Introduction
Esophageal cancer is a common digestive malignant
tumor, the main types of which are esophageal squamous cell
carcinoma (ESCC) and esophageal adenocarcinoma (1). China has the highest incidence and
mortality rates of ESCC in the world, constituting ~50% of the
global prevalence (2). Since early
ESCC has no apparent symptoms, the cancer is usually in the middle
and late stages when clinical symptoms appear. Notably, the 5-year
survival rate of patients with middle-to-late stage esophageal
cancer is as low as 6–15% (3–5). To
improve the long-term survival rate of patients with ESCC, it is
necessary to explore new biomarkers and molecular mechanisms.
Lipid metabolism serves a vital role in cancer
progression, and excessive lipid uptake, storage and lipogenesis
result in rapid tumor growth (6).
The subtilisin-like proprotein convertase proprotein convertase
subtilisin/kexin type 9 (PCSK9) has a critical role in regulating
plasma cholesterol homeostasis and fatty acid metabolism (7). PCSK9 regulates not only lipid
transport but also viral infection and insulin resistance, as well
as tumor progression and apoptosis. PCSK9 has been reported to be
associated with tumor progression in various types of cancer
(8,9). In hepatocellular carcinoma and colon
cancer, PCSK9 has been shown to promote tumor proliferation
(10,11). It has also been found to facilitate
tumor invasion and suppress apoptosis in gastric cancer (12). However, it is unclear how PCSK9
contributes to ESCC development.
The dynamic interplay between neoplastic cells and
their adjacent stromal microenvironment facilitates the initiation,
advancement and dissemination of cancer, and the acquisition of
resistance to therapeutic agents in solid tumor development
(13). Tumor stroma facilitates
tumor metastasis; for example, TGF-β secreted by stromal cells
surrounding the tumor induces epithelial-mesenchymal transition
(EMT), thereby facilitating tumor cell infiltration (14). In cancer cells, EMT is associated
with tumor growth, metastatic tumor formation and enhanced
resistance to numerous therapeutic regimens. EMT has been
associated with tumor development in a number of studies. Breast
tumors induced by receptor tyrosine-protein kinase erbB-2 have been
shown to spontaneously express Snail and exhibit EMT
characteristics; in prostate cancer, tumor progression is
significantly associated with the switch from E-cadherin to
N-cadherin (15). In addition, the
presence of EMT markers is associated with high-grade and -stage
bladder cancer (16). As evidenced
by current research, EMT mechanisms are integral components of
cancer progression (17,18).
Tumors express high levels of chemokines and their
receptors, which function as growth factors for tumor cells,
promote their development and facilitate their metastasis (19). Previous studies have reported that
chemokine (C-C motif) ligand 25 (CCL25) is upregulated in various
malignancies, where it is associated with organ-specific tumor
metastasis. In non-small cell lung cancer, breast cancer and
hepatocellular carcinoma, CCL25 has been shown to promote migration
and invasion (20–22).
The present study aimed to investigate the functions
of the PCSK9 in human ESCC progression. The present study may
provide novel theoretical perspectives regarding ESCC diagnosis and
treatment.
Materials and methods
Patients and specimens
A total of 60 ESCC tissues and 20 normal esophageal
tissues were collected from patients (41 men and 19 women; age
range, 49–76 years; mean age, 68 years) who received esophagectomy
between January and May 2020 at The Fourth Hospital of Hebei
Medical University (Shijiazhuang, China). A total of 100 esophageal
cancer tissue samples were also obtained between October 2017 and
January 2018 from patients (48 men and 19 women; age range, 48–84
years; mean age, 65 years) who had undergone esophagectomy at The
Fourth Hospital of Hebei Medical University (Shijiazhuang, China).
5 Paired carcinomatous and para-carcinomatous tissues were from
patients with ESCC (3 men and 2 women; age range, 63–77 years; mean
age, 71 years) who had undergone esophagectomy at The Fourth
Hospital of Hebei Medical University (Shijiazhuang, China). All
patients did not undergo preoperative adjuvant chemotherapy and
radiotherapy. All patients signed informed consent forms, and the
study was approved (no. 2018MEC015) by the Medical Ethics Committee
of Hebei Medical University.
Cell culture
In vitro experiments were conducted using
TE1, KYSE30, KYSE150 and KYSE170 human ESCC cell lines grown in
RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing
10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
All cell lines were purchased from Procell Life Science &
Technology Co., Ltd. The cells were maintained in a humidified
atmosphere containing 5% CO2 at 37°C.
Transient transfection
To establish cell lines with PCSK9 overexpression,
transfection was performed when the cell density reached ~90%. The
PCSK9 plasmid (cat. no. RC220000; Origene Technologies, Inc.) was
employed to induce overexpression of PCSK9, whereas the empty
pCMV6-Entry plasmid (cat. no. PS100001; Origene Technologies, Inc.)
served as a control. A total of 2 µg plasmid was used for
transfection of each well of a 6-well culture plate for 6 h at
37°C. According to the manufacturer's protocol, transient
transfection was conducted using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) and subsequent
functional experiments were conducted 24 h post-transfection.
Small interfering RNA (siRNA)
transfection
KYSE-150 cells were subjected to transfection with a
double-stranded siRNA obtained from Guangzhou RiboBio Co., Ltd.
using Hi-perfect Transfection Reagent (Qiagen GmbH) according to
the manufacturer's protocol. To establish cell lines with PCSK9
knockdown, transfection was performed when the cell density reached
~50% of a 6-well culture plate. Cells were transfected with 50 nM
siRNAs for 6 h at 37°C and the knockdown gene effect was assessed
48 h post-transfection via western blot analysis and RT-qPCR. The
sequences were as follows: si-PCSK9-1 sense,
5′-GCACCCUCAUAGGCCUGGAGUUUAU-3′; and antisense,
5′-AUAAACUCCAGGCCUAUGAGGGUGC-3′; si-PCSK9-2 sense,
5′-GACAUCAUUGGUGCCUCCAGCGACU-3′; and antisense,
5′-AGUCGCUGGAGGCACCAAUGAUGUC-3′; si-PCSK9-3 sense,
5′-CAUAGGCCUGGAGUUUAUUCGAAA-3′; and antisense,
5′-UUUCGAAUAAACUCCAGGCCUAUG-3′; si-negative control (NC) sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′.
Cytokine treatment
CCL25 was purchased from PeproTech EC Ltd. In the
rescue experiment, after transfection of si-PCSK9, ESCC cells were
cultured with CCL25 (150 ng/ml) for 24 h at 37°C in a humidified
atmosphere containing 5% CO2.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA from ESCC tissues and cells was extracted
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.), and cDNA was prepared using GoScript Reverse Transcriptase
according to the manufacturer's protocol (Promega Corporation).
GoTaq qPCR Master Mix (Promega Corporation) was used to perform
qPCR amplification. RT-qPCR was conducted by using an SYBR Green
PCR Kit (Promega Corporation) with a real-time PCR System (ABI
7500). The thermocycling conditions of qPCR were as follows:
Initial denaturation, 70°C for 5 min; annealing, 25°C for 5 min;
extension, 42°C for 60 min; and denaturation, 70°C for 15 min
(23). The primer sequences are as
follows: PCSK9 forward, 5′-GCTGAGCTGCTCCAGTTTCT-3′, reverse,
5′-AATGGCGTAGACACCCTCAC-3′; GAPDH forward,
5′-AAGGTGAAGGTCGGAGTCAAC-3′ and reverse,
5′-GGGGTCATTGATGGCAACAATA-3′. Relative expression levels were
calculated using the 2−∆∆Cq method (24).
