Introduction
More than 500,000 patients around the world succumb
to complications associated with hepatocellular carcinoma (HCC),
which is one of the most prevalent cancers and the third leading
cause of cancer-related death (1,2).
Despite advances in medical therapy, such as regorafenib and
metronomic capecitabine treatment, prognosis of HCC remains poor
(3,4). Even with immunotherapy under
evaluation in patients with HCC, agents such as programmed cell
death-1 inhibitor have so far not brought the expected results
(5–8). Thus, there is an urgent need to
understand the underlying disease pathogenesis to inform and
improve current therapeutic approaches.
MicroRNAs (miRNAs) are a class of small, conserved,
non-coding RNA molecules with an average length of 22 nucleotides
that negatively regulate protein expression by interacting with
their target mRNAs (9). Previous
reports have shown the significance of miRNAs function in a variety
of biological processes, such as cell division, death, migration
and invasion (10,11). A growing number of miRNAs have been
reported to be closely related to a variety of cancer types,
including HCC (12). For instance,
Yin et al (13) demonstrated
the tumor suppressor role of miR-361-5p in inhibiting viability,
migration and invasion in HCC cell lines through targeting Twist1.
Jiang et al (14) showed
that miR-874, a tumor suppressor, inhibited metastasis and
epithelial-mesenchymal transition in HCC by targeting
sex-determining region Y-box 12. By contrast, Li et al
(15) reported that miR-155 serves
as an oncogene by increasing the invasiveness and metastasis of HCC
cells. In several studies, a number of miRNAs have been linked to
the clinical outcomes of patients with HCC (16,17).
miR-342-3p upregulation was a factor in the poorer overall survival
(OS) of patients with HCC (16).
Therefore, examining the role of miRNAs in HCC may help to find
potential treatment targets for this condition.
Long non-coding RNAs (lncRNAs) are a class of
non-protein coding transcripts >200 nucleotides in length; they
are the largest subclass of the non-coding transcriptome in humans
(18). Previous studies have
revealed a regulatory mechanism for lncRNAs as a competitive
endogenous RNA (ceRNA) to miRNA, thus releasing the suppressive
effects of miRNAs on their target mRNAs (19–21).
For example, lncRNA PP7080 promoted HCC cell viability, migration
and invasion through the miR-601/SIRT1 signaling axis (22). Guan et al (23) showed that lncRNA TP73-AS1 promoted
cell proliferation, migration and invasion of cervical cancer cell
lines by targeting miR-329-3p to regulate the expression of the
SMAD2 gene. Zhuang et al (24) demonstrated that miR-92b promoted HCC
progression by targeting Smad7, and this was mediated by lncRNA
XIST. Therefore, the focus of the present study was the interaction
between miRNAs and lncRNAs in the development of HCC.
In the present study, miR-206, and its expression in
HCC tissues and its association with the clinicopathological
parameters of patients with HCC was investigated. Then, the
influence of miR-206 upon HCC cell function was explored, and the
connection between miR-206 and lncRNA MALAT1 was studied. Moreover,
the underlying molecular mechanisms of miR-206 in HCC were also
studied in vitro. Results suggested that miR-206 may be
chosen as a new treating target and is a potential biomarker for
HCC.
Materials and methods
Clinical specimens
A total of 50 resected HCC and matched
tumor-adjacent tissues (>5 cm away from the tumor) were obtained
from patients admitted to the First Affiliated Hospital of Henan
University of Science and Technology (Luoyang, China) between
January 2019 and May 2020. The inclusion and exclusion criteria
were strictly controlled. The details are as follows. Inclusion
criteria are: i) Accurate pathological diagnosis of primary HCC;
ii) Obtain complete clinicopathological and follow-up data; iii)
Mainly surgical resection; iv) Willing to sign informed consent.
The exclusion criteria are (1)
perioperative death; (2) undergo
palliative surgery; (3) have been
treated with chemotherapy drugs; (4) combined with other cancers. Patient
clinicopathological data is shown in Table I. Written informed consent was
obtained from all patients. This study was approved by the Ethics
Committee of the First Affiliated Hospital of Henan University of
Science and Technology (Ethics approval number: 2019-006).
 | Table I.Association between miR-206 and
clinicopathological features of patients with hepatocellular
carcinoma. |
Table I.
