Cancer is not only a major public health problem
worldwide, but also one of the leading causes of human mortality.
Host-microbe immune interactions profoundly influence cancer
development, progression and treatment outcomes (1–11).
Fungi and bacteria co-colonize the mammalian skin epithelium,
respiratory tract, gastrointestinal tract and reproductive organs,
forming a complex ecosystem of microbe-microbe and host-microbe
interactions that significantly impact human health (12–21).
Although only ~0.1% of microbial DNA is present in the gut
(22), fungal infections cause more
than 1.5 million deaths worldwide each year (23).
There is growing evidence linking the human
microbiome (bacteria, fungi and viruses) to cancer and cancer
outcomes (24,25). In recent years, several bacteria
have been observed to be associated with cancer development and
progression. Helicobacter pylori infection is the strongest
risk factor for the development of malignant tumors in the stomach,
and epidemiologic studies have determined that the attributable
risk of gastric cancer due to Helicobacter pylori is ~75%
(26). At the same time, in the
lower gastrointestinal tract, genotoxic Escherichia coli,
Bacteroides fragilis, Streptococcus bovis and Fusobacterium
nucleatum are associated with the pathogenesis of colorectal
cancer (27). Among these
cancer-associated bacteria, they can modulate host immunity and
cause chronic inflammation, which is thought to have oncogenic
effects. Recent reports have also shown that bacterial DNA
circulating in the blood of cancer patients can be used as a
predictive biomarker for tumors (28,29)
and intracellular bacteria have been found in numerous tumor types
(30).
In recent times, scientists have been exploring the
link between cancer and bacteria and viruses, but few studies have
focused on the relationship between fungi and cancer. Although the
human fungal biome is less abundant than the bacterial group, it
can still significantly affect human health (31–33).