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 kit (Dojindo Laboratories, Inc.) was used to
evaluate cell viability. First, 4×103/cells well were
inoculated into 96-well plates (100 µm/well) and incubated at 37°C
overnight. Subsequently, 10 µl CCK-8 solution was added to each
well and incubated for 1.5 h at 37°C. Finally, absorbance was
measured at 450 nm using a microplate reader at 24, 48, 72 and 96
h.
Colony formation assay
ESCC cells (~1,000) were isolated and seeded in a
6-well plate. After 10–14 days, the colonies were fixed by 4%
paraformaldehyde for 15 min at room temperature and stained with
0.1% crystalline violet-stained for 10 min at room temperature. The
colonies (>50 cells) were then counted using ImageJ (version
1.8.0_172).
Transwell migration and invasion
assays
Transwell migration and invasion assays were
conducted using empty (migration) or precoated (30 µl Matrigel at
37°C for 1 h) (invasion) Transwell chambers (BD Biosciences).
Briefly, 4×104 cells were plated into the upper chamber
in serum-free culture medium, whereas complete culture medium was
added to the lower chamber. After 36–48 h incubation (dependent on
cell type) in a 37°C incubator, cells that were attached to the
lower compartment of the filter were identified by crystal violet
staining (0.1%; 37°C for 15 min) and were counted under a light
microscope.
Wound healing assay
A previously described method was used to conduct
the wound healing assay (23).
Briefly, 3×105 transfected cells were evenly distributed
onto a 6-well plate. Once the cells reached 80% confluence in
serum-free medium, they were scratched using a 200-µl pipette tip.
Subsequently, images were captured at 0 and 24 h to determine the
wound healing rate. The wound healing rate was calculated as
follows: (Original wound area-non-healing wound area)/original
wound area ×100.
Western blotting
Following PCSK9 overexpression or knockdown, total
protein was extracted using RIPA lysis buffer (Beyotime Institute
of Biotechnology) and was denatured by boiling. Protein
concentrations were detected by using a BCA protein assay kit
(Thermo Fisher Scientific, Inc.). Proteins (40 µg/lane) were then
separated on 10% gels using SDS-PAGE and were transferred onto PVDF
membranes (MilliporeSigma). After blocking the proteins with 5%
non-fat dried milk at 37°C for 1 h (Beijing Solarbio Science &
Technology Co., Ltd.), incubation was performed at 4°C overnight
with primary antibodies at a 1:1,000 dilution. Rabbit polyclonal
antibodies against E-cadherin, N-cadherin and vimentin (cat. nos.
20874-1-AP, 22018-1-AP and 10366-1-AP; 1:1,000; Proteintech Group,
Inc.) were used to detect the corresponding proteins. Anti-PCSK9
antibody (cat. no. 27882-1-AP; 1:1,000; Proteintech Group, Inc.)
was used to detect the transfection effect. The loading control was
β-actin (cat. no. 20536-1-AP; 1:1,000; Proteintech Group, Inc.)
Membranes were then washed with Tris-buffered saline plus Tween (1%
Tween-20) and incubated with secondary HRP-conjugated antibodies
(1:10,000) for 2 h at room temperature. The secondary antibodies
were purchased from Proteintech Group, Inc. (cat. no. PR30012).
Enhanced chemiluminescence SuperSignal™ West Atto
reagent (cat. no. A38554; Thermo Fisher Scientific, Inc.) was used
to visualize the proteins according the protocol described by
Scherbakov et al (25).
ImageJ software (version 1.8.0_172; National Institutes of Health)
was used to analyze the gray value of the western blot.
Immunohistochemistry
ESCC samples and normal esophageal epithelial
tissues were dehydrated in 4% paraformaldehyde in PBS (4°C), made
transparent, dipped in wax, embedded, and sliced into 5-µm thick
serial sections using a microtome (RM2235; Leica Microsystems
GmbH). Slides were deparaffinized in xylene, rehydrated using a
decreasing alcohol gradient, and washed three times for 5 min in 1X
PBS. Following antigen retrieval, the sections were heat-treated
for 5 min in 10 mmol/l Na-citrate buffer (pH 6.0) and washed again
in 1X PBS. For antigen retrieval, the sections were heated with
EDTA buffer (pH 9.0) in a microwave oven for 5 min. Subsequently,
the sections were incubated for 45 min at 37°C in 5% normal goat
serum (Proteintech Group, Inc.), after which, the sections were
incubated with an anti-PCSK9 antibody (cat. no. 27882-1-AP; 1:100;
Proteintech Group, Inc.) at 4°C overnight. A
streptavidin-biotinylated HRP-based detection system was used to
reveal specific binding after incubation with a secondary antibody
(cat. no. PR30011; 1:100; Proteintech Group, Inc.) at 37°C for 1 h.
The sections were then incubated with 3,3′-diaminobenzidine
chromogen for 1 h at 37°C. Sections were observed at ×200
magnification using a Leica DM4000 B LED microscope (Leica
Microsystems GmbH). ImageJ (version 1.8.0_172; National Institutes
of Health) was used to calculate the average optical density of
positive expression (26). The
expression was ranked on the sum of intensity and area from 0 to 7:
0–2, negative expression; 3–7, positive expression (3–4, weak
positive expression; 5–7, strong positive expression). Staining
intensity was graded as follows: 0, no staining; 1, mild staining;
2, moderate staining; and 3, intense staining. The staining area
was scored as follows: 0, no staining; 1, 1–25% area; 2, 26–50%
area; 3, 51–75% area; and 4, 76–100% area.
Enzyme-linked immunosorbent assay
(ELISA)
Cell supernatants were collected for ELISA. ELISA
assays were performed in 96-well ELISA plates using a CCL25 ELISA
kit (cat. no. ab256624; Abcam), according to the manufacturer's
instructions.
Bioinformatics analysis
The Cancer Genome Atlas (TCGA; http://portal.gdc.com) was used to obtain
RNA-sequencing expression profiles (level 3) for ESCC (tumor,
n=163; adjacent paracancerous tissue, n=11). Statistical analysis
was performed using R program v4.0.3 (URL http://www.R-project.org/), with ggplot2 (v3.3.2) used
to detect the expression level of PCSK9 in normal tissues adjacent
to ESCC and cancer tissues. Differential expression analysis was
performed using DESeq2 (bioconductor.org/packages/DESeq2/) for PCSK9 high and
low expression groups in ESCC. P<0.05 was considered to indicate
a statistically significant difference.
Statistical analysis
For the in vitro studies, experiments were
conducted with at least three biological repeats. SPSS 25.0 (IBM
Corp.) was used for statistical analysis. Paired and unpaired
Student's t-tests were used to compare continuous data between two
groups. For comparisons between multiple groups, ANOVA (parametric)
and the Kruskal-Wallis test (nonparametric) were used.
Subsequently, if the obtained results were deemed significant, a
post hoc test (LSD/SNK) was conducted for ANOVA, while Dunn's test
was employed for the Kruskal-Wallis test. The Kaplan-Meier method
and Cox regression analysis were used to evaluate cumulative
survival, and a χ2 test was used to examine the
association between PCSK9 expression and clinicopathological
findings. Kaplan-Meier survival curves were analyzed by log-rank
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
PCSK9 expression in ESCC tissues is
associated with poor prognosis
To investigate the role of PCSK9 in ESCC, 20 normal
esophageal and 60 ESCC tissues were assessed. At the protein level,
immunohistochemical staining results suggested that PCSK9
expression was markedly higher in ESCC tissues compared with that
in normal tissues (Fig. 1A). In
addition, TCGA database revealed that PCSK9 was significantly
elevated in ESCC tissues compared with that in normal tissues
(Fig. 1B). The results of RT-qPCR
analysis verified this finding in ESCC samples (Fig. 1C). Furthermore, analysis of PCSK9
expression levels was conducted in freshly obtained esophageal
cancer tissues compared with adjacent paracancerous tissues in a
cohort of 5 patients. The findings indicated a significant
upregulation of PCSK9 expression in ESCC tissues when compared to
para-carcinomatous tissues (Fig.
