Introduction
Liver cancer is the third leading cause of
cancer-related death worldwide and has a poor prognosis, and thus
the development of more effective treatments is strongly desired
(1). It is estimated that over 1
million people will be affected by liver cancer annually by 2025
(2), and hepatocellular carcinoma
(HCC), as the most common form of liver cancer, accounts for
approximately 90% of cases (3). In
most cases, due to the lack of tumor-specific symptoms, HCC is
diagnosed at an advanced stage (Barcelona Clinic Liver Cancer
classification C), and the mainstay of treatment is chemotherapy
(4). In addition, the recurrence
rate in the remnant liver, even after R0 resection, has been
reported to be up to 80% (5), and
the main treatment is shifted to chemotherapy within 2 years from
diagnosis in over 50% of cases diagnosed as early-stage HCC and
treated radically with surgery or ablation (6). For unresectable advanced HCC,
molecular-targeted therapy using lenvatinib (7), sorafenib (8,9),
atezolizumab, and bevacizumab (10)
is reported to be effective. In particular, oral molecular targeted
drugs such as lenvatinib and sorafenib are used as crucial drugs in
the first-line chemotherapy of HCC.
Lenvatinib is an oral multi-kinase inhibitor that
inhibits mainly vascular endothelial growth factor receptor 1–3,
fibroblast growth factor receptor (FGFR) 1–4, platelet-derived
growth factor receptor (PDGFR) α, c-KIT, and RET (11). In terms of the disease control rate
(complete response + partial response + stable disease ratio),
response rate (complete response + partial response ratio), median
progression-free survival, and median time to progression,
lenvatinib shows significantly better results compared with
sorafenib, another oral multi-kinase inhibitor (75.5 vs. 60.5%,
24.1 vs. 9.2%, 7.4 vs. 3.7 months, 8.9 vs. 3.7 months,
respectively) (7). In contrast,
tumor growth or metastasis occurs in more than 50% of patients who
receive lenvatinib therapy within 1 year (7), and the development of acquired
resistance is a crucial clinical problem to be resolved (12).
The factors underlying primary resistance to
lenvatinib have been investigated widely and identified, and
include low protein levels of fibroblast growth factor 19 (FGF19),
which is a ligand of FGFR4, the main reaction pathway of
lenvatinib, and Kelch-like ECH-associated protein 1 (13–15);
activation of the hepatocyte growth factor/c-MET pathway (16); and high protein levels of
stomatin-like protein 2 and epidermal growth factor receptor
(17,18).
Few studies have investigated acquired resistance to
lenvatinib. Possible mechanisms for developing acquired resistance
to anti-cancer drugs include mutations of genes encoding proteins
that are the targets of drugs or are present in downstream pathways
of proteins inhibited by drugs as well as the activation of
collateral pathways (19,20). Genetic variations associated with
developing resistance to many anti-cancer drugs have also been
investigated. However, the genetic variations responsible for
developing acquired resistance to lenvatinib have yet to be
elucidated. In 2021, Myojin et al (14) established a cell line that acquired
resistance to lenvatinib from a lenvatinib-sensitive cell line,
Hep3B, by continuous exposure to lenvatinib, and revealed that
FGF19 overexpression restored lenvatinib susceptibility to this
cell line. However, in their study, no significant changes in FGF19
protein levels were observed with the acquisition of resistance,
and we cannot conclude that the reduced expression of FGF19 is the
cause of acquired resistance to lenvatinib.
In order to investigate the drug resistance
mechanism with the activation of alternative pathways in detail, a
comprehensive analysis of proteins derived from cancer cells and
identification of signaling pathways whose activity significantly
changes before and after the development of resistance to
anti-cancer drugs should be performed. Recently, the usefulness of
a comprehensive protein analytic approach has been revealed in many
studies such as the analysis of predictive markers for the efficacy
of erlotinib in non-small cell lung cancer (21–25).
In the present study, we established a lenvatinib-resistant HCC
cell line, JHH-7_LR, from a lenvatinib-sensitive HCC cell line by
continuous exposure to lenvatinib. Then, to clarify the factors
related to the acquisition of lenvatinib resistance, we identified
the proteins with significant changes in their expression before
and after the development of resistance. In addition, we analyzed
the signaling pathways composed by the proteins with significant
changes in expression. Furthermore, we assessed the impact of
inhibiting those signaling pathways on the efficacy of lenvatinib
in the JHH-7_LR cell line.
Materials and methods
Reagents
A lenvatinib-sensitive human hepatocellular
carcinoma cell line, JHH-7, was purchased from the Japanese
Collection of Research Bioresources Cell Bank (Osaka, Japan).
William's E Medium and GlutaMAX™ Supplement were purchased from
Thermo Fisher Scientific K.K. Lenvatinib and dasatinib were
purchased from LC Laboratories and Cayman Chemical, respectively. A
Cell Proliferation Kit I was purchased from Merck. The other cell
culture and sample preparation reagents were purchased from
Fujifilm Wako Pure Chemical Corporation. All other reagents were
obtained from commercial sources, and those used for proteomic
analysis were graded for high-performance liquid chromatography or
liquid chromatography-mass spectrometry (LC/MS).
Establishment of a lenvatinib-acquired
resistance cell line
JHH-7 cells were cultured in a humidified incubator
at 37°C with 5% CO2. A lenvatinib-acquired resistance
cell line (JHH-7_LR) was generated upon continuous exposure of
JHH-7 cells to lenvatinib; the exposure concentration started at
0.5 µmol/l and was increased to 40 µmol/l for over 1 year.
The acquisition of lenvatinib resistance was
verified by a cytotoxicity assay using a Cell Proliferation Kit I.
