The cytotoxicity of gefitinib on patient‑derived induced pluripotent stem cells reflects gefitinib‑induced liver injury in the clinical setting
- Authors:
- Published online on: October 18, 2023 https://doi.org/10.3892/ol.2023.14108
- Article Number: 520
-
Copyright: © Fujisaka et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Lung cancer is the most frequently diagnosed cancer with an estimated 2.20 million new cases and is the leading cause of cancer-related deaths with 1.79 million deaths worldwide (1). Gefitinib is an oral epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI), recommended as the first-choice treatment for patients with advanced stage disease (2), and was associated with a significantly higher frequency of grade 3 or greater hepatotoxicity than the other EGFR-TKIs, erlotinib or afatinib, although severe rash and diarrhea were less frequent than afatinib (3). Drug-induced hepatotoxicity is one of the major causes of approved drugs being removed from the market (4).
Human induced pluripotent stem cells (iPSCs) have opened new doors in biology and regenerative medicine (5). One advantage of iPSCs is that they can be generated from human tissues of subjects with genetic variations or various traits, such as drug sensitivity (6,7). Remodeling the pathogenesis, iPSCs have been generated for various diseases, such as severe combined immunodeficiency (ADA-SCID) (8), type 1 diabetes (9), and Parkinson's disease (10). iPSCs could be also an ideal platform for drug discovery or evaluation (11).
Various protocols for hepatic-differentiation from iPSCs have been developed (12–15). Despite differences among iPSC-derived hepatocytes (iPSC-heps), hepatocyte cell lines, and primary hepatocytes (16,17), iPSC-heps have been shown to have the potential to predict drug toxicity and improve drug screening (7,18,19).
In this study, we generated iPSCs by reprogramming peripheral blood mononuclear cells (PBMC) obtained from 2 groups of gefitinib-treated patients who either had grade 3 or greater hepatotoxicity (Toxicity group, T) or grade 1 or less hepatotoxicity (No clinical toxicity group, N) in the Common Terminology Criteria for Adverse Events (CTCAE v5.0). iPSCs were differentiated into hepatocytes (iPSC-heps). We examined gefitinib-induced cytotoxicity in both iPSCs and iPSC-heps from both the T and N groups and evaluated the correlation between in vitro cytotoxicity and clinical hepatocytotoxicity to compare the cells from both groups.
Materials and methods
Antibodies
Anti-EGFR antibodies (sheep polyclonal, Upstate®, Merck, Darmstadt, DE), phospho-EGFR (Y1068) [mouse monoclonal (m), Abcam, Cambridge, UK], GAPDH (m, MBL, Nagoya, Japan), human albumin (goat polyclonal, Bethyl Laboratories, Montgomery, TX), α-fetoprotein [rabbit monoclonal (r, mAb), Abcam, Cambridge, UK], and hepatocyte nuclear factor 4 alpha (HNF4α) (r, mAb, Abcam, Cambridge, UK) were used for our experiments.
Generation of iPSCs
Peripheral blood mononuclear cells (PBMCs) culture and ReprogrammingiPSCs were established according to the protocol described by Okita et al (20) with minor modifications. Peripheral blood was obtained from the patients according to the Osaka Medical and Pharmaceutical University Review Board's guidelines. PBMCs were isolated using a BD Vacutainer®CPT™ mononuclear cell preparation tube with sodium citrate (BD Biosciences, Franklin Lakes, NJ, USA), according to the manufacturer's instructions. Half of the collected cells were suspended in a STEM-CELL Banker® (ZENOEN PHARMA, Fukushima, Japan), aliquoted into cryovials at >2.5×106 cells/500 µl/vial, frozen, and stored at −80°C. The rest of the cells were plated in 6-well plates in PBMC culture medium: Stem Span-ACF (STEMCELL Technologies, Vancouver, BC, Canada) with 10 ng/ml IL-3, 100 ng/ml IL-6, 300 ng/ml SCF, 300 ng/ml TPO, and 300 ng/ml Flt3 ligand, and cultured for 1 week with more medium added as appropriate. Unless otherwise stated, cytokines were purchased from FUJIFILM Wako Pure Chemical Corp. (Osaka, Japan).
The plasmids pCXLE-hOCT3/4-shp53-F, pCXLE-hSK, pCXLE-hUL, and pCXLE-EBANA required for reprogramming were electroporated into 3–5×106 PBMCs using a Nucleofector 2b Device (Lonza, Basel, Switzerland) with an Amaxa human CD34+ cell Nucleofector kit (Lonza, Basel, Switzerland) according to the manufacturer's instructions. The transfected cells were transferred into iMatrix 511 silk (TaKaRa, Shiga, Japan)-coated 6-well plates in PBMC culture medium using a sequential dilution. The PBMC medium was replaced with Stem Fit AK03N w/o C solution (Ajinomoto, Tokyo, Japan) by adding 1.5 ml of Stem Fit every other day, three times. The medium was replaced with Stem Fit w/o C solution on the eighth day of culture. The cells were cultured continuously, and iPSC colonies were picked when they were approximately 1 mm in diameter. The iPSCs reprogramed from patients (iPSCs) were used for further characterization.