1D).
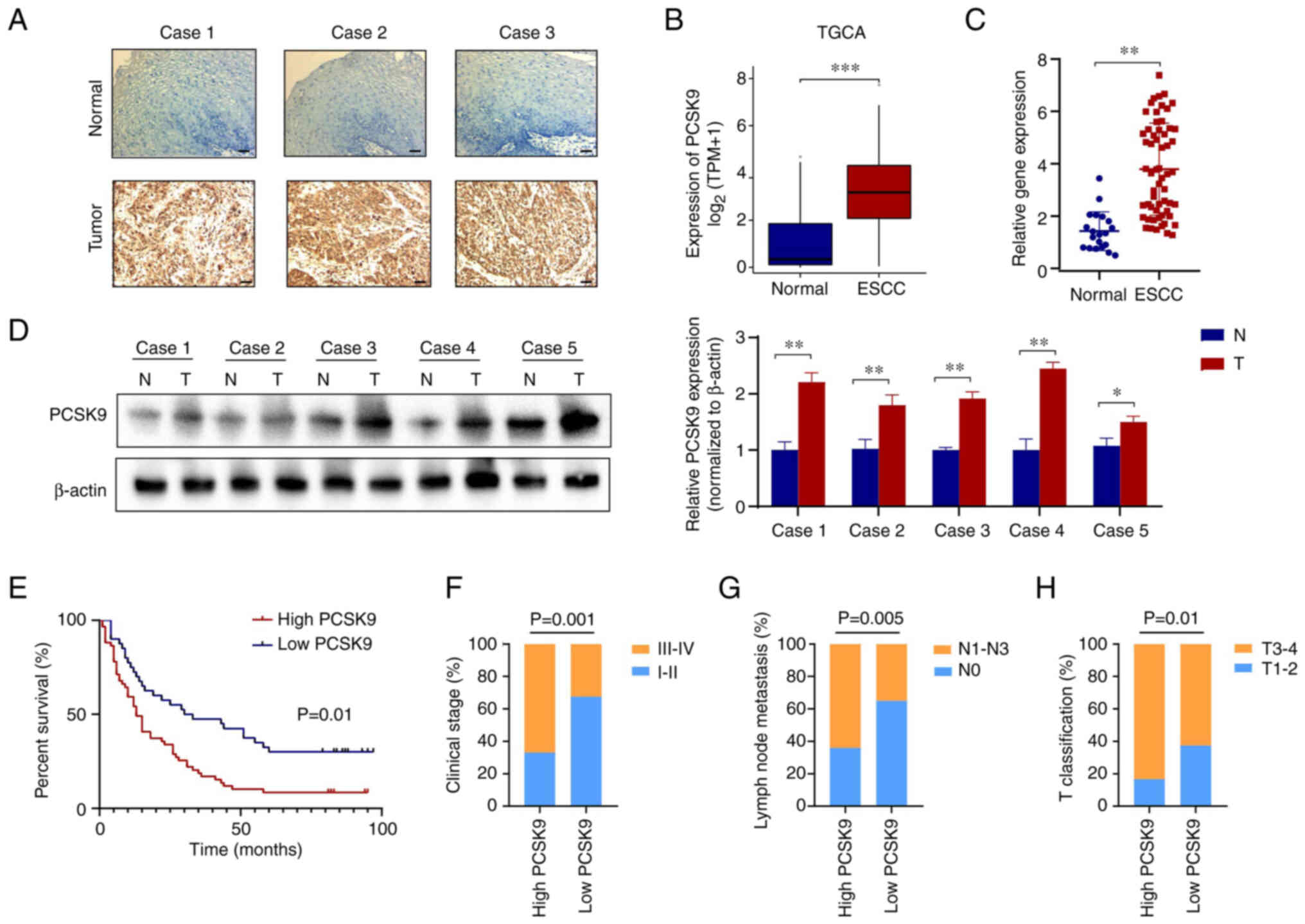 | Figure 1.High PCSK9 expression is associated
with poor prognosis in patients with ESCC. (A) Immunohistochemistry
of normal esophageal epithelial and ESCC tissues from patients with
ESCC. Typical cases with PCSK9 expression are shown. Scale bar, 100
µm. (B) PCSK9 expression levels in an ESCC validation set obtained
from The Cancer Genome Atlas database. (C) Differential expression
of PCSK9 in ESCC tissues and normal esophageal epithelial tissues
from 60 patients. (D) PCSK9 expression levels in freshly obtained
esophageal cancer tissues compared with adjacent paracancerous
tissues in five patients. (E) Based on the expression of PCSK9,
Kaplan-Meier analysis showed that high PCSK9 was associated with a
shorter overall survival in 100 patients with ESCC. (F) In the
PCSK9-high expression group, the clinical stage was higher than
that in the PCSK9-low expression group. (G) PCSK9-high expression
groups demonstrated a higher rate of lymph node metastasis. (H) A
higher T classification was observed in the PCSK9-high expression
group than that in the PCSK9-low expression group. Scale bars, 100
µm. *P<0.05, **P<0.01, ***P<0.001. PCSK9, proprotein
convertase subtilisin/kexin type 9; ESCC, esophageal squamous cell
carcinoma; N, normal; T, tumor. |
A total of 100 patients with ESCC were analyzed to
explore the role of PCSK9 in ESCC. Based on PCSK9 expression
levels, 60 patients were categorized as having high PCSK9
expression and 40 as having low PCSK9 expression. The PCSK9
expression cut-off value was identified as the median IHC score.
Based on the survival analysis, the PCSK9-high expression group had
a shorter overall survival (OS) than the PCSK9-low expression group
(Fig. 1E). In addition, it was
observed that high levels of PCSK9 expression were associated with
clinical stage, lymph node metastasis and T classification in
patients with ESCC, but not with their age or sex (Fig. 1F-H; Table I). Based on univariate analysis, OS
was associated with clinical stage, T classification, lymph node
metastasis and PCSK9 expression. Multivariate analysis, however,
showed that clinical stage, T classification, lymph node metastasis
and PCSK9 expression were associated with OS (Table II). Overall, the expression of
PCSK9 was revealed to be increased in ESCC tissue and was
associated with poor prognosis.
 | Table I.Relationship between the expression
levels of PCSK9 and clinical pathological features in patients with
esophageal squamous cell carcinoma. |
Table I.
Relationship between the expression
levels of PCSK9 and clinical pathological features in patients with
esophageal squamous cell carcinoma.
|
|
| PCSK9
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | n | High | Low | χ2 | P-value |
|---|
| Age, years |
|
|
| 3.038 | 0.053 |
|
≤60 | 34 | 18 | 16 |
|
|
|
>60 | 66 | 42 | 24 |
|
|
| Sex |
|
|
| 0.554 | 0.457 |
|
Male | 74 | 46 | 28 |
|
|
|
Female | 26 | 14 | 12 |
|
|
| T stage |
|
|
| 6.645 | 0.013 |
|
T1-2 | 25 | 10 | 15 |
|
|
|
T3-4 | 75 | 50 | 25 |
|
|
| Lymph node
metastasis |
|
|
| 7.719 | 0.005 |
| No | 51 | 22 | 29 |
|
|
|
Yes | 49 | 38 | 11 |
|
|
| Clinical stage |
|
|
| 10.635 | 0.001 |
| I and
II | 47 | 20 | 27 |
|
|
|
III | 53 | 40 | 13 |
|
|
 | Table II.Univariate and multivariate analyses
of prognostic factors in esophageal squamous cell carcinoma. |
Table II.