The cells were plated in a 96-well plate at 1.0×103
cells/well in medium supplemented with 10% fetal bovine serum.
After overnight incubation, the medium was changed to 100 µl fresh
medium containing 0 or 0.005–50 µmol/l of lenvatinib and cultured
for another 5 days. Then, 10 µl of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
solution (5 mg/ml) was added to each well. After incubation for 4
h, 10% SDS solution was added to stop the reaction, and absorbance
at 550 and 690 nm of each well was measured immediately. The growth
inhibition ratio (Ir) was calculated by equation 1:
Ir = (AbsLB -
AbsS)/(AbsLB - AbsM) (1)
where AbsLB, AbsS, and
AbsM represent the values of Abs550-Abs690 nm of the
sample incubated without lenvatinib, sample incubated with
lenvatinib, and medium, respectively.
The calculated Ir and lenvatinib concentrations were
applied to equation 2, and the nonlinear least-squares MULTI
program was used to estimate the inhibition of 50% cancer cell
growth (IC50):
Ir = 100 x Imax/(1 + Exp (-S x (C -
IC50) (2)
where S, C, and Imax represent the
sigmoid variable, lenvatinib concentration, and maximum inhibition
rate, respectively.
Proteome analysis
Cytoplasmic proteins of each cell line were
extracted using a Minute Plasma Membrane Protein Isolation Kit
(Invent Biotechnologies), and the concentrations of the extracted
proteins were measured using a DC™ Protein Assay Kit (Bio-Rad
Laboratories). The extracts were diluted to 0.25 mg/ml with
phosphate-buffered saline, and 100 µl of the diluted samples was
incubated at 37°C for 60 min with 100 mg urea and 10 µl of 650
mmol/l dithiothreitol in 8 mol/l urea/0.5 mol/l Tris HCL (pH 8.5).
After spiking with 10 µl of 1 mol/l iodoacetamide in 8 mol/l
urea/0.5 mol/l Tris HCL (pH 8.5), the samples were incubated at
37°C for 30 min. Subsequently, to digest the proteins, 12 µl of 1
mg/ml trypsin in 20 mmol/l acetic acid was added and incubated at
37°C for 3 h. Finally, the trypsinized samples were desalted and
concentrated using an ISOLUTE C18 (EC) column (Biotage Japan Ltd.,
Tokyo, Japan) and applied to LC/MS.
LC/MS analysis was performed with an EksigentNanoLC
425 coupled to a Triple TOF 6600 (AB Sciex). First, 10 µl of the
sample was loaded on a trap column (Acclaim PepMap 100 C18, 5 µm,
0.2 mm I.D. ×10 mm; Thermo Fisher Scientific K.K.) and then
separated using an analytical column (Acclaim PepMap 100 C18, 3 µm,
0.075 mm I.D. ×250 mm; Thermo Fisher Scientific K.K.) with a
gradient from 2 to 32% solvent B at a flow rate of 300 nl/min for
120 min (solvent A: 0.1% formic acid in water; solvent B: 0.1%
formic acid in acetonitrile). Ion source parameters were set as
follows: ion source voltage, 2,350 V; ion source gas (GS1 and GS2),
5 and 0; interface heater temperature, 150°C; declustering
potential, 80 V.
Sequential window acquisition of all theoretical
fragment ion spectra (SWATH) was acquired using the 100 SWATH
variable window method (AB Sciex Pte. Ltd.) from m/z
100–1,800 with each 25 ms accumulation time. Library samples were
prepared by mixing all samples to be analyzed equally and measured
three times by data-dependent acquisition, selecting the top 25
highest peaks. UniProt (uniprot_sprot.fasta) was used for library
data preparation. The ion chromatograms were analyzed for five
transitions per peptide and five peptides per protein and then
processed with a peptide confidence threshold of 99% and a false
discovery rate of <1%. ProteinPilot ver. 5.0.1, SWATH
Acquisition Micro App ver. 2.0 (AB Sciex), and Peak View ver. 2.2
(AB Sciex) were used for analysis.
Pathway analysis
The differences in protein expression levels between
the lenvatinib-sensitive and -resistant cell lines were analyzed by
a t-test, and the P-value was adjusted by the Benjamini-Hochberg
method (q-value). Using the UniProt code, expression level
ratio, P-value, and q-value of proteins with
q<0.05 and those with a change of expression in the
acquired resistance cell line of more than 2- or 0.5-fold compared
to the original cell line, core analysis was performed with
bioinformatics software (Ingenuity Pathway Analysis ver. Winter
2021; Qiagen, Venlo, Netherlands).
Effect of dasatinib on lenvatinib
sensitivity
The JHH-7_LR and JHH-7 cell lines were plated in
96-well plates at 1.0×103 cells/well in medium
supplemented with 10% fetal bovine serum. After overnight
incubation, the medium was changed to 100 µl fresh medium
containing lenvatinib (0 or 0.005–50 µmol/l) with or without
dasatinib (2.5 or 5 µmol/l). After incubation for another 5 days,
IC50 was estimated as described earlier.
Statistical analysis
Statistical analysis was performed on the difference
between two groups with unpaired t-test. For multiple comparisons,
one-way ANOVA was first performed, followed by Dunnett's test.
Multiple comparisons were controlled for the false discovery rate
based on the Benjamini-Hochberg method. SPSS ver. 28 (IBM Japan
Co., Ltd.) was used for statistical analysis.
Results
Establishment of a
lenvatinib-resistant cell line
We established a lenvatinib-resistant cell line,
JHH-7_LR, after exposing the parental HCC cell line JHH-7 to
step-wise increasing concentrations of lenvatinib up to 40 µmol/l.
