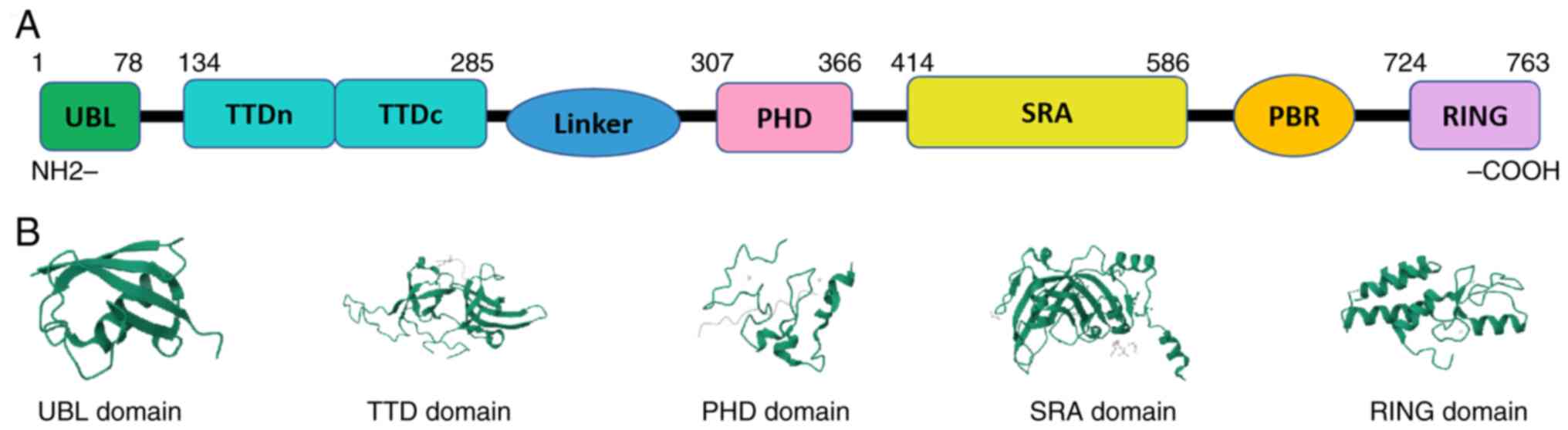
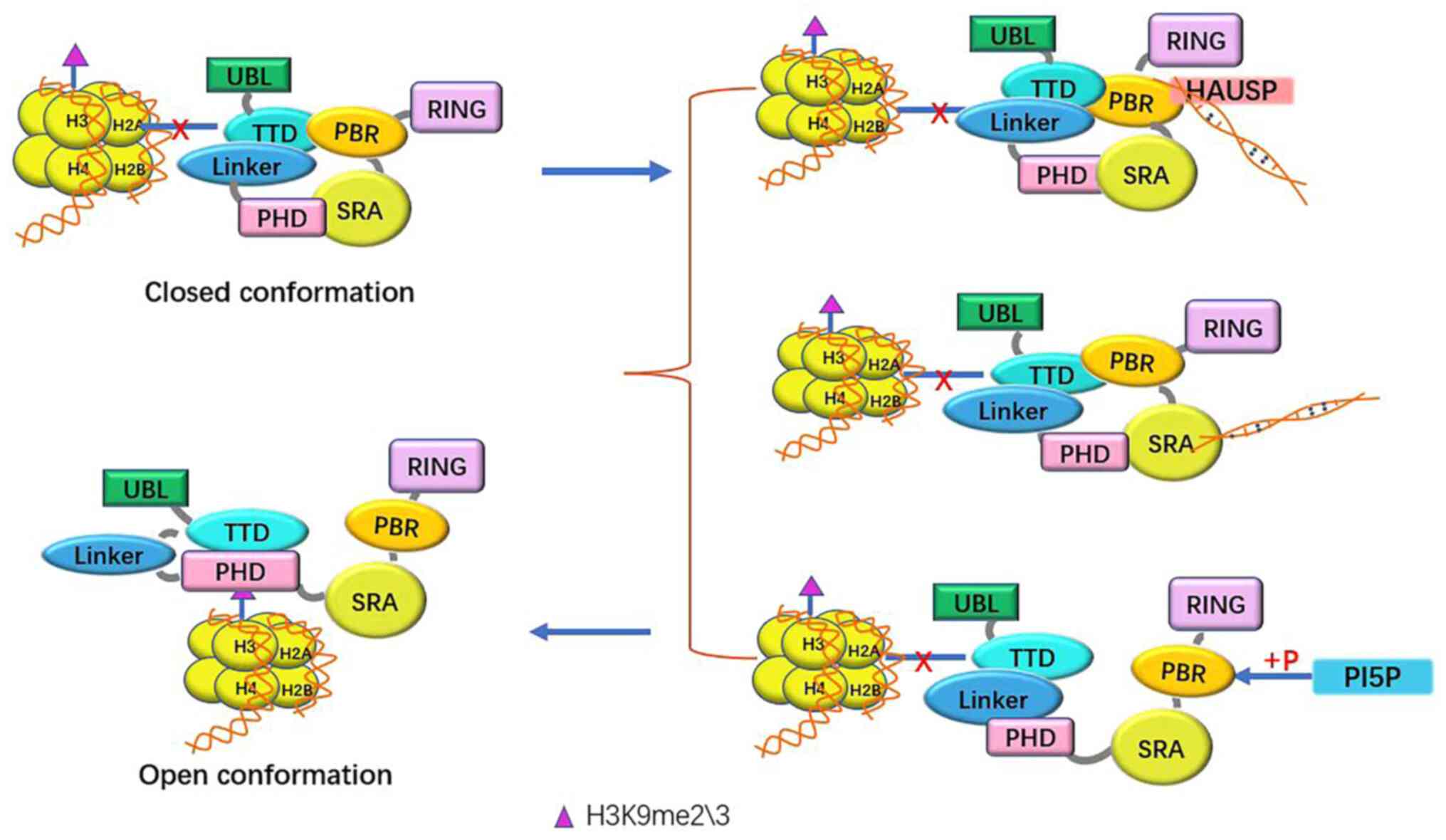
|
1
|
Probst AV, Dunleavy E and Almouzni G:
Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell
Biol. 10:192–206. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zafon C, Gil J, Pérez-González B and Jordà
M: DNA methylation in thyroid cancer. Endocr Relat Cancer.
26:R415–R439. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abhishek S, Nakarakanti NK, Deeksha W and
Rajakumara E: Mechanistic insights into recognition of symmetric
methylated cytosines in CpG and non-CpG DNA by UHRF1 SRA. Int J
Biol Macromol. 170:514–522. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bronner C, Krifa M and Mousli M:
Increasing role of UHRF1 in the reading and inheritance of the
epigenetic code as well as in tumorogenesis. Biochem Pharmacol.
86:1643–1649. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sheng Y, Wang H, Liu D and Zhang C, Deng
Y, Yang F, Zhang T and Zhang C: Methylation of tumor suppressor
gene CDH13 and SHP1 promoters and their epigenetic regulation by
the UHRF1/PRMT5 complex in endometrial carcinoma. Gynecol Oncol.
140:145–151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Daskalos A, Oleksiewicz U, Filia A,
Nikolaidis G, Xinarianos G, Gosney JR, Malliri A, Field JK and
Liloglou T: UHRF1-mediated tumor suppressor gene inactivation in
nonsmall cell lung cancer. Cancer. 117:1027–1037. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mudbhary R, Hoshida Y, Chernyavskaya Y,
Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson
RT, et al: UHRF1 overexpression drives DNA hypomethylation and
hepatocellular carcinoma. Cancer Cell. 25:196–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhuo H, Tang J, Lin Z, Jiang R, Zhang X,
Ji J, Wang P and Sun B: The aberrant expression of MEG3 regulated
by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol
Carcinog. 55:209–219. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reardon ES, Shukla V, Xi S, Gara SK, Liu
Y, Straughan D, Zhang M, Hong JA, Payabyab EC, Kumari A, et al:
UHRF1 is a novel druggable epigenetic target in malignant pleural
mesothelioma. J Thorac Oncol. 16:89–103. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nakamura K, Baba Y, Kosumi K, Harada K,
Shigaki H, Miyake K, Kiyozumi Y, Ohuchi M, Kurashige J, Ishimoto T,
et al: UHRF1 regulates global DNA hypomethylation and is associated
with poor prognosis in esophageal squamous cell carcinoma.