Univariate and multivariate analyses
of prognostic factors in esophageal squamous cell carcinoma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | P-value | 95% CI | HR | P-value | 95% CI |
|---|
| Expression of
PCSK9, high vs. low | 2.113 | 0.002 | 1.328–3.364 | 1.755 | 0.030 | 1.057–2.915 |
| Sex, male vs.
female | 1.555 | 0.126 | 0.884–2.732 |
|
|
|
| Age, <60 vs. ≥60
years | 1.208 | 0.443 | 0.745–1.958 |
|
|
|
| T stage, T1-2 vs.
T3-4 | 2.012 | 0.015 | 1.146–3.565 | 1.855 | 0.036 | 1.042–3.301 |
| Lymph node
metastasis, no vs. yes | 1.545 | 0.001 | 1.216–1.962 | 1.332 | 0.032 | 1.025–1.729 |
| Clinical stage, I
and II vs. III | 2.333 | <0.001 | 1.482–3.671 |
|
|
|
PCSK9 promotes ESCC cell
proliferation, invasion and migration in vitro
To explore the biological function of PCSK9 in
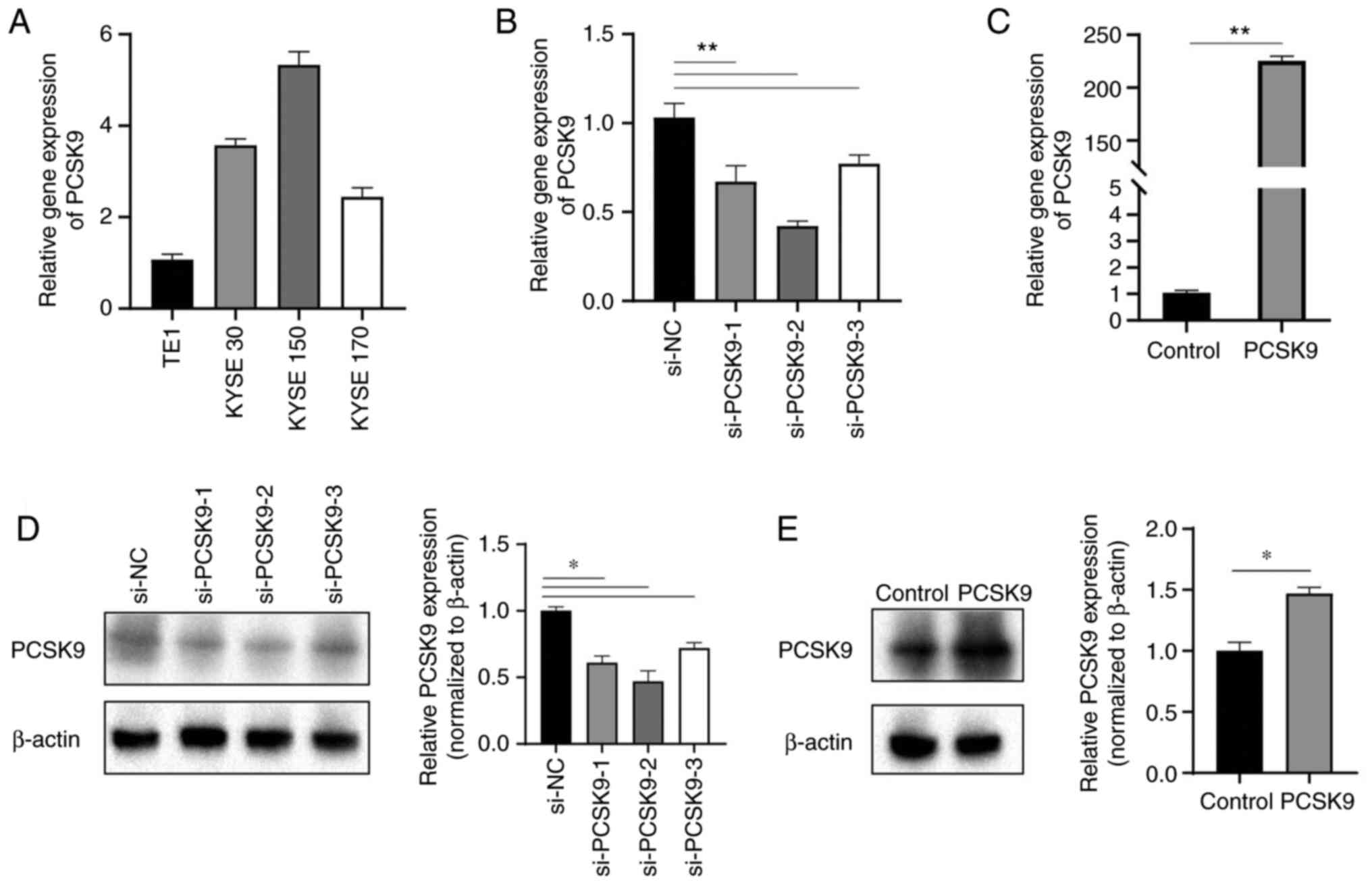
vitro, its expression level was first measured in four ESCC
cell lines using RT-qPCR. The expression levels of PCSK9 were
lowest in TE1 cells and highest in KYSE150 cells (Fig. 2A). Therefore, biological function
experiments were conducted in TE1 and KYSE150 cells. A PCSK9
overexpression vector was used for the overexpression experiment in
TE1 cells, whereas PCSK9 siRNA was used for the knockdown
experiment in KYSE150 cells. RT-qPCR was used to determine the
efficiency of overexpression and knockdown (Fig. 2B and C). si-PCSK9-2 with the
greatest efficiency was then selected for further experiments.
Additionally, western blotting was conducted to analyze alterations
in the protein expression levels of PCSK9 subsequent to its
overexpression and knockdown. The overexpression of PCSK9 resulted
in an upregulation of its protein expression level, while the
knockdown of PCSK9 led to the inhibition of its expression
(Fig. 2D and E).
Following PCSK9 overexpression, the proliferation of
TE1 cells was increased, as determined by CCK-8 assay (Fig. 3A). Similar results were determined
using the colony formation assay (Fig.
3B). In addition, PCSK9 affected the migration and invasion of
ESCC cells. The Transwell assay showed that PCSK9 overexpression
promoted the migration and invasion of TE1 cells (Fig. 3C). The wound healing assay also
confirmed that PCSK9 overexpression could promote the migration of
TE1 cells (Fig. 3D). Conversely,
following PCSK9 knockdown, the proliferation of KYSE150 cells was
inhibited, as determined by CCK-8 and colony formation assays
(Fig. 3E and F). Furthermore, the
migration and invasion of the cells were inhibited (Fig. 3G and H). These findings suggested
that PCSK9 could facilitate the proliferation, migration and
invasion of ESCC cells in vitro.