Association between miR-206 and
clinicopathological features of patients with hepatocellular
carcinoma.
|
|
| miR-206
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
feature | Total (n=50) | High | Low | P-value |
|---|
| Sex |
|
|
| 0.8045 |
|
Male | 34 | 14 | 20 |
|
|
Female | 16 | 6 | 10 |
|
| Age, years |
|
|
| 0.7256 |
|
≥50 | 29 | 11 | 18 |
|
|
<50 | 21 | 9 | 12 |
|
| Tumor size, cm |
|
|
| 0.1220 |
| ≥5 | 19 | 5 | 14 |
|
|
<5 | 31 | 15 | 16 |
|
| Tumor nodule
number |
|
|
| 0.0445a |
|
Multiple (≥2) | 38 | 12 | 26 |
|
|
Solitary | 12 | 8 | 4 |
|
| TNM stage |
|
|
| 0.0088a |
|
I–II | 10 | 8 | 2 |
|
|
III–IV | 40 | 12 | 28 |
|
| Venous
infiltration |
|
|
| 0.0223a |
|
Negative | 18 | 11 | 7 |
|
|
Positive | 32 | 9 | 23 |
|
|
Differentiation |
|
|
| 0.0771 |
| Well
and moderate | 30 | 9 | 21 |
|
|
Poorly | 20 | 11 | 9 |
|
| Serum AFP level,
µg/l |
|
|
| 0.2971 |
|
≤400 | 27 | 9 | 18 |
|
|
>400 | 23 | 11 | 12 |
|
miRNA expression profile data from
GEO
Data on miRNA expression profiles were downloaded
from the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo) using the accession
number GSE10694. Based on interactive web tool GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r),
differentially expressed miRNAs between HCC tumor samples and
adjacent normal tissues were evaluated through the R ‘limma’
package (Version 4.2), which is a widely used tool that can be used
to analyze data from any GEO series and significance analysis of
microarray (SAM), to determine the differential expression of
miRNAs among groups. The differentially expressed miRNAs were
identified with general linear model, and a fold change >2 and
P-value <0.05 were recognized as significantly differentially
expressed miRNAs. Data were visualized as heat maps using the
online tool Morpheus (a web-based tool,
software.broadinstitute.org/morpheus/). Subsequently, reverse
transcription-quantitative PCR (RT-qPCR) was used to confirm the
miRNAs that were the most significantly differentially expressed
(P<0.05).
Reverse transcription-quantitative PCR
(RT-qPCR)
miRNA was extracted from HCC tissue or cell lines
using a miRNeasy mini kit (cat no. 217004; Qiagen Sciences, Inc.)
and total RNA was extracted with the TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to
manufacturer's protocols. cDNA was synthesized using a miScript II
RT kit (Qiagen Sciences, Inc.). For mRNA reverse transcription,
cDNA was synthesized using the PrimeScript™ First Strand cDNA
Synthesis kit (Takara Biotechnology Co., Ltd.). On an ABI PRISM
7500 Real-time PCR equipment, miRNA and mRNA real-time qPCR were
carried out according to a standard procedure from the SYBR Green
PCR kit (Toyobo Life Science; Thermo Fisher Scientific, Inc.). The
expression the level of miR-206 was normalized to small nuclear RNA
U6, while c-Met and MALAT1 was normalized to GAPDH. The primers for
qPCR analysis were as follows: miR-206 forward,
5′-GCGTGGAATGTAAGGAAGT-3′; miR-206 universal reverse,
5′-GCAGGGTCCGAGGTATTC-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; c-Met forward,
5′-CATCTCAGAACGGTTCATGCC-3′ and reverse,
5′-TGCACAATCAGGCTACTGGG-3′; MALAT1 forward,
5′-ATGCGAGTTGTTCTCCGTCT-3′ and reverse, 5′-TATCTGCGGTTTCCTCAAGC-3′;
GAPDH forward, 5′-TCAACGACCCCTTCATTGACC-3′ and reverse,
5′-CTTCCCGTTGATGACAAGCTTC-3′. qPCR amplification protocol was as
follows: Initial denaturation at 95°C for 5 min; followed by 40
cycles of denaturation at 94°C for 15 sec, annealing at 55°C for 30
sec and extension at 70°C for 30 sec. The RT-qPCR assays were run
in triplicate and the change in expression was calculated using the
2−ΔΔCq method (25).
Cell lines and cultures
The HCC cell lines MHCC97-H, Huh-7, Hep3B and HCCLM3
were acquired from the American Type Culture Collection (ATCC).
293T cells were acquired from the cell bank of Chinese Academic of
Sciences, Shanghai, China. The immortalized non-tumorigenic human
hepatocyte cell line, MIHA, was obtained from the Cell Bank of Type
Culture Collection of Chinese Academy of Sciences. All cells were
cultured in DMEM containing 10% fetal bovine serum (FBS, Gibco) and
1% penicillin/streptomycin (Thermo Fisher Scientific, Inc.) at 37°C
under 5% CO2.
Cell transfection
miR-206 mimics, mimics negative control (NC),
miR-206 inhibitor and inhibitor NC were purchased from Chang Jing
Bio-Tech, Ltd. For MALAT1 upregulation, lncRNA MALAT1 (NCBI
reference sequence, NR_002819.4) was inserted into pcDNA3.1 vector
(cat no. V87020; Thermo Fisher Scientific, Inc.) with the
XbaI and EcoRI restriction sites. Similarly, the open
reading frame (ORF) of c-Met (NCBI GenBank accession number,
J02958.1) was cloned into pcDNA3.1 vector with the HindIII
and BamHI restriction sites. In addition, MALAT1-targeted
small interfering (si)RNA (si-MALAT1) and NC Scramble siRNA
(si-Scramble) were also purchased from Chang Jing Bio-Tech, Ltd.