Oncotarget. 7:57821–57831. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hervouet E, Lalier L, Debien E, Cheray M,
Geairon A, Rogniaux H, Loussouarn D, Martin SA, Vallette FM and
Cartron PF: Disruption of Dnmt1/PCNA/UHRF1 interactions promotes
tumorigenesis from human and mice glial cells. PLoS One.
5:e113332010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kan G, He H, Zhao Q, Li X, Li M, Yang H
and Kim JK: Functional dissection of the role of UHRF1 in the
regulation of retinoblastoma methylome. Oncotarget. 8:39497–39511.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Obata Y, Furusawa Y, Endo TA, Sharif J,
Takahashi D, Atarashi K, Nakayama M, Onawa S, Fujimura Y, Takahashi
M, et al: The epigenetic regulator Uhrf1 facilitates the
proliferation and maturation of colonic regulatory T cells. Nat
Immunol. 15:571–579. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Elia L, Kunderfranco P, Carullo P,
Vacchiano M, Farina FM, Hall IF, Mantero S, Panico C, Papait R,
Condorelli G and Quintavalle M: UHRF1 epigenetically orchestrates
smooth muscle cell plasticity in arterial disease. J Clin Invest.
128:2473–2486. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Unoki M, Nishidate T and Nakamura Y:
ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG
through its SRA domain. Oncogene. 23:7601–7610. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jenkins Y, Markovtsov V, Lang W, Sharma P,
Pearsall D, Warner J, Franci C, Huang B, Huang J, Yam GC, et al:
Critical role of the ubiquitin ligase activity of UHRF1, a nuclear
RING finger protein, in tumor cell growth. Mol Biol Cell.
16:5621–569. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Unoki M, Brunet J and Mousli M: Drug
discovery targeting epigenetic codes: The great potential of UHRF1,
which links DNA methylation and histone modifications, as a drug
target in cancers and toxoplasmosis. Biochem Pharmacol.
78:1279–1288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mousli M, Hopfner R, Abbady AQ, Monté D,
Jeanblanc M, Oudet P, Louis B and Bronner C: ICBP90 belongs to a
new family of proteins with an expression that is deregulated in
cancer cells. Br J Cancer. 89:120–127. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Unoki M, Kelly JD, Neal DE, Ponder BA,
Nakamura Y and Hamamoto R: UHRF1 is a novel molecular marker for
diagnosis and the prognosis of bladder cancer. Br J Cancer.
101:98–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Crnogorac-Jurcevic T, Gangeswaran R,
Bhakta V, Capurso G, Lattimore S, Akada M, Sunamura M, Prime W,
Campbell F, Brentnall TA, et al: Proteomic analysis of chronic
pancreatitis and pancreatic adenocarcinoma. Gastroenterology.
129:1454–1463. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lorenzato M, Caudroy S, Bronner C, Evrard
G, Simon M, Durlach A, Birembaut P and Clavel C: Cell cycle and/or
proliferation markers: What is the best method to discriminate
cervical high-grade lesions? Hum Pathol. 36:1101–1107. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Y, Cheng D, Wang Y, Xi S, Wang T, Sun
W, Li G, Ma D, Zhou S, Li Z and Ni C: UHRF1-mediated ferroptosis
promotes pulmonary fibrosis via epigenetic repression of GPX4 and
FSP1 genes. Cell Death Dis. 13:10702022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fang L, Hao Y, Yu H, Gu X, Peng Q, Zhuo H,
Li Y, Liu Z, Wang J, Chen Y, et al: Methionine restriction promotes
cGAS activation and chromatin untethering through demethylation to
enhance antitumor immunity. Cancer Cell. 41:1118–1133. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Achour M, Mousli M, Alhosin M, Ibrahim A,
Peluso J, Muller CD, Schini-Kerth VB, Hamiche A, Dhe-Paganon S and
Bronner C: Epigallocatechin-3-gallate up-regulates tumor suppressor
gene expression via a reactive oxygen species-dependent
down-regulation of UHRF1. Biochem Biophys Res Commun. 430:208–212.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alhosin M, Abusnina A, Achour M, Sharif T,
Muller C, Peluso J, Chataigneau T, Lugnier C, Schini-Kerth VB,
Bronner C and Fuhrmann G: Induction of apoptosis by thymoquinone in
lymphoblastic leukemia Jurkat cells is mediated by a p73-dependent
pathway which targets the epigenetic integrator UHRF1. Biochem
Pharmacol. 79:1251–1260. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alhosin M, Sharif T, Mousli M,
Etienne-Selloum N, Fuhrmann G, Schini-Kerth VB and Bronner C:
Down-regulation of UHRF1, associated with re-expression of tumor
suppressor genes, is a common feature of natural compounds
exhibiting anti-cancer properties. J Exp Clin Cancer Res.