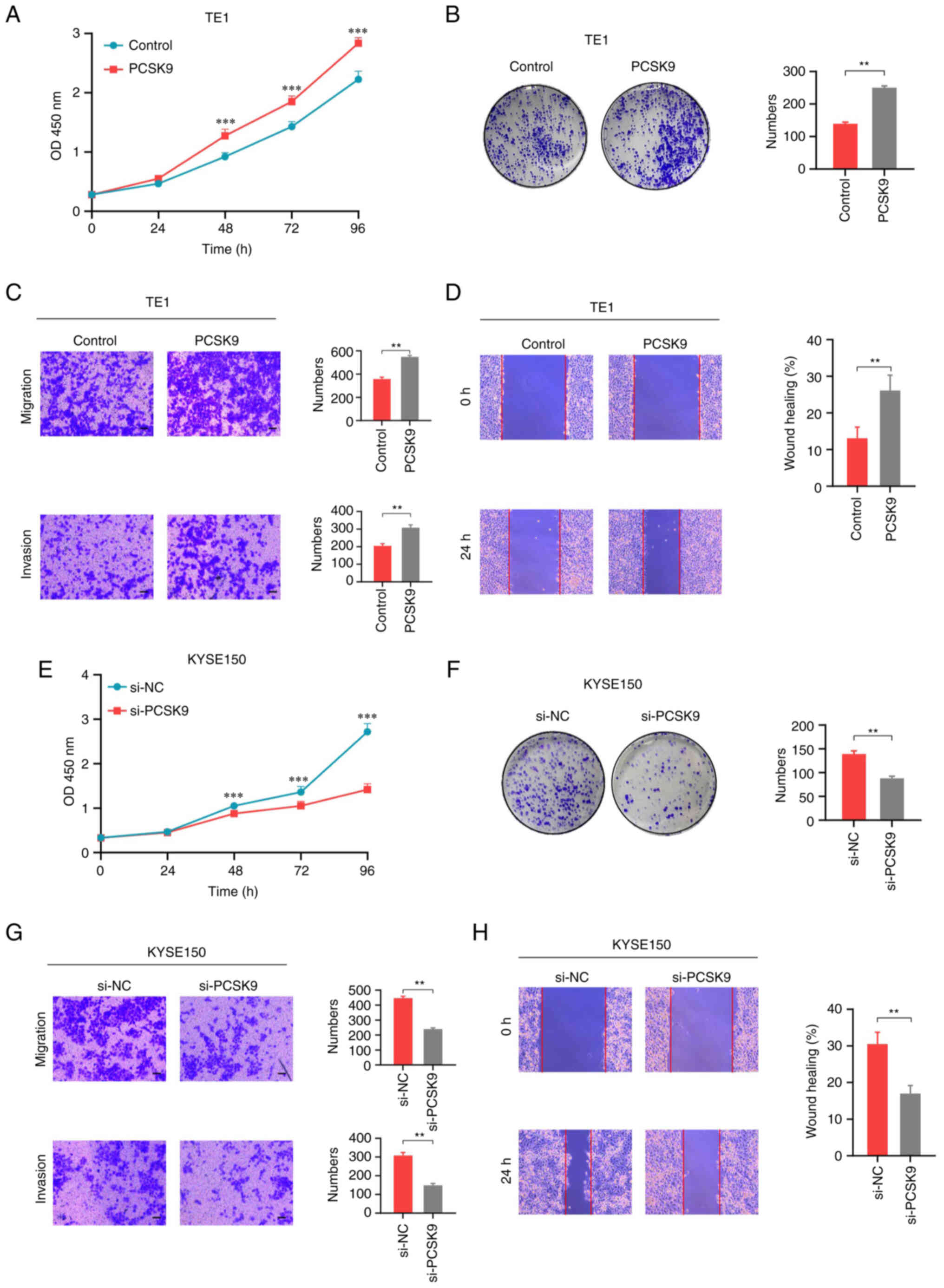 | Figure 3.PCSK9 promotes cell proliferation,
migration and invasion in vitro. (A) Proliferation of TE1
cells overexpressing PCSK9 and those transfected with a vector
control, as determined using CCK-8 assay. PCSK9 promoted ESCC cell
proliferation. ***P<0.001 vs. Control. (B) Colony formation
assay of TE1 cells; PCSK9 overexpression promoted colony formation.
Migration and invasion of TE1 cells with PCSK9 overexpression were
detected using (C) Transwell migration and invasion assays, and (D)
wound healing assays. PCSK9 promoted the invasion and migration of
ESCC cells. (E) Proliferation of KYSE150 cells with PCSK9 knockdown
and those transfected with si-NC, as determined using CCK-8 assay.
(F) Colony formation assay of KYSE150 cells. (G) Transwell
migration and invasion, and (H) wound healing assays detected
migration and invasion of KYSE150 cells. Scale bars, 100 µm.
**P<0.01. ***P<0.001. PCSK9, proprotein convertase
subtilisin/kexin type 9; CCK-8, Cell Counting Kit-8; ESCC,
esophageal squamous cell carcinoma; si, small interfering; NC,
negative control. |
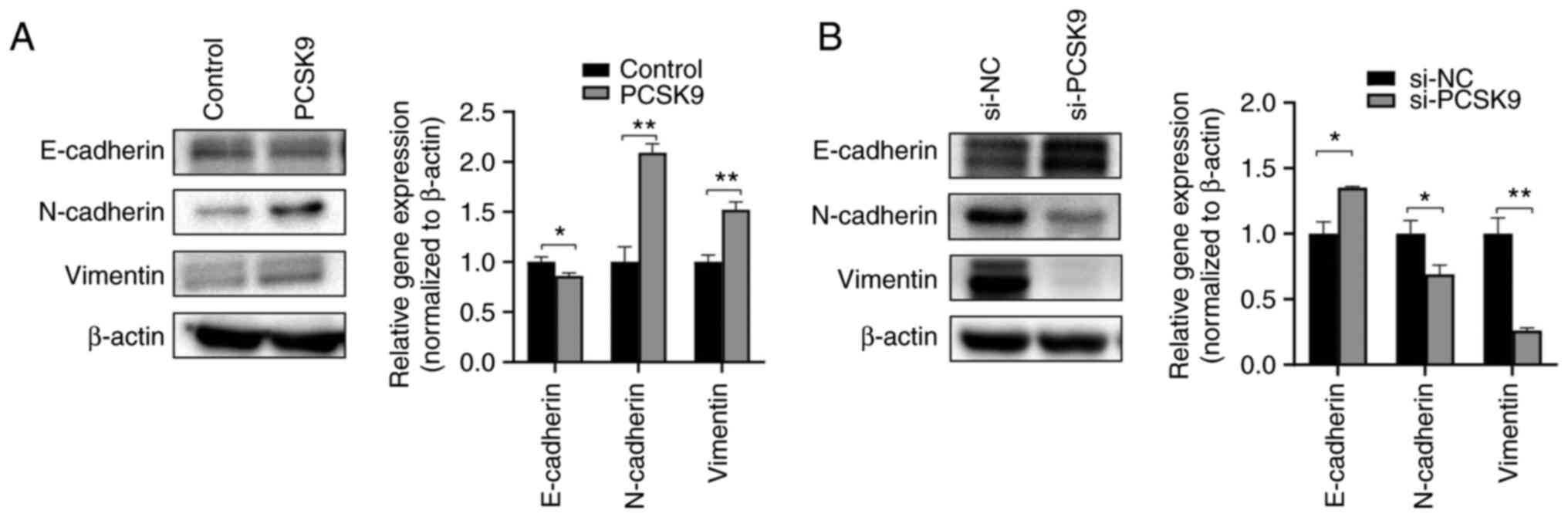
PCSK9 modulates the EMT of ESCC
cells
EMT is the process of epithelial cells acquiring
mesenchymal characteristics. Tumor initiation, invasion, metastasis
and resistance to cancer treatment are all affected by the EMT
process (18). The key proteins
involved in EMT were analyzed to better understand the effects of
PCSK9 on ESCC. Following PCSK9 overexpression, the protein
expression levels of E-cadherin were suppressed; however, the
expression levels of N-cadherin and vimentin were increased
(Fig. 4A). PCSK9 knockdown
increased the protein expression levels of E-cadherin, while
inhibiting those of vimentin and N-cadherin (Fig. 4B). These data indicated that PCSK9
was involved in EMT progression, and may regulate ESCC migration
and invasion through EMT.
