Introduction
Recent advances in cellular, molecular, and genomic
technologies and approaches have markedly improved disease
diagnosis and treatment (1).
However, the early prediction and diagnosis of certain diseases,
including cancer, remains to be successfully achieved. In 2018, the
WHO estimated the occurrence of ≥9.6 million deaths worldwide,
which is expected to be <21.4 million by 2030 (2,3).
Female breast cancer (BC) is a highly prevalent type of cancer
among young and aged women in the Kingdom of Saudi Arabia, with an
incidence rate of 29.7% (4).
Regardless of the effective and globally available BC therapies for
the treatment of BC, diagnosis at the earliest stage of the disease
remains a challenge (5,6). Consequently, early detection followed
by prompt referral to the patient BC clinics are crucial for
overcoming many of the challenges in disease management (7); thus there is an urgent need for the
discovery of powerful BC biomarkers.
It has been well documented that multiple signaling
pathways are involved in BC pathophysiology, including development,
proliferation, differentiation, motility and metastasis (8,9). These
mainly include the WNT (10), NOTCH
(11), bone morphogenetic protein-2
(12), STAT3 (13), estrogen receptor (ER) (14), human epidermal growth factor
receptor 2 (HER2) (15), MAPK
(9,16), PI3K/Akt/NF-κB (17), TGF-β (18), Hedgehog (19) and HIPPO (20) pathways. In addition, inflammation
has been strongly linked to BC progression through TLR activities
(21–23). Furthermore, microRNAs (miRNAs/miRs)
have been demonstrated to be critical in orchestrating and
fine-tuning the signaling during BC stem cell self-renewal,
proliferation, tumor metastasis and drug resistance (8,24).
Frizzled receptors (FZDs) and their WNT ligands have
been revealed to regulate several cellular processes during
embryonic development and tumorigenesis (25). Mutations in the WNT components
pathway can trigger diseases, specifically cancer (25–27).
Several FZDs have been reported to play a crucial role in cancer
progression (28–31). FZD8 receptor is included in this
category, cloned and characterized in humans on chromosome 10pll.2
(32). FZD8 has been demonstrated
in several studies to play differential roles in various types of
cancer (33–37). In BC, FZD8 has been demonstrated to
mediate resistance to chemotherapy in patients with triple-negative
(TN)BC, thus rendering it an important candidate as a therapeutic
target (33). In gastric cancer
(34), FZD8 has been demonstrated
to promote metastasis through WNT/β-catenin. It also exhibits the
same function in prostate cancer, in addition to the activation of
TGF-β signaling (36). In lung
cancer, FZD8 has been reported as a novel powerful prognostic
marker (37).
Regardless of the important role of FZD8 in
different types of cancer, a limited number of studies have
previously analyzed its role in BC (33,38).
Therefore, the aim of the present study was to assess the
prognostic value of FZD8 expression in a BC cohort using
immunohistochemistry (IHC), multivariate Cox analysis and
Kaplan-Meier univariate survival analysis. The association between
the FZD8 expression pattern and patient clinicopathological
parameters was determined. In addition, using BC microarray
datasets in the Gene Expression Omnibus (GEO) and miRNA target
prediction bioinformatics tools, the silencing machinery involving
FZD8 in BC was investigated, and potential miRNAs targeting FZD8
expression were identified.
Materials and methods
Patients
The present study was reviewed and approved by the
Ethics Committee of the Center of Excellence in Genomic Medicine
Research (CEGMR), King Abdulaziz University (Jeddah, Saudi Arabia;
approval no. 012-CEGMR-ETH). All patients who participated in the
study provided written informed consent in accordance with The
Declaration of Helsinki (39). The
procedures for collecting patient specimens adhered to the King
Abdulaziz University Hospital (KAUH) guidelines. The BC specimens
used in the present study were obtained from the Pathology
Department, KAUH, and covered the period between January, 1995 and
December, 2017. A total of 562 patients aged between 25–70 years
with an age median of 50 years were enrolled in the present study.
The surgery was performed on almost all patients, usually in the
form of a lumpectomy, or a radical or modified radical mastectomy
with axillary clearance. Per patient, one sample/biopsy was
included in the study and in all further analyses. The
clinicopathological data of the patients were obtained from the
KAUH medical records. The inclusion criteria that were applied
included all female breast cancer patients diagnosed, monitored and
followed-up at KAUH. Only patients with available clinical records
and retrospective samples were included in the study. Patients with
other concomitant diseases or with no medical records or available
samples were excluded from the study. The present study did not
include patients who had received neoadjuvant therapy. Following
surgery, the BC and lymph node specimens of the patients were
immediately fixed in 10% formalin buffer overnight at 4°C with
shaking and processed for the typical formalin-fixed
paraffin-embedded (FFPE) blocks, followed by paraffin sectioning at
a thickness of 4 µm. For the assessment of the histopathological
characteristics, histological grading, and tumor, node and
metastasis-based staging of the biopsies, the paraffin sections
were stained at room temperature according to the manufacturer's
protocol of the hematoxylin and eosin kit (ab245880, Abcam), and
then examined and analyzed by pathologists. For the BC grading
system, the method published by Schauer et al (40) was followed. Briefly, upon
histological examination, tumor cells were classified based on
their similarity or dissimilarity to the normal cells, as well as
their mitotic activity: i) When tumor cells were closely similar to
normal cells or well-differentiated with lower mitotic count, they
were classified as grade 1; ii) when tumor cells were
moderately-differentiated with moderate mitotic activity, they were
considered as grade 2; iii) when tumor cells were
poorly-differentiated with high mitotic activity, they were
classified as grade 3; and iv) undifferentiated tumor cells with
very high mitotic activity were classified as grade 4.
Treatment and follow-up
For post-operative early adjuvant systemic therapy,
75, 59 and 34% of the patients received chemotherapy, radiation
therapy and hormone therapy, respectively. Patients who completed
the therapy were followed-up every 6–12 months until they either
succumbed or reached the end of follow-up by December, 2017. During
the follow-up, some patients did not survive. The mean follow-up
time for the entire series was 99 months (range, 2–630 months). In
addition to routine clinical check-ups throughout the follow-up
period, the patients were also subjected to any necessary chest,
abdominal-pelvic and bone isotope scans. In total, ~19% of the
patients experienced recurrence and 23% succumbed to the disease.
Disease-specific survival (DSS) was determined as the interval
between the time of diagnosis and the disease-related mortality or
the last time a patient was seen alive. Patients who succumbed due
to unspecified or unrelated causes were not included in the DSS
calculation. Based on the clinical evidence alone, the causes of
mortality were typically clear. In those cases, no autopsy was
carried out. The date on which patients were last seen disease-free
was used to calculate disease-free survival (DFS), which was also
determined as the interval between the diagnosis and the occurrence
of disease recurrence.
Tissue microarray and IHC
Tissue microarray (TMA) slides (41) were prepared using a total number of
562 BC FFPE blocks. In brief, BC tissue cores were extracted from
the donor block and placed into a recipient paraffin block using a
computerized TMA platform (TMA Master 1.14 SP3; 3DHISTECH, Ltd.).
FZD8 protein expression patterns in BC and lymph node tissues were
detected by staining the TMA slides with anti-FZD8 antibody using a
fully automated Ventana IHC-staining system (BenchMark XT automated
slide preparation system; Roche Tissue Diagnostics). The Ventana
protocol included paraffin removal from the 4-µm-thick sections
with Ventana EZ-Prep at 75°C for 15 min and treatment with human
anti-FZD8 monoclonal primary antibody (cat. no. ab155093, Abcam;
dilution, 1:100) for 16 min at 37°C. Chromogen color staining was
developed at room temperature using the iView DAB Detection Kit
(cat. no. 760–091, Ventana Medical Systems), which was carried out
as previously reported (42). The
TMA-stained sections were counterstained with Hematoxylin II (Roche
Tissue Diagnostics) for 3–5 min at room temperature and treated
with running tap water for bluing for 4 min, followed by washing
with PBS for 1 min. This was followed by immersion in ascending
grades of ethanol buffer (50, 70, 80, 90, 95 and 100%) for 3 min
for each change. The stained sections were mounted with Tissue-Tek
xylene-based mounting media (Sakura Finetek USA, Inc.) and covered
with a glass coverslip (Corning, Inc.).