iPSC culture
iPSCs were cultured in a feeder-free system. Briefly, 6-well plates were pre-incubated in 1.5 ml/wells of Stem Fit AK03N (Ajinomoto, Tokyo, Japan) containing 0.25% iMatrix 511 silk (TaKaRa, Shiga, Japan) and 10 µM Y-27632, a rho-associated coiled-coil kinase (ROCK) inhibitor (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan), in a CO2 incubator. iPSCs (1–1.4×104 cells), suspended in 500 µl of Stem Fit AK03N (Ajinomoto, Tokyo, Japan) and then plated onto a pre-incubated 6-well plates. The following day, the medium was replaced with Stem Fit AK03N (Ajinomoto, Tokyo, Japan) alone and changed every other day.
iPSC passage
At confluency, iPSCs were washed twice with PBS (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan) and incubated with Accutase™ (Innovative Cell Technologies, Inc., San Diego, CA) for approximately 5 min at 37°C, after which the supernatant was removed. The cells were washed with PBS, suspended in Stem Fit AK03N (Ajinomoto, Tokyo, Japan), and counted using an automated cell counter (TC20, Bio-Rad, Hercules, CA, USA). The cells were cultured as described above.
Hepatocyte differentiation of iPSC
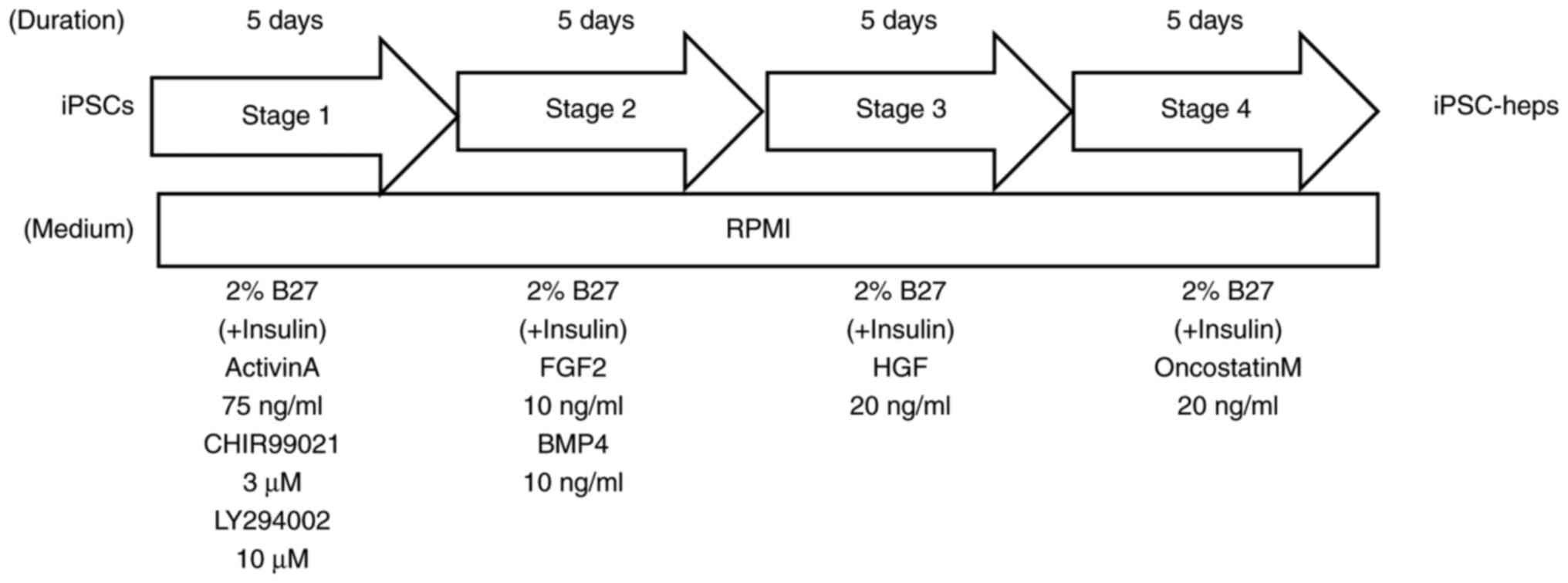
iPSCs were plated at 60–80% of confluence with 1 ml/well of Stem Fit AK03N (Ajinomoto, Tokyo, Japan) containing 0.25% iMatrix 511 silk (TaKaRa, Shiga, Japan) and 10 µM Y-27632 in a 12-well plate. Hepatic differentiation was performed using 4-step protocol. (STAGE 1) The next day, the medium was changed to RPMI1640 (Thermo Fisher Scientific, Waltham, MA) with 2% B27 (Thermo Fisher Scientific, Amarillo, TX) with insulin, 75 ng/ml Activin A (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan), 3 µM CHIR99021 (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan), GSK3β inhibitor, and 10 µM LY294002 (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan), PI3K inhibitor. The cells were then incubated for 5 days and the medium changed daily. (STAGE 2) The medium was replaced with RPMI1640 (Thermo Fisher Scientific, Waltham, MA) containing 2% B27 (Thermo Fisher Scientific, Amarillo, TX) with insulin, 10 ng/ml BMP4 (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan), and 10 ng/ml FGF2 (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan). The cells were then incubated for 5 days and the medium changed daily. (STAGE 3) At day 11, the cells were incubated with RPMI1640 (Thermo Fisher Scientific, Waltham, MA) containing 2% B27 (Thermo Fisher Scientific, Amarillo, TX) with insulin, and 20 ng/ml HGF (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan) for 5 days with daily medium change. (STAGE 4). On day 16, the medium was again replaced with RPMI1640 (Thermo Fisher Scientific, Waltham, MA) containing 2% B27 (Thermo Fisher Scientific, Amarillo, TX) with insulin, 20 ng/ml Oncostatin M (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan) for a further 5 days with daily medium change, after which the iPSC-derived hepatocytes (iPSC-heps) were characterized.