PCSK9 promotes ESCC progression by
upregulating CCL25
To further investigate how PCSK9 promoted the
progression of ESCC, ESCC data were extracted from the selected
TCGA public database, and divided into high and low expression
groups by PCSK9 expression The data were organized based on the
levels of expression exhibited by the respective molecules, with
the lower 50% being classified as the low expression group, and the
upper 50–100% as the high expression group (high expression group,
n=82; low expression group, n=81). Based on standard procedures,
raw count matrices from the public data were compared using DESeq2.
Among the differentially expressed genes, CCL25 was highly
associated with PCSK9 (Fig. 5A).
Therefore, it was hypothesized that PCSK9 could promote ESCC
progression by affecting the secretion of CCL25. To validate this
hypothesis, ELISA experiments were performed to determine the
effect of PCSK9 on CCL25 secretion. The results suggested that
PCSK9 overexpression promoted CCL25 secretion, whereas PCSK9
knockdown had the opposite effect (Fig.
5B). Subsequently, rescue experiments were performed. As
determined by the CCK-8 assay, PCSK9 knockdown inhibited cell
proliferation, whereas the exogenous addition of CCL25 suppressed
this effect (Fig. 5C). The colony
formation assay also confirmed that the addition of CCL25 inhibited
the diminished cell proliferation induced by PCSK9 knockdown
(Fig. 5D). Furthermore, the
migration and invasion of cells were inhibited following PCSK9
knockdown, whereas CCL25 reversed this effect (Fig. 5E and F).
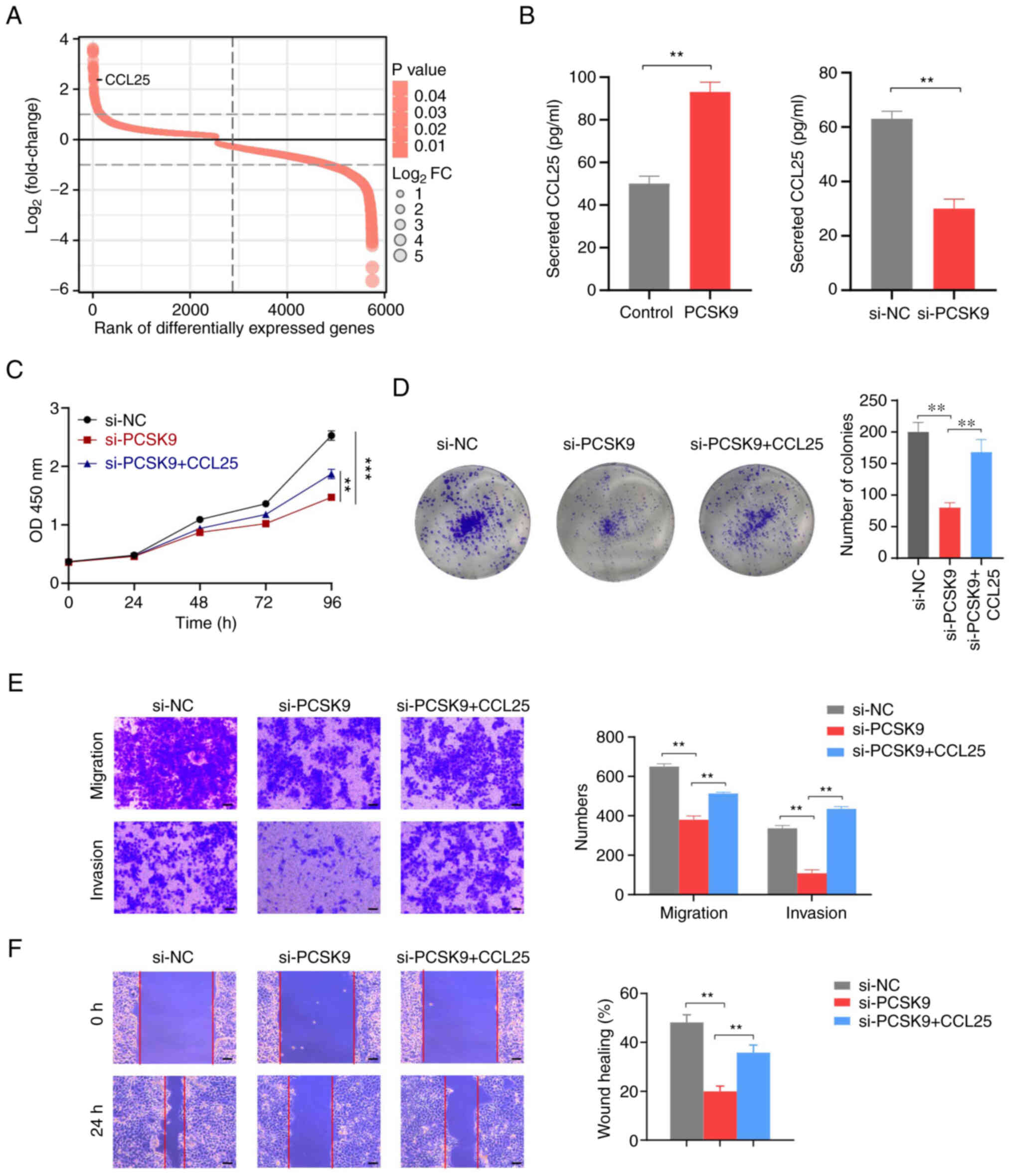 | Figure 5.PCSK9 promotes ESCC progression
through CCL25 upregulation. (A) PCSK9 data were extracted from The
Cancer Genome Atlas public database and divided into high and low
expression groups, according to the expression of the PCSK9. The
original count matrix of the selected public data was analyzed for
differences according to the standard process using the DESeq2
package. CCL25 expression was also low in the PCSK9-low expression
group. (B) As determined by ELISA, PCSK9 overexpression increased
CCL25 secretion, whereas PCSK9 knockdown inhibited CCL25 secretion.
(C) Cell Counting Kit-8 assay showed that the exogenous addition of
CCL25 inhibited PCSK9 knockdown-induced inhibition of cell
proliferation. (D) Colony formation assay showed that the exogenous
addition of CCL25 rescued PCSK9 knockdown-induced colony formation
inhibition. (E) Transwell migration and invasion, and (F) wound
healing assays were performed to examine whether the exogenous
addition of CCL25 could reverse PCSK9 knockdown-induced inhibition
of the migration and invasion of ESCC cells. Scale bars, 100 µm.
**P<0.01, ***P<0.001. PCSK9, proprotein convertase
subtilisin/kexin type 9; ESCC, esophageal squamous cell carcinoma;
CCL25, chemokine (C-C motif) ligand 25; si, small interfering; NC,
negative control. |
Discussion
ESCC is a common malignant gastrointestinal tumor
that seriously endangers human health. Although surgery, endoscopy
and radiotherapy techniques have improved and complications have
decreased, the overall treatment outcome of ESCC remains
unsatisfactory (2). Therefore, it
is essential to identify more effective tumor markers and discover
new therapeutic targets for ESCC to improve the long-term survival
rate of patients.