Previous studies have demonstrated that fungal DNA is present in
most human cancer tissues. However, the role and impact of the
tumor mycobiome on cancer pathogenesis remain largely unknown.
Fungal biome imbalance was found in patients with
gastric cancer. The fungal biome characteristics of patients with
gastric cancer differed significantly from controls, with reduced
diversity and enrichment of Candida and Alternaria in
gastric cancer tissues (36).
Colorectal fungi were altered in patients with colorectal cancer
compared to normal subjects. Patients with colorectal cancer had
increased Malasseziomycetes, decreased
Saccharomycetes and a distorted ratio of
Basidiomycota to Ascomycota (37). The distribution of the fungal genera
Aspergillus, Malassezia, Rhodotorula, Pseudogymnoascus,
Kwoniella, Talaromyces, Debaryomyces, Moniliophthora,
Pneumocystis and Nosemia was altered in colorectal
cancer, as verified in independent Chinese and European cohorts
(38).
Analysis of the tumor mycobiome in various body
parts showed a maximum of 1 fungal cell per 10,000 tumor cells
(39). In The Cancer Genome Atlas
(TCGA) primary tumors, the average relative abundance of bacteria
and fungi was 96 and 4%, respectively, which confirms the lower
abundance of fungi compared to bacteria (34). It has been found that there is a
symbiotic rather than competitive ecological interaction between
fungal and bacterial biomes in the tumor microenvironment (35). However, this differs from the
manifestation of alternating fungal and bacterial populations in
the gastrointestinal tract (9,40).
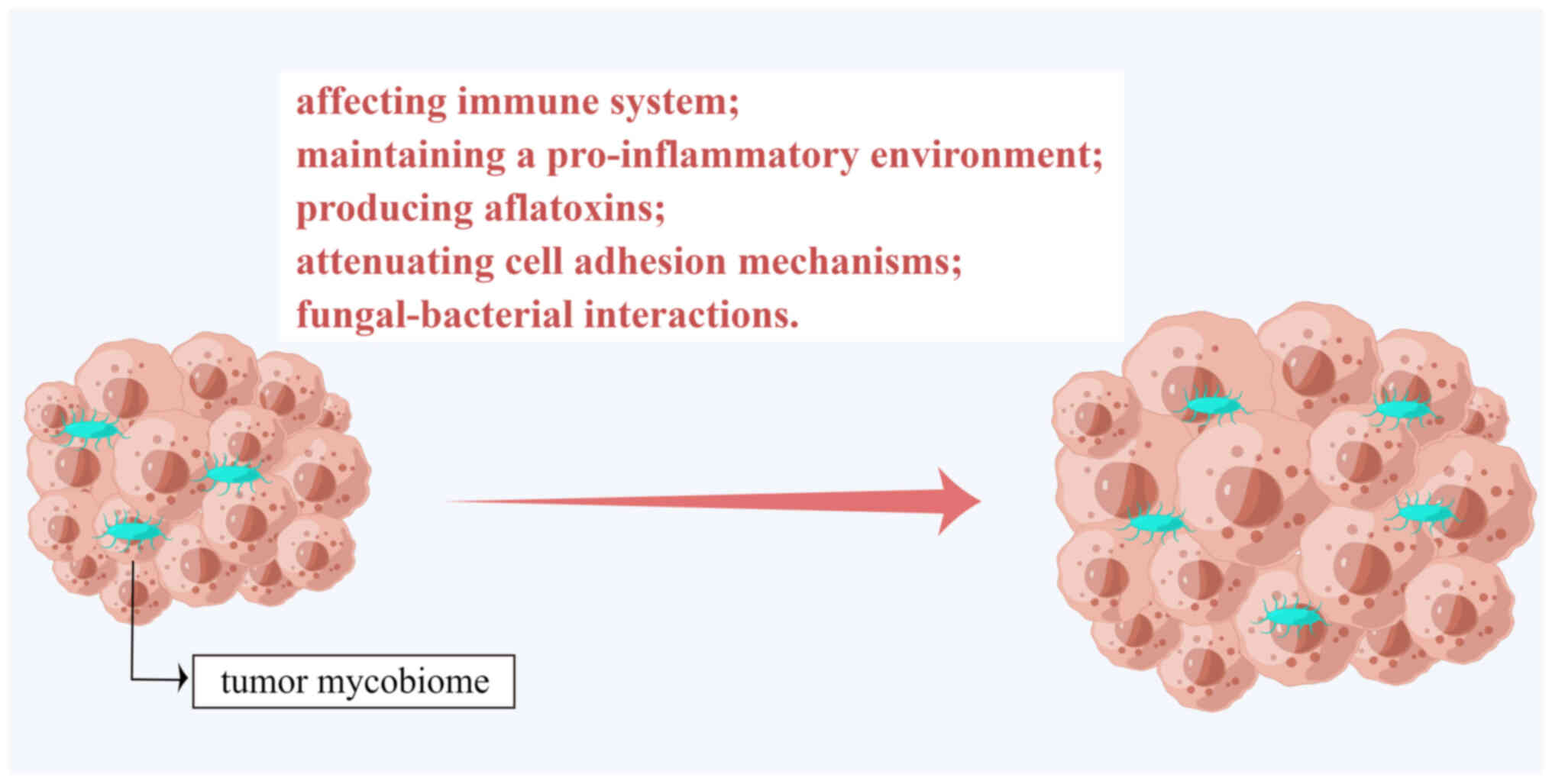
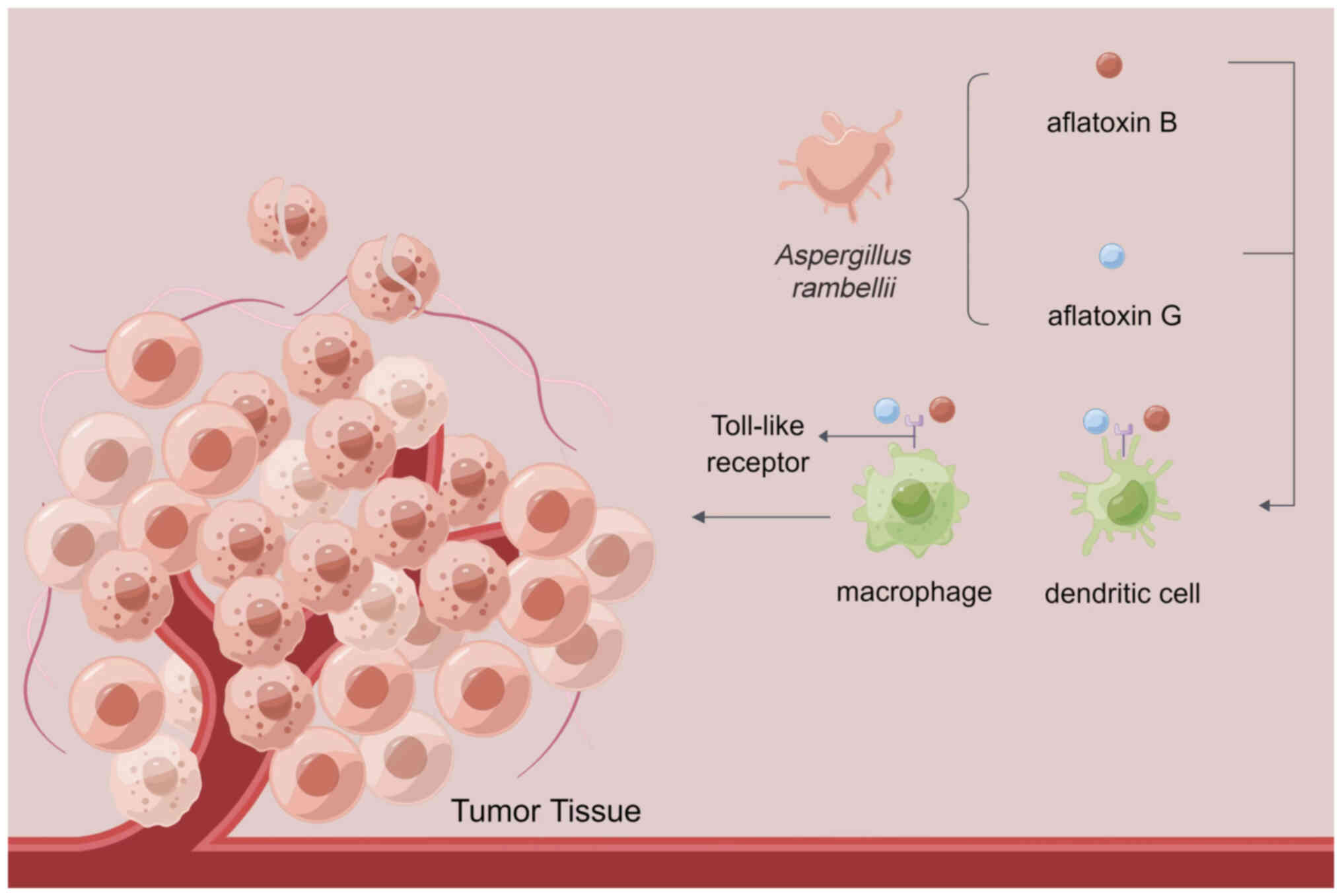
The tumor mycobiome not only resides in tumors but
also promotes tumor progression and metastasis and spreads
systemically by affecting the human immune system, maintaining a
pro-inflammatory environment, producing aflatoxins, attenuating
cell adhesion mechanisms and fungal-bacterial interactions
(Fig. 1).