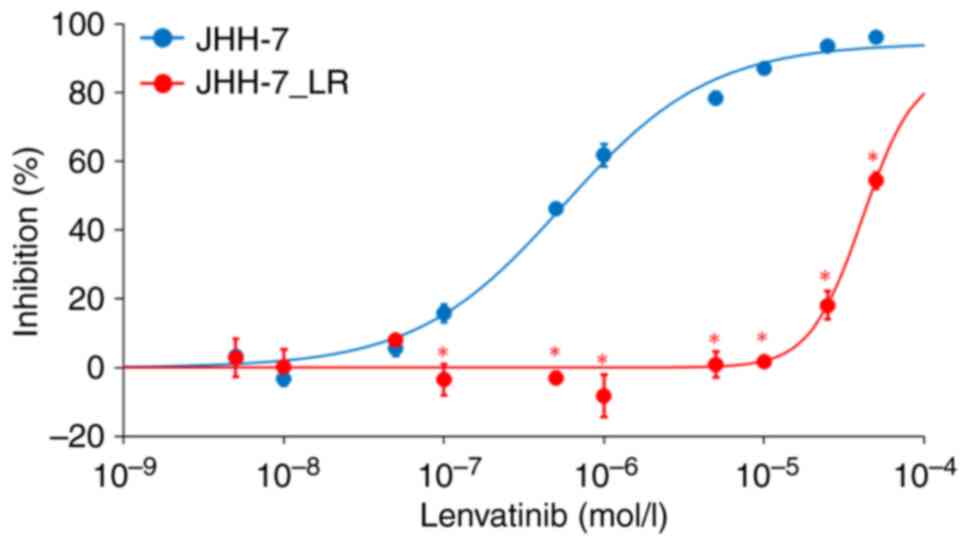
Lenvatinib inhibited the proliferation of both the JHH-7 and
JHH-7_LR HCC cells in a concentration-dependent manner. The
IC50 of JHH-7_LR cells to lenvatinib was approximately
70 times higher than that of JHH-7 cells (41.3 and 0.56 µmol/l,
respectively) (Fig. 1).
Proteome analysis
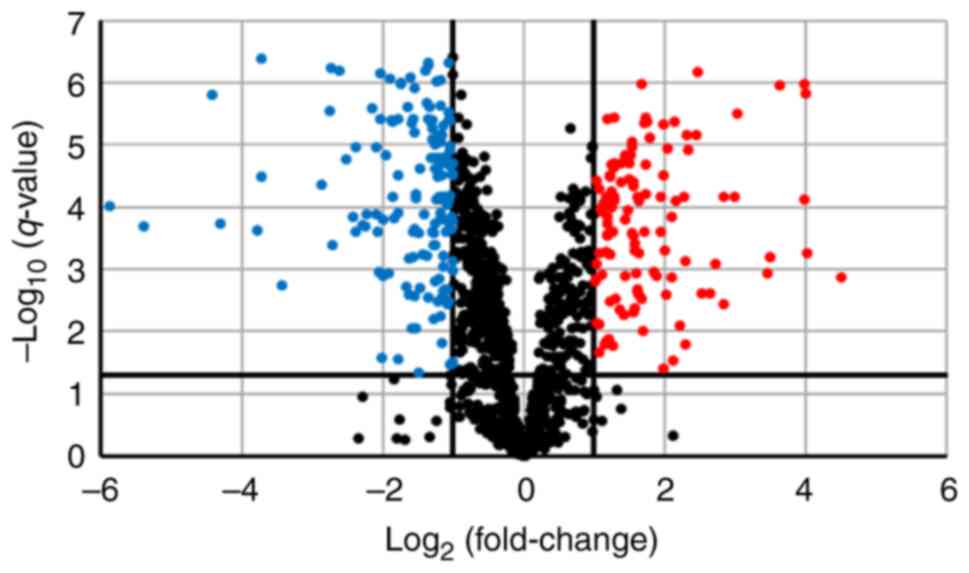
We performed SWATH analysis to investigate the
changes in protein expression levels associated with lenvatinib
resistance. SWATH analysis detected 4,013 peptides and identified
1,323 proteins. Among them, 1,321 proteins for which quantitative
values were obtained in all samples were analyzed. Along with the
development of resistance, the expression levels of 115 proteins
were more than doubled and significantly increased
(q<0.05). However, the expression levels of 152 proteins
were reduced by half and significantly decreased (q<0.05)
(Fig. 2).