Transfections of the miRNA mimics (10 nM), miRNA inhibitors (10
nM), pcDNA vectors (2 µg) and 25 nM of siRNAs, including the
respective NCs, were performed using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.) at 37°C for 24 h, according to
manufacturer's instructions. Non-transfected cells are used as
control group (Blank). Sequences of miR-206 mimics, inhibitor,
MALAT1 siRNA and corresponding controls are as follows: miR-206
mimics, 5′-UGGAAUGUAAGGAAGUGUGUGG-3′; mimics NC,
5′-UAAGGGGAUGAGUGGAUAUGUG-3′; miR-206 inhibitor,
5′-CCACACACUUCCUUACAUUCCA-3′; inhibitor NC,
5′-CUCUAACACUCUACCUCACCUA-3′; si-MALAT1,
5′-CAGAAGGUCUGAAGCUCAUACCUAA-3′; si-Scramble,
5′-AGGCAUCAUAAGUGAACUCGUACCA-3′. All subsequent experiments were
performed 24 h after transfection.
Cell viability assay
Cell viability was assessed by MTT assay. HCCLM3 and
MHCC97-H cells (2×104) were seeded in 96-well plates at
24-h post-transfection. At 0, 24, 48, 72 and 96 h, 20 µl MTT
reagent (5 mg/ml) was added to each well and further incubated at
37°C for 4 h. Then the culture medium was removed from each well,
and 150 µl DMSO was added to each well to dissolve the purple
MTT-formazan crystals for 10 min at 37°C. The absorbance of each
well at 490 nm was determined using a microplate reader.
Apoptosis assay
HCCLM3 and MHCC97-H cells (5×105
cells/well) were seeded in 6-well plates overnight at 37°C,
transfected for 24 h with miR-206 mimics, miR-206 mimics +
pcDNA-c-Met, or mimics NC. Apoptotic rates were determined using
the Annexin V-FITC Apoptosis Detection Kit (Abcam). Cells were
digested with 0.25% trypsin (MilliporeSigma; Merck KGaA),
centrifuged at 300 g for 5 min, resuspended in 20 µl binding buffer
and incubated with 5 µl Annexin V-FITC and 1 µl PI in a dark room
at room temperature for 20 min. The stained cells were then
analyzed using BD FACSCalibur Flow Cytometer System (BD
BioSciences). Data analysis was performed using BD FACSuite™
software (Version 6.0, BD Biosciences).
Caspase-3 activity
Caspase-3 activity assay was performed using
Caspase-3 colorimetric assay kit (BD Biosciences) according to the
manufacturer's protocol. The results were evaluated using a
microplate reader (Bio-Rad, California, USA) at 405 nm.
Wound-healing assay
HCCLM3 and MHCC97-H cells were seeded into six-well
plates at a density of 3×105 cells/well overnight at
37°C, the cells were transfected for 24 h with miR-206 mimics,
miR-206 mimics + pcDNA-c-Met, or NC oligonucleotides. A new 1 ml
pipette tip was used to scratch the center of the cell layer once
it had reached 70–80% confluence. Afterward, serum-free medium was
added and cultivated for 24 h at 37°C and 5% CO2. The
distances between the wound sides were captured in the same fields
at the baseline and 48 h later using an inverted microscope
(magnification, ×200). The percentage of the wound healing was
quantified using the ImageJ Software (version 1.49; National
Institutes of Health).
Matrigel invasion assays
Cell invasive ability was examined using 24-well
Transwell chambers with an 8 µm pore size (Corning, Inc.) at 37°C.
Briefly, The HCCLM3 and MHCC97-H cells at 1×105 density
were put in the top chamber with 200 µl serum-free DMEM.
Furthermore, 500 µl complete DMEM and 10% FBS were added to the
bottom chamber. The cells was washed with PBS twice 48 h after
transfection, and stained with 0.1% crystal violet (Baomanbio
Biomart Co, Shanghai, China) for 15 min at 37°C. The cells that
invaded the bottom chamber were counted under a light microscope
(magnification, ×200; Olympus Corporation).
Luciferase reporter assay
The biological prediction website microRNA.org was employed to analyze and predict the
target genes of miR-206. To create the wild-type (wt) c-Met-3′-UTR
vector and the mutant c-Met-3′-UTR vector, respectively, the 3′-UTR
of c-Met and the fragment of Met 3′-UTR mutant were separately
inserted into the pGL3 control vector (Promega Corporation,
Madison, WI, USA). The 293T cells (1×105) were cultured
in 24-well plates and transfected for 48 h at 37°C using
Lipofectamine 2000 with 0.5 µg wild type c-Met 3′ UTR and mutant
c-Met 3′UTR reporter plasmid, together with 50 nM miR-206 mimics,
100 nM miR-206 inhibitor or the respective NC. The relative firefly
luciferase activity normalized with Renilla luciferase was measured
48 h after transfection by using the Dual-Luciferase Reporter Assay
system (Promega Corporation), according to the manufacturer's
instructions. Three duplicates of each experiment were carried
out.