As numerous cancer patients are immunosuppressed,
they are more susceptible to fungal infections, which may further
aggravate their condition (41). It
has been demonstrated that fungal-driven pancreatic cancer occurs
through complement cascade activation and IL-33 secretion (35). Aykut et al (31) have shown that Malassezia can
secrete hydrolases to release host lipids and activate the C3
complement mannose-binding lectin pathway to promote an
immunosuppressive tumor environment in pancreatic cancer. The tumor
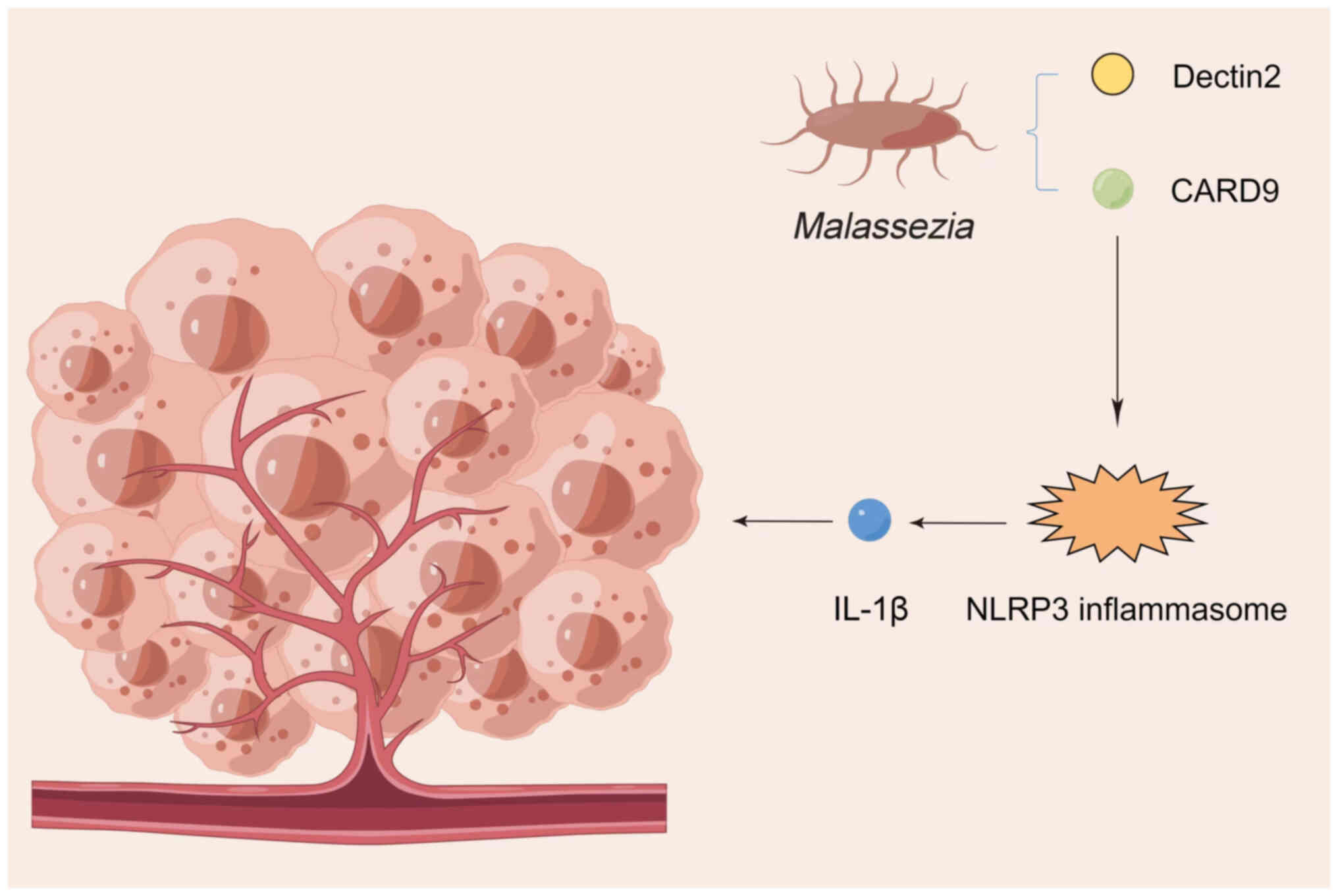
mycobiome activates dectin-1-mediated Src-Syk-caspase recruitment
domain family member 9 (CARD9) signaling in the pancreas, leading
to IL-33 secretion and tumor growth, and thus, this may be the
mechanism by which the tumor mycobiome promotes pancreatic cancer
progression (42).