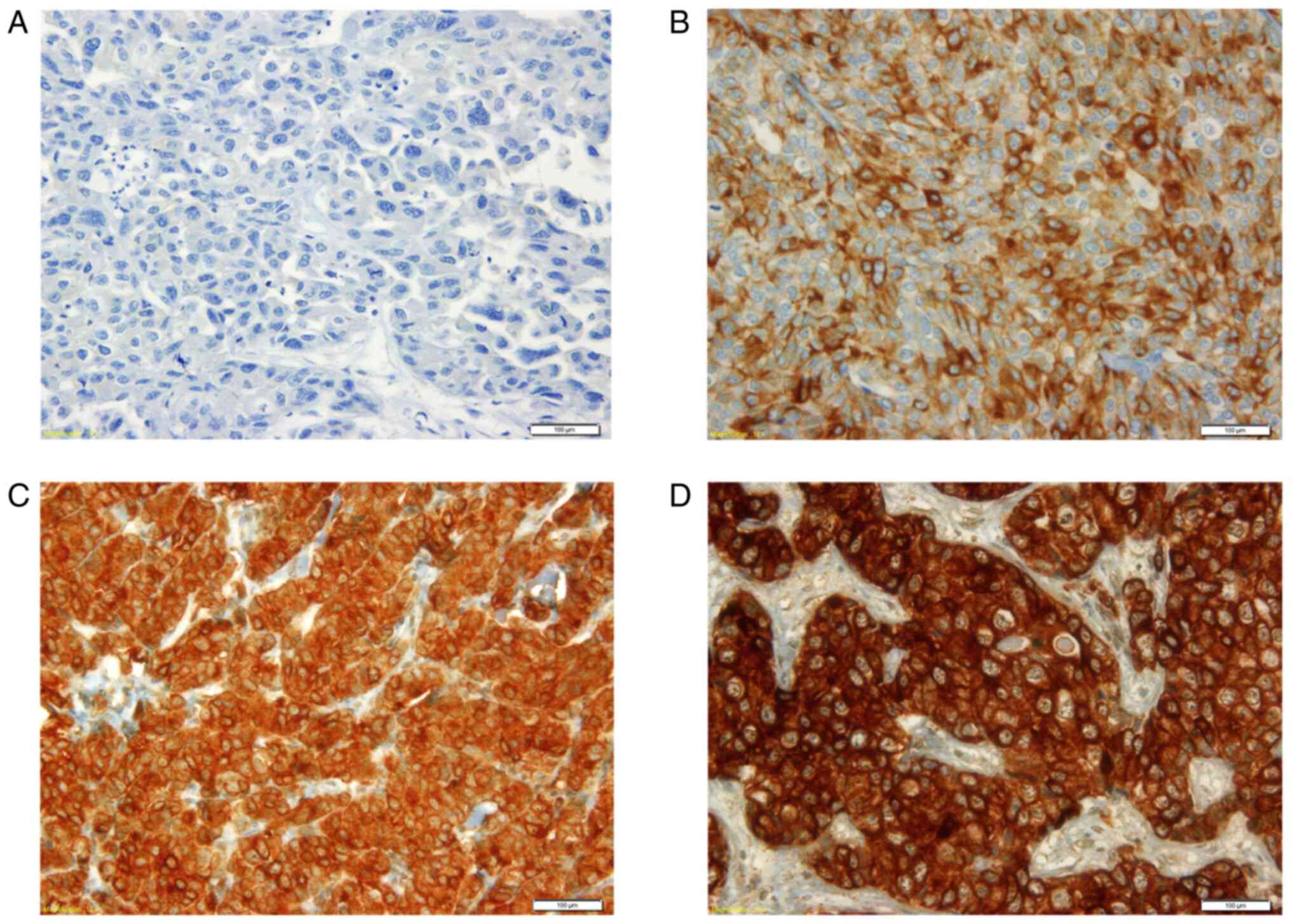
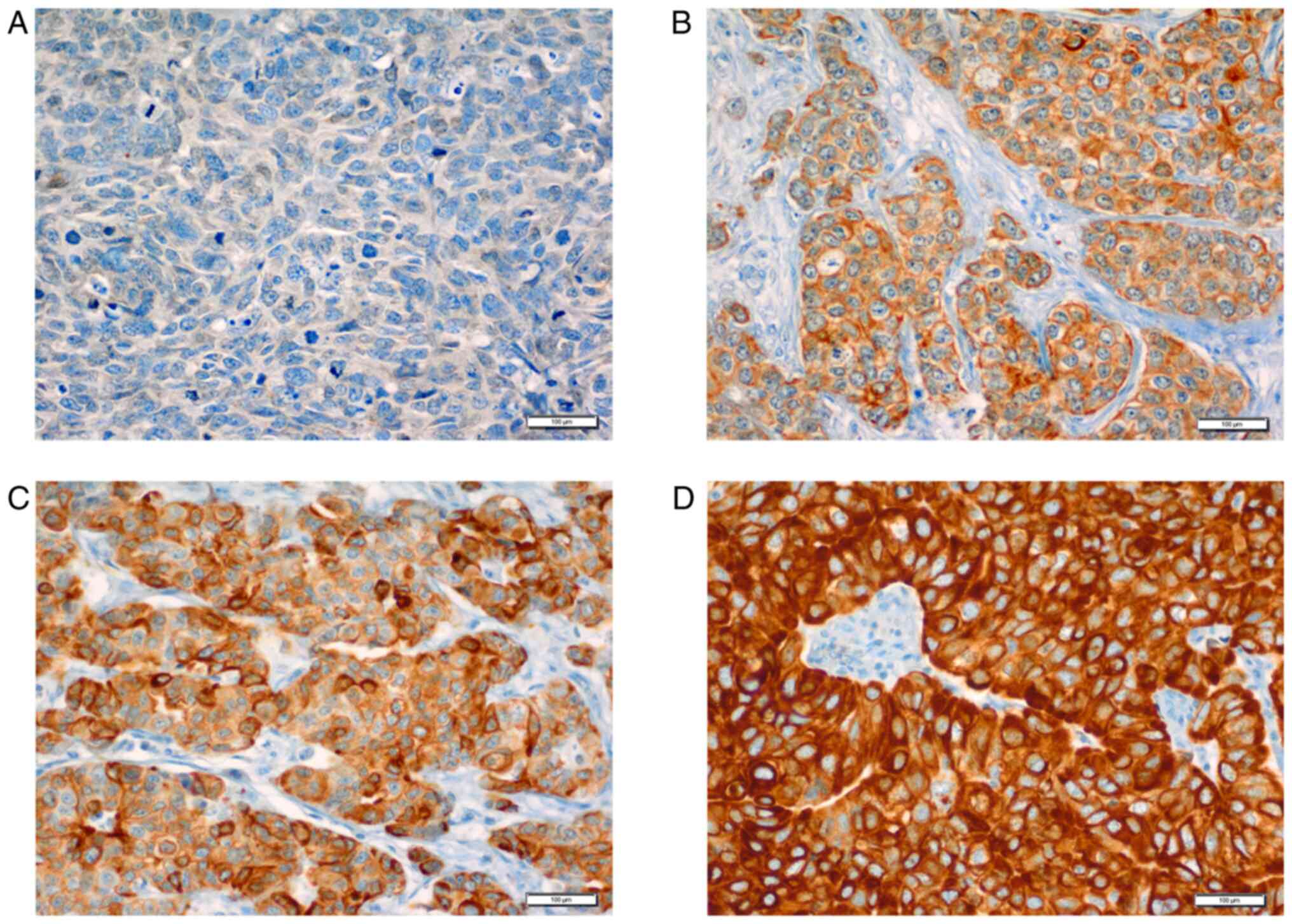
FZD8 protein expression scoring
The evaluation and scoring of the IHC-stained FZD8
expression in BC and lymph node TMA sections was performed by two
experienced pathologists blinded to the clinical data. The IHC
Index Score System, which has already been previously validated
(43,44), was used for scoring. The BC tumor
cells with FZD8 cytoplasmic staining were classified into four
groups as follows: i) 0, negative or no detectable staining; ii)
1+, weak but still detectable staining; iii) 2+, moderate or
clearly positive; and iv) 3+, strong or highly strong (heavy
staining). The cytoplasmic index was determined using the following
formula, considering the intensity of the staining, as well as the
fraction of the positively stained cells (45): I=0×f0 + 1×f1 + 2×f2 + 3×f3, where
(I) is the staining index and (f0-f3) are the proportions of cells
exhibiting a given staining intensity (0–3+). The index value may
vary between 0 and 300. Different cut-off points were used to test
their potential value as prognostic indicators. The FZD8 expression
pattern was examined using a Nikon light microscope (model no.
6132; Nikon Corporation) at a magnification of ×40 and imaged using
a Coolsnap Pro Color camera equipped with Image Pro Plus software,
v6 (Media Cybernetics, Inc.).
miRNA target-prediction analysis
Microarray datasets from the GEO (accession no.
GSE65194) (46) were used for in
silico analysis. GEO2R was used to identify the differentially
expressed genes (DEGs) in TNBC patient samples compared with
healthy breast tissue. Herein, the raw P-value was calculated using
unpaired Student's t-test (47).
The DEGs were subsequently investigated using the iPathwayGuide
high-throughput knowledge discovery tool (Advaita Corporation)
using a cut-off fold change of 1.5 and a cut-off of P-value at
<0.05 based on unpaired Student's t-test, as previously
described (48). TargetScanHuman
(version 7.2, http://www.targetscan.org/vert_72/) was used to
validate the miRNAs acquired from the iPathwayGuide study for FZD8
and WNT ligand-target prediction (49) and then validated further using the
new interface of miRabel miRNAs target-prediction platform
(http://bioinfo.univ-rouen.fr/mirabel/) (50). In the miRabel platform, four miRNA
target-prediction bioinformatics tools (miRanda, PITA, SVmicrO and
TargetScan) are merged into one database.
Statistical analysis
SPSS, Inc. (v19, IBM SPSS Statistics for Windows)
software packages were used to conduct the statistical analysis. To
determine the significance of the association between the various
categorical variables, Fisher's exact test was used. For the
univariate survival analysis of the survival outcome measures (DSS
and DFS), the Kaplan-Meier method with a log-rank (Mantel-Cox)
comparison test was used. P<0.05 was considered to indicate a
statistically significant difference.
