preoperative chemotherapy. The accuracy of the model
is shown in Table SII. The
post-chemotherapy model had a sensitivity of 40.5% (17/42),
specificity of 56.1% (23/41), accuracy of 48.2% (40/83) and AUC
value of 0.592 (Fig. S1C). The
post-chemotherapy deep learning model had inferior predictive
performance to the model based on pre-chemotherapy endoscopic
images.
Discussion
AI technology has achieved unprecedented success in
various fields. Deep learning, which is a subset of machine
learning that focuses on deep artificial neural networks (21,25),
has been used in numerous areas of oncology, ranging from cancer
detection and classification to the molecular characterization of
tumors and their microenvironment (33,34),
drug discovery (35) and prediction
of distant metastasis (36). In the
context of predicting the efficacy of preoperative treatment for
RC, deep learning based on MRI images and hematoxylin-eosin (HE)
staining images has been reportedly useful in predicting complete
response to preoperative chemoradiotherapy (37). The colonoscopy that was the source
of images in the current study was an examination conducted for
nearly all patients with RC who received preoperative treatment,
meaning it was relatively straightforward to implement a deep
learning model using this modality in clinical practice.
The present study examined the ability of deep
learning models based on colonoscopy images to predict the response
of RC to preoperative chemotherapy. This study did not include the
patients who were provided neoadjuvant radiation therapy or
neoadjuvant chemoradiotherapy because the mechanisms of antitumor
effects of chemotherapy and radiation therapy differ. The model
based on colonoscopy images taken before preoperative chemotherapy
was able to differentiate PR-RC from GR-RC with a sensitivity of
77.6% and accuracy of 71.4% with an AUC of 0.713. Furthermore, the
present study created a program to visualize the parts of the image
that the AI recognized and diagnosed using a color map (38–40).
AI-based image recognition detects the location of the tumor as
well as the information around the tumor, including the bleeding
area, which might influence the prediction accuracy. Implementing a
program that accurately recognizes the tumor surface may improve
the accuracy of the prediction. The analyses in the study did not
exclude patients who used molecularly targeted drugs. The present
study also attempted to create another model using pre-treatment
images except for the patients who used molecularly targeted drugs
and the accuracy was assessed. The sensitivity and accuracy of the
model that excluded the patient on targeted therapy were 71.4%
(35/49) and 65.5% (55/84), respectively.
The current study defined MRI-detected clinical
response as the outcome of deep learning models because MRI
assessments of tumor regression grade are imaging markers that
predict survival outcomes (15).
Pathologic tumor regression grading is based on the percentage of
fibrosis in the surgical specimen and correlates with survival at a
greater statistical significance (15,16);
however, by applying a similar approach with MRI, the present study
was able to assess tumor response and predict the outcome of
patients before surgery (15).
Future validation studies are needed to determine the tumor
response and survival outcome before chemotherapy using deep
learning and endoscopic images.
To the best of our knowledge, the present study was
the first to report on the utility of deep learning based on
pretreatment endoscopic images to predict the treatment response to
neoadjuvant chemotherapy. The current results had several important
implications. The first was the possibility of improving the model
by combining the modality with other modalities, such as MRI images
and clinicopathological factors. Recently, deep learning prediction
models have combined multiple modalities to predict the clinical
efficacy of preoperative RC treatment. Predictive models combining
multiple modalities significantly outperform predictive models
involving individual modalities (37). Secondly, the application of deep
learning to pretreatment endoscopic images could serve as a
potential method for predicting the complete response to
preoperative chemoradiotherapy. In the future, if deep learning
models based on endoscopic images demonstrate improved predictive
accuracy compared with models based on histological images, their
significance must be further emphasized through future validation.
Thirdly, pre-chemotherapy endoscopic images may be more useful
compared with post-chemotherapy images to build models predicting
the effect of chemotherapy. The present study revealed that the
deep learning model based on pre-chemotherapy images was more
accurate in predicting chemotherapy efficacy compared with that
using post-chemotherapy images. Furthermore, the present study
validated a combination model; however, the sensitivity (20.0%) and
accuracy (40.0%) were so low that the model was not considered
useful. The deep learning model based on pre-chemotherapy images
may capture features of tumor surface structures that reflect
chemotherapy responsiveness.