The Colorectal Cancer Risk Factor Assessment report
indicated that the human body has an inadequate immune response to
fungi, such as inflammatory bowel disease (43) and ulcerative colitis (12). Antifungal treatment has been
reported to exacerbate colitis and colorectal cancer, while colonic
fungi enhance azoxymethane/dextran sodium sulfate-induced
inflammatory vesicle activation in colitis (44).
There are numerous hypotheses about tumor
pathogenesis, among which the theory of an inflammatory mechanism
is a widely accepted hypothesis. Inflammation is usually the basis
for resistance to harmful stimuli, accelerating wound recovery and
maintaining normal tissue function, and its role involves
endothelial cells, immune cells and inflammatory factors (45).
Colorectal cancer is the third most common cancer
type worldwide, with >500,000 related deaths per year (59,60).
The contribution of the intestinal flora to colorectal cancer
progression has been widely recognized (61–63).
Intestinal fungi constitute a significant component of the human
intestinal flora, but their role in colorectal cancer has remained
elusive (64). Several studies have
confirmed a correlation between intestinal fungi and colon cancer
(38,44,65–67).
Through extensive genomic analysis, tumor
fungal-bacterial biome interactions can promote colorectal
carcinogenesis through the upregulation of D-arginine and
D-ornithine metabolic pathways and stimulation of the butanoate
metabolic pathway (77). Liu et
al (77) demonstrated that two
marker genes, oraS and oraE, in the D-arginine and D-ornithine
metabolic pathways were upregulated in colorectal cancer. The
butanoate metabolic pathway, which is strongly activated in
colorectal cancer but less studied, has also been identified
(78). Tumor fungal-bacterial biome
interactions promote colorectal cancer progression through
upregulation of bdhA and bdhB gene expression in the
butanoate metabolic pathway (77).
Therefore, butanoate in the butanoate metabolic pathway is crucial
in supporting the tumor microenvironment (79). Tumor fungal-bacterial biome
interactions are being explored as an effective means of
maintaining homeostasis in the gut.
Numerous studies have indicated the potential of
bacteria as biomarkers for the diagnosis of colorectal cancer
(64). Wirbel et al
(80) and Thomas et al
(81) performed meta-analyses to
identify several bacteria enriched in colorectal cancer with
utility as diagnostic biomarkers for colorectal cancer.
By contrast, with the study of the tumor microbiome,
fungi, in addition to bacteria, may be used as biomarkers for the
noninvasive diagnosis of tumor patients. In TCGA cohorts, fungal
biome richness varied significantly among cancers (35). For instance, Candida,
Saccharomyces cerevisiae and Cyberlindnera jadinii are
highly abundant in the gastrointestinal tumor fungal organism
community and Blastomyces and Malassezia are highly
abundant in lung cancer and breast cancer, respectively (39).
A further study performed qualitative and
quantitative analyses of 20 different fungal DNAs released into the
bloodstream and the results suggested that they may be used to
distinguish patients with cancer from healthy individuals, even in
early-stage disease (35). This
suggests that the tumor fungal biome has utility in cancer
diagnosis.
The use of antimicrobial agents that target known
pathogenic microorganisms effectively prevents the onset and
progression of the disease. The use of targeted antifungal agents
is helpful in the prevention or treatment of gastrointestinal
cancers (37). In a mouse model of
human pancreatic ductal adenocarcinoma, Malassezia
infiltrates and accelerates the progression of human pancreatic
ductal adenocarcinoma, a condition that can be reversed by
antifungal treatment (31).
Antifungal treatment targeting Malassezia resulted in a 40%
reduction in the incidence of pancreatic cancer in mice (31). In a mouse model of esophageal
cancer, the oral fungus Dictyostelium significantly
increased the severity of esophageal squamous cell carcinoma, a
condition that could be reversed by antifungal treatment (82).
In addition, the use of fungal probiotics can
prevent and treat gastrointestinal cancers. Probiotics are
microorganisms that improve health when consumed in the correct
amounts. The most common probiotics are bacteria that have been
shown to inhibit the proliferation of pathogenic intestinal
microorganisms and to prevent carcinogenic inflammation in the
esophagus, stomach, pancreas and colorectum (83–85).
By contrast, fungi can also be ingested as probiotics and have been
reported to be used to alleviate gastrointestinal cancers (37). The Helicobacter pylori
eradication rate improved when Saccharomyces boulardii was
combined with Lactobacillus gasseri, Lactobacillus reuteri,
Lactobacillus acidophilus, Streptococcus faecalis, Bacillus
subtilis and Bifidobacterium (86).
The use of fungal probiotics in treating
gastrointestinal cancers is of great importance. Because of their
characteristic cellular structure, they can survive in the
unfavorable environment of the gastrointestinal tract (87,88).
As more research on the efficacy and safety of fungal probiotics is
conducted, they may directly or indirectly modulate the tumor
microbiome to prevent and treat gastrointestinal cancers (37).
With the above summary and analysis, the fungal
biome that targets and attacks tumors is likely to be a good way to
treat cancer.