30:412011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krifa M, Alhosin M, Muller CD, Gies JP,
Chekir-Ghedira L, Ghedira K, Mély Y, Bronner C and Mousli M:
Limoniastrum guyonianum aqueous gall extract induces apoptosis in
human cervical cancer cells involving p16 INK4A re-expression
related to UHRF1 and DNMT1 down-regulation. J Exp Clin Cancer Res.
32:302013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abusnina A, Alhosin M, Keravis T, Muller
CD, Fuhrmann G, Bronner C and Lugnier C: Down-regulation of cyclic
nucleotide phosphodiesterase PDE1A is the key event of p73 and
UHRF1 deregulation in thymoquinone-induced acute lymphoblastic
leukemia cell apoptosis. Cell Signal. 23:152–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang C, Wang Y, Zhang F, Sun G, Li C, Jing
S, Liu Q and Cheng Y: Inhibiting UHRF1 expression enhances
radiosensitivity in human esophageal squamous cell carcinoma. Mol
Biol Rep. 40:5225–5235. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao L, Tan XF, Zhang S, Wu T, Zhang ZM, Ai
HW and Song J: An intramolecular interaction of uhrf1 reveals dual
control for its histone association. Structure. 26:304–311. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kong X, Chen J, Xie W, Brown SM, Cai Y, Wu
K, Fan D, Nie Y, Yegnasubramanian S, Tiedemann RL, et al: Defining
UHRF1 domains that support maintenance of human colon cancer DNA
methylation and oncogenic properties. Cancer Cell. 35:633–648.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bronner C, Achour M, Arima Y, Chataigneau
T, Saya H and Schini-Kerth VB: The UHRF family: Oncogenes that are
drugable targets for cancer therapy in the near future? Pharmacol
Therap. 115:419–434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bronner C, Alhosin M, Hamiche A and Mousli
M: Coordinated dialogue between UHRF1 and DNMT1 to ensure faithful
inheritance of methylated DNA patterns. Genes (Basel). 10:652019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schultz MD, He Y, Whitaker JW, Hariharan
M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, et
al: Human body epigenome maps reveal noncanonical DNA methylation
variation. Nature. 523:212–216. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ming X, Zhang Z, Zou Z, Lv C, Dong Q, He
Q, Yi Y, Li Y, Wang H and Zhu B: Kinetics and mechanisms of mitotic
inheritance of DNA methylation and their roles in aging-associated
methylome deterioration. Cell Res. 30:980–996. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Harrison JS, Cornett EM, Goldfarb D,
DaRosa PA, Li ZM, Yan F, Dickson BM, Guo AH, Cantu DV, Kaustov L,
et al: Hemi-methylated DNA regulates DNA methylation inheritance
through allosteric activation of H3 ubiquitylation by UHRF1. ELife.