Research has shown that lipid metabolism is critical
to cancer progression (27–29). PCSK9 is a subtilisin-like proprotein
convertase family member that serves a critical role in maintaining
plasma cholesterol levels and fatty acid metabolism. Aside from
regulating lipid transport, PCSK9 also affects cell functions,
including viral infection and insulin resistance, and tumor
progression and tumor apoptosis (30,31).
Furthermore, PCSK9 has been implicated in cancer biology in several
studies (9). In hepatocellular
carcinoma, PCSK9 promotes tumor growth by inhibiting tumor cell
apoptosis through the FASN/Bax/Bcl-2/caspase 9/caspase 3 pathway
(10). PCSK9 also promotes colon
cancer cell progression and metastasis by regulating EMT and
PI3K/AKT signaling (11). In
gastric cancer, PCSK9 promotes invasion and suppresses apoptosis by
promoting the MAPK signaling pathway through the upregulation of
heat shock protein 70 (12).
Notably, it has been reported that serum PCSK9 antigen levels are
not associated with esophageal cancer (32); this may be due to the fact that
PCSK9 is mainly produced and metabolized by the liver. In the
present study, ESCC tissues exhibited higher expression levels of
PCSK9 than non-ESCC tissues, and PCSK9 expression was associated
with T classification, clinical stage and lymph node metastasis. It
was also identified as an independent risk factor for the OS of
patients with ESCC. In vitro experiments revealed that PCSK9
stimulated the proliferation, invasion and migration of ESCC cells.
The present findings on the role of PCSK9 in ESCC are consistent
with findings in other types of cancer, suggesting that PCSK9 is an
oncogenic gene that has a vital role in cancer development.
EMT refers to the transformation of epithelial cells
into mesenchymal cells (33). The
epithelial cells lose polarity and intercellular adhesion junctions
during this process and become mesenchymal cells, with increased
migratory and invasive abilities (18). Studies have shown that EMT serves a
crucial role in the development of malignancies, cell invasion and
tumor metastasis. During EMT, the cell phenotype is transformed
from epithelial to mesenchymal, and cellular markers change. The
expression of epithelial markers, such as E-cadherin and
cytokeratin, is decreased; the expression of mesenchymal markers,
such as N-cadherin, vimentin and smooth muscle actin proteins, are
increased (14,16). E-cadherin is a vital adhesion
molecule for maintaining the epithelial phenotype, and has a
decisive role in intercellular adhesion and cell polarity (15,17,34).
The tumor microenvironment also plays an important role in EMT. In
the tumor microenvironment, mesenchymal stem cells release growth
factors that promote EMT, such as IL-6 and TNF-α. In non-small cell
lung cancer, IL-6 and TNF-α levels have been shown to be positively
correlated with the protein levels of vimentin and N-cadherin, and
negatively correlated with the protein levels of E-cadherin in
tumor tissues. These results suggested that IL-6 and TNF-α may have
an essential role in tumor EMT development (35). Decreased or absent E-cadherin
expression in tumor cells is considered the gold standard for the
loss of epithelial features in cancer cells in EMT and a critical
manifestation of tumor metastasis (36). In the present study, the
overexpression of PCSK9 inhibited the protein expression levels of
E-cadherin, and promoted those of N-cadherin and vimentin. During
PCSK9 knockdown, the opposite was observed. By knocking down PCSK9
in colorectal cancer, colon cancer cells have been shown to display
reduced EMT and PI3K/AKT activation (11). Thus, PCSK9 may promote tumor cell
metastasis and invasion by facilitating EMT.
Chemokines and their receptors are closely
associated with tumor cell genesis, development and metastasis, and
appear highly expressed in several tumors. Thymus-expressed
chemokine 25, also known as CCL25, is the sole ligand of C-C
chemokine receptor type 9. Various studies have shown that CCL25 is
involved in tumor metastasis and tumorigenesis, and is
overexpressed in malignant tumors (21,22,37).
VEGF and MMPs play essential roles in tumor neovascularization and
metastasis. Notably, tumor size and the number of metastasized
lymph nodes are positively correlated with the serum levels of
MMP-2 and MMP-9 in breast cancer (38), whereas in non-small cell lung
cancer, CCL25 can promote migration and invasion by regulating VEGF
and MMPs (21). Breast cancer cells
may also express MMPs through CCL25 (20).
In addition, CCL25 plays a vital role in leukemia
invasion and osteosarcoma metastasis (39,40).
The present study showed that PCSK9 could promote the development
of ESCC by promoting the secretion of CCL25. The rescue experiment
showed that PCSK9 knockdown inhibited cell migration and invasion,
whereas these effects could be reversed through the addition of
CCL25.
The present results demonstrated that PCSK9 may act
as a tumor promoter in ESCC. PCSK9 could facilitate the
proliferation, migration and invasion of ESCC in vitro.
Mechanistically, PCSK9 could promote EMT by secreting CCL25. Hence,
the present study could provide novel theoretical perspectives
regarding ESCC diagnosis and treatment.
In conclusion, a comprehensive in vitro cell
experiment and tissue sample analysis was conducted in the present
study to assess the involvement of PCSK9 in ESCC progression and
metastasis. In ESCC tissues, PCSK9 protein was highly expressed and
associated with poor prognosis. In addition, it was suggested that
PCSK9 may promote tumor progression through EMT in ESCC,
potentially by secreting CCL25. Overall, the present data
demonstrated the oncogenic activity of PCSK9 in ESCC. Future
studies should investigate PCSK9 inhibition as a potential
treatment for ESCC.
Acknowledgements
Not applicable.
Funding
This work was supported by the Specific Project of the National
Cancer Center of China on the Scientific Research of Oncology
(grant no. NCC2017A24).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZT and HW were involved in study conception and
design. ZT provided administrative support. HW provided study
materials or patients. HW and QG performed the experiments. QG and
MW collected and assembled data. HW and CL analyzed and interpreted
data. ZT and HW confirm the authenticity of all the raw data. All
authors wrote the manuscript, and read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of The Fourth Hospital of Hebei Medical University.
Written informed consent was obtained from each patient included
and this study was performed in accordance with The Declaration of
Helsinki.
Patient consent for publication
The patients provided written informed consent for
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Klingelhöfer D, Zhu Y, Braun M, Brüggmann
D, Schöffel N and Groneberg DA: A world map of esophagus cancer
research: A critical accounting. J Transl Med. 17:1502019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kelly RJ: Emerging multimodality
approaches to treat localized esophageal cancer. J Natl Compr Canc
Netw. 17:1009–1014. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gronnier C and Collet D: New trends in
esophageal cancer management. Cancers (Basel). 13:30302021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martin-Perez M, Urdiroz-Urricelqui U,
Bigas C and Benitah SA: The role of lipids in cancer progression
and metastasis. Cell Metab. 34:1675–1699. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lambert G, Sjouke B, Choque B, Kastelein
JJP and Hovingh GK: The PCSK9 decade. J Lipid Res. 53:2515–2524.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mahboobnia K, Pirro M, Marini E, Grignani
F, Bezsonov EE, Jamialahmadi T and Sahebkar A: PCSK9 and cancer:
Rethinking the link. Biomed Pharmacother. 140:1117582021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bonaventura A, Vecchié A, Ruscica M,
Grossi F and Dentali F: PCSK9 as a new player in cancer: New
opportunity or red herring? Curr Med Chem. 29:960–969. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang SZ, Zhu XD, Feng LH, Li XL, Liu XF,
Sun HC and Tang ZY: PCSK9 promotes tumor growth by inhibiting tumor
cell apoptosis in hepatocellular carcinoma. Exp Hematol Oncol.