HaCaT cells culture
HaCaT cells, an immortalized human keratinocyte cell line, obtained from COSMO BIO CO., Ltd. (300493-ACADEMIC) were cultured in MCDB 153 medium containing 5% FBS and 10 µg epidermal growth factor (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan). To collect the cell lysates, the cells were grown until they reached 80–100% confluence.
Western blot analysis
The samples were prepared with RIPA buffer and quantified using a Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). The quantified samples were electrophoresed and electroblotted using a semi-dry system (Trans-Blot SD Semi-Dry Transfer cell, Bio-Rad, Hercules, CA, USA) onto a PVDF membrane (Merck Millipore, Burlington, MA). The membrane was then incubated with the appropriate primary antibodies followed by HRP-conjugated anti-mouse or -rabbit secondary antibodies (SouthernBiotech, Birmingham, AL) and developed using Luminata™ Crescendo (Merck Millipore, Burlington, MA). The images were captured using a FUSION SYSTEM FX7 (VILBER LOURMAT, Marne-la-Vallée Cedex 3, France).
qPCR analysis
Total RNA was prepared using the RNAzol®RT Reagent (Molecular Research Center, Inc., Cincinnati, OH, USA) and then transcribed into cDNA with a PrimeScript™ RT reagent kit (Perfect Real Time) (TaKaRa, Shiga, Japan), 1 µg of total RNA. TaqMan® probes for AFP (Hs01040598_m1), ALB (Hs00609411_m1), NANOG (Hs02387400_g1), and POU5F1 (Hs04260367_gH) were obtained from Thermo Fisher Scientific (Waltham, MA), and qPCR was performed using the StepOnePlus real-time PCR system (Thermo Fisher Scientific, Waltham, MA). The data were analyzed using the ΔΔCq method and expressed as relative quantities (RQ) according to the manufacturer's instructions.
Lactate dehydrogenase (LDH) release assay
iPSCs were plated in Stem Fit AK03N (Ajinomoto, Tokyo, Japan) containing 0.25% iMatrix 511 silk (TaKaRa, Shiga, Japan) and 10 µM Y-27632 (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan) in a 12-well plate. as described above. The following day, the medium was replaced with Stem Fit AK03N (Ajinomoto, Tokyo, Japan) containing gefitinib (0, 3, 6, and 10 µM). The medium was collected on days 0, 1, and 2. The medium was not replaced during gefitinib treatment. For iPSC-heps, gefitinib (0, 3, 6, and 10 µM) was added on day 2 of STAGE 4, and the conditioned medium and iPSCs were collected. LDH was measured using the LDH-Glo™ Cytotoxicity Assay combined with the GloMax Multi/Luminescence System (Promega, Madison, WI, USA) according to the manufacturer's instructions.
Statistical analysis
All analyses were performed using ‘MEPHAS’ (http://www.gen-info.osaka-u.ac.jp/testdocs/tomocom/). One-way ANOVA with Dunnett's test or Tukey's test, and Student's-t test were employed for statistical analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
Generation of iPSCs and iPSC-heps
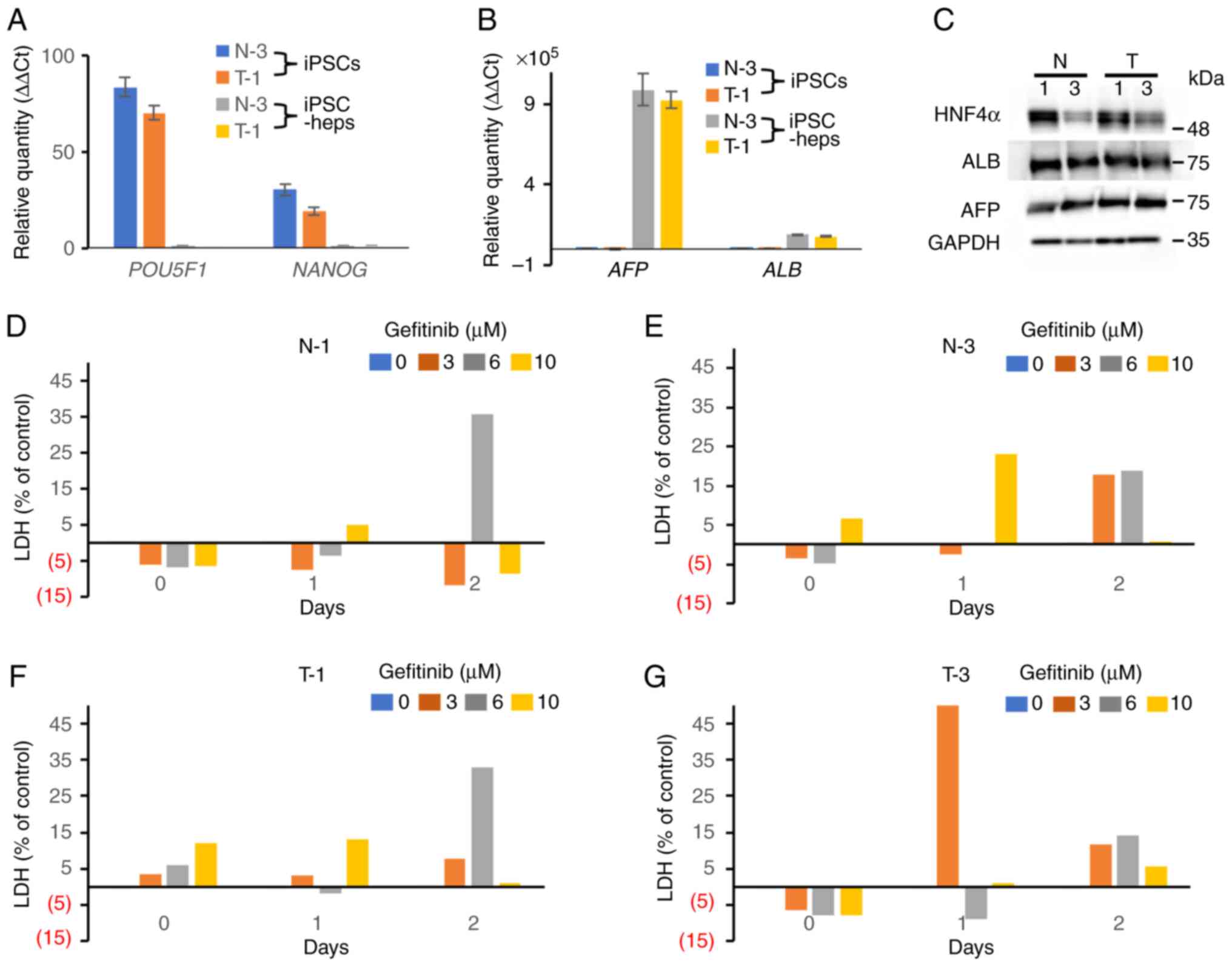