Western blot
Total protein was extracted from cells using the
RIPA lysis buffer 48 h after transfection (Beyotime Institute of
Biotechnology). A BCA protein assay reagent kit was used to measure
the protein contents in the supernatant after cell lysates were
centrifuged at 12,000 × g for 20 min at 4°C (Beyotime Institute of
Biotechnology). Proteins (40 µg/lane) were separated by SDS-PAGE
using 8% gels, and then electroblotted on PVDF membranes. Primary
antibodies against c-Met (1:1,000; BF8218, Affinity Biosciences,
Ltd.), E-cadherin (1:1,000, sc-8426, Santa Cruz Biotechnology,
Inc.), N-cadherin (1:1,000, sc-8424, Santa Cruz Biotechnology,
Inc.), fibronectin (1:1,000, sc-8422, Santa Cruz Biotechnology,
Inc.), vimentin (1:2,000, sc-6260, Santa Cruz Biotechnology, Inc.),
phosphorylated (p)-c-Met (1:1,000, AF8121, Affinity Biosciences,
Ltd.), p-mTOR [1:1,000; cat no. 5536, Cell Signaling Technology,
Inc. (CST)], mTOR (1:1,000; cat no. 4517; CST), p-AKT (1:1,000; cat
no. 5106, CST), AKT (1:1,000; cat no. 2920, CST) and β-actin
(1:2,000; sc-47778, Santa Cruz Biotechnology, Inc.) were added to
membranes and incubated at 4°C overnight. Subsequently, the
membranes were incubated with secondary antibody (Horseradish
Peroxidase-conjugated IgG, 1:5,000, sc-516102, Santa Cruz
Biotechnology, Santa Cruz, CA) for 2 h at 37°C. The protein bands
were visualized using an enhanced chemiluminescence kit (Thermo
Fisher Scientific Inc., Waltham, USA), and the relative gray value
was analyzed were quantified using the ImageJ Software (version
1.49; National Institutes of Health).
Statistical analysis
Statistical analysis was performed using SPSS
(version 18.0; SPSS, Inc.). Data are presented as mean ± SD. Paired
or unpaired Student's t-test was used to compare differences
between two groups; one-way ANOVA followed by Tukey's post hoc test
was used to compare differences between multiple groups.
χ2 test and Fisher's exact test were used to investigate
the relationships between miR-206 levels and clinicopathological
characteristics. Pearson's correlation coefficient was used to
examine the correlation between the expression levels of miR-206
and lncRNA MALAT1 or c-Met mRNA. Kaplan-Meier survival curves were
analyzed by log-rank. P≤0.05 was considered to indicate a
statistically significant difference.
Results
miR-206 expression is downregulated in