5:e171012016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xue B, Zhao J, Feng P, Xing J, Wu H and Li
Y: Epigenetic mechanism and target therapy of UHRF1 protein complex
in malignancies. Onco Targets Ther. 12:549–559. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gelato KA, Tauber M, Ong MS, Winter S,
Hiragami-Hamada K, Sindlinger J, Lemak A, Bultsma Y, Houliston S,
Schwarzer D, et al: Accessibility of different histone H3-binding
domains of UHRF1 is allosterically regulated by
phosphatidylinositol 5-phosphate. Mol Cell. 54:905–919. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Greiner VJ, Kovalenko L, Humbert N,
Richert L, Birck C, Ruff M, Zaporozhets OA, Dhe-Paganon S, Bronner
C and Mély Y: Site-selective monitoring of the interaction of the
SRA domain of UHRF1 with target DNA sequences labeled with
2-aminopurine. Biochemistry. 54:6012–6020. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hashimoto H, Horton JR, Zhang X, Bostick
M, Jacobsen SE and Cheng X: The SRA domain of UHRF1 flips
5-methylcytosine out of the DNA helix. Nature. 455:826–829. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Avvakumov GV, Walker JR, Xue S, Li Y, Duan
S, Bronner C, Arrowsmith CH and Dhe-Paganon S: Structural basis for
recognition of hemi-methylated DNA by the SRA domain of human
UHRF1. Nature. 455:822–825. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Arita K, Ariyoshi M, Tochio H, Nakamura Y
and Shirakawa M: Recognition of hemi-methylated DNA by the SRA
protein UHRF1 by a base-flipping mechanism. Nature. 455:818–821.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang H, Liu Y, Xie Y, Zhu Y, Liu J and Lu
F: H3K27me3 shapes DNA methylome by inhibiting UHRF1-mediated H3
ubiquitination. Sci China Life Sci. 65:1685–1700. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xie S and Qian C: The growing complexity
of UHRF1-mediated maintenance DNA methylation. Genes (Basel).
9:6002018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ferry L, Fournier A, Tsusaka T, Adelmant
G, Shimazu T, Matano S, Kirsh O, Amouroux R, Dohmae N, Suzuki T, et
al: Methylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to
replicating DNA and regulates DNA methylation. Mol Cell.
67:550–565. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kori S, Ferry L, Matano S, Jimenji T,
Kodera N, Tsusaka T, Matsumura R, Oda T, Sato M, Dohmae N, et al:
Structure of the UHRF1 tandem tudor domain bound to a methylated
non-histone protein, LIG1, reveals rules for binding and
regulation. Structure. 27:485–496. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li T, Wang L, Du Y, Xie S, Yang X, Lian F,
Zhou Z and Qian C: Structural and mechanistic insights into
UHRF1-mediated DNMT1 activation in the maintenance DNA methylation.
Nucl Acids Res. 46:3218–3231. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Povlsen LK, Beli P, Wagner SA, Poulsen SL,
Sylvestersen KB, Poulsen JW, Nielsen ML, Bekker-Jensen S, Mailand N
and Choudhary C: Systems-wide analysis of ubiquitylation dynamics
reveals a key role for PAF15 ubiquitylation in DNA-damage bypass.
Nat Cell Biol. 14:1089–1098. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Karg E, Smets M, Ryan J, Forné I, Qin W,
Mulholland CB, Kalideris G, Imhof A, Bultmann S and Leonhardt H:
Ubiquitome analysis reveals PCNA-associated factor 15 (PAF15) as a
specific ubiquitination target of UHRF1 in embryonic stem cells. J
Mol Biol. 429:3814–3824. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
González-Magaña A, de Opakua AI, Merino N,
Monteiro H, Diercks T, Murciano-Calles J, Luque I, Bernadó P,
Cordeiro TN, Biasio A and Blanco FJ: Double monoubiquitination
modifies the molecular recognition properties of p15(PAF) promoting
binding to the reader module of Dnmt1. ACS Chem Biol. 14:2315–2326.
2019.PubMed/NCBI
|
|
51
|
Miyashita R, Nishiyama A, Qin W, Chiba Y,
Kori S, Kato N, Konishi C, Kumamoto S, Kozuka-Hata H, Oyama M, et
al: The termination of UHRF1-dependent PAF15 ubiquitin signaling is
regulated by USP7 and ATAD5. Elife. 12:e790132023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bronner C, Krifa M and Mousli M:
Increasing role of UHRF1 in the reading and inheritance of the
epigenetic code as well as in tumorogenesis. Biochem Pharmacol.
86:1643–1649. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kori S, Shibahashi Y, Ekimoto T, Nishiyama
A, Yoshimi S, Yamaguchi K, Nagatoishi S, Ohta M, Tsumoto K,
Nakanishi M, et al: Structure-based screening combined with
computational and biochemical analyses identified the inhibitor
targeting the binding of DNA Ligase 1 to UHRF1. Bioorg Med Chem.