To prevent dose reduction or cessation of gefitinib treatment caused by hepatotoxicity, it is important to establish a cell-based assay system that can predict gefitinib-induced toxicity before clinical use. We hypothesized that iPSC-heps may predict gefitinib-induced hepatotoxicity in vitro and thus generated iPSCs from two groups of gefitinib-treated patients, those with grade 3 or greater hepatotoxicity (T group), and those who had grade 1 or less hepatotoxicity (N group) in the CTCAE v5.0. iPSCs were generated from 6 patients, 3 each from T and N groups. The patient characteristics are displayed in Table SI. We established 3 clones from a patient and got 18 clones in total. To confirm the pluripotency, we examined the mRNA expression levels of POU5F1 and NANOG, stem cell markers, showing that the clones were in a pluripotent state (Fig. 1A). In addition, the other stem cell markers, alkaline phosphatase (ALP) and SOX2, were highly expressed in the clones, whereas they were barely detected in the differentiated cells, HaCaT cells, in western blot analyses (Fig. S1). Since all clones were in a pluripotent state, we randomly chose 1 clone from each of the patients (6 clones), and was subjected to further analysis including the cyto/hepatotoxicity assay. We then differentiated iPSCs into iPSC-heps. A qPCR analysis showed that the expression levels of AFP and ALB, hepatocyte-specific markers, in iPSC-heps were much higher than those in iPSCs (Fig. 1B). In contrast, NANOG and POU5F1 were not expressed in iPSC-heps (Fig. 1A). A western blot analysis also showed that HNF4α, ALBUMIN (ALB), and AFP, hepatocyte-specific markers, were comparably expressed in iPSC-heps (Fig. 1C). These data suggest that iPSCs had differentiated into cells in a hepatocyte lineage. The four-step protocol we used for differentiation of iPSCs into iPSC-heps was depicted in Fig. 2. Collectively, we concluded that the iPSC clones were successfully differentiated into the iPSC-heps.
Evaluation of hepatotoxicity by gefitinib via LDH-release assay with iPSC-heps
To evaluate the hepatotoxicity of gefitinib, we employed a chemiluminescence-based LDH release assay using iPSC-heps. iPSCs (N-1, N-3, T-1, and T-3) were simultaneously differentiated into iPSC-heps. Gefitinib (0, 3, 6, and 10 µM) was administered on day 2 of STAGE 4 (Fig. 2). We confirmed that AFP and ALB, hepatocyte-specific markers, were already expressed at the beginning of STAGE 4 (data not shown). As hepatic maturation proceeded, evidence of naturally dying cells appeared. To differentiate natural cell death from gefitinib-related cytotoxicity as much as possible, day 2 of STAGE 4 was selected as the starting point. Each iPSC-heps were treated with 0, 2, 6, and 10 µM gefitinib, and conditioned medium was collected on days 0, 1, and 2 after administration. However, we could not find dose- or group (N and T)-dependency (Fig. 1D-G). These data suggest that our experimental design may need to be improved to be a good platform for gefitinib-induced hepatotoxicity assays.