The present results were limited by the potential
risk of overfitting due to the use of data from a single-center and
small sample size. Additionally, the current results are limited by
uncertain reproducibility due to the lack of prospective
validation; therefore, additional validation is required.
In conclusion, deep learning based on endoscopic
images may allow for enhanced accuracy when predicting the response
of RC to neoadjuvant chemotherapy. There is a need to collect more
images and investigate additional image features to improve
prediction accuracy.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Mr. Hiroyuki Hishida
and Ms. Misa Taguchi (The MathWorks Japan, Inc., Tokyo, Japan) for
their technical assistance and support in program development.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SK, NM, SF, MU, YD and HE conceived and designed the
study. Material preparation and data collection were performed by
SK, SM, AN and RH. Data analyses were performed by SK, NM, YS, TH,
AH, TO, HT, MU, HY, MT and YK. The first draft of the manuscript
was written by SK, and all the authors commented on previous
versions of the manuscript. SK and NM confirm the authenticity of
all the raw data All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was conducted in accordance with the
principles of the Declaration of Helsinki. Written informed consent
was obtained from all participants. This study was approved by the
Institutional Review Board of Osaka University (approval no.
19020).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fong Y, Cohen AM, Fortner JG, Enker WE,
Turnbull AD, Coit DG, Marrero AM, Prasad M, Blumgart LH and Brennan
MF: Liver resection for colorectal metastases. J Clin Oncol.
15:938–946. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rajput A and Bullard Dunn K: Surgical
management of rectal cancer. Semin Oncol. 34:241–249. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weiser MR, Landmann RG, Wong WD, Shia J,
Guillem JG, Temple LK, Minsky BD, Cohen AM and Paty PB: Surgical
salvage of recurrent rectal cancer after transanal excision. Dis
Colon Rectum. 48:1169–1175. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wiig JN, Larsen SG and Giercksky KE:
Operative treatment of locally recurrent rectal cancer. Recent
Results Cancer Res. 165:136–147. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schrag D, Weiser MR, Goodman KA, Gonen M,
Hollywood E, Cercek A, Reidy-Lagunes DL, Gollub MJ, Shia J, Guillem
JG, et al: Neoadjuvant chemotherapy without routine use of
radiation therapy for patients with locally advanced rectal cancer:
A pilot trial. J Clin Oncol. 32:513–518. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Glynne-Jones R, Hava N, Goh V, Bosompem S,
Bridgewater J, Chau I, Gaya A, Wasan H, Moran B, Melcher L, et al:
Bevacizumab and Combination Chemotherapy in rectal cancer Until
Surgery (BACCHUS): A phase II, multicentre, open-label, randomised
study of neoadjuvant chemotherapy alone in patients with high-risk
cancer of the rectum. BMC Cancer. 15:7642015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kamiya T, Uehara K, Nakayama G, Ishigure
K, Kobayashi S, Hiramatsu K, Nakayama H, Yamashita K, Sakamoto E,
Tojima Y, et al: Early results of multicenter phase II trial of
perioperative oxaliplatin and capecitabine without radiotherapy for
high-risk rectal cancer: CORONA I study. Eur J Surg Oncol.
42:829–835. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Folkesson J, Birgisson H, Pahlman L,
Cedermark B, Glimelius B and Gunnarsson U: Swedish Rectal Cancer
Trial: Long lasting benefits from radiotherapy on survival and
local recurrence rate. J Clin Oncol. 23:5644–5650. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peeters KC, Marijnen CA, Nagtegaal ID,
Kranenbarg EK, Putter H, Wiggers T, Rutten H, Pahlman L, Glimelius
B, Leer JW, et al: The TME trial after a median follow-up of 6
years: Increased local control but no survival benefit in
irradiated patients with resectable rectal carcinoma. Ann Surg.