This review provides a comprehensive summary of the
impact of the tumor mycobiome on cancer pathogenesis. The tumor
mycobiome promotes tumor progression and metastasis by affecting
the human immune system, maintaining a pro-inflammatory
environment, producing aflatoxins, attenuating cellular adhesion
mechanisms and fungal-bacterial interactions. Furthermore, the
tumor mycobiome also has tremendous potential for cancer
prevention, diagnosis and treatment.
Although studies on the effect of the tumor
mycobiome on cancer pathogenesis have become more frequent, the
results are not uniform because of the differences between
different populations and inconsistent standards for metagenomic
data generation and processing. The future development of
standardized and low-cost sequencing technologies and pipeline
analysis methods to improve the quality of data collection and
analytical processing, and the initiation of longitudinal studies
with large sample sizes in different populations to clarify the
specific mechanistic relationships, will be crucial for research in
this field.
Not applicable.
This study was supported by a grant from the Natural Science
Foundation of Shandong (grant no. ZR2017LH050).
Not applicable.
WL wrote the manuscript, searched the literature and
prepared the figures. ZL was involved in the design of the study
and revised the manuscript. BH provided ideas to improve the
article, modified the figures and revised the manuscript. XL, HC,
HJ and QN performed the literature search and selected relevant
articles. Data authentication is not applicable. All authors have
read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Davar D, Dzutsev AK, McCulloch JA,
Rodrigues RR, Chauvin JM, Morrison RM, Deblasio RN, Menna C, Ding
Q, Pagliano O, et al: Fecal microbiota transplant overcomes
resistance to anti-PD-1 therapy in melanoma patients. Science.
371:595–602. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dzutsev A, Badger JH, Perez-Chanona E, Roy
S, Salcedo R, Smith CK and Trinchieri G: Microbes and cancer. Annu
Rev Immunol. 35:199–228. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Finlay BB, Goldszmid R, Honda K,
Trinchieri G, Wargo J and Zitvogel L: Can we harness the microbiota
to enhance the efficacy of cancer immunotherapy? Nat Rev Immunol.
20:522–528. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garrett WS: The gut microbiota and colon
cancer. Science. 364:1133–1135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iida N, Dzutsev A, Stewart CA, Smith L,
Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S,
et al: Commensal bacteria control cancer response to therapy by
modulating the tumor microenvironment. Science. 342:967–970. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Routy B, Le Chatelier E, Derosa L, Duong
CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C,
Roberti MP, et al: Gut microbiome influences efficacy of PD-1-based
immunotherapy against epithelial tumors. Science. 359:91–97. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sharma P, Hu-Lieskovan S, Wargo JA and
Ribas A: Primary, adaptive, and acquired resistance to cancer
immunotherapy. Cell. 168:707–723. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shiao SL, Kershaw KM, Limon JJ, You S,
Yoon J, Ko EY, Guarnerio J, Potdar AA, McGovern DPB, Bose S, et al:
Commensal bacteria and fungi differentially regulate tumor
responses to radiation therapy. Cancer cell. 39:1202–1213.e6. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spencer CN, McQuade JL, Gopalakrishnan V,
McCulloch JA, Vetizou M, Cogdill AP, Khan MAW, Zhang X, White MG,
Peterson CB, et al: Dietary fiber and probiotics influence the gut
microbiome and melanoma immunotherapy response. Science.
374:1632–1640. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tanoue T, Morita S, Plichta DR, Skelly AN,
Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, et
al: A defined commensal consortium elicits CD8 T cells and
anti-cancer immunity. Nature. 565:600–605. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sokol H, Leducq V, Aschard H, Pham HP,
Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier
I, et al: Fungal microbiota dysbiosis in IBD. Gut. 66:1039–1048.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Findley K, Oh J, Yang J, Conlan S, Deming
C, Meyer JA, Schoenfeld D, Nomicos E, Park M; NIH Intramural
Sequencing Center Comparative Sequencing Program, ; et al:
Topographic diversity of fungal and bacterial communities in human
skin. Nature. 498:367–370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hoarau G, Mukherjee PK, Gower-Rousseau C,
Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J,
Colombel JF, et al: Bacteriome and mycobiome interactions
underscore microbial dysbiosis in familial Crohn's disease. mBio.
7:e01250–16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leonardi I, Paramsothy S, Doron I, Semon
A, Kaakoush NO, Clemente JC, Faith JJ, Borody TJ, Mitchell HM,
Colombel JF, et al: Fungal trans-kingdom dynamics linked to
responsiveness to fecal microbiota transplantation (FMT) therapy in
ulcerative colitis. Cell Host Microbe. 27:823–829.e3. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Doron I, Mesko M, Li XV, Kusakabe T,
Leonardi I, Shaw DG, Fiers WD, Lin WY, Bialt-DeCelie M, Román E, et
al: Mycobiota-induced IgA antibodies regulate fungal commensalism
in the gut and are dysregulated in Crohn's disease. Nat Microbiol.
6:1493–1504. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lewis JD, Chen EZ, Baldassano RN, Otley
AR, Griffiths AM, Lee D, Bittinger K, Bailey A, Friedman ES,
Hoffmann C, et al: Inflammation, antibiotics, and diet as
environmental stressors of the gut microbiome in pediatric Crohn's
disease. Cell Host Microbe. 18:489–500. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liguori G, Lamas B, Richard ML, Brandi G,
da Costa G, Hoffmann TW, Di Simone MP, Calabrese C, Poggioli G,
Langella P, et al: Fungal dysbiosis in mucosa-associated microbiota
of Crohn's disease patients. J Crohns Colitis. 10:296–305. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tipton L, Müller CL, Kurtz ZD, Huang L,
Kleerup E, Morris A, Bonneau R and Ghedin E: Fungi stabilize
connectivity in the lung and skin microbial ecosystems. Microbiome.