EGFR expression, and the phosphorylation following gefitinib treatment in the clones of iPSCs or iPSC-heps from the N or T group
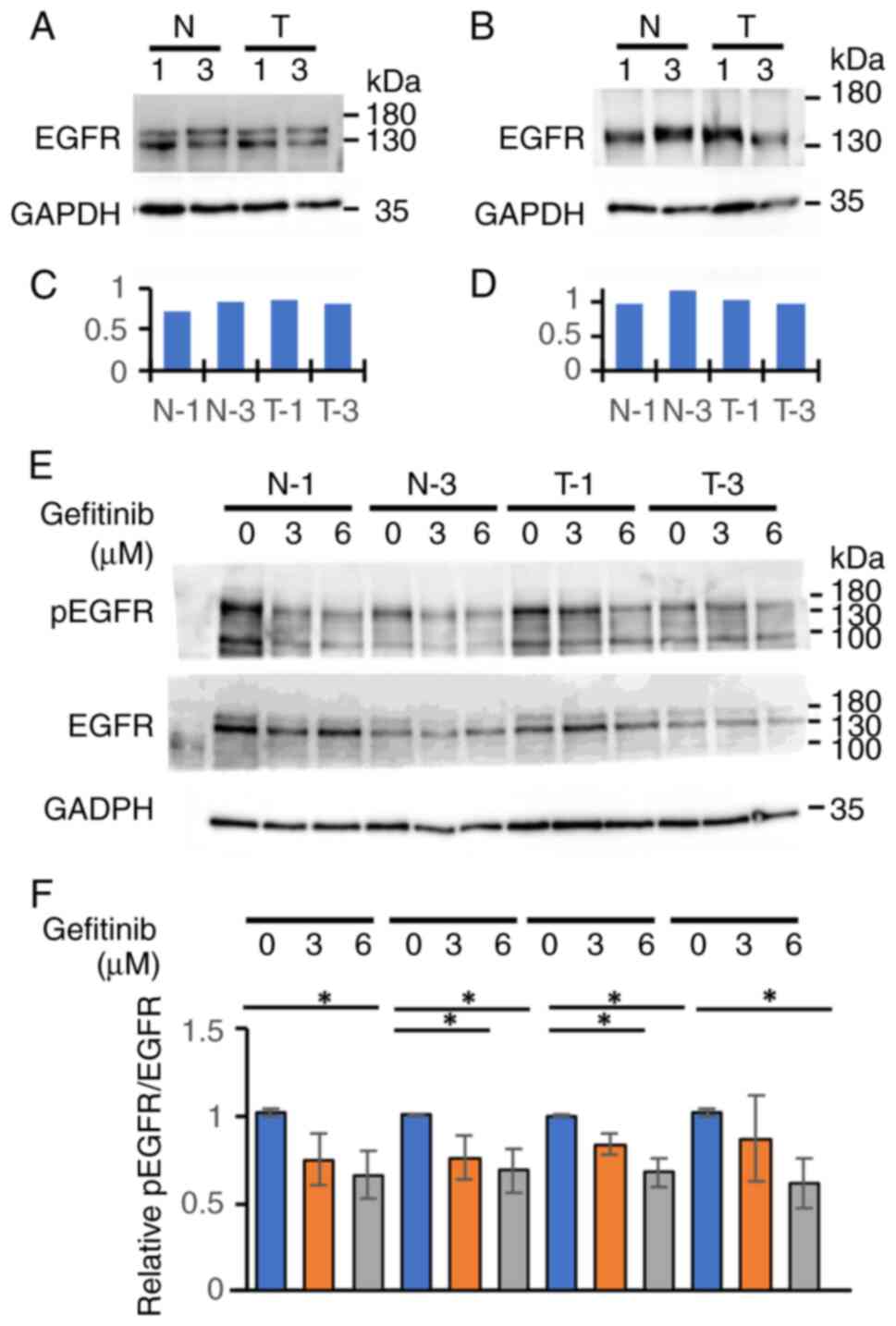
Since gefitinib is an EGFR-TKI, we examined the expression level of EGFR and the phosphorylation following gefitinib treatment in the clones of iPSCs and iPSC-heps from the N or T group. Western blot analyses showed that they were expressed similarly among the clones of iPSCs (Fig. 3A and C) and iPSC-heps (Fig. 3B and D) from each group, irrespective of whether those were from the N or T group. EGFR phosphorylation status (the ratio of pEGFR to EGFR) were also similar among the clones of iPSCs from the two groups following gefitinib treatment (Fig. 3E and F). Although treatment significantly suppressed EGFR phosphorylation in a dose-dependent manner, we did not find any differences in phosphorylation status between the two groups following gefitinib treatment. Fig. 3E and F shows that gefitinib treatment inhibited the EGFR signaling pathway equally in each iPSCs. Taken together, an LDH-release assay using iPSCs would potentially be a good predictor of gefitinib-induced hepatotoxicity.