Pathway analysis
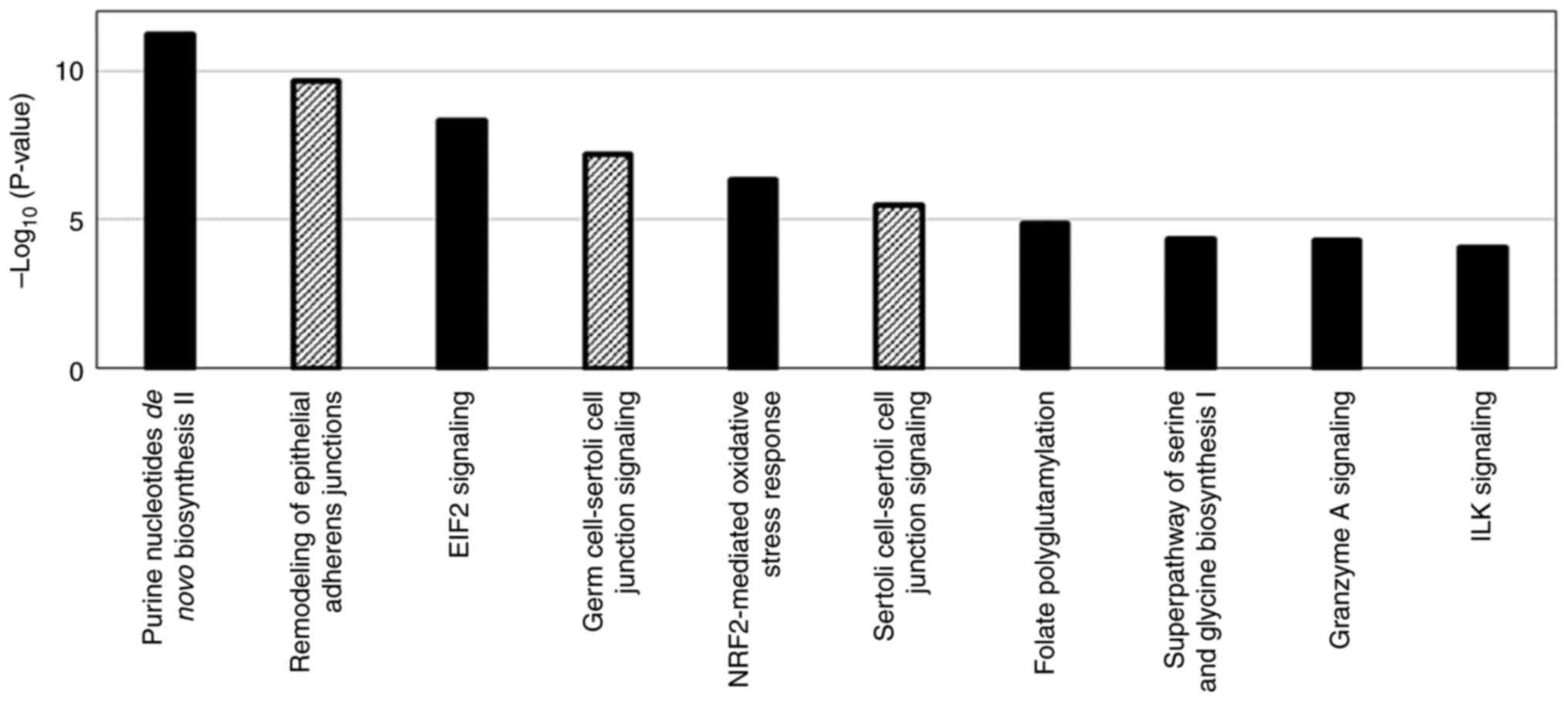
Pathway analysis was performed using 267 proteins
with substantial changes in expression with the acquisition of
resistance to lenvatinib as well as 124 signaling pathways were
detected in which activity was affected by changes in the
expression of those proteins (Fig.
3). For the proteins involved in the top 10 signaling pathways
with the highest -log(P-value), calculated based on the number and
proportion of proteins contained in the signaling pathway, the top
10 proteins with the most significant change of expression with the
acquisition of resistance to lenvatinib were extracted as
lenvatinib resistance-related protein candidates (Tables I and II). Among those proteins, c-SRC,
Ras-related C3 botulinum toxin substrate 1, nucleoside diphosphate
kinase A, serine hydroxymethyltransferase (cytosolic), and serine
hydroxymethyltransferase (mitochondrial) were found to have
enzymatic activity and are involved in multiple pathways. The
changes in their expression with the acquisition of resistance were
5.44-, 0.45-, 0.34-, 0.42-, and 2.0-fold, respectively. The changes
in cadherin-2, NAD(P)H dehydrogenase [quinone] 1, and Filamin-A
expressions were more significant than those in c-SRC expression
upon acquisition of resistance. However, those proteins were
contained in only one signaling pathway extracted by pathway
analysis.
 | Table I.Top 10 proteins exhibiting increased
expression following the development of lenvatinib resistance. |
Table I.
Top 10 proteins exhibiting increased
expression following the development of lenvatinib resistance.
|
|
| Expression
levelsa | Ratio of expression
levelsb |
|
|---|
|
|
|
|
|
|
|---|
| UniProt code | Name | JHH-7 | JHH-7_LR | JHH-7_LR/JHH-7 |
P-valuec |
|---|
| P19022 | Cadherin-2 | 13,995 | 222,657 | 15.91 |
2.4×10−8 |
| P12931 | Proto-oncogene
tyrosine-protein kinase Src | 103,614 | 563,152 | 5.44 |
3.2×10−7 |
| P04179 | Superoxide
dismutase [Mn], mitochondrial | 45,934 | 226,645 | 4.93 |
3.1×10−7 |
| P07305 | Histone H1.0 | 21,414 | 105,396 | 4.92 |
8.5×10−3 |
| P78330 | Phosphoserine
phosphatase | 3,982,655 | 13,190,082 | 3.31 |
10.0×10−8 |
| O43707 | α-actinin-4 | 1,885,027 | 5,750,919 | 3.05 |
7.7×10−6 |
| P35221 | Catenin α-1 | 51,001 | 150,172 | 2.94 |
2.1×10−3 |
| P35222 | Catenin β-1 | 26,094 | 76,507 | 2.93 |
4.8×10−6 |
| Q14651 | Plastin-1 | 97,393 | 274,947 | 2.82 |
3.5×10−6 |
| P08263 | Glutathione
S-transferase A1 | 1,866,497 | 5,041,783 | 2.70 |
2.9×10−5 |
 | Table II.Top 10 proteins exhibiting decreased
expression following the development of lenvatinib resistance. |
Table II.