246:693–701. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sauer R, Liersch T, Merkel S, Fietkau R,
Hohenberger W, Hess C, Becker H, Raab HR, Villanueva MT, Witzigmann
H, et al: Preoperative versus postoperative chemoradiotherapy for
locally advanced rectal cancer: Results of the German
CAO/ARO/AIO-94 randomized phase III trial after a median follow-up
of 11 years. J Clin Oncol. 30:1926–1933. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ciseł B, Pietrzak L, Michalski W, Wyrwicz
L, Rutkowski A, Kosakowska E, Cencelewicz A, Spałek M, Polkowski W,
Jankiewicz M, et al: Long-course preoperative chemoradiation versus
5x5 Gy and consolidation chemotherapy for clinical T4 and fixed
clinical T3 rectal cancer: Long-term results of the randomized
Polish II study. Ann Oncol. 30:1298–1303. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bahadoer RR, Dijkstra EA, van Etten B,
Marijnen CAM, Putter H, Kranenbarg EM, Roodvoets AGH, Nagtegaal ID,
Beets-Tan RGH, Blomqvist LK, et al: Short-course radiotherapy
followed by chemotherapy before total mesorectal excision (TME)
versus preoperative chemoradiotherapy, TME, and optional adjuvant
chemotherapy in locally advanced rectal cancer (RAPIDO): A
randomised, open-label, phase 3 trial. Lancet Oncol. 22:29–42.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jin J, Tang Y, Hu C, Jiang LM, Jiang J, Li
N, Liu WY, Chen SL, Li S, Lu NN, et al: Multicenter, randomized,
Phase III trial of short-term radiotherapy plus chemotherapy versus
long-term chemoradiotherapy in locally advanced rectal cancer
(STELLAR). J Clin Oncol. 40:1681–1692. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Patel UB, Taylor F, Blomqvist L, George C,
Evans H, Tekkis P, Quirke P, Sebag-Montefiore D, Moran B, Heald R,
et al: Magnetic resonance imaging-detected tumor response for
locally advanced rectal cancer predicts survival outcomes: MERCURY
experience. J Clin Oncol. 29:3753–3760. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fokas E, Liersch T, Fietkau R, Hohenberger
W, Beissbarth T, Hess C, Becker H, Ghadimi M, Mrak K, Merkel S, et
al: Tumor regression grading after preoperative chemoradiotherapy
for locally advanced rectal carcinoma revisited: Updated results of
the CAO/ARO/AIO-94 trial. J Clin Oncol. 32:1554–1562. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ghadimi BM, Grade M, Difilippantonio MJ,
Varma S, Simon R, Montagna C, Füzesi L, Langer C, Becker H, Liersch
T, et al: Effectiveness of gene expression profiling for response
prediction of rectal adenocarcinomas to preoperative
chemoradiotherapy. J Clin Oncol. 23:1826–1838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cercek A, Dos Santos Fernandes G, Roxburgh
CS, Ganesh K, Ng S, Sanchez-Vega F, Yaeger R, Segal NH,
Reidy-Lagunes DL, Varghese AM, et al: Mismatch repair-deficient
rectal cancer and resistance to neoadjuvant chemotherapy. Clin
Cancer Res. 26:3271–3279. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lynch ML and Brand MI: Preoperative
evaluation and oncologic principles of colon cancer surgery. Clin
Colon Rectal Surg. 18:163–173. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arteaga-González I, Martín-Malagón A,
Fernández EM, Arranz-Durán J, Parra-Blanco A, Nicolas-Perez D,
Quintero-Carrión E, Luis HD and Carrillo-Pallares A: The use of
preoperative endoscopic tattooing in laparoscopic colorectal cancer
surgery for endoscopically advanced tumors: A prospective
comparative clinical study. World J Surg. 30:605–611. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
LeCun Y, Bengio Y and Hinton G: Deep
learning. Nature. 521:436–444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Duch W, Swaminathan K and Meller J:
Artificial intelligence approaches for rational drug design and
discovery. Curr Pharm Des. 13:1497–1508. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ehteshami Bejnordi B, Veta M, Johannes van
Diest P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak J,
Hermsen M, Manson QF, Balkenhol M, et al: Diagnostic assessment of
deep learning algorithms for detection of lymph node metastases in
women with breast cancer. JAMA. 318:2199–2210. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Khosravi P, Kazemi E, Imielinski M,
Elemento O and Hajirasouliha I: Deep Convolutional neural networks
enable discrimination of heterogeneous digital pathology images.