HCC tissues and is associated with clinicopathological
features
To explore the role of miRNAs in HCC, differentially
expressed miRNAs from the GEO dataset GSE10694 were analyzed.
Cluster analysis based on miRNA expression demonstrated a
significant difference between HCC and adjacent normal tissue; 29
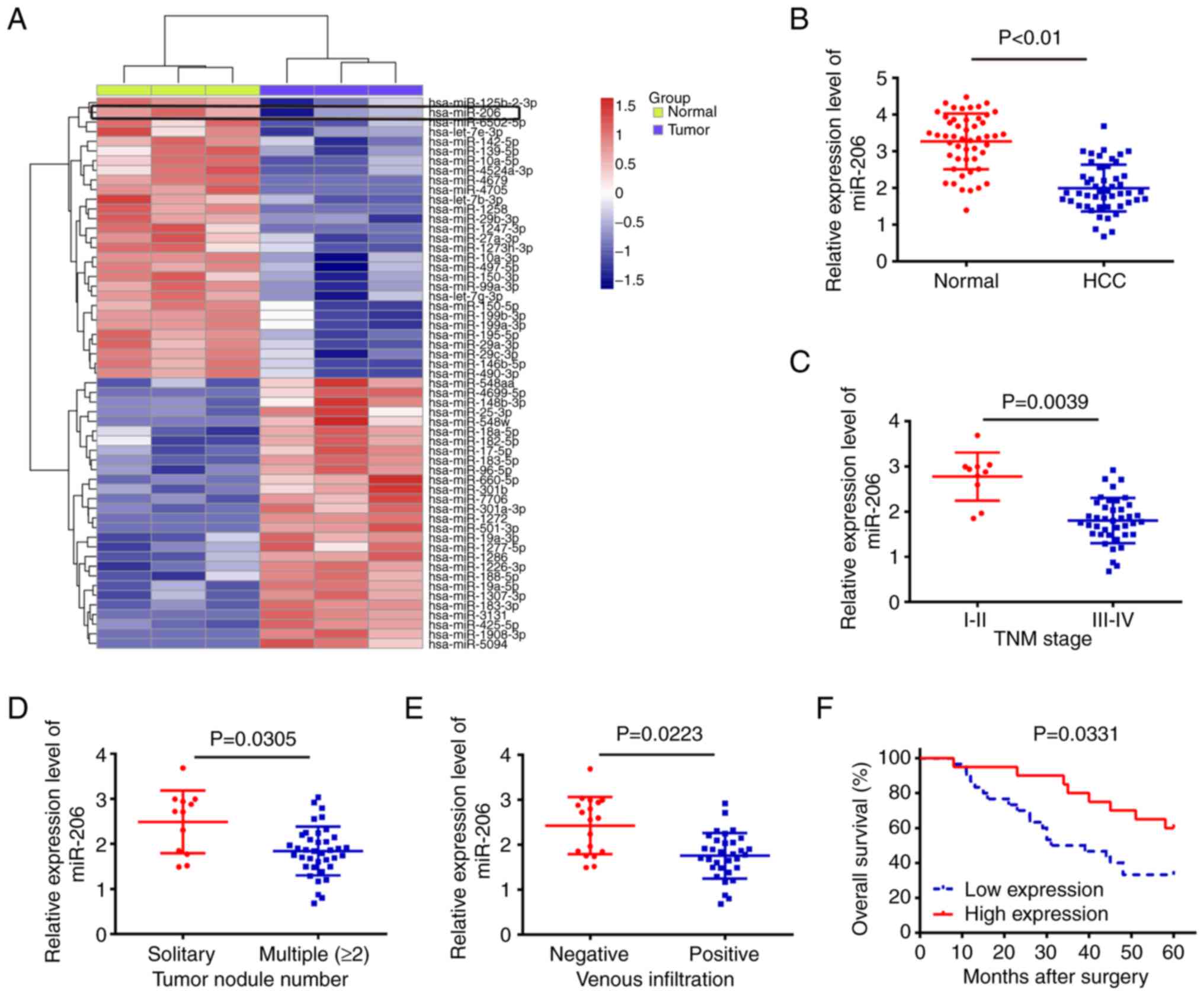
miRNAs were upregulated and 27 miRNAs were downregulated (Fig. 1A). Among aberrantly expressed
miRNAs, miR-206 had the lowest expression level in the HCC group.
Notably, miR-206 has previously been reported to function as a
tumor suppressor in various types of cancers, including lung
adenocarcinoma, breast cancer and colorectal cancer (26–28).
Although miR-206 has been reported to function as tumor suppressor
in various tumors including HCC, we conducted this study to further
determine the roles of miR-206 in HCC, especially in clinical
pathological characteristics. To validate the expression of miR-206
identified from the miRNA microarray assay, RT-qPCR was conducted
to investigate the expression of miR-206 in 50 pairs of HCC tissue.
The results showed that the expression of miR-206 in HCC tissues
was significantly lower compared with that of their matched
adjacent normal tissues (Fig.
1B).
Subsequently, the 50 patients with HCC were divided
into two groups, the miR-206 high-expression group and the miR-206
low-expression group, using the mean expression level of miR-206 as
a cut-off value. The association between miR-206 expression and
clinicopathological characteristics of the patients is summarized
on Table I. Low miR-206 expression
was shown to be associated with advanced tumor nodularity, TNM
stage and venous infiltration (Fig.
1C-E). The Kaplan-Meier survival curve revealed that patients
in the low miR-206 expression group had a worse 5-year OS rate
compared with those in the high miR-206 expression group (Fig. 1F). These findings suggested that
miR-206 may be useful as a biomarker for predicting the clinical
prognosis of patients with HCC.
Overexpression of miR-206 suppresses
cell viability and induces cell apoptosis
To determine the effects of miR-206 in HCC, the
expression of miR-206 was investigated by RT-qPCR in the HCC cell
lines MHCC97-H, Huh-7, Hep3B and HCCLM3; the immortalized
non-tumorigenic human hepatocyte cell line, MIHA, was used as a NC.