Results
Frequency of FZD8 protein expression
pattern profiling in BC
FZD8 cytoplasmic expression pattern analysis
revealed that 37.5% of the primary samples exhibited a high
expression profile (2+ and 3+), whereas almost 63% of the samples
exhibited a low expression profile (0 and 1+) (Table I). Furthermore, ~46% of the
malignant tissues from the lymph node-positive samples exhibited a
high FZD8 cytoplasmic expression (2+ and 3+), with 54% exhibiting a
decreased expression in the cytoplasm (0 and 1+) (Table I). The cytoplasmic protein
expression patterns in the primary BC samples (Fig. 1), as well as those in the lymph
nodes (Fig. 2), varied in
intensity, ranging from no expression (0), to weak (1+), moderate
(2+) and strong expression (3+).
 | Table I.FZD8 cytoplasmic expression pattern
intensities and corresponding percentages in the primary BC tissue
and secondary lymph node samples. |
Table I.
FZD8 cytoplasmic expression pattern
intensities and corresponding percentages in the primary BC tissue
and secondary lymph node samples.
|
| BC tissues | Lymph nodes |
|---|
|
|
|
|
|---|
| FZD8 cytoplasmic
expression intensity | No. of samples | Percentage | No. of samples | Percentage |
|---|
| Negative (0) | 133 | 23 | 23 | 6.5 |
| Weak (1+) | 229 | 39.5 | 166 | 47.3 |
| Moderate (2+) | 119 | 20.5 | 73 | 20.8 |
| Strong (3+) | 98 | 17 | 89 | 25.4 |
Association between the FZD8
expression pattern and the patient clinicopathological
characteristics
In this analysis, a cut-off point of a low
cytoplasmic expression (0 and 1+ scores) compared to a high
expression in the cytoplasm (2+ and 3+ scores) was used (Table II). There was a significant
association between the FZD8 expression pattern and various
clinicopathological features. Tumors with vascular invasion
exhibited a stronger FZD8 expression (P<0.03) than the tumors
without invasion, while low/intermediate grade tumors exhibited a
stronger FZD8 expression (P<0.003). Of note, and consistent with
the molecular dichotomy of BC [triple-positive (TP) vs. TN], TP
tumors [ER-, progesterone receptor (PR)- and HER2-positive]
exhibited a higher FZD8 expression compared with TN tumors
(P<0.001). However, the HER2-enriched BC molecular subtype was
very rare in the present study cohort and hence, the analysis of
its association with FZD8 expression was not possible. A marginally
significant association was identified between tumor size and FZD8
cytoplasmic expression (P=0.06), whereas small tumors exhibited a
high expression of FZD8. However, there was no significant
association between FZD8 cytoplasmic expression, and patient age
(P=0.41), lymph node status (P=0.23) and histopathological type
(P=0.26).
 | Table II.Associations between FZD8 protein
expression patterns and the clinicopathological characteristics of
patients with BC. |
Table II.
Associations between FZD8 protein
expression patterns and the clinicopathological characteristics of
patients with BC.
|
|
| FZD8 protein
expression pattern (%) |
|
|---|
|
|
|
|
|
|---|
| Features | No. of cases
(%) | Low expression (0,
1+) (%) | High expression
(2+, 3+) (%) | P-value |
|---|
| Age (years) |
|
|
| 0.41 |
|
<50 | 353 (63) | 193 (64) | 110 (36) |
|
|
>50 | 209 (37) | 160 (62) | 99 (38) |
|
| Tumor invasion |
|
|
| 0.11 |
|
Positive | 15 (3) | 6 (40) | 9 (60) |
|
|
Negative | 499 (97) | 315 (63) | 184 (37) |
|
| Lymph node
status |
|
|
| 0.23 |
|
Positive | 183 (39) | 122 (67) | 61 (33) |
|
|
Negative | 289 (61) | 183 (63) | 106 (37) |
|
| Vascular
invasion |
|
|
| 0.03 |
|
Positive | 236 (59) | 160 (68) | 76 (32) |
|
|
Negative | 164 (41) | 103 (63) | 61 (37) |
|
| Tumor size
(cm) |
|
|
| 0.06 |
|
0-3 | 189 (39) | 116 (61) | 73 (39) |
|
|
3-6 | 231 (48) | 150 (65) | 81 (35) |
|
|
>7 | 62 (13) | 45 (73) | 17 (27) |
|
| Tumor grade |
|
|
| 0.003 |
| Grade
1 | 88 (18) | 44 (50) | 44 (50) |
|
| Grade
2 | 239 (50) | 151 (63) | 88 (37) |
|
| Grade
3 | 151 (32) | 101 (67) | 50 (33) |
|
| Histopathological
type |
|
|
| 0.26 |
| Ductal
carcinoma | 500 (90) | 309 (62) | 191 (38) |
|
|
Others | 56 (10) | 39 (70) | 17 (30) |
|
| Molecular
subtypes |
|
|
| 0.001 |
|
Triple-positive | 62 (41) | 57 (92) | 5 (8) |
|
|
Triple-negative | 88 (59) | 42 (48) | 46 (52) |
|
Association between FZD8 expression
status and survival outcomes
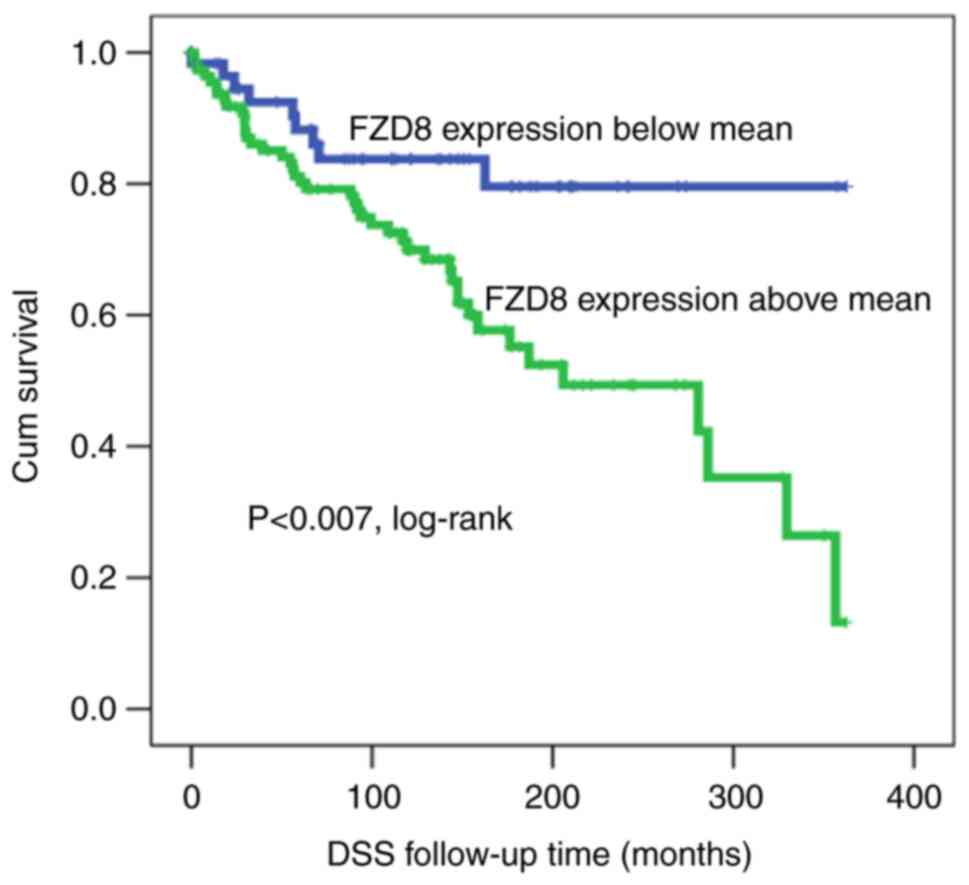
The Kaplan-Meier survival analysis revealed a
significant (log-rank test, P<0.007) association between the
FZD8 cytoplasmic expression and DSS. In this case, patients
exhibiting a low expression of FZD8 survived for a significantly
longer period of time (longer DSS) than those with a higher FZD8
expression (Fig. 3). Of note, the
present study demonstrated that at the median follow-up time (200
months), ~50% of all patients exhibiting FZD8 overexpression were
deceased, as compared with only 18% of those exhibiting a low
expression of FZD8, suggesting that patients with a low FZD8
expression had a better prognosis.