EBioMedicine. 27:317–328. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bhinder B, Gilvary C, Madhukar NS and
Elemento O: Artificial intelligence in cancer research and
precision medicine. Cancer Discov. 11:900–915. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
TNM Classifcation of Maligant Tumours,
Eighth Edition. Wiley Blackwell; Hoboken: 2016
|
|
27
|
Yukimoto R, Uemura M, Tsuboyama T, Sekido
Y, Hata T, Ogino T, Miyoshi N, Takahashi H, Kida A, Furuyashiki M,
et al: Efficacy of PET/CT in diagnosis of regional lymph node
metastases in patients with colorectal cancer: Retrospective cohort
study. BJS Open. 6:2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Japanese Classification of Colorectal,
Appendiceal and Anal Carcinoma: The 3d English Edition [Secondary
Publication]. J Anus Rectum Colon. 3:175–195. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Benson AB, Venook AP, Al-Hawary MM, Azad
N, Chen YJ, Ciombor KK, Cohen S, Cooper HS, Deming D,
Garrido-Laguna I, et al: Rectal cancer, version 2.2022, NCCN
clinical practice guidelines in oncology. J Natl Compr Canc Netw.
20:1139–1167. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Krizhevsky AS and Hinton GEI: Imagenet
classification with deep convolutional neural networks. Adv Neural
Inf Process Syst. 25:1097–1105. 2012.PubMed/NCBI
|
|
32
|
Minami S, Saso K, Miyoshi N, Fujino S,
Kato S, Sekido Y, Hata T, Ogino T, Takahashi H, Uemura M, et al:
Diagnosis of depth of submucosal invasion in colorectal cancer with
AI using deep learning. Cancers (Basel). 14:53612022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yamashita R, Long J, Longacre T, Peng L,
Berry G, Martin B, Higgins J, Rubin DL and Shen J: Deep learning
model for the prediction of microsatellite instability in
colorectal cancer: A diagnostic study. Lancet Oncol. 22:132–141.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiao Y, Li J, Qian C and Fei S: Deep
learning-based tumor microenvironment analysis in colon
adenocarcinoma histopathological whole-slide images. Comput Methods
Programs Biomed. 204:1060472021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gupta R, Srivastava D, Sahu M, Tiwari S,
Ambasta RK and Kumar P: Artificial intelligence to deep learning:
Machine intelligence approach for drug discovery. Mol Divers.
25:1315–1360. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu X, Zhang D, Liu Z, Li Z, Xie P, Sun K,
Wei W, Dai W, Tang Z, Ding Y, et al: Deep learning radiomics-based
prediction of distant metastasis in patients with locally advanced
rectal cancer after neoadjuvant chemoradiotherapy: A multicentre
study. EBioMedicine. 69:1034422021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Feng L, Liu Z, Li C, Li Z, Lou X, Shao L,
Wang Y, Huang Y, Chen H, Pang X, et al: Development and validation
of a radiopathomics model to predict pathological complete response
to neoadjuvant chemoradiotherapy in locally advanced rectal cancer:
A multicentre observational study. Lancet Digit Health. 4:e8–e17.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Miyoshi N: AI application for surgery. J
Jpn Soc Precis Eng. 88:9–11. 2022. View Article : Google Scholar
|
|
39
|
Lu L, Dercle L, Zhao B and Schwartz LH:
Deep learning for the prediction of early on-treatment response in
metastatic colorectal cancer from serial medical imaging. Nat
Commun. 12:66542021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Horvat N, Veeraraghavan H, Khan M, Blazic
I, Zheng J, Capanu M, Sala E, Garcia-Aguilar J, Gollub MJ and
Petkovska I: MR Imaging of rectal cancer: Radiomics analysis to
assess treatment response after neoadjuvant therapy. Radiology.
287:833–843. 2018. View Article : Google Scholar : PubMed/NCBI
|