Top 10 proteins exhibiting decreased
expression following the development of lenvatinib resistance.
|
|
| Expression
levelsa | Ratio of expression
levelsb |
|
|---|
|
|
|
|
|
|
|---|
| UniProt code | Name | JHH-7 | JHH-7_LR | JHH-7_LR/JHH-7 |
P-valuec |
|---|
| P15559 | NAD(P)H
dehydrogenase [quinone] 1 | 1,145,340 | 27,193 | 0.02 |
4.1×10−5 |
| P21333 | Filamin-A | 1,611,699 | 221,586 | 0.14 |
4.4×10−6 |
| Q99615 | DnaJ homolog
subfamily C member 7 | 323,245 | 93,505 | 0.29 |
2.8×10−6 |
| P11586 |
C-1-tetrahydrofolate synthase,
cytoplasmic | 2,096,222 | 627,168 | 0.30 |
1.6×10−8 |
| Q9Y3U8 | 60S ribosomal
protein L36 | 405,738 | 121,794 | 0.30 |
1.2×10−8 |
| P35579 | Myosin-9 | 773,819 | 251,904 | 0.33 |
9.7×10−4 |
| P18206 | Vinculin | 1,547,743 | 505,010 | 0.33 |
7.4×10−9 |
| Q13885 | Tubulin beta-2A
chain | 491,799 | 165,287 | 0.34 |
1.7×10−4 |
| Q06830 |
Peroxiredoxin-1 | 3,870,952 | 1,313,386 | 0.34 |
5.5×10−5 |
| P15531 | Nucleoside
diphosphate kinase A | 276,722 | 94,293 | 0.34 |
2.7×10−7 |
Effect of dasatinib on lenvatinib
sensitivity
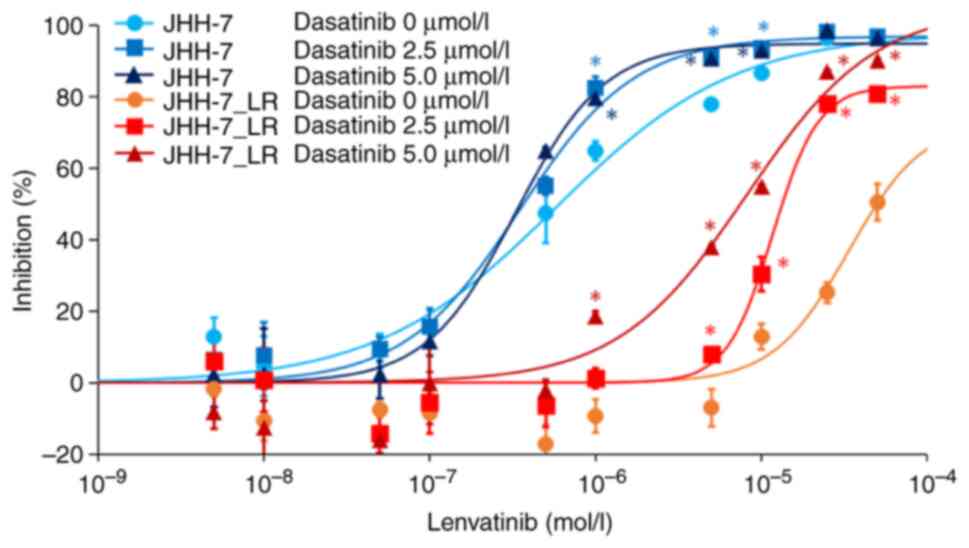
We examined the involvement of c-SRC in lenvatinib
sensitivity. The concomitant use of dasatinib, a c-SRC inhibitor,
increased the sensitivity of JHH-7_LR cells to lenvatinib in a
dose-dependent manner. However, dasatinib had no effect on the
lenvatinib sensitivity of JHH-7 cells (Fig. 4).
Discussion
In the present study, we found that multiple
signaling pathways were associated with the development of
resistance to lenvatinib. Among them, the expression levels of many
proteins that constitute the signaling pathways linked with c-SRC
changed significantly with the acquisition of resistance to
lenvatinib, and the expression level of c-SRC increased by
approximately 5-fold with the acquisition of resistance.
c-SRC is a non-receptor tyrosine kinase encoded by
SRC, which is homologous to the Rous sarcoma virus
proto-oncogene γ-Src (26–28).
c-SRC is located downstream of receptor tyrosine kinases such as
epidermal growth factor receptor (29–31),
FGFR (32), PDGFR (33), HER2/neu (29), and c-MET (34), and it is involved in cell
proliferation and metastasis in various carcinomas (35–37) by
activation of the RAS/RAF/MEK/ERK pathway (38), PI3K/Akt/mTOR pathway (39), integrin/FAK pathway (40,41),
and JAK/STAT pathway (39,40). In addition, c-SRC is reported to be
involved in the acquisition of resistance to other tyrosine kinase
inhibitors. Yoshida et al (42) found that multiple phosphorylation
reactions change with the acquisition of resistance, and c-SRC was
identified as a factor related to these phosphorylation reactions
in vitro using a gefitinib-resistant cell line, established
from a gefitinib-sensitive non-small cell lung cancer cell line,
PC-9. In our data, changes in cadherin-2 expression were more
significant than those in c-SRC expression upon acquisition of
resistance. However, it has been reported that the expression of
N-cadherin encoded by CDH2 is affected by c-SRC (43), and we speculated that the variation
in its expression level was affected by the variation in c-SRC
activity.
Given that our data suggested the possibility that
acquired resistance to lenvatinib is induced by enhancement of
c-SRC-related pathways, we assessed the effect of a c-SRC inhibitor
on the sensitivity of JHH-7_LR cells to lenvatinib. Of the drugs
used in the clinical setting, dasatinib, which is widely used for
treating chronic myelogenous leukemia and Philadelphia
chromosome-positive acute lymphoblastic leukemia, is known to
inhibit c-SRC activity (44,45).