52:1165002021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Senisterra G, Zhu HY, Luo X, Zhang H, Xun
G, Lu C, Xiaon W, Hajian T, Loppnau P, Chau I, et al: Discovery of
small-molecule antagonists of the H3K9me3 binding to UHRF1 tandem
tudor domain. SLAS Discov. 23:930–940. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Choudalakis M, Kungulovski G, Mauser R,
Bashtrykov P and Jeltsch A: Refined read-out: The hUHRF1
Tandem-Tudor domain prefers binding to histone H3 tails containing
K4me1 in the context of H3K9me2/3. Protein Sci. 32:e47602023.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hata K, Kobayashi N, Sugimura K, Qin W,
Haxholli D, Chiba Y, Yoshimi S, Hayashi G, Onoda H, Ikegami T, et
al: Structural basis for the unique multifaceted interaction of
DPPA3 with the UHRF1 PHD finger. Nucleic Acids Res. 50:12527–12542.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jain K, Fraser CS, Marunde MR, Parker MM,
Sagum C, Burg JM, Hall N, Popova IK, Rodriguez KL, Vaidya A, et al:
Characterization of the plant homeodomain (PHD) reader family for
their histone tail interactions. Epigenetics Chromatin. 13:32020.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Musselman CA and Kutateladze TG:
Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res.
39:9061–9071. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xie S, Jakoncic J and Qian C: UHRF1 double
tudor domain and the adjacent PHD finger act together to recognize
K9me3-containing histone H3 tail. J Mol Biol. 415:318–328. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Morinière J, Rousseaux S, Steuerwald U,
Soler-López M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K,
Hart DJ, et al: Cooperative binding of two acetylation marks on a
histone tail by a single bromodomain. Nature. 461:664–668. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Rajakumara E, Wang Z, Ma H, Hu L, Chen H,
Lin Y, Guo R, Wu F, Li H, Lan F, et al: PHD finger recognition of
unmodified histone H3R2 links UHRF1 to regulation of euchromatic
gene expression. Mol Cell. 43:275–284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cheng J, Yang Y, Fang J, Xiao J, Zhu T,
Chen F, Wang P, Li Z, Yang H and Xu Y: Structural insight into
coordinated recognition of trimethylated histone H3 lysine 9
(H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain
(TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger
domains, 1) protein. J Biol Chem. 288:1329–1339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Rothbart SB, Krajewski K, Nady N, Tempel
W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT,
Fuchs SM, et al: Association of UHRF1 with methylated H3K9 directs
the maintenance of DNA methylation. Nat Struct Mol Biol.
19:1155–1160. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Arita K, Isogai S, Oda T, Unoki M, Sugita
K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M, et al:
Recognition of modification status on a histone H3 tail by linked
histone reader modules of the epigenetic regulator UHRF1. Proc Natl
Acad Sci USA. 109:12950–1295. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Rothbart SB, Dickson BM, Ong MS, Krajewski
K, Houliston S, Kireev DB, Arrowsmith CH and Strahl BD: Multivalent
histone engagement by the linked tandem Tudor and PHD domains of
UHRF1 is required for the epigenetic inheritance of DNA
methylation. Genes Dev. 27:1288–1298. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Unoki M: Current and potential anticancer
drugs targeting members of the UHRF1 complex including epigenetic
modifiers. Recent Pat Anticancer Drug Discov. 6:116–130. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Du W, Dong Q, Zhang Z, Liu B, Zhou T, Xu
RM, Wang H, Zhu B and Li Y: Stella protein facilitates DNA
demethylation by disrupting the chromatin association of the RING
finger-type E3 ubiquitin ligase UHRF1. J Biol Chem. 294:8907–8917.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Vaughan RM, Rothbart SB and Dickson BM:
The finger loop of the SRA domain in the E3 ligase UHRF1 is a
regulator of ubiquitin targeting and is required for the
maintenance of DNA methylation. J Biol Chem. 294:15724–15732. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Alhosin M, Omran Z, Zamzami MA, Al-Malki
AL, Choudhry H, Mousli M and Bronner C: Signalling pathways in
UHRF1-dependent regulation of tumor suppressor genes in cancer. J