6:122018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhai B, Ola M, Rolling T, Tosini NL,
Joshowitz S, Littmann ER, Amoretti LA, Fontana E, Wright RJ,
Miranda E, et al: High-resolution mycobiota analysis reveals
dynamic intestinal translocation preceding invasive candidiasis.
Nat Med. 26:59–64. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zuo T, Wong SH, Cheung CP, Lam K, Lui R,
Cheung K, Zhang F, Tang W, Ching JYL, Wu JCY, et al: Gut fungal
dysbiosis correlates with reduced efficacy of fecal microbiota
transplantation in Clostridium difficile infection. Nat Commun.
9:36632018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qin J, Li R, Raes J, Arumugam M, Burgdorf
KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al: A
human gut microbial gene catalogue established by metagenomic
sequencing. Nature. 464:59–65. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Brown GD, Denning DW, Gow NA, Levitz SM,
Netea MG and White TC: Hidden killers: Human fungal infections. Sci
Transl Med. 4:165rv132012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Helmink BA, Khan MAW, Hermann A,
Gopalakrishnan V and Wargo JA: The microbiome, cancer, and cancer
therapy. Nat Med. 25:377–388. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vogtmann E and Goedert JJ: Epidemiologic
studies of the human microbiome and cancer. Br J Cancer.
114:237–242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Polk DB and Peek RM Jr: Helicobacter
pylori: Gastric cancer and beyond. Nat Rev Cancer. 10:403–414.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sepich-Poore GD, Zitvogel L, Straussman R,
Hasty J, Wargo JA and Knight R: The microbiome and human cancer.
Science. 371:eabc45522021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Poore GD, Kopylova E, Zhu Q, Carpenter C,
Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ,
et al: Microbiome analyses of blood and tissues suggest cancer
diagnostic approach. Nature. 579:567–574. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dohlman AB, Arguijo Mendoza D, Ding S, Gao
M, Dressman H, Iliev ID, Lipkin SM and Shen X: The cancer
microbiome atlas: A pan-cancer comparative analysis to distinguish
tissue-resident microbiota from contaminants. Cell Host Microbe.
29:281–298.e5. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nejman D, Livyatan I, Fuks G, Gavert N,
Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E,
et al: The human tumor microbiome is composed of tumor
type-specific intracellular bacteria. Science. 368:973–980. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Aykut B, Pushalkar S, Chen R, Li Q,
Abengozar R, Kim JI, Shadaloey SA, Wu D, Preiss P, Verma N, et al:
The fungal mycobiome promotes pancreatic oncogenesis via activation
of MBL. Nature. 574:264–267. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Elaskandrany M, Patel R, Patel M, Miller
G, Saxena D and Saxena A: Fungi, host immune response, and
tumorigenesis. Am J Physiol Gastrointest Liver Physiol.
321:G213–G222. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Iliev ID and Leonardi I: Fungal dysbiosis:
Immunity and interactions at mucosal barriers. Nat Rev Immunol.
17:635–646. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Narunsky-Haziza L, Sepich-Poore GD,
Livyatan I, Asraf O, Martino C, Nejman D, Gavert N, Stajich JE,
Amit G, González A, et al: Pan-cancer analyses reveal
cancer-type-specific fungal ecologies and bacteriome interactions.
Cell. 185:3789–3806.e17. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X and Saxena D: The tumor mycobiome: A
paradigm shift in cancer pathogenesis. Cell. 185:3648–3651. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhong M, Xiong Y, Zhao J, Gao Z, Ma J, Wu
Z, Song Y and Hong X: Candida albicans disorder is
associated with gastric carcinogenesis. Theranostics. 11:4945–4956.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Coker OO: Non-bacteria microbiome (virus,
fungi, and archaea) in gastrointestinal cancer. J Gastroenterol
Hepatol. 37:256–262. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong
SH, Ng SC, Chan FKL, Sung JJY and Yu J: Enteric fungal microbiota
dysbiosis and ecological alterations in colorectal cancer. Gut.
68:654–662. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dohlman AB, Klug J, Mesko M, Gao IH,
Lipkin SM, Shen X and Iliev ID: A pan-cancer mycobiome analysis
reveals fungal involvement in gastrointestinal and lung tumors.
Cell. 185:3807–3822.e12. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Seelbinder B, Chen J, Brunke S,
Vazquez-Uribe R, Santhaman R, Meyer AC, de Oliveira Lino FS, Chan
KF, Loos D, Imamovic L, et al: Antibiotics create a shift from
mutualism to competition in human gut communities with a
longer-lasting impact on fungi than bacteria. Microbiome.
8:1332020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Viscoli C, Castagnola E and Machetti M:
Antifungal treatment in patients with cancer. J Intern Med Suppl.