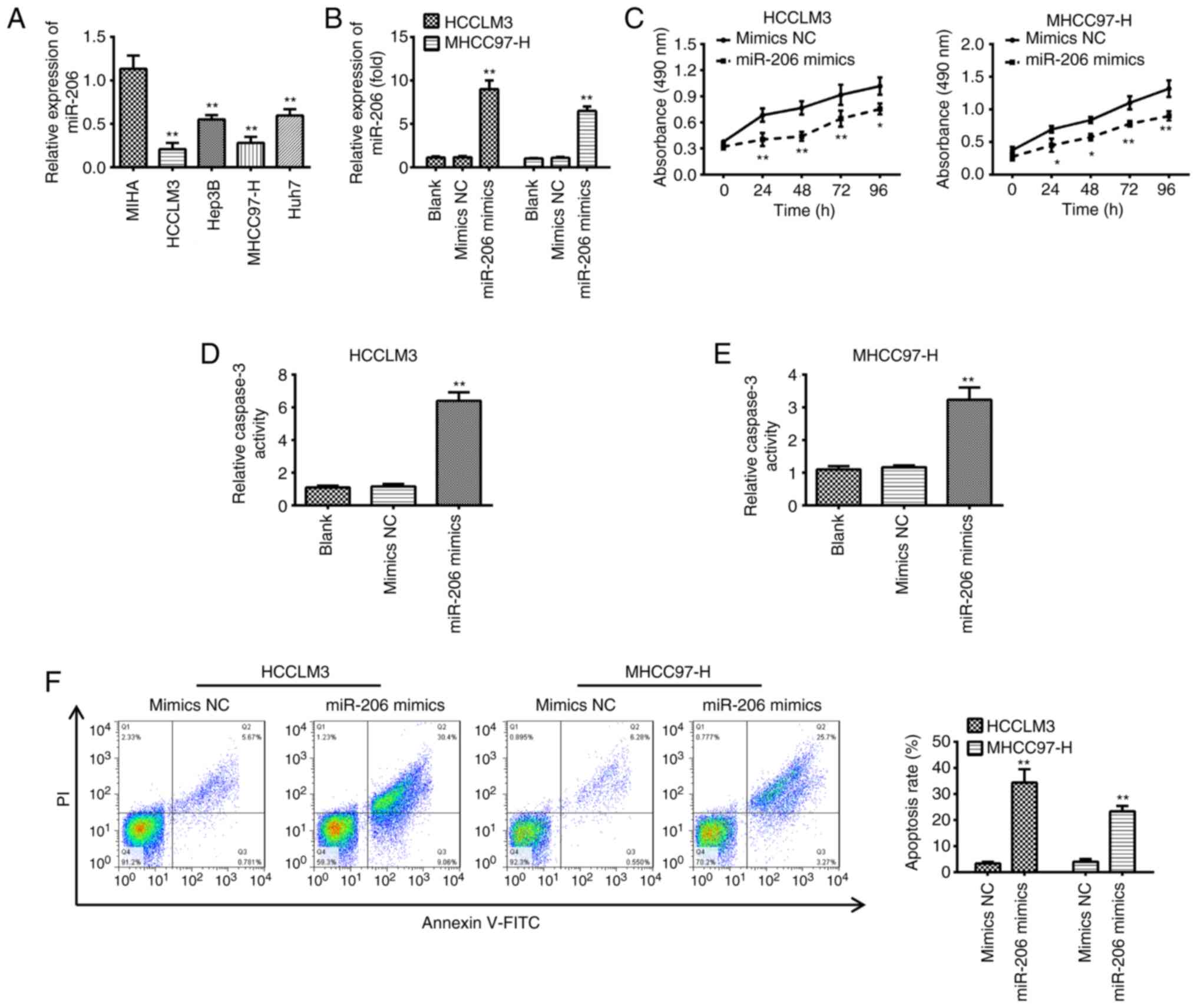
RT-qPCR demonstrated that all HCC cell lines had significantly
lower levels of miR-206 compared with MIHA cells, which is similar
to the findings in HCC tissues (Fig.
2A). miR-206 mimics were transfected into MHCC97-H and HCCLM3
cells because they exhibited the lowest expression levels of
miR-206 compared with the other cell lines; the results showed a
significant increase in miR-206 expression (Fig. 2B). miR-206 overexpression resulted
in a significant reduction in cell viability of MHCC97-H and HCCLM3
cells compared with cells transfected with mimics NC (Fig. 2C). Furthermore, the effects of
overexpression of miR-206 on cell apoptosis were determined by
caspase-3 activity and flow cytometry. Overexpression of miR-206
significantly promoted caspase-3 activity and induced apoptosis in
both MHCC97-H and HCCLM3 cells compared with the respective mimics
NC groups (Fig. 2D-F). These
results demonstrated that miR-206 inhibited HCC cell viability and
induced apoptosis.