Exp Clin Cancer Res. 35:1742016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hahm JY, Park JW, Kang JY, Park J, Kim CH,
Kim JY, Ha NC, Kim JW and Seo SB: Acetylation of UHRF1 regulates
hemi--methylated DNA binding and maintenance of Genome-wide DNA
methylation. Cell Rep. 32:1079582020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Achour M, Jacq X, Rondé P, Alhosin M,
Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A,
Hughes AD, et al: The interaction of the SRA domain of ICBP90 with
a novel domain of DNMT1 is involved in the regulation of VEGF gene
expression. Oncogene. 27:2187–2197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bashtrykov P, Jankevicius G, Jurkowska RZ,
Ragozin S and Jeltsch A: The UHRF1 protein stimulates the activity
and specificity of the maintenance DNA methyltransferase DNMT1 by
an allosteric mechanism. J Biol Chem. 289:4106–4115. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Taniue K, Hayashi T, Kamoshida Y, Kurimoto
A, Takeda Y, Negishi L, Iwasaki K, Kawamura Y, Goshima N and
Akiyama T: UHRF1-KAT7-mediated regulation of TUSC3 expression via
histone methylation/acetylation is critical for the proliferation
of colon cancer cells. Oncogene. 39:1018–1030. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Luo G, Li Q, Yu M, Wang T, Zang Y, Liu Z,
Niu Z, Yang H and Lai J: UHRF1 modulates breast cancer cell growth
via estrogen signaling. Med Oncol. 39:1112022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Awal MA, Nur SM, Al Khalaf AK, Rehan M,
Ahmad A, Hosawi SBI, Choudhry H and Khan MI: Structural-guided
identification of small molecule inhibitor of UHRF1
methyltransferase activity. Front Gene. 13:9288842022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zaayter L, Mori M, Ahmad T, Ashraf W,
Boudier C, Kilin V, Gavvala K, Richert L, Eiler S, Ruff M, et al: A
molecular tool targeting the base-flipping activity of human UHRF1.
Chemistry. 25:13363–13375. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Ciaco S, Mazzoleni V, Javed A, Eiler S,
Ruff M, Mousli M, Mori M and Mély Y: Inhibitors of UHRF1 base
flipping activity showing cytotoxicity against cancer cells. Bioorg
Chem. 137:1066162023. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Hu CL, Chen BY, Li Z, Yang T, Xu CH, Yang
R, Yu PC, Zhao J, Liu T, Liu N, et al: Targeting UHRF1-SAP30-MXD4
axis for leukemia initiating cell eradication in myeloid leukemia.
Cell Res. 32:1105–1123. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Baell JB and Holloway GA: New substructure
filters for removal of pan assay interference compounds (PAINS)
from screening libraries and for their exclusion in bioassays. J
Med Chem. 53:2719–2740. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Baell J and Walters MA: Chemistry:
Chemical con artists foil drug discovery. Nature. 513:481–483.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Nady N, Lemak A, Walker JR, Avvakumov GV,
Kareta MS, Achour M, Xue S, Duan S, Allali-Hassani A, Zuo X, et al:
Recognition of multivalent histone states associated with
heterochromatin by UHRF1 protein. J Biol Chem. 286:24300–24311.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
DaRosa PA, Harrison JS, Zelter A, Davis
TN, Brzovic P, Kuhlman B and Klevit RE: A bifunctional role for the
UHRF1 UBL domain in the control of hemi-methylated DNA-dependent
histone ubiquitylation. Mol Cell. 72:753–765. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Foster BM, Stolz P, Mulholland CB, Montoya
A, Kramer H, Bultmann S and Bartke T: Critical role of the UBL
domain in stimulating the E3 ubiquitin ligase activity of UHRF1
toward chromatin. Mol Cell. 72:739–752. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Abdullah O, Omran Z, Hosawi S, Hamiche A,
Bronner C and Alhosin M: Thymoquinone is a multitarget single
epidrug that inhibits the UHRF1 protein complex. Genes (Basel).
12:6222021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Alhosin M, Ibrahim A, Boukhari A, Sharif
T, Gies JP, Auger C and Schini-Kerth VB: Anti-neoplastic agent
thymoquinone induces degradation of α and β tubulin proteins in
human cancer cells without affecting their level in normal human
fibroblasts. Invest New Drugs. 30:1813–1819. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Peng Y, Tang R, Ding L, Zheng R, Liu Y,
Yin L, Fu Y, Deng T and Li X: Diosgenin inhibits prostate cancer
progression by inducing UHRF1 protein degradation. Eur J Pharmacol.