Evaluation of gefitinib-induced hepatotoxicity via LDH-release assay with iPSCs
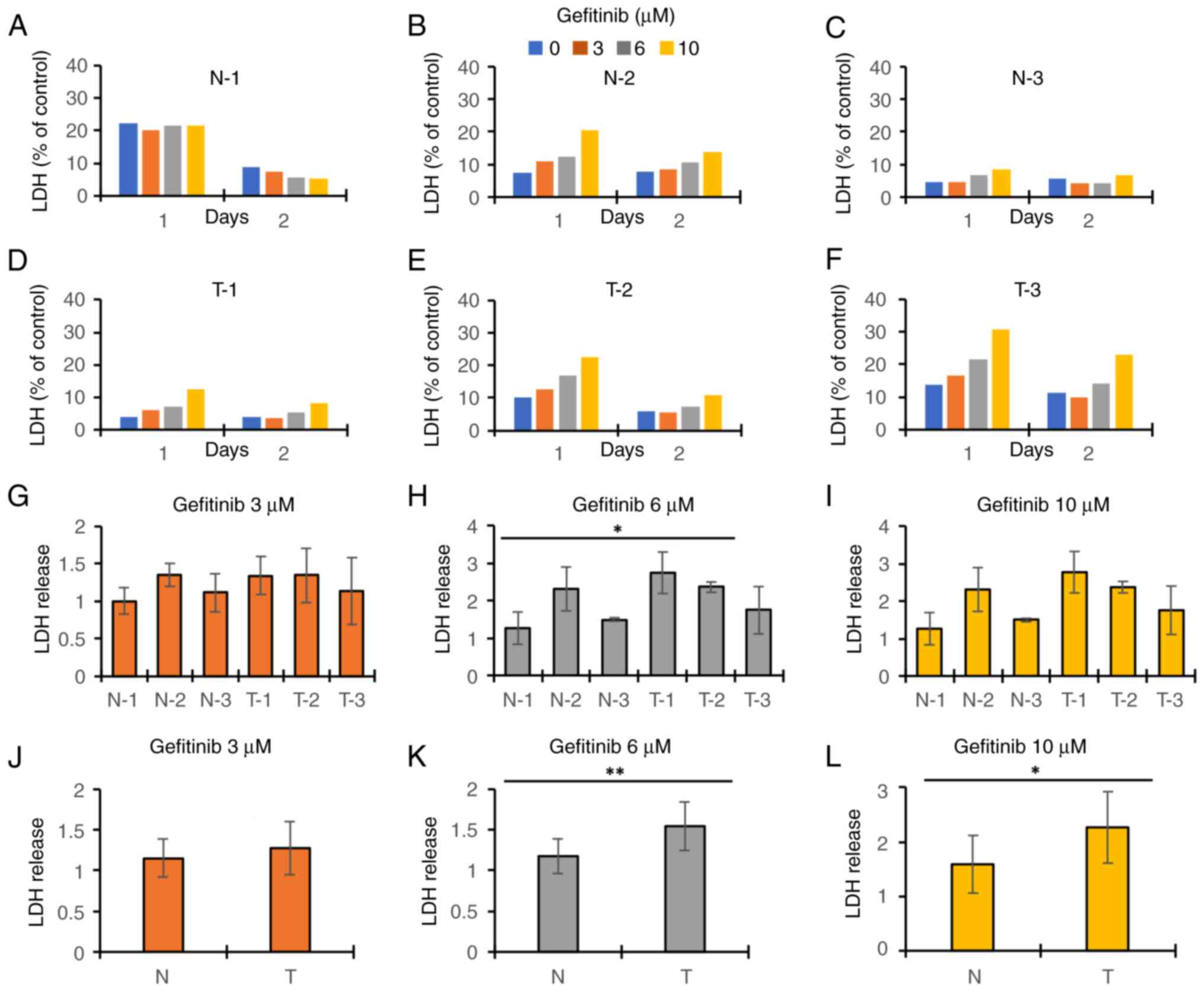
While we evaluated the gefitinib-induced hepatotoxicity with the iPSC-heps, we used iPSCs as a control in the assays. We found that iPSCs in T group released LDH more than those in N group after gefitinib treatment, and the increase was dose dependent (Fig. 4A-F). Because the difference between the groups at day 1 after gefitinib treatment seemed to be the largest among the time points analyzed, we focused on the toxicity at day 1. T-2 was significantly more sensitive to gefitinib than N-1 after 6 µM gefitinib treatment (Fig. 4H). There were no significant differences between the T and N groups in any other concentrations or combinations except that T-2 was significantly more sensitive to gefitinib than N-1 after 6 µM gefitinib treatment (Fig. 4G-I). However, iPSCs in T group tended to have the cytotoxicity more than those in N group. Therefore, we combined the data of each group and compared the 2 groups (Fig. 4J-L). The result showed that iPSCs in T group had higher cytotoxicity than those in N group after 6 or 10 µM gefitinib treatment (Fig. 4K and L), whereas there were no significant differences between N and T groups after 3 µM treatment (Fig. 4J). These results were consistent with the data of the cell viability assay (Fig. S2) and the cell morphology (Fig. S3). The cell viability in T group at day 2 after gefitinib treatment was significantly aggravated, except for T-2 and T-3 at 3 µM and T-2 at 6 µM, whereas no significant damages were observed in N group, except for N-1 and N-3 at 10 µM (Fig. S2). In the cell morphology at day 2 after gefitinib treatment, gefitinib decreased the number and size of attached iPSCs in the T group in a dose-dependent manner, whereas no significant damages were observed in N group, except for N-2 that seemed to be slightly damaged at 10 µM treatment (Fig. S3).