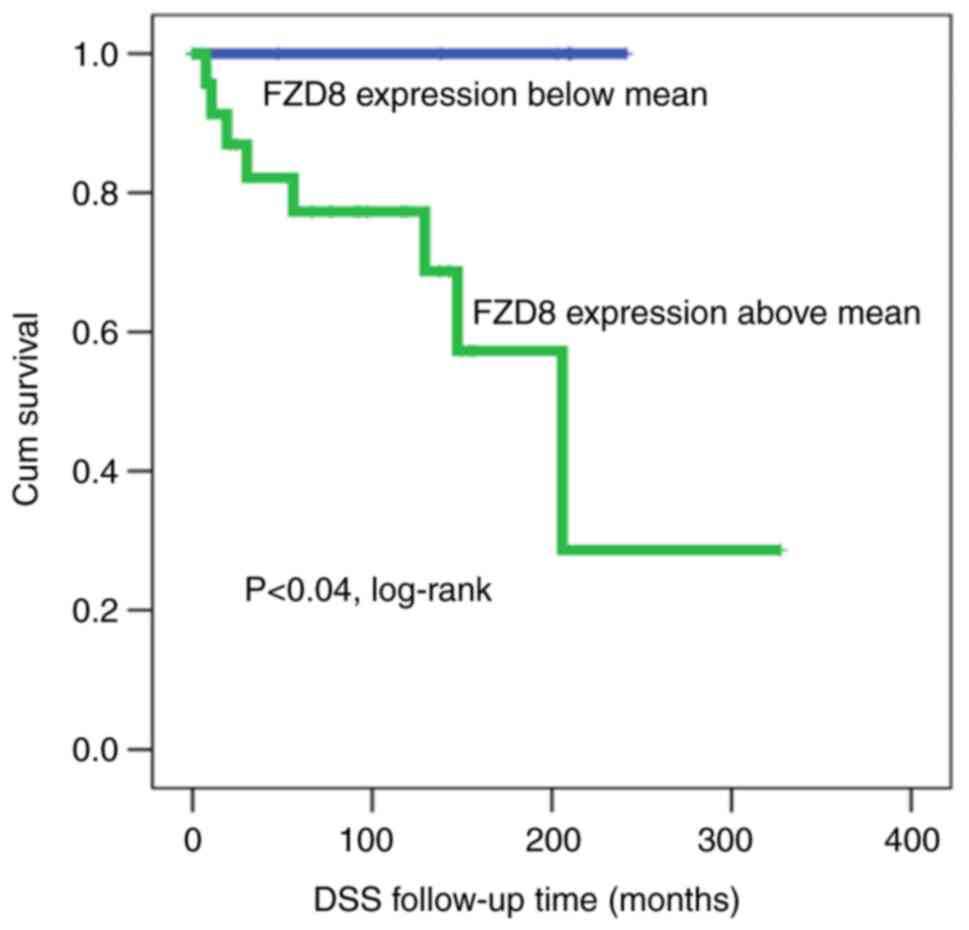
When the cohort was classified and analyzed
according to TP and TN molecular subtypes (ER+,
PR+, HER2+ vs. ER−,
PR−, HER2−), the prognostic value of FZD8
protein expression was reserved for patients with TPBC, whereas it
was completely lost in those with TN tumors. In that regard,
patients with TPBC and lower FZD8 expression patterns survived for
longer than those with higher FZD8 expression patterns (Fig. 4). Following a 5-year follow-up
period, 20% of the patients with BC and a high FZD8 protein
expression were deceased, whereas none of the patients with a low
FZD8 expression were deceased, suggesting that TP patients
exhibiting a reduced FZD8 expression had a better prognosis and a
longer survival (log-rank test, P<0.04).
It is worth mentioning that DSS was calculated as
the time from the diagnosis of the disease to mortality (due to
cancer) or to the date patients were last recorded alive. In
general, patients were followed-up at 3- to 6-month intervals until
they succumbed or did not pursue medical follow-up (i.e., who were
not admitted for hospitalization). When follow-up was not available
(patients censored or when the exact death time of the patient was
not known/not appeared), the patients were excluded from the
statistical survival analysis since the outcome (deceased or alive)
was not disclosed. Overall, the present study revealed a stable
trend toward the worse survival of patients with BC exhibiting an
increased FZD8 expression, as compared with that of those with a
decreased FZD8 expression pattern.
Cox regression analysis
According to a multivariate Cox regression analysis,
a reduced FZD8 cytoplasmic protein expression pattern was found to
be an independent predictor of a poor survival, along with age,
lymph node status, tumor grade, and vascular and tumor invasion
(P<0.008). Tumor invasion is one of the most considerable
histopathological variables and is often used as a potential
prognostic indicator. The association of FZD8 expression with tumor
invasion may reflect the tumorigenic effects of the FZD protein
family in general, including FZD8 in particular. In addition, Cox
regression analysis revealed that patients with BC with a high
expression of FZD8 exhibited an ~5-fold higher risk of
cancer-related mortality compared with those with a low expression
of FZD8 (Table III).
 | Table III.Cox regression analysis of the
prognostic values of FZD8, age at diagnosis, tumor grade and lymph
node status. |
Table III.
Cox regression analysis of the
prognostic values of FZD8, age at diagnosis, tumor grade and lymph
node status.
| Feature | P-value | Standard error
value | Relative risk | 95% CI |
|---|
| FZD 8 expression
(low vs. high) | 0.008 | 0.614 | 5.13 | 0.058-0.649 |
| Tumor grade (low
vs. high) | 0.05 | 0.627 | 0.29 | 0.994-11.614 |
| Age at diagnosis
(<50 vs. >50 years) | 0.12 | 0.389 | 1.70 | 0.274-1.260 |
| Lymph node status
(Neg vs. Pos) | 0.47 | 0.409 | 1.20 | 0.372-1.848 |
miRNA target prediction analysis
In this target prediction analysis, 3,923 DEGs were
identified from a total of 20,150 genes whose expression had been
measured. These were identified using a statistical significance
(P-value) threshold of 0.05 and a log-fold change in expression
with an absolute value of a minimum of 1.5. The analysis of
upstream regulatory miRNAs using iPathwayGuide revealed 374 miRNAs
predicted to regulate DEGs in TNBC (Table SI). Further filtering based on the
association between regulatory upstream miRNAs and FZD8 yielded
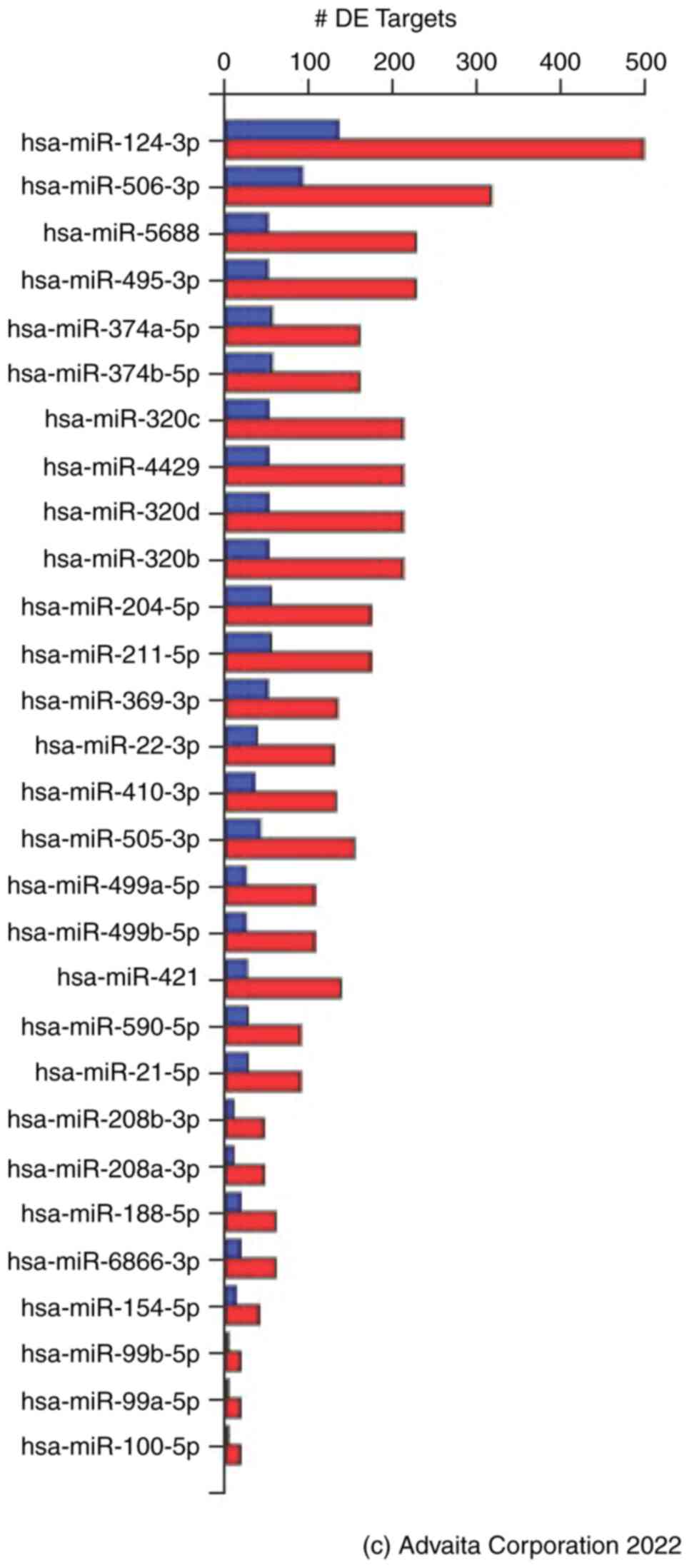
29/374 miRNAs (Fig. 5). Filtering
of the 29 miRNAs based on the prediction score revealed that eight
miRNAs, (hsa-miR-124-3p, hsa-miR-506-3p, hsa-miR-495-3p,
hsa-miR-410-3p, hsa-miR-208b-3p, hsa-miR-208a-3p, hsa-miR-99b-5p
and hsa-miR-99a-5p) were significantly associated with FZD8
signaling in TNBC (Table IV). Of
note, the same sets of the aforementioned miRNAs were found when
the TNBCs were compared with the non-TNBCs, and similar to the
TNBCs compared with healthy tissues. The DEGs potentially regulated
by these miRNAs are listed in Table
SII.