Dasatinib inhibits SRC family kinases, BCR-ABL, PDGFRβ, and c-Kit
among tyrosine kinases and is reported as a potent inhibitor of
c-SRC (IC50: 0.5 nmol/l) (46). In our study, dasatinib increased the
sensitivity of JHH-7_LR cells to lenvatinib in a dose-dependent
manner. Similarly, Murakami et al (47) reported that dasatinib partially
restored the afatinib sensitivity of afatinib-resistant non-small
cell lung cancer cell lines established by continuous exposure to
afatinib. However, although the concentration of dasatinib in the
medium was set at sufficiently high levels (2.5 and 5.0 µmol/l),
which were more than 10-fold higher than the IC50 value
of dasatinib against c-SRC (0.5 nmol/l) and the maximum blood
concentration of dasatinib (approximately 0.2 µmol/l) after
continuous oral administration, dasatinib did not completely
restore the sensitivity of acquired drug resistance cell lines to
lenvatinib and afatinib. There are two possible reasons why
dasatinib is unable to fully restore lenvatinib sensitivity. First,
because the concentration of dasatinib in the medium required for
inhibiting c-SRC phosphorylation varies depending on the cell line
(48), it may not have been able to
inhibit c-SRC in JHH-7_LR cells sufficiently. For this point,
evaluating the impact of a more potent c-SRC inhibitor on the
lenvatinib sensitivity of JHH-7_LR cells is required. Next, it is
possible that pathways other than the c-SRC-related pathways
simultaneously influence the development of resistance to
lenvatinib. For this point, the involvement of other pathways needs
to be studied in more detail.
This study has some limitations. First, in the
proteome analysis, we were unable to detect changes in FGF19
expression, which has been reported as one of the factors affecting
the development of resistance in previous studies, nor were we able
to assess its effects on the lenvatinib sensitivity of the JHH-7_LR
cell line. On the other hand, the expression level of N-cadherin,
which has been reported to be decreased by knocking down FGF19 in
hepatocellular carcinoma-derived cell lines (49), was confirmed to increase with the
development of resistance in our study. In addition, Myojin et
al (14) also reported that
protein levels of FGF19 did not change with the development of
resistance, unlike the change in its mRNA level. Thus, although we
could not detect FGF19 in our study, we considered that the
decrease in FGF19 expression did not cause the development of
resistance to lenvatinib. Next, we used only one type of HCC cell
line; therefore, conducting similar studies using multiple cell
lines and clinical samples is necessary. In the future, we aim to
construct a lenvatinib-resistant cell line derived from other
multi-cell lines. In addition, we should assess the contribution of
the change in c-SRC expression to developing resistance to
lenvatinib in the clinical setting in detail. Thirdly, we could not
analyze the change of expression of proteins, which could not be
detected by comprehensive protein expression analysis using
LC-MS/MS. For this point, additional analysis, such as
comprehensive mRNA analysis, could provide helpful information. In
the subsequent study, we will analyze this point in detail and
elucidate the factors that cause resistance to lenvatinib in more
detail.
In conclusion, the activation of c-SRC may be an
essential factor in the development of lenvatinib resistance
induced by long-term exposure to lenvatinib. We also found that
exposure to dasatinib, a c-SRC inhibitor, could partially eliminate
the resistance of the JHH-7_LR cell line to lenvatinib. We believe
that detailed examinations of the mechanism of c-SRC activation
associated with lenvatinib resistance will lead to the development
of a more effective method for overcoming lenvatinib
resistance.
Acknowledgements
Not applicable.
Funding
This work was supported by JSPS KAKENHI (grant no.
JP18K06743).
Availability of data and materials
The raw mass spectrometry data and peptide/protein
identification results generated and/or analyzed during the current
study are available in the jPOST repository, https://repository.jpostdb.org/entry/JPST002307.
The other datasets used and/or analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
KY, TA, DN, HY and AN conceived and designed the
experiments. MT performed the experiments. MT and TA analyzed the
data and wrote the paper. MT and TA confirm the authenticity of all
the raw data. KY, TA, DN, HY and AN revised the paper. All authors
have read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Villanueva A: Hepatocellular carcinoma. N
Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Akinyemiju T, Abera S, Ahmed M, Alam N,
Alemayohu MA, Allen C, Al-Raddadi R, Alvis-Guzman N, Amoako Y,
Artaman A, et al: The burden of primary liver cancer and underlying
etiologies From 1990 to 2015 at the global, regional, and national
level: Results from the global burden of disease study 2015. JAMA
Oncol. 3:1683–1691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park JW, Chen M, Colombo M, Roberts LR,
Schwartz M, Chen PJ, Kudo M, Johnson P, Wagner S, Orsini LS and
Sherman M: Global patterns of hepatocellular carcinoma management
from diagnosis to death. Liver Int. 35:2155–2166. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kokudo N, Takemura N, Hasegawa K, Takayama
T, Kubo S, Shimada M, Nagano H, Hatano E, Izumi N, Kaneko S, et al:
Clinical practice guidelines for hepatocellular carcinoma: The
Japan society of hepatology 2017 (4th JSH-HCC guidelines) 2019
update. Hepatol Res. 49:1109–1113. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tabrizian P, Jibara G, Shrager B, Schwartz
M and Roayaie S: Recurrence of hepatocellular cancer after
resection: Patterns, treatments, and prognosis. Ann Surg.