miR-206 overexpression suppresses cell
migration and invasion
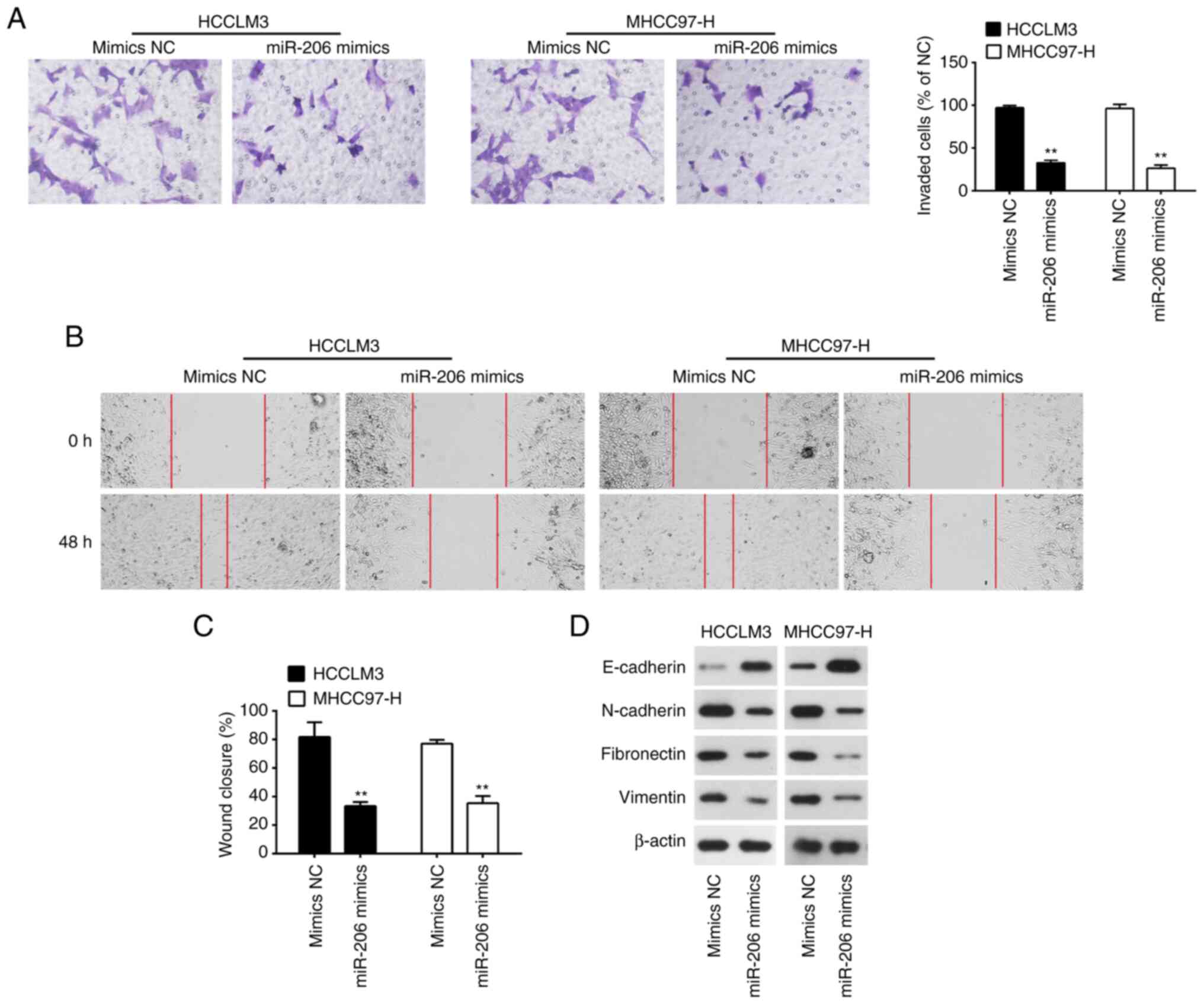
Whether miR-206 overexpression could modify the
metastatic potential of HCC was investigated, as the ability of HCC
to spread is a significant determinant in the poor prognosis of
patients (29). Overexpression of
miR-206 significantly reduced the migratory and invasive capacity
of MHCC97-H and HCCLM3 cells (Fig.
3A-C). Epithelial-mesenchymal transition (EMT) is an important
contributor in the early stage of the metastatic cascade (30). To determine if miR-206 affects EMT
in HCC cells, Western blot was performed to examine the expression
of EMT related proteins. The findings showed that overexpression of
miR-206 in MHCC97-H and HCCLM3 cells increased the expression of
E-cadherin, a known epithelial marker, and decreased the levels of
the mesenchymal-markers N-cadherin, Vimentin and Fibronectin
(Fig. 3D). These results suggested
that miR-206 overexpression inhibited cell migration and invasion
by regulating the process of EMT.