 | Table IV.miRNA target-prediction for FZD8
expression, highlighting predicted WNT ligands for each miRNA. |
Table IV.
miRNA target-prediction for FZD8
expression, highlighting predicted WNT ligands for each miRNA.
| MicroRNA | No. of
differentially expressed targets (−/all) | No. of targets
(−/all) | P-value | Prediction
score | Predicted WNT
ligands for the microRNA |
|---|
|
hsa-miR-124-3p | 134/631 | 1065/2110 | 1 | 0.010 | WNT2B, 4, 5A,
5B, 7B, 9A, 9B, 11, 16 |
|
hsa-miR-506-3p | 91/407 | 622/1262 | 1 | 0.005 | WNT2B, 4, 5A,
5B, 7B, 9A, 9B, 10B, 11, 16 |
|
hsa-miR-374a-5p | 55/214 | 396/717 | 1 | 0.581 | WNT2, 2B, 3, 3A,
5A, 5B, 11, 16 |
|
hsa-miR-374b-5p | 55/214 | 396/717 | 1 | 0.105 | WNT2, 2B, 3, 3A,
5A, 5B, 11, 16 |
| hsa-miR-204-5p | 54/227 | 391/744 | 1 | 0.211 | WNT2, 2B, 3, 3A, 4,
5A, 8B, 9B, 10B, 11, 16 |
| hsa-miR-211-5p | 54/227 | 391/744 | 1 | 0.202 | WNT2, 2B, 3, 3A, 4,
5A, 8B, 9B, 10B, 11 |
| hsa-miR-320c | 51/262 | 394/803 | 1 | 0.994 | WNT2, 2B, 5A, 8A,
8B, 9A, 9B, 10B, 11, 16 |
| hsa-miR-4429 | 51/262 | 394/803 | 1 | 0.999 | WNT2B, 8A, 8B, 9A,
9B, 10B |
| hsa-miR-320d | 51/262 | 394/803 | 1 | 0.994 | WNT2, 2B, 8A, 10B,
11, 5A, 8B, 9A, 9B, 16 |
| hsa-miR-320b | 51/262 | 394/803 | 1 | 0.994 | WNT2, 2B, 5A, 8A,
8B, 9A, 9B, 10B, 11, 16 |
| hsa-miR-5688 | 50/276 | 411/831 | 1 | 0.980 | WNT2B |
|
hsa-miR-495-3p | 50/276 | 411/831 | 1 | 0.040 | WNT2B, 4, 5B,
8A, 8B, 9B, 11, 16 |
| hsa-miR-369-3p | 50/183 | 338/600 | 1 | 0.807 | WNT2, 2B, 3, 5A,
5B, 16 |
| hsa-miR-505-3p | 41/194 | 282/586 | 1 | 0.851 | WNT2B, 4, 5A, 7A,
7B, 8B, 16 |
| hsa-miR-22-3p | 37/166 | 323/584 | 1 | 0.991 | WNT1, 2B, 3, 3A, 4,
5A, 8A, 9B, 10B, 11 |
|
hsa-miR-410-3p | 34/165 | 295/581 | 1 | 0.013 | WNT2, 2B, 3A, 4,
5A, 5B, 7B, 9B, 11, 16 |
| hsa-miR-21-5p | 26/115 | 187/365 | 1 | 0.989 | WNT2B, 4, 3A, 5A,
9B |
| hsa-miR-590-5p | 26/115 | 188/366 | 1 | 0.989 |
|
| hsa-miR-421 | 25/162 | 196/436 | 1 | 0.597 | WNT2B, 5A, 7A, 7B,
8B, 9B, 16 |
Discussion
BC accounts of at least 53% of all female patient
cancer cases in Saudi Arabia, rendering it one of the most frequent
malignancies among Saudi women (4).
Such increased incidence of BC in Saudi Arabia may be mainly
attributed to the lack of early, frequent and effective BC
screening programs (51).
Therefore, the development of such programs and the identification
of BC biomarkers are critical for the early detection of the
disease. BC is controlled by crucial complexities of signaling
cascades (52), which, following
decades of focused research are not yet fully understood. Wnt/FZD
signaling has been demonstrated to regulate numerous cellular
events during embryonic development and cancer. Despite having a
significant impact on embryonic development and disease, FZD8 is a
critical WNT receptor that has received minimal attention in BC
investigations. Of note, FZD8 expression has been detected during
mammary stem cell development (53–55).
In the present study, the clinicopathological characteristics of
Saudi patients with BC and their association with the FZD8
expression pattern were examined. The majority of the patients
exhibited medium-to-high expression levels, either in BC or lymph
node tissues. A previous study (33) revealed that increased expression
levels of FZD8 were detected in the breast squamous cell
carcinoma-derived TNBC cell line. Elevated FZD8 expression levels
have also been observed in various other malignancies, including
lung cancer (56), medulloblastoma
(57), renal cancer (58) and osteosarcoma of the spine
(59). Taken together, the findings
of the present study suggest that targeting FZD8 expression in BC
may be an effective therapeutic approach.
Herein, Cox regression analysis results revealed
that FZD8 was associated with tumor invasion. An increased
expression of FZD8 has been reported to be associated with other
significant cellular events during carcinogenesis in different
types of cancer. For instance, in spinal osteosarcoma, the
downregulation of FZD8 has been shown to suppress the invasion,
migration and proliferation of osteosarcoma cells (59). Increased FZD8 expression has been
linked to proliferation and metastasis in renal cell cancer
(58). Similarly, in prostate
(35) and colorectal (60) cancer, an increased expression of
FZD8 has been revealed to promote metastasis.
The results of the present study revealed a
significant association between low expression levels of FZD8 and
tumor aggressiveness, including vascular invasion, tumor size and
grade, molecular subtype and survival outcomes in the BC cohort
studied. A higher proportion of patients with grade III cancer and
lymphovascular invasion also exhibited a decreased FZD8 expression.
Notably, these clinicopathological features were mainly observed in
the TP molecular subtype. It is known that the TPBC subtype is
associated with a good prognosis, as compared with the TN molecular
subtype, mainly due to the multimodality of treatment options,
which leads to a better prognosis and longer survival (61,62).
Overall, these results suggest a powerful prognostic value of FZD8
expression in BC. To the best of our knowledge, this is the first
report revealing the association between an increased expression of
FZD8 and these clinicopathological characteristics of patients with
BC. In other types of cancer, including gastric cancer, an
increased expression of FZD8 has been reported and demonstrated to
indicate a poor prognosis (34,63).
One of the limitations of the survival analysis performed during
the present study was the limited sample size of the patient
cohort. However, in future studies, the authors aim to expand the
sample size, launching a national sample network termed Saudi
Cancer Tissue Array-based Network (SCTAN) that could aim towards
the collection of additional tumor samples.
The present study attempted to unravel several
complex signaling mechanisms through which FZD8 may function.
miRNAs have been shown to mediate BC tumorigenesis through WNT
signaling (64). In the present
study, microarray datasets were analyzed in TNBC patient samples
from the GEO database and compared with healthy BC samples, using
the iPathwayGuide high-throughput knowledge discovery platform. A
total of 29 miRNAs were predicted to target FZD8 expression in BC,
eight of which exhibited significant prediction scores. Previous
reports have revealed that several of these eight miRNAs, including
hsa-miR-124-3p, hsa-miR-506-3p, hsa-miR-410-3p and hsa-miR-99a-5p
function as tumor suppressors (65–68).
Additionally, hsa-miR-495-3p has been reported to be a predictor of
a poor prognosis of patients with TNBC (69), hsa-miR-208a-3p has been found to be
a TNBC promoter (70), while
hsa-miR-99b-5p has been found to be a promoter of tumor
aggressiveness (71). The miRNA
prediction analysis of the present study suggested that these
miRNAs could target FZD8 expression and hence, they may play a
pivotal role in the miRNA silencing of WNT signaling in BC.
However, validation analysis of the miRNAs with a significantly
high prediction score was not performed in the present study. In
vivo and/or in vitro validation assays would further
elucidate the mechanisms through which this silencing functions.
This could include the use of total RNA from BC biopsies to
investigate whether the expression of the predicted miRNAs was
simultaneously increased with FZD8 expression, using RT-qPCR.
Another approach could include using miRNA antagomirs to knock down
the predicted miRNA function in BC cell line(s) and also
investigate the increase in FZD8 expression.
One important miRNA, hsa-miR-100, has been reported
to inhibit migration and invasion, and regulate apoptosis and
metastasis in BC (38,72,73).
The present study investigated whether FZD8 was a predicted target
for this miRNA, and it was revealed that FZD8 could be a target for
hsa-miR-100 with a prediction score of 0.075. In addition, two
other miRNAs expressed in BC were investigated in the present study
that were previously reported to target FZD8 in other types of
cancer, including hsa-miR-375 [colorectal cancer (60)] and hsa-miR-520b [spinal osteosarcoma
(59)]. The miRNA prediction
analysis performed herein revealed a prediction score of 0.017 for
hsa-miR-375 and 0.974 for hsa-miR-520. However, these results need
to be further investigated in future validation research in BC
experimental models.