942:1755222023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Nishiyama A, Yamaguchi L, Sharif J,
Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T,
Ishikawa F, et al: Uhrf1-dependent H3K23 ubiquitylation couples
maintenance DNA methylation and replication. Nature. 502:249–253.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Qin W, Wolf P, Liu N, Link S, Smets M, La
Mastra F, Forné I, Pichler G, Hörl D, Fellinger K, et al: DNA
methylation requires a DNMT1 ubiquitin interacting motif (UIM) and
histone ubiquitination. Cell Res. 25:911–929. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Liang T, Zhang Q, Wu Z, Chen P, Huang Y,
Liu S and Li L: UHRF1 suppresses HIV-1 transcription and promotes
HIV-1 latency by competing with p-TEFb for
ubiquitination-proteasomal degradation of tat. mBio.
12:e01625212021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yang J, Liu K, Yang J, Jin B, Chen H, Zhan
X, Li Z, Wang L, Shen X, Li M, et al: PIM1 induces cellular
senescence through phosphorylation of UHRF1 at Ser311. Oncogene.
36:4828–4842. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chu J, Loughlin EA, Gaur NA, SenBanerjee
S, Jacob V, Monson C, Kent B, Oranu A, Ding Y, Ukomadu C and Sadler
KC: UHRF1 phosphorylation by cyclin A2/cyclin-dependent kinase 2 is
required for zebrafish embryogenesis. Mol Biol Cell. 23:59–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhang ZM, Rothbart SB, Allison DF, Cai Q,
Harrison JS, Li L, Wang Y, Strahl BD, Wang GG and Song J: An
allosteric interaction links USP7 to deubiquitination and chromatin
targeting of UHRF1. Cell Rep. 12:1400–1406. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Fang J, Cheng J, Wang J, Zhang Q, Liu M,
Gong R, Wang P, Zhang X, Feng Y, Lan W, et al: Hemi-methylated DNA
opens a closed conformation of UHRF1 to facilitate its histone
recognition. Nat Commun. 7:111972016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhang J, Gao Q, Li P, Liu X, Jia Y, Wu W,
Li J, Dong S, Koseki H and Wong J: S phase-dependent interaction
with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA
methylation maintenance. Cell Res. 21:1723–1739. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Geng Y, Gao Y, Ju H and Yan F: Diagnostic
and prognostic value of plasma and tissue ubiquitin-like,
containing PHD and RING finger domains 1 in breast cancer patients.
Cancer Sci. 104:194–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chen X, Cheung ST, So S, Fan ST, Barry C,
Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al: Gene expression
patterns in human liver cancers. Mol Biol Cell. 13:1929–1939. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhu M, Xu Y, Ge M, Gui Z and Yan F:
Regulation of UHRF1 by microRNA-9 modulates colorectal cancer cell
proliferation and apoptosis. Cancer Sci. 106:833–839. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yang H, Cheung S and Churg A: UHRF1
Immunohistochemical staining separates benign reactive spindle cell
mesothelial proliferations from sarcomatoid mesotheliomas. Am J
Surg Pathol. 46:840–845. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Cai Y, Tsai HC, Yen RC, Zhang YW, Kong X,
Wang W, Xia L and Baylin SB: Critical threshold levels of DNA
methyltransferase 1 are required to maintain DNA methylation across
the genome in human cancer cells. Genome Res. 27:533–544. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Niinuma T, Kitajima H, Kai M, Yamamoto E,
Yorozu A, Ishiguro K, Sasaki H, Sudo G, Toyota M, Hatahira T, et
al: UHRF1 depletion and HDAC inhibition reactivate epigenetically
silenced genes in colorectal cancer cells. Clin Epigenetics.
11:702019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Fu Y, Cao T, Zou X, Ye Y, Liu Y, Peng Y,
Deng T, Yin L and Li X: AKT1 regulates UHRF1 protein stability and
promotes the resistance to abiraterone in prostate cancer.
Oncogenesis. 12:12023. View Article : Google Scholar : PubMed/NCBI
|