740:89–94. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Alam A, Levanduski E, Denz P,
Villavicencio HS, Bhatta M, Alhorebi L, Zhang Y, Gomez EC, Morreale
B, Senchanthisai S, et al: Fungal mycobiome drives IL-33 secretion
and type 2 immunity in pancreatic cancer. Cancer Cell.
40:153–167.e11. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qiu X, Zhang F, Yang X, Wu N, Jiang W, Li
X, Li X and Liu Y: Changes in the composition of intestinal fungi
and their role in mice with dextran sulfate sodium-induced colitis.
Sci Rep. 5:104162015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Malik A, Sharma D, Malireddi RKS, Guy CS,
Chang TC, Olsen SR, Neale G, Vogel P and Kanneganti TD: SYK-CARD9
signaling axis promotes gut fungi-mediated inflammasome activation
to restrict colitis and colon cancer. Immunity. 49:515–530.e5.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Medzhitov R: Origin and physiological
roles of inflammation. Nature. 454:428–435. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bannenberg GL, Chiang N, Ariel A, Arita M,
Tjonahen E, Gotlinger KH, Hong S and Serhan CN: Molecular circuits
of resolution: Formation and actions of resolvins and protectins. J
Immunol. 174:4345–4355. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gordon S: Phagocytosis: An immunobiologic
process. Immunity. 44:463–475. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bishehsari F, Engen PA, Preite NZ, Tuncil
YE, Naqib A, Shaikh M, Rossi M, Wilber S, Green SJ, Hamaker BR, et
al: Dietary fiber treatment corrects the composition of gut
microbiota, promotes SCFA production, and suppresses colon
carcinogenesis. Genes (Basel). 9:1022018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Singh N, Baby D, Rajguru JP, Patil PB,
Thakkannavar SS and Pujari VB: Inflammation and cancer. Ann Afr
Med. 18:121–126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Naylor MS, Stamp GW, Foulkes WD, Eccles D
and Balkwill FR: Tumor necrosis factor and its receptors in human
ovarian cancer. Potential role in disease progression. J Clin
Invest. 91:2194–2206. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Haghnegahdar H, Du J, Wang D, Strieter RM,
Burdick MD, Nanney LB, Cardwell N, Luan J, Shattuck-Brandt R and
Richmond A: The tumorigenic and angiogenic effects of MGSA/GRO
proteins in melanoma. J Leukoc Biol. 67:53–62. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Thun MJ, Namboodiri MM, Calle EE, Flanders
WD and Heath CW Jr: Aspirin use and risk of fatal cancer. Cancer
Res. 53:1322–1327. 1993.PubMed/NCBI
|
|
54
|
Yang Q, Ouyang J, Pi D, Feng L and Yang J:
Malassezia in inflammatory bowel disease: Accomplice of
evoking tumorigenesis. Front Immunol. 13:8464692022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wolf AJ, Limon JJ, Nguyen C, Prince A,
Castro A and Underhill DM: Malassezia spp. induce
inflammatory cytokines and activate NLRP3 inflammasomes in
phagocytes. J Leukoc Biol. 109:161–172. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang YJ, Han Y, Sun YZ, Jiang HH, Liu M,
Qi RQ and Gao XH: Extracellular vesicles derived from
Malassezia furfur stimulate IL-6 production in keratinocytes
as demonstrated in in vitro and in vivo models. J Dermatol Sci.
93:168–175. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Berti M and Vindigni A: Replication
stress: Getting back on track. Nat Struct Mol Biol. 23:103–109.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kawanishi S, Ohnishi S, Ma N, Hiraku Y and
Murata M: Crosstalk between DNA damage and inflammation in the
multiple steps of carcinogenesis. Int J Mol Sci. 18:18082017.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lin Y, Wang G, Yu J and Sung JJY:
Artificial intelligence and metagenomics in intestinal diseases. J
Gastroenterol Hepatol. 36:841–847. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wong SH and Yu J: Gut microbiota in
colorectal cancer: Mechanisms of action and clinical applications.
Nat Rev Gastroenterol Hepatol. 16:690–704. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Perrone G and Gallo A: Aspergillus
species and their associated mycotoxins. Methods Mol Biol.
1542:33–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao
L, Chen Z, Chan FKL, Kristiansen K, Sung JJY, Wong SH and Yu J:
Multi-cohort analysis of colorectal cancer metagenome identified
altered bacteria across populations and universal bacterial
markers. Microbiome. 6:702018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lin Y, Lau HC, Liu Y, Kang X, Wang Y, Ting
NL, Kwong TN, Han J, Liu W, Liu C, et al: Altered mycobiota
signatures and enriched pathogenic Aspergillus rambellii are
associated with colorectal cancer based on multicohort fecal
metagenomic analyses. Gastroenterology. 163:908–921. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Luan C, Xie L, Yang X, Miao H, Lv N, Zhang
R, Xiao X, Hu Y, Liu Y, Wu N, et al: Dysbiosis of fungal microbiota
in the intestinal mucosa of patients with colorectal adenomas. Sci
Rep. 5:79802015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Gao R, Kong C, Li H, Huang L, Qu X, Qin N
and Qin H: Dysbiosis signature of mycobiota in colon polyp and
colorectal cancer. Eur J Clin Microbiol Infect Dis. 36:2457–2468.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Richard ML, Liguori G, Lamas B, Brandi G,
da Costa G, Hoffmann TW, Pierluigi Di Simone M, Calabrese C,
Poggioli G, et al: Mucosa-associated microbiota dysbiosis in
colitis associated cancer. Gut Microbes. 9:131–142. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cary JW, Ehrlich KC, Beltz SB,
Harris-Coward P and Klich MA: Characterization of the
Aspergillus ochraceoroseus aflatoxin/sterigmatocystin
biosynthetic gene cluster. Mycologia. 101:352–362. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Frisvad JC, Skouboe P and Samson RA:
Taxonomic comparison of three different groups of aflatoxin
producers and a new efficient producer of aflatoxin B1,
sterigmatocystin and 3-O-methylsterigmatocystin, Aspergillus
rambellii sp. nov. Syst Appl Microbiol. 28:442–453. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Navale V, Vamkudoth KR, Ajmera S and Dhuri
V: Aspergillus derived mycotoxins in food and the
environment: Prevalence, detection, and toxicity. Toxicol Rep.