261:947–955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kudo M, Finn RS, Qin S, Han KH, Ikeda K,
Piscaglia F, Baron A, Park JW, Han G, Jassem J, et al: Lenvatinib
versus sorafenib in first-line treatment of patients with
unresectable. Lancet. 391:1163–1173. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
et al: Sorafenib in advanced hepatocellular carcinoma. N Engl J
Med. 359:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S,
Kim JS, Luo R, Feng J, Ye S, Yang TS, et al: Efficacy and safety of
sorafenib in patients in the Asia-pacific region with. Lancet
Oncol. 10:25–34. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Finn RS, Qin S, Ikeda M, Galle PR, Ducreux
M, Kim TY, Kudo M, Breder V, Merle P, Kaseb AO, et al: Atezolizumab
plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J
Med. 382:1894–1905. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matsui J, Yamamoto Y, Funahashi Y,
Tsuruoka A, Watanabe T, Wakabayashi T, Uenaka T and Asada M: E7080,
a novel inhibitor that targets multiple kinases, has potent
antitumor activities against stem cell factor producing human small
cell lung cancer H146, based on angiogenesis inhibition. Int J
Cancer. 122:664–671. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ao J, Chiba T, Shibata S, Kurosugi A,
Qiang N, Ma Y, Kan M, Iwanaga T, Sakuma T, Kanzaki H, et al:
Acquisition of mesenchymal-like phenotypes and overproduction of
angiogenic factors in lenvatinib-resistant hepatocellular carcinoma
cells. Biochem Biophys Res Commun. 549:171–178. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Matsuki M, Hoshi T, Yamamoto Y,
Ikemori-Kawada M, Minoshima Y, Funahashi Y and Matsui J: Lenvatinib
inhibits angiogenesis and tumor fibroblast growth factor signaling
pathways in human hepatocellular carcinoma models. Cancer Med.
7:2641–2653. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Myojin Y, Kodama T, Maesaka K, Motooka D,
Sato Y, Tanaka S, Abe Y, Ohkawa K, Mita E, Hayashi Y, et al:
ST6GAL1 is a novel serum biomarker for lenvatinib-susceptible
FGF19-driven hepatocellular carcinoma. Clin Cancer Res.
27:1150–1161. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng A, Chevalier N, Calderoni M, Dubuis
G, Dormond O, Ziros PG, Sykiotis GP and Widmann C: CRISPR/Cas9
genome-wide screening identifies KEAP1 as a sorafenib, lenvatinib,
and regorafenib sensitivity gene in hepatocellular carcinoma.
Oncotarget. 10:7058–7070. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu R, Jiang S, Li J, Chen H and Zhang X:
Activation of the HGF/c-MET axis promotes lenvatinib resistance in
hepatocellular carcinoma cells with high c-MET expression. Med
Oncol. 37:242020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng Y, Huang C, Lu L, Yu K, Zhao J, Chen
M, Liu L, Sun Q, Lin Z, Zheng J, et al: STOML2 potentiates
metastasis of hepatocellular carcinoma by promoting PINK1-mediated
mitophagy and regulates sensitivity to lenvatinib. J Hematol Oncol.
14:162021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin H, Shi Y, Lv Y, Yuan S, Ramirez CFA,
Lieftink C, Wang L, Wang S, Wang C, Dias MH, et al: EGFR activation
limits the response of liver cancer to lenvatinib. Nature.
595:730–734. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vasan N, Baselga J and Hyman DM: A view on
drug resistance in cancer. Nature. 575:299–309. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chatterjee N and Bivona TG: Polytherapy
and targeted cancer drug resistance. Trends Cancer. 5:170–182.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chung CH, Seeley EH, Roder H, Grigorieva
J, Tsypin M, Roder J, Burtness BA, Argiris A, Forastiere AA,
Gilbert J, et al: Detection of tumor epidermal growth factor
receptor pathway dependence by serum mass spectrometry in cancer
patients. Cancer Epidemiol Biomarkers Prev. 19:358–365. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pitteri SJ, Amon LM, Buson TS, Zhang Y,
Johnson MM, Chin A, Kennedy J, Wong CH, Zhang Q, Wang H, et al:
Detection of elevated plasma levels of epidermal growth factor
receptor before breast cancer diagnosis among hormone therapy
users. Cancer Res. 70:8598–8606. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Garrisi VM, Bongarzone I, Mangia A,
Cremona M, De Bortoli M, Vaghi E, Galetta D, Pastorino U, Quaranta
M, Abbate I and Paradiso A: Characterization of a serum protein
pattern from NSCLC patients treated with Gefitinib. Clin Biochem.
44:936–940. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lazzari C, Spreafico A, Bachi A, Roder H,
Floriani I, Garavaglia D, Cattaneo A, Grigorieva J, Viganò MG,
Sorlini C, et al: Changes in plasma mass-spectral profile in course
of treatment of non-small cell lung cancer patients with epidermal
growth factor receptor tyrosine kinase inhibitors. J Thorac Oncol.
7:40–48. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gregorc V, Novello S, Lazzari C, Barni S,
Aieta M, Mencoboni M, Grossi F, De Pas T, de Marinis F, Bearz A, et
al: Predictive value of a proteomic signature in patients with
non-small-cell lung cancer treated with second-line erlotinib or
chemotherapy (PROSE): A biomarker-stratified, randomised phase 3
trial. Lancet Oncol. 15:713–721. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rous P: A sarcoma of the fowl
transmissible by an agent separable from the tumor cells. J Exp
Med. 13:397–411. 1911. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Stehelin D, Varmus HE, Bishop JM and Vogt
PK: DNA related to the transforming gene(s) of avian sarcoma
viruses is present in normal avian DNA. Nature. 260:170–173. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Czernilofsky AP, Levinson AD, Varmus HE,
Bishop JM, Tischer E and Goodman HM: Nucleotide sequence of an
avian sarcoma virus oncogene (src) and proposed amino acid sequence
for gene product. Nature. 287:198–203. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luttrell DK, Lee A, Lansing TJ, Crosby RM,
Jung KD, Willard D, Luther M, Rodriguez M, Berman J and Gilmer TM:
Involvement of pp60c-src with two major signaling pathways in human
breast cancer. Proc Natl Acad Sci USA. 91:83–87. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Maa MC, Leu TH, McCarley DJ, Schatzman RC
and Parsons SJ: Potentiation of epidermal growth factor
receptor-mediated oncogenesis by c-Src: Implications for the
etiology of multiple human cancers. Proc Natl Acad Sci USA.