Discussion
Drug-induced hepatotoxicity is a major concern in drug development and clinical therapy (21,22). Severe hepatotoxicity caused by gefitinib treatment often leads to acute/chronic liver injury, drug discontinuation, and further treatment failure (3,23,24). To develop preclinical tests to predict hepatotoxicity of gefitinib, we evaluated a cell-based assay system using iPSCs or iPSC-heps generated from patients receiving gefitinib therapy. We first tried iPSC-heps for the gefitinib-induced hepatotoxicity assay. We estimated the hepatotoxicity by the measurement of LDH amount released into the culture medium from iPSC-heps. The hepatotoxicity assay using iPSC-heps did not reflect gefitinib-induced liver injury in the clinical setting; there were no significant differences between the 2 groups, although gefitinib treatment increased LDH release in both groups in a dose- and time-dependent manner (Fig. 1D-G). This may be because of the high variability of gefitinib-related cytotoxicity among the clones in the same group.
We next tried undifferentiated iPSCs for the cytotoxicity assay. The result showed that iPSCs from the T group had significantly higher cytotoxicity after gefitinib treatment than that from the N group (Fig. 4). Although we considered to use cell lines for the positive determination and feasibility of the assay, we did not try cell lines for the assay. That is because the reactivity to drugs in cell lines was quite different from that in primary cultured cells in our experiences. Taken together, we concluded that the cytotoxicity of gefitinib on patient-derived-iPSCs reflects gefitinib-induced liver injury in the clinical setting more than the hepatotoxicity of gefitinib on patient-derived-iPSC-heps, showing that iPSCs may have a possibility to become a platform for preclinical testing to predict gefitinib-induced hepatotoxicity.
The possibility of applying iPSCs in disease modeling and drug evaluation has been recently proposed by Sano et al (2021) (25). They used iPS cells transfected with ACE2, a SARS-CoV2 receptor (ACE2-iPS cells) to recapitulate SARS-CoV2 infection and assess anti-COVID-19 drug sensitivity in a pluripotent state and noticed that ACE-iPS cells could be infected by the virus, and that the virus could replicate in the cells. In addition, replication was strongly inhibited by the approved anti-COVID19 drug, remdesivir, but not by chloroquine (25), findings consistent with clinical results (26,27). Intriguingly, ACE2-iPS cells generated from men are more sensitive to SARS-CoV2 infection than those from women (25). These results indicate that if a molecular mechanism of a given clinical phenotype stems from a fundamental biological event encoded in the genome, iPSCs can model the phenotype. This might be the case in our gefitinib-induced hepatotoxicity study.
To determine the molecular mechanism by which gefitinib exerted higher toxicity against iPSCs from the T group compared to those from the N group, we examined various molecules in possible pathways such as pNFκB/NFκB and pTank-binding kinase (TBK)1/TBK1 related to inflammation. However, we could not identify gefitinib-induced changes for the pathways between N and T groups (data not shown). The expression levels of the major metabolizing enzyme of gefitinib, cytochrome p450 3A4 (CYP 3A4) (28–30), in iPSCs from both groups (Fig. S4) were also examined. It was shown that there were no significant differences on the expression levels among the cells. Interestingly, all the patients in the N group have an exon 19 deletion mutation in EGFR, while all the patients in the T group have an Exon 21 L585R mutation (Table SI). There is a possibility that different EGFR mutations have an impact on gefitinib-induced hepatotoxicity. We are now investigating the molecular mechanism of gefitinib-induced hepatotoxicity.
Collectively, these results suggest that a cell-based assay system using iPSCs may become a platform for preclinical tests to predict gefitinib-induced hepatotoxicity.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was partly supported by the grant-in-Aid for Scientific Research© (grant no. 19K07179) from the Japan Society for the Promotion of Science (to YF) and the OMC Internal Research Grant (to MA).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YF, TN, KT and MA participated in the conception and design of the study. YF and MW performed all the experiments with help from TN, KT, NM, YT, SI and AI. TN and MA wrote the manuscript. TN and MW performed the statistical analyses. YF, TN, KT, NM, YT, SI, AI and MA evaluated the results. YF, TN, MW and MA confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was conducted according to the guidelines of The Declaration of Helsinki and was approved by the Ethical Committee of the Osaka Medical College (now Osaka Medical and Pharmaceutical University) (approval no. 2100 on 10.03.2017). Written informed consent was obtained from all study subjects.
Patient consent for publication
Written informed consent has been obtained from all the study subjects to publish this paper.
Competing interests
The authors declare that they have no competing interests.