8:1008–1030. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Uka V, Cary JW, Lebar MD, Puel O, De
Saeger S and Diana Di Mavungu J: Chemical repertoire and
biosynthetic machinery of the Aspergillus flavus secondary
metabolome: A review. Compr Rev Food Sci Food Saf. 19:2797–2842.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
McCullough AK and Lloyd RS: Mechanisms
underlying aflatoxin-associated mutagenesis-implications in
carcinogenesis. DNA Repair (Amst). 77:76–86. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Bianco G, Russo R, Marzocco S, Velotto S,
Autore G and Severino L: Modulation of macrophage activity by
aflatoxins B1 and B2 and their metabolites aflatoxins M1 and M2.
Toxicon. 59:644–650. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Mohammadi A, Mehrzad J, Mahmoudi M and
Schneider M: Environmentally relevant level of aflatoxin B1
dysregulates human dendritic cells through signaling on key
toll-like receptors. Int J Toxicol. 33:175–186. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Soler AP, Miller RD, Laughlin KV, Carp NZ,
Klurfeld DM and Mullin JM: Increased tight junctional permeability
is associated with the development of colon cancer. Carcinogenesis.
20:1425–1431. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Martin TA and Jiang WG: Loss of tight
junction barrier function and its role in cancer metastasis.
Biochim Biophys Acta. 1788:872–891. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Liu NN, Jiao N, Tan JC, Wang Z, Wu D, Wang
AJ, Chen J, Tao L, Zhou C, Fang W, et al: Multi-kingdom microbiota
analyses identify bacterial-fungal interactions and biomarkers of
colorectal cancer across cohorts. Nat Microbiol. 7:238–250. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Gmeiner WH, Hellmann GM and Shen P:
Tissue-dependent and -independent gene expression changes in
metastatic colon cancer. Oncol Rep. 19:245–251. 2008.PubMed/NCBI
|
|
79
|
Tjalsma H, Boleij A, Marchesi JR and
Dutilh BE: A bacterial driver-passenger model for colorectal
cancer: Beyond the usual suspects. Nat Rev Microbiol. 10:575–582.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wirbel J, Pyl PT, Kartal E, Zych K,
Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R,
et al: Meta-analysis of fecal metagenomes reveals global microbial
signatures that are specific for colorectal cancer. Nat Med.
25:679–689. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Thomas AM, Manghi P, Asnicar F, Pasolli E,
Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, et
al: Metagenomic analysis of colorectal cancer datasets identifies
cross-cohort microbial diagnostic signatures and a link with
choline degradation. Nat Med. 25:667–678. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhu F, Willette-Brown J, Song NY, Lomada
D, Song Y, Xue L, Gray Z, Zhao Z, Davis SR, Sun Z, et al:
Autoreactive T cells and chronic fungal infection drive esophageal
carcinogenesis. Cell Host Microbe. 21:478–493.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Azad MAK, Sarker M, Li T and Yin J:
Probiotic species in the modulation of gut microbiota: An overview.
Biomed Res Int. 2018:94786302018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Mozaffari Namin B, Daryani NE, Mirshafiey
A, Yazdi MKS and Dallal MMS: Effect of probiotics on the expression
of Barrett's oesophagus biomarkers. J Med Microbiol. 64:348–354.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Rosania R, Minenna MF, Giorgio F,
Facciorusso A, De Francesco V, Hassan C, Panella C and Ierardi E:
Probiotic multistrain treatment may eradicate Helicobacter
pylori from the stomach of dyspeptics: A placebo-controlled
pilot study. Inflamm Allergy Drug Targets. 11:244–249. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhu R, Chen K, Zheng YY, Zhang HW, Wang
JS, Xia YJ, Dai WQ, Wang F, Shen M, Cheng P, et al: Meta-analysis
of the efficacy of probiotics in Helicobacter pylori
eradication therapy. World J Gastroenterol. 20:18013–18021. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Kumar V, Yadav AN, Verma P, Sangwan P,
Saxena A, Kumar K and Singh B: β-Propeller phytases: Diversity,
catalytic attributes, current developments and potential
biotechnological applications. Int J Biol Macromol. 98:595–609.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Lipke PN and Ovalle R: Cell wall
architecture in yeast: New structure and new challenges. J
Bacteriol. 180:3735–3740. 1998. View Article : Google Scholar : PubMed/NCBI
|