In conjunction to the miRNA prediction analysis
carried out in the present study, the expression of another WNT
receptor (FZD6) in BC was previously analyzed by the authors
(74). It was demonstrated that the
FZD6 prognostic value was more potent in younger patients with BC,
which indeed is not the case for FZD8 expression. In addition, at
least 18 miRNAs were predicted to silence FZD6 expression,
presented with a high prediction score. Specifically, the miRNAs
were hsa-miR-101-3p, hsa-miR-302b-3p, hsa-miR-302d-3p,
hsa-miR-372-3p, hsa-miR-373-3p, hsa-miR-520c-3p, hsa-miR-519a-3p,
hsa-miR-519b-3p, hsa-miR-568, hsa-miR-545-3p, hsa-miR-130a-3p,
hsa-miR-130b-3p, hsa-miR-301a-3p, hsa-miR-301b-3p, hsa-miR-454-3p,
hsa-miR-3121-3p, hsa-miR-19a-3p, and hsa-miR-19b-3p (74). In this previous study by the
authors, the predicted miRNAs that could target FZD6 expression
were compared with those predicted to target FZD8 expression in BC.
The comparison analysis revealed that there were no common miRNAs
to target both FZD6 and FZD8, indicating that the prediction
database tools used in both studies and the analysis carried out
were gene specific.
The therapeutic potential of FZD8 in TNBC has been
previously reported to play a crucial role in mediating
chemotherapy resistance through WNT signaling (33). The biological mechanism through
which FZD8 exerts this function remains unclear. It is suggested
that FZD8 expression may be either downregulated/fine-tuned or
silenced by one or more of the predicted miRNAs revealed in the
analysis of the present study to activate this chemotherapy
resistance process. Overall, the miRNAs identified in the present
study, particularly those with the top prediction scores, are
highly recommended for further validation analyses (Fig. 6).
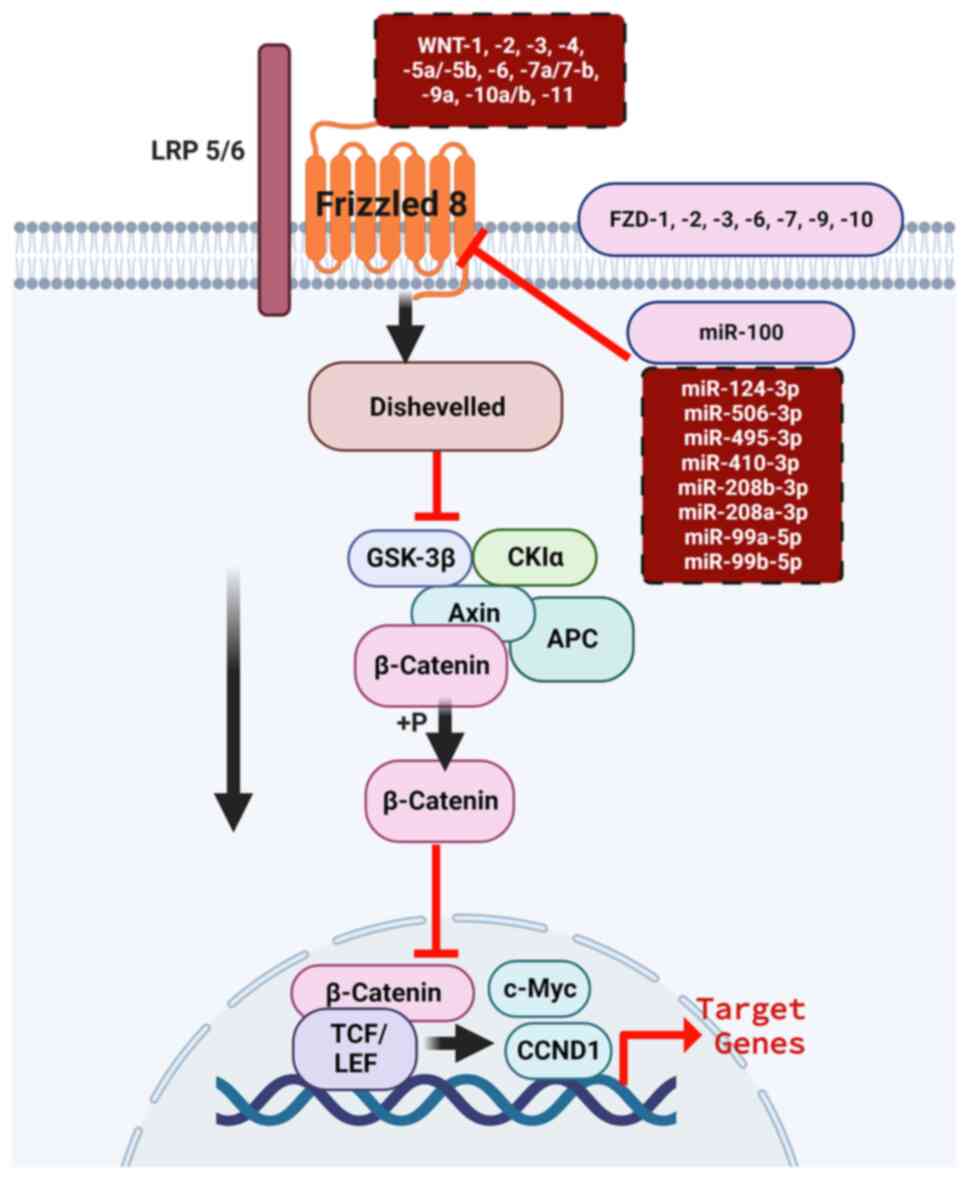 | Figure 6.Predicted WNT ligands and miRNA
regulation of FZD8 expression in breast cancer through the
canonical (β-catenin-dependent) pathway. Potential WNT ligands,
including WNT1, WNT2, WNT3, WNT4, WNT5a/b, WNT6, WNT7a/b, WNT9a,
WNT10a/b and WNT11 (top brown dotted box), bind to the FZD
receptors, including FZD1, FZD2, FZD3, FZD6, FZD7, FZD8, FZD9 and
FZD10. Several potential miRNAs (lower brown dotted brown box,
right), including hsa-miR-100, hsa-miR-124-3p, hsa-miR-506-3p,
hsa-miR-495-3p, hsa-miR-410-3p, hsa-miR-208a/b-3p and
hsa-miR-99a/b-5p silence FZD8 expression to recruit Dvl, which in
turn suppresses protein complex containing β-catenin, GSK-3β, Axin,
APC and CKIa. Upon phosphorylation, β-catenin translocates to the
nucleus and binds to the TCF/LEF complex to drive the transcription
of the target genes. LRP5/6 are the FZD co-receptors. miRNA,
microRNA; FZD8, frizzled receptor 8; Dvl, Dishevelled; TCF/LEF,
T-cell factor/lymphoid enhancer factor; LRP, low-density
lipoprotein receptor-related protein; FZD, FZD8, frizzled receptor;
GSK-3β, glycogen synthase kinase-3 β; APC, adenomatous polyposis
coli; CKIα, cyclin-dependent kinase inhibitory protein α. |
The ability of FZD8 to activate the non-canonical or
canonical WNT/β-catenin pathways during tumorigenesis in various
cancer types such as renal cell carcinoma (58) and BC (75) has been previously reported. In BC,
FZD8 has been revealed to play a crucial role in TNBC drug
resistance through canonical WNT/β-catenin (75). The miRNA analysis performed herein
predicted several WNT ligands to be involved in FZD8 silencing in
BC. These included WNT1, WNT2, WNT3, WNT4, WNT5a/b, WNT6, WNT7a/b,
WNT9a, WNT10a/b and WNT11 (Fig. 6).
These potential ligands could bind to several FZD receptors,
including FZD1, FZD2, FZD3, FZD6, FZD7, FZD8, FZD9 and FZD10. This
activates the downstream targets of the canonical pathways of
Dishevelled, which in turn blocks several downstream protein
targets (β-catenin, glycogen synthase kinase-3, Axin, adenomatous
polyposis coli and cyclin-dependent kinase inhibitory protein a;
Fig. 6). Once β-catenin is
phosphorylated, it translocates to the nucleus to bind to the
T-cell factor/lymphoid enhancer factor complex, following which the
transcription of the target genes is activated (Fig. 6). The miRNAs identified in the
present study could play a role in fine-tuning the expression of
FZDs, including that of FZD8, to activate the WNT downstream
targets (Fig. 6). These results
support those of previous research, demonstrating that these WNT
ligands are highly expressed and play critical functional roles in
the development of BC through the canonical WNT/β-catenin pathway,
as previously reviewed by Xu et al (10). It is also suggested that FZD8 in the
BC cohort of the present study was more likely to function through
the canonical WNT/β-catenin pathway than the non-canonical pathway;
however, further validation studies are required in the future.
In conclusion, female BC has been identified in
multiple studies and in the Saudi Cancer Registry as the most
frequent type of cancer among female patients in Saudi Arabia. The
lack of early BC biomarkers to aid the early detection of the
disease further aggravates the existing difficulties of the health
system and individual patients. Herein, the expression of the WNT
signaling receptor, FZD8, was investigated in a Saudi BC cohort.