References
|
Thai AA, Solomon BJ, Sequist LV, Gainor JF and Heist RS: Lung cancer. Lancet. 398:535–554. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Ettinger DS, Bepler G, Bueno R, Chang A, Chang JY, Chirieac LR, D'Amico TA, Demmy TL, Feigenberg SJ, Grannis FW Jr, et al: Non-small cell lung cancer clinical practice guidelines in oncology. J Natl Compr Canc Netw. 4:548–582. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Takeda M, Okamoto I and Nakagawa K: Pooled safety analysis of EGFR-TKI treatment for EGFR mutation-positive non-small cell lung cancer. Lung Cancer. 88:74–79. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Lee WM: Acute liver failure in the United States. Semin Liver Dis. 23:217–226. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Sharma A, Mücke M and Seidman CE: Human induced pluripotent stem cell production and expansion from blood using a non-integrating viral reprogramming vector. Curr Protoc Mol Biol. 122:e582018. View Article : Google Scholar : PubMed/NCBI | |
|
Corbett JL and Duncan SA: iPSC-derived hepatocytes as a platform for disease modeling and drug discovery. Front Med (Lausanne). 6:2652019. View Article : Google Scholar : PubMed/NCBI | |
|
Davidson MD, Ware BR and Khetani SR: Stem cell-derived liver cells for drug testing and disease modeling. Discov Med. 19:349–358. 2015.PubMed/NCBI | |
|
Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K and Daley GQ: Disease-specific induced pluripotent stem cells. Cell. 134:877–886. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Maehr R, Chen S, Snitow M, Ludwig T, Yagasaki L, Goland R, Leibel RL and Melton DA: Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci USA. 106:15768–15773. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al: Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 136:964–977. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Ellis J and Bhatia M: iPSC technology: Platform for drug discovery. Point. Clin Pharmacol Ther. 89:639–641. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Carpentier A, Nimgaonkar I, Chu V, Xia Y, Hu Z and Liang TJ: Hepatic differentiation of human pluripotent stem cells in miniaturized format suitable for high-throughput screen. Stem Cell Res. 16:640–650. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Varghese DS, Alawathugoda TT and Ansari SA: Fine tuning of hepatocyte differentiation from human embryonic stem cells: Growth factor vs small molecule-based approaches. Stem Cells Int. 2019:59682362019. View Article : Google Scholar : PubMed/NCBI | |
|
Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S and Duncan SA: Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 51:297–305. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Hay DC, Zhao D, Fletcher J, Hewitt ZA, McLean D, Urruticoechea-Uriguen A, Black JR, Elcombe C, Ross JA, Wolf R and Cui W: Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells. 26:894–902. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Gao X and Liu Y: A transcriptomic study suggesting human iPSC-derived hepatocytes potentially offer a better in vitro model of hepatotoxicity than most hepatoma cell lines. Cell Biol Toxicol. 33:407–421. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Yamaguchi T, Matsuzaki J, Katsuda T, Saito Y, Saito H and Ochiya T: Generation of functional human hepatocytes in vitro: Current status and future prospects. Inflamm Regen. 39:132019. View Article : Google Scholar : PubMed/NCBI | |
|
Ware BR, Berger DR and Khetani SR: Prediction of drug-induced liver injury in micropatterned co-cultures containing iPSC-derived human hepatocytes. Toxicol Sci. 145:252–262. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou W, Graham K, Lucendo-Villarin B, Flint O, Hay DC and Bagnaninchi P: Combining stem cell-derived hepatocytes with impedance sensing to better predict human drug toxicity. Expert Opin Drug Metab Toxicol. 15:77–83. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Okita K, Yamakawa T, Matsumura Y, Sato Y, Amano N, Watanabe A, Goshima N and Yamanaka S: An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 31:458–466. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Ballet F: Hepatotoxicity in drug development: Detection, significance and solutions. J Hepatol. 26 (Suppl 2):S26–S36. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
Dirven H, Vist GE, Bandhakavi S, Mehta J, Fitch SE, Pound P, Ram R, Kincaid B, Leenaars CHC, Chen M, et al: Performance of preclinical models in predicting drug-induced liver injury in humans: A systematic review. Sci Rep. 11:64032021. View Article : Google Scholar : PubMed/NCBI | |
|
Takeda M, Okamoto I, Tsurutani J, Oiso N, Kawada A and Nakagawa K: Clinical impact of switching to a second EGFR-TKI after a severe AE related to a first EGFR-TKI in EGFR-mutated NSCLC. Jpn J Clin Oncol. 42:528–533. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Ho C, Davis J, Anderson F, Bebb G and Murray N: Side effects related to cancer treatment: CASE 1. Hepatitis following treatment with gefitinib. J Clin Oncol. 23:8531–8533. 2005. View Article : Google Scholar : PubMed/NCBI | |
|
Sano E, Deguchi S, Sakamoto A, Mimura N, Hirabayashi A, Muramoto Y, Noda T, Yamamoto T and Takayama K: Modeling SARS-CoV-2 infection and its individual differences with ACE2-expressing human iPS cells. iScience. 24:1024282021. View Article : Google Scholar : PubMed/NCBI | |
|
Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, et al: Remdesivir for the treatment of Covid-19-final report. N Engl J Med. 383:1813–1826. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Axfors C, Schmitt AM, Janiaud P, van't Hooft J, Abd-Elsalam S, Abdo EF, Abella BS, Akram J, Amaravadi RK, Angus DC, et al: Mortality outcomes with hydroxychloroquine and chloroquine in COVID-19 from an international collaborative meta-analysis of randomized trials. Nat Commun. 12:23492021. View Article : Google Scholar : PubMed/NCBI | |
|
Culy CR and Faulds D: Gefitinib. Drugs. 62:2237–2250. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Cohen MH, Williams GA, Sridhara R, Chen G, McGuinn WD Jr, Morse D, Abraham S, Rahman A, Liang C, Lostritto R, et al: United states food and drug administration drug approval summary: Gefitinib (ZD1839; Iressa) tablets. Clin Cancer Res. 10:1212–1218. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
McKillop D, McCormick AD, Millar A, Miles GS, Phillips PJ and Hutchison M: Cytochrome P450-dependent metabolism of gefitinib. Xenobiotica. 35:39–50. 2005. View Article : Google Scholar : PubMed/NCBI |