10:252021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang L, Li S, Luo H, Lu Q and Yu S: PCSK9
promotes the progression and metastasis of colon cancer cells
through regulation of EMT and PI3K/AKT signaling in tumor cells and
phenotypic polarization of macrophages. J Exp Clin Cancer Res.
41:3032022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu B, Li S, Fang Y, Zou Y, Song D, Zhang S
and Cai Y: Proprotein convertase subtilisin/kexin type 9 promotes
gastric cancer metastasis and suppresses apoptosis by facilitating
mapk signaling pathway through HSP70 up-regulation. Front Oncol.
10:6096632020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jurisic V: Multiomic analysis of cytokines
in immuno-oncology. Expert Rev Proteomics. 17:663–674. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moody SE, Perez D, Pan TC, Sarkisian CJ,
Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD and
Chodosh LA: The transcriptional repressor Snail promotes mammary
tumor recurrence. Cancer Cell. 8:197–209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gravdal K, Halvorsen OJ, Haukaas SA and
Akslen LA: A switch from E-cadherin to N-cadherin expression
indicates epithelial to mesenchymal transition and is of strong and
independent importance for the progress of prostate cancer. Clin
Cancer Res. 13:7003–7011. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baumgart E, Cohen MS, Silva Neto B, Jacobs
MA, Wotkowicz C, Rieger-Christ KM, Biolo A, Zeheb R, Loda M,
Libertino JA and Summerhayes IC: Identification and prognostic
significance of an epithelial-mesenchymal transition expression
profile in human bladder tumors. Clin Cancer Res. 13:1685–1694.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ye X, Tam WL, Shibue T, Kaygusuz Y,
Reinhardt F, Ng Eaton E and Weinberg RA: Distinct EMT programs
control normal mammary stem cells and tumour-initiating cells.
Nature. 525:256–260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rhim AD, Mirek ET, Aiello NM, Maitra A,
Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK,
Vonderheide RH, et al: EMT and dissemination precede pancreatic
tumor formation. Cell. 148:349–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Propper DJ and Balkwill FR: Harnessing
cytokines and chemokines for cancer therapy. Nat Rev Clin Oncol.
19:237–253. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Johnson-Holiday C, Singh R, Johnson E,
Singh S, Stockard CR, Grizzle WE and Lillard JW Jr: CCL25 mediates
migration, invasion and matrix metalloproteinase expression by
breast cancer cells in a CCR9-dependent fashion. Int J Oncol.
38:1279–1285. 2011.PubMed/NCBI
|
|
21
|
Niu Y, Tang D, Fan L, Gao W and Lin H:
CCL25 promotes the migration and invasion of non-small cell lung
cancer cells by regulating VEGF and MMPs in a CCR9-dependent
manner. Exp Ther Med. 19:3571–3580. 2020.PubMed/NCBI
|
|
22
|
Zhang Z, Sun T, Chen Y, Gong S, Sun X, Zou
F and Peng R: CCL25/CCR9 signal promotes migration and invasion in
hepatocellular and breast cancer cell lines. DNA Cell Biol.
35:348–357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng Y, Sang M, Liu F, Gu L, Li J, Wu Y
and Shan B: Aprepitant inhibits the progression of esophageal
squamous cancer by blocking the truncated neurokinin-1 receptor.
Oncol Rep. 50:1312023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Scherbakov AM, Vorontsova SK, Khamidullina
AI, Mrdjanovic J, Andreeva OE, Bogdanov FB, Salnikova DI, Jurisic
V, Zavarzin IV and Shirinian VZ: Novel pentacyclic derivatives and
benzylidenes of the progesterone series cause anti-estrogenic and
antiproliferative effects and induce apoptosis in breast cancer
cells. Invest New Drugs. 41:142–152. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Prasad K and Prabhu GK: Image analysis
tools for evaluation of microscopic views of immunohistochemically
stained specimen in medical research-a review. J Med Syst.
36:2621–2631. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng C, Geng F, Cheng X and Guo D: Lipid
metabolism reprogramming and its potential targets in cancer.
Cancer Commun (Lond). 38:272018.PubMed/NCBI
|
|
28
|
Diao XY and Lin T: Progress in therapeutic
strategies based on cancer lipid metabolism. Thorac Cancer.
10:1741–1743. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nickels JT Jr: New links between lipid
accumulation and cancer progression. J Biol Chem. 293:6635–6636.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fasolato S, Pigozzo S, Pontisso P, Angeli
P, Ruscica M, Savarino E, De Martin S, Lupo MG and Ferri N: PCSK9
levels are raised in chronic HCV patients with hepatocellular
carcinoma. J Clin Med. 9:31342020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mbikay M, Sirois F, Gyamera-Acheampong C,
Wang GS, Rippstein P, Chen A, Mayne J, Scott FW and Chrétien M:
Variable effects of gender and Western diet on lipid and glucose
homeostasis in aged PCSK9-deficient C57BL/6 mice CSK9PC57BL/6. J
Diabetes. 7:74–84. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ito M, Hiwasa T, Oshima Y, Yajima S,
Suzuki T, Nanami T, Sumazaki M, Shiratori F, Funahashi K, Li SY, et
al: Association of serum anti-PCSK9 antibody levels with favorable
postoperative prognosis in esophageal cancer. Front Oncol.
11:7080392021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu W and Kang Y: Epithelial-mesenchymal
plasticity in cancer progression and metastasis. Dev Cell.
49:361–374. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Y, Donaher JL, Das S, Li X,
Reinhardt F, Krall JA, Lambert AW, Thiru P, Keys HR, Khan M, et al:
Genome-wide CRISPR screen identifies PRC2 and KMT2D-COMPASS as
regulators of distinct EMT trajectories that contribute
differentially to metastasis. Nat Cell Biol. 24:554–564. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jurisic V, Srdic-Rajic T, Konjevic G,
Bogdanovic G and Colic M: TNF-α induced apoptosis is accompanied
with rapid CD30 and slower CD45 shedding from K-562 cells. J Membr
Biol. 239:115–122. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu X and Tian X: Long noncoding RNA
TCONS_00068220 promotes breast cancer progression by regulating
epithelial-mesenchymal transition marker E-cadherin. Med Sci Monit.
27:e9298322021.PubMed/NCBI
|
|
37
|
Sharma PK, Singh R, Novakovic KR, Eaton
JW, Grizzle WE and Singh S: CCR9 mediates PI3K/AKT-dependent
antiapoptotic signals in prostate cancer cells and inhibition of
CCR9-CCL25 interaction enhances the cytotoxic effects of etoposide.
Int J Cancer. 127:2020–2030. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stankovic S, Konjevic G, Gopcevic K, Jovic
V, Inic M and Jurisic V: Activity of MMP-2 and MMP-9 in sera of
breast cancer patients. Pathol Res Pract. 206:241–247. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li J, Muhammad J, Xie T, Sun J, Lei Y, Wei
Z, Pan S, Qin H, Shao L, Jiang D and Zhang Q: LINC00853 restrains T
cell acute lymphoblastic leukemia invasion and infiltration by
regulating CCR9/CCL25. Mol Immunol. 140:267–275. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li J, Zhao C, Wang D, Wang S, Dong H, Wang
D, Yang Y, Li J, Cui F, He X and Qin J: ZIM3 activation of CCL25
expression in pulmonary metastatic nodules of osteosarcoma recruits
M2 macrophages to promote metastatic growth. Cancer Immunol
Immunother. 72:903–916. 2023. View Article : Google Scholar : PubMed/NCBI
|