The majority of the patients of the cohort exhibited
moderate-to-high FZD8b cytoplasmic expression. Patients exhibiting
an increased expression of FZD8 had a low survival rate, and vice
versa. Increased levels of FZD8 expression were consistently
associated with a poor prognosis. This refers to several
clinicopathological features, including tumor vascular invasion,
size, grade, molecular subtypes and survival outcomes. miRNA target
prediction analysis using microarray TNBC datasets revealed that
FZD8 was a target for 29 miRNAs expressed in BC, among which eight
miRNAs exhibited significant prediction scores. This miRNA analysis
would benefit from further validation assays. The results reported
in the present study suggest the necessity of future functional
analyses, particularly by applying in vivo and in
vitro gain- and loss-of-function approaches to further decipher
FZD8 biological functions in BC. The results of the present study
suggest that FZD8 may be a powerful prognostic BC marker and
partially elucidate the mechanistic complexity of the involvement
of WNT/FZD8/miRNA silencing in BC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to express their gratitude to
Dr Habiba Abu-Elmagd from Bristol Dental School, University of
Bristol, UK for proofreading the manuscript.
Funding
The authors extend their appreciation to the Deputyship for
Research and Innovation, Ministry of Education in Saudi Arabia for
funding this research work through the project number
(IFPRC-088-247-2020) and King Abdulaziz University, Deanship of
Scientific Research, DSR, Jeddah, Saudi Arabia.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
MAE, AB, JAM and MHAZ designed the study. MAE, AB,
MA, AA and PNP performed the histopathological, statistical and
bioinformatics analyses. MAE, AB, JAM and AZ were responsible for
collecting breast cancer tumor samples and the preparation of the
tissue microarray slides. MAE, AB, MA, AA and PNP conducted data
curation. MAE, AB and MA drafted the manuscript. MAE, PNP and AZ
reviewed and edited the manuscript. MAE was responsible for study
supervision and project administration. All authors read and
approved the final manuscript. AB and PNP confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the Center of Excellence in Genomic Medicine Research, King
Abdulaziz University (Approval no. 012-CEGMR-ETH), and was
conducted in accordance with The Declaration of Helsinki. Written
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Abu-Elmagd M, Assidi M, Alrefaei AF and
Rebai A: Editorial: Advances in genomic and genetic tools, and
their applications for understanding embryonic development and
human diseases. Front Cell Dev Biol. 10:10164002022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
World Health Organization, . Global status
report on noncommunicable diseases 2010, 2010. https://www.who.int/nmh/publications/ncd_report_full_en.pdf
|
|
3
|
World Health Organization, . Fact sheets
2018, 2018. https://www.who.int/news-room/factsheets/detail/cancer
|
|
4
|
Alqahtani WS, Almufareh NA, Domiaty DM,
Albasher G, Alduwish MA, Alkhalaf H, Almuzzaini B, Al-Marshidy SS,
Alfraihi R, Elasbali AM, et al: Epidemiology of cancer in Saudi
Arabia thru 2010–2019: A systematic review with constrained
meta-analysis. AIMS Public Health. 7:679–696. 2020.PubMed/NCBI
|
|
5
|
Unger-Saldaña K: Challenges to the early
diagnosis and treatment of breast cancer in developing countries.
World J Clin Oncol. 5:465–477. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barrios CH: Global challenges in breast
cancer detection and treatment. Breast. 62 (Suppl 1):S3–S6. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
World Health Organization, . Cancer
control: Knowledge into action: WHO guide for effective programmes:
Module 3: Early detection. Geneva: World Health Organization;
2007
|
|
8
|
Song K and Farzaneh M: Signaling pathways
governing breast cancer stem cells behavior. Stem Cell Res Ther.
12:2452021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yousefnia S, Seyed Forootan F, Seyed
Forootan S, Nasr Esfahani MH, Gure AO and Ghaedi K: Mechanistic
pathways of malignancy in breast cancer stem cells. Front Oncol.
10:4522020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu X, Zhang M, Xu F and Jiang S: Wnt
signaling in breast cancer: Biological mechanisms, challenges and
opportunities. Mol Cancer. 19:1652020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kontomanolis EN, Kalagasidou S, Pouliliou
S, Anthoulaki X, Georgiou N, Papamanolis V and Fasoulakis ZN: The
notch pathway in breast cancer progression.
TScientificWorldJournal. 2018:24154892018.
|
|
12
|
Arnold SF, Tims E and McGrath BE:
Identification of bone morphogenetic proteins and their receptors
in human breast cancer cell lines: Importance of BMP2. Cytokine.
11:1031–1037. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Furth PA: STAT signaling in different
breast cancer sub-types. Mol Cell Endocrinol. 382:612–615. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yip CH and Rhodes A: Estrogen and
progesterone receptors in breast cancer. Future Oncol.
10:2293–2301. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Krishnamurti U and Silverman JF: HER2 in
breast cancer: A review and update. Adv Anat Pathol. 21:100–107.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mirzoeva OK, Das D, Heiser LM,
Bhattacharya S, Siwak D, Gendelman R, Bayani N, Wang NJ, Neve RM,
Guan Y, et al: Basal subtype and MAPK/ERK kinase
(MEK)-phosphoinositide 3-kinase feedback signaling determine
susceptibility of breast cancer cells to MEK inhibition. Cancer
Res. 69:565–572. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mohapatra P, Preet R, Das D, Satapathy SR,
Siddharth S, Choudhuri T, Wyatt MD and Kundu CN: The contribution
of heavy metals in cigarette smoke condensate to malignant
transformation of breast epithelial cells and in vivo initiation of
neoplasia through induction of a PI3K-AKT-NFκB cascade. Toxicol
Appl Pharmacol. 274:168–179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu X, Zhang L, He X, Zhang P, Sun C, Xu X,
Lu Y and Li F: TGF-β plays a vital role in triple-negative breast
cancer (TNBC) drug-resistance through regulating stemness, EMT and
apoptosis. Biochem Biophys Res Commun. 502:160–165. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bhateja P, Cherian M, Majumder S and
Ramaswamy B: The Hedgehog signaling pathway: A viable target in
breast cancer? Cancers (Basel). 11:11262019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu L and Yang X: Targeting the hippo
pathway for breast cancer therapy. Cancers (Basel). 10:4222018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bhatelia K, Singh K and Singh R: TLRs:
Linking inflammation and breast cancer. Cell Signal. 26:2350–2357.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
González-Reyes S, Marín L, González L,
González LO, del Casar JM, Lamelas ML, González-Quintana JM and
Vizoso FJ: Study of TLR3, TLR4 and TLR9 in breast carcinomas and
their association with metastasis. BMC Cancer. 10:6652010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Roychowdhury A, Jondhale M, Saldanha E,
Ghosh D, Kumar Panda C, Chandrani P and Mukherjee N: Landscape of
toll-like receptors expression in tumor microenvironment of triple
negative breast cancer (TNBC): Distinct roles of TLR4 and TLR8.
Gene. 792:1457282021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Niu T, Zhang W and Xiao W: MicroRNA
regulation of cancer stem cells in the pathogenesis of breast
cancer. Cancer Cell Int. 21:312021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Taciak B, Pruszynska I, Kiraga L, Bialasek
M and Krol M: Wnt signaling pathway in development and cancer. J
Physiol Pharmacol. 69:2018.PubMed/NCBI
|
|
26
|
Klaus A and Birchmeier W: Wnt signalling
and its impact on development and cancer. Nat Rev Cancer.
8:387–398. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yin P, Wang W, Zhang Z, Bai Y, Gao J and
Zhao C: Wnt signaling in human and mouse breast cancer: Focusing on
Wnt ligands, receptors and antagonists. Cancer Sci. 109:3368–3375.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang X, Zhang R, Hou C, He R, Wang QS,
Zhou TH, Li XQ, Zhai QL and Feng YM: FOXF2 oppositely regulates
stemness in luminal and basal-like breast cancer cells through the
Wnt/beta-catenin pathway. J Biol Chem. 298:1020822022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang C, Ye Z, Wan J, Liang J, Liu M, Xu X
and Li L: Secreted frizzled-related protein 2 is associated with
disease progression and poor prognosis in breast cancer. Dis
Markers. 2019:61493812019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Klopocki E, Kristiansen G, Wild PJ, Klaman
I, Castanos-Velez E, Singer G, Stöhr R, Simon R, Sauter G, Leibiger
H, et al: Loss of SFRP1 is associated with breast cancer
progression and poor prognosis in early stage tumors. Int J Oncol.
25:641–649. 2004.PubMed/NCBI
|
|
32
|
Saitoh T, Hirai M and Katoh M: Molecular
cloning and characterization of human frizzled-8 gene on chromosome
10p11.2. Int J Oncol. 18:991–996. 2001.PubMed/NCBI
|
|
33
|
Yin S, Xu L, Bonfil RD, Banerjee S, Sarkar
FH, Sethi S and Reddy KB: Tumor-initiating cells and FZD8 play a
major role in drug resistance in triple-negative breast cancer. Mol
Cancer Ther. 12:491–498. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen W, Liu Z, Mai W, Xiao Y, You X and
Qin L: FZD8 indicates a poor prognosis and promotes gastric cancer
invasion and metastasis via B-catenin signaling pathway. Ann Clin
Lab Sci. 50:13–23. 2020.PubMed/NCBI
|
|
35
|
Li Q, Ye L, Zhang X, Wang M, Lin C, Huang
S, Guo W, Lai Y, Du H, Li J, et al: FZD8, a target of p53, promotes
bone metastasis in prostate cancer by activating canonical
Wnt/β-catenin signaling. Cancer Lett. 402:166–176. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Murillo-Garzón V, Gorroño-Etxebarria I,
Åkerfelt M, Puustinen MC, Sistonen L, Nees M, Carton J, Waxman J
and Kypta RM: Frizzled-8 integrates Wnt-11 and transforming growth
factor-β signaling in prostate cancer. Nat Commun. 9:17472018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang K, Li Y, Wang J, Chen R and Li J: A
novel 12-gene signature as independent prognostic model in stage IA
and IB lung squamous cell carcinoma patients. Clin Transl Oncol.