92:6981–6985. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mao W, Irby R, Coppola D, Fu L, Wloch M,
Turner J, Yu H, Garcia R, Jove R and Yeatman TJ: Activation of
c-Src by receptor tyrosine kinases in human colon cancer cells with
high metastatic potential. Oncogene. 15:3083–3090. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
LaVallee TM, Prudovsky IA, McMahon GA, Hu
X and Maciag T: Activation of the MAP kinase pathway by FGF-1
correlates with cell proliferation induction while activation of
the Src pathway correlates with migration. J Cell Biol.
141:1647–1658. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Courtneidge SA, Fumagalli S, Koegl M,
Superti-Furga G and Twamley-Stein GM: The Src family of protein
tyrosine kinases: Regulation and functions. Dev Suppl. 57–64.
1993.PubMed/NCBI
|
|
34
|
Rahimi N, Hung W, Tremblay E, Saulnier R
and Elliott B: c-Src kinase activity is required for hepatocyte
growth factor-induced motility and anchorage-independent growth of
mammary carcinoma cells. J Biol Chem. 273:33714–33721. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Irby RB and Yeatman TJ: Role of Src
expression and activation in human cancer. Oncogene. 19:5636–5642.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao R, Wu Y, Wang T, Zhang Y, Kong D,
Zhang L, Li X, Wang G, Jin Y, Jin X and Zhang F: Elevated Src
expression associated with hepatocellular carcinoma metastasis in
northern Chinese patients. Oncol Lett. 10:3026–3034. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ito Y, Kawakatsu H, Takeda T, Sakon M,
Nagano H, Sakai T, Miyoshi E, Noda K, Tsujimoto M, Wakasa K, et al:
Activation of c-Src gene product in hepatocellular carcinoma is
highly correlated with the indices of early stage phenotype. J
Hepatol. 35:68–73. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Haas M, Askari A and Xie Z: Involvement of
Src and epidermal growth factor receptor in the signal-transducing
function of Na+/K+-ATPase. J Biol Chem. 275:27832–27837. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu H and Jove R: The STATs of cancer-new
molecular targets come of age. Nat Rev Cancer. 4:97–105. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yeatman TJ: A renaissance for SRC. Nat Rev
Cancer. 4:470–480. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lau GM, Lau GM, Yu GL, Gelman IH, Gutowski
A, Hangauer D and Fang JW: Expression of Src and FAK in
hepatocellular carcinoma and the effect of Src inhibitors on
hepatocellular carcinoma in vitro. Dig Dis Sci. 54:1465–1475. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yoshida T, Zhang G, Smith MA, Lopez AS,
Bai Y, Li J, Fang B, Koomen J, Rawal B, Fisher KJ, et al: Tyrosine
phosphoproteomics identifies both codrivers and cotargeting
strategies for T790M-related EGFR-TKI resistance in non-small cell
lung cancer. Clin Cancer Res. 20:4059–4074. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Debiais F, Lemonnier J, Hay E, Delannoy P,
Caverzasio J and Marie PJ: Fibroblast growth factor-2 (FGF-2)
increases N-cadherin expression through protein kinase C and
Src-kinase pathways in human calvaria osteoblasts. J Cell Biochem.
81:68–81. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Talpaz M, Shah NP, Kantarjian H, Donato N,
Nicoll J, Paquette R, Cortes J, O'Brien S, Nicaise C, Bleickardt E,
et al: Dasatinib in imatinib-resistant Philadelphia
chromosome-positive leukemias. N Engl J Med. 354:2531–2541. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ottmann O, Dombret H, Martinelli G,
Simonsson B, Guilhot F, Larson RA, Rege-Cambrin G, Radich J,
Hochhaus A, Apanovitch AM, et al: Dasatinib induces rapid
hematologic and cytogenetic responses in adult patients with
Philadelphia chromosome positive acute lymphoblastic leukemia with
resistance or intolerance to imatinib: Interim results of a phase 2
study. Blood. 110:2309–2315. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lombardo LJ, Lee FY, Chen P, Norris D,
Barrish JC, Behnia K, Castaneda S, Cornelius LA, Das J, Doweyko AM,
et al: Discovery of N-(2-chloro-6-methyl-
phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino)
thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase
inhibitor with potent antitumor activity in preclinical assays. J
Med Chem. 47:6658–6661. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Murakami Y, Sonoda K, Abe H, Watari K,
Kusakabe D, Azuma K, Kawahara A, Akiba J, Oneyama C, Pachter JA, et
al: The activation of SRC family kinases and focal adhesion kinase
with the loss of the amplified, mutated EGFR gene contributes to
the resistance to afatinib, erlotinib and osimertinib in human lung
cancer cells. Oncotarget. 8:70736–70751. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chang AY and Wang M: Molecular mechanisms
of action and potential biomarkers of growth inhibition of
dasatinib (BMS-354825) on hepatocellular carcinoma cells. BMC
Cancer. 13:2672013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao H, Lv F, Liang G, Huang X, Wu G,
Zhang W, Yu L, Shi L and Teng Y: FGF19 promotes
epithelial-mesenchymal transition in hepatocellular carcinoma cells
by modulating the GSK3β/β-catenin signaling cascade via FGFR4
activation. Oncotarget. 7:13575–13586. 2016. View Article : Google Scholar : PubMed/NCBI
|