23:2368–2381. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang Q, He M, Guan S, Ma M, Wu H, Yu Z,
Jiang L, Wang Y, Zong X, Jin F and Wei M: MicroRNA-100 suppresses
the migration and invasion of breast cancer cells by targeting
FZD-8 and inhibiting Wnt/β-catenin signaling pathway. Tumour Biol.
37:5001–5011. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
World Medical Association, . World medical
association declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schauer A, Korabiowska M, Kellner S,
Schumacher M, Sauer R, Bojar H and Rauschecker H: Grading system
for breast cancer. Anticancer Res. 18:2139–2144. 1998.PubMed/NCBI
|
|
41
|
Al-Maghrabi J, Emam E, Gomaa W, Saggaf M,
Buhmeida A, Al-Qahtani M and Al-Ahwal M: c-MET immunostaining in
colorectal carcinoma is associated with local disease recurrence.
BMC Cancer. 15:6762015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nedjadi T, Al-Maghrabi J, Assidi M, Dallol
A, Al-Kattabi H, Chaudhary A, Al-Sayyad A, Al-Ammari A, Abuzenadah
A, Buhmeida A and Al-Qahtani M: Prognostic value of HER2 status in
bladder transitional cell carcinoma revealed by both IHC and BDISH
techniques. BMC Cancer. 16:6532016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lipponen PK and Collan Y: Simple
quantitation of immunohistochemical staining positivity in
microscopy for histopathology routine. Acta Stereol. 11:125–132.
1992.
|
|
44
|
Buhmeida A, Elzagheid A, Algars A, Collan
Y, Syrjänen K and Pyrhönen S: Expression of the cell-cell adhesion
molecule beta-catenin in colorectal carcinomas and their
metastases. APMIS. 116:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lipponen P and Collan Y: Simple
quantitation of immuno-histochemical staining positivity in
microscopy. Acta Stereol. 11:125–132. 1992.
|
|
46
|
Maubant S, Tesson B, Maire V, Ye M,
Rigaill G, Gentien D, Cruzalegui F, Tucker GC, Roman-Roman S and
Dubois T: Transcriptome analysis of Wnt3a-treated triple-negative
breast cancer cells. PLoS One. 10:e01223332015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Maire V, Baldeyron C, Richardson M, Tesson
B, Vincent-Salomon A, Gravier E, Marty-Prouvost B, De Koning L,
Rigaill G, Dumont A, et al: TTK/hMPS1 is an attractive therapeutic
target for triple-negative breast cancer. PLoS One. 8:e637122013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pushparaj PN, Kalamegam G, Wali Sait KH
and Rasool M: Decoding the role of astrocytes in the entorhinal
cortex in Alzheimer's disease using high-dimensional single-nucleus
RNA sequencing data and next-generation knowledge discovery
methodologies: Focus on drugs and natural product remedies for
dementia. Front Pharmacol. 12:7201702022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Quillet A, Saad C, Ferry G, Anouar Y,
Vergne N, Lecroq T and Dubessy C: Improving bioinformatics
prediction of microRNA targets by ranks aggregation. Front Genet.
10:13302020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
AlSaleh KA: Efficacy of breast cancer
screening program in Kingdom of Saudi Arabia. Saudi Med J.
43:428–430. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Katoh M: Networking of WNT, FGF, Notch,
BMP, and Hedgehog signaling pathways during carcinogenesis. Stem
Cell Rev. 3:30–38. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Oakes SR, Gallego-Ortega D and Ormandy CJ:
The mammary cellular hierarchy and breast cancer. Cell Mol Life
Sci. 71:4301–4324. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kendrick H, Regan JL, Magnay FA,
Grigoriadis A, Mitsopoulos C, Zvelebil M and Smalley MJ:
Transcriptome analysis of mammary epithelial subpopulations
identifies novel determinants of lineage commitment and cell fate.
BMC Genomics. 9:5912008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Alexander CM, Goel S, Fakhraldeen SA and
Kim S: Wnt signaling in mammary glands: Plastic cell fates and
combinatorial signaling. Cold Spring Harb Perspect Biol.
4:a0080372012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang HQ, Xu ML, Ma J, Zhang Y and Xie CH:
Frizzled-8 as a putative therapeutic target in human lung cancer.
Biochem Biophys Res Commun. 417:62–66. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Salsano E, Paterra R, Figus M, Menghi F,
Maderna E, Pollo B, Solero CL, Massimi L and Finocchiaro G:
Expression profile of frizzled receptors in human medulloblastomas.
J Neurooncol. 106:271–280. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yang Q, Wang Y, Pan X, Ye J, Gan S, Qu F,
Chen L, Chu C, Gao Y and Cui X: Frizzled 8 promotes the cell
proliferation and metastasis of renal cell carcinoma. Oncotarget.
8:78989–79002. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang J, Pang W, Zuo Z, Zhang W and He W:
MicroRNA-520b suppresses proliferation, migration, and invasion of
spinal osteosarcoma cells via downregulation of frizzled-8. Oncol
Res. 25:1297–1304. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xu L, Wen T, Liu Z, Xu F, Yang L, Liu J,
Feng G and An G: MicroRNA-375 suppresses human colorectal cancer
metastasis by targeting frizzled 8. Oncotarget. 7:40644–40656.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Bianchini G, De Angelis C, Licata L and
Gianni L: Treatment landscape of triple-negative breast
cancer-expanded options, evolving needs. Nat Rev Clin Oncol.
19:91–113. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kirikoshi H, Sekihara H and Katoh M:
Expression profiles of 10 members of frizzled gene family in human
gastric cancer. Int J Oncol. 19:767–771. 2001.PubMed/NCBI
|
|
64
|
Hu X, Zhang Q, Xing W and Wang W: Role of
microRNA/lncRNA intertwined with the Wnt/β-catenin axis in
regulating the pathogenesis of triple-negative breast cancer. Front
Pharmacol. 13:8149712022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang Y, Chen L, Wu Z, Wang M, Jin F, Wang
N, Hu X, Liu Z, Zhang CY, Zen K, et al: miR-124-3p functions as a
tumor suppressor in breast cancer by targeting CBL. BMC Cancer.
16:8262016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhao X, Bai X, Li W, Gao X, Wang X and Li
B: microRNA-506-3p suppresses the proliferation of triple negative
breast cancer cells via targeting SNAI2. Mol Cell Toxicol.
17:513–522. 2021. View Article : Google Scholar
|
|
67
|
Zhang YF, Yu Y, Song WZ, Zhang RM, Jin S,
Bai JW, Kang HB, Wang X and Cao XC: miR-410-3p suppresses breast
cancer progression by targeting Snail. Oncol Rep. 36:480–486. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Long X, Shi Y, Ye P, Guo J, Zhou Q and
Tang Y: MicroRNA-99a suppresses breast cancer progression by
targeting FGFR3. Front Oncol. 9:14732020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lin S, Zhao M, Lv Y, Mao G, Ding S and
Peng F: The lncRNA GATA3-AS1/miR-495-3p/CENPU axis predicts poor
prognosis of breast cancer via the PLK1 signaling pathway. Aging
(Albany NY). 13:13663–13679. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zou Y, Zheng S, Xiao W and Xie X, Yang A,
Gao G, Xiong Z, Xue Z, Tang H and Xie X: circRAD18 sponges
miR-208a/3164 to promote triple-negative breast cancer progression
through regulating IGF1 and FGF2 expression. Carcinogenesis.
40:1469–1479. 2019.PubMed/NCBI
|
|
71
|
Gujrati H, Ha S, Waseem M and Wang BD:
Downregulation of miR-99b-5p and upregulation of nuclear mTOR
cooperatively promotes the tumor aggressiveness and drug resistance
in African American prostate cancer. Int J Mol Sci. 23:96432022.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Gong Y, He T, Yang L, Yang G, Chen Y and
Zhang X: The role of miR-100 in regulating apoptosis of breast
cancer cells. Sci Rep. 5:116502015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang W, Liu Y, Guo J, He H, Mi X, Chen C,
Xie J, Wang S, Wu P, Cao F, et al: miR-100 maintains phenotype of
tumor-associated macrophages by targeting mTOR to promote tumor
metastasis via Stat5a/IL-1ra pathway in mouse breast cancer.
Oncogenesis. 7:972018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Assidi M, Buhmeida A, Al-Zahrani MH,
Al-Maghrabi J, Rasool M, Naseer MI, Alkhatabi H, Alrefaei AF, Zari
A, Elkhatib R, et al: The prognostic value of the developmental
gene FZD6 in young Saudi breast cancer patients: A biomarkers
discovery and cancer inducers OncoScreen approach. Front Mol
Biosci. 9:7837352022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zeng CM, Chen Z and Fu L: Frizzled
receptors as potential therapeutic targets in human cancers. Int J
Mol Sci. 19:15432018. View Article : Google Scholar : PubMed/NCBI
|