c-Met is a direct target of
miR-206
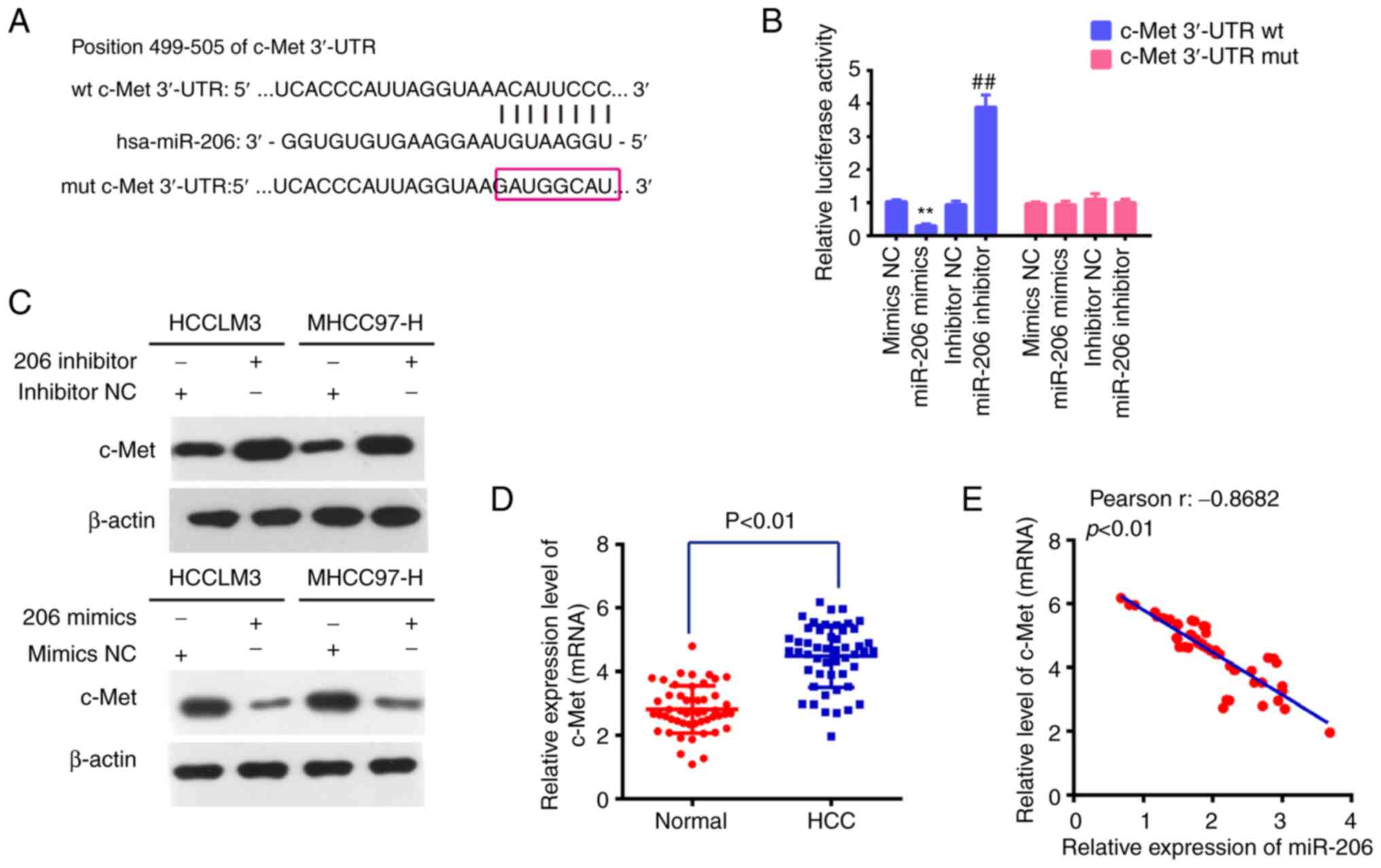
To identify specific gene targets of miR-206 by
which it might anti-oncogenic behavior in vitro, the public
algorithm microRNA.org (microrna.org) was used.
Among hundreds of predicted targets, c-Met was chosen for the
reason that it was not only identified as an oncogene, but also as
a direct target of miR-206 in various types of cancers (31–33).
The target sites for miR-206 in the c-Met sequence are shown in
Fig. 4A. To validate whether c-Met
was a direct target of miR-206, a dual-luciferase reporter assay
was conducted in 293T cells, which showed that overexpression of
miR-206 significantly reduced luciferase activity of wt
c-Met-3′-UTR when compared with negative control, whereas
inhibition of miR-206 promoted the luciferase activity of wt
c-Met-3 3′-UTR (Fig. 4B); the
activity of the mutant-type c-Met-3 3′-UTR displayed no obvious
change. In addition, the protein expression level of c-Met in
transfected HCC cells was investigated by western blotting. The
results showed that c-Met was downregulated in MHCC97-H and HCCLM3
cells transfected with miR-206 mimics and upregulated in cells
transfected with the miR-206 inhibitor (Fig. 4C). Moreover, RT-qPCR was used to
assess the levels of c-Met in the 50 pairs of HCC tumor and matched
tumor-adjacent tissues. When compared with matched tumor-adjacent
tissues, the data revealed that c-Met was considerably elevated in
HCC tissues (Fig. 4D). Pearson's
correlation analysis revealed an inverse relationship between the
levels of miR-206 and c-Met in 50 HCC tissues (Fig. 4E). These findings indicated that
miR-206 suppressed the expression of the oncogene c-Met and may
serve as a tumor suppressor.
c-Met reverses the inhibitory effects
of miR-206 overexpression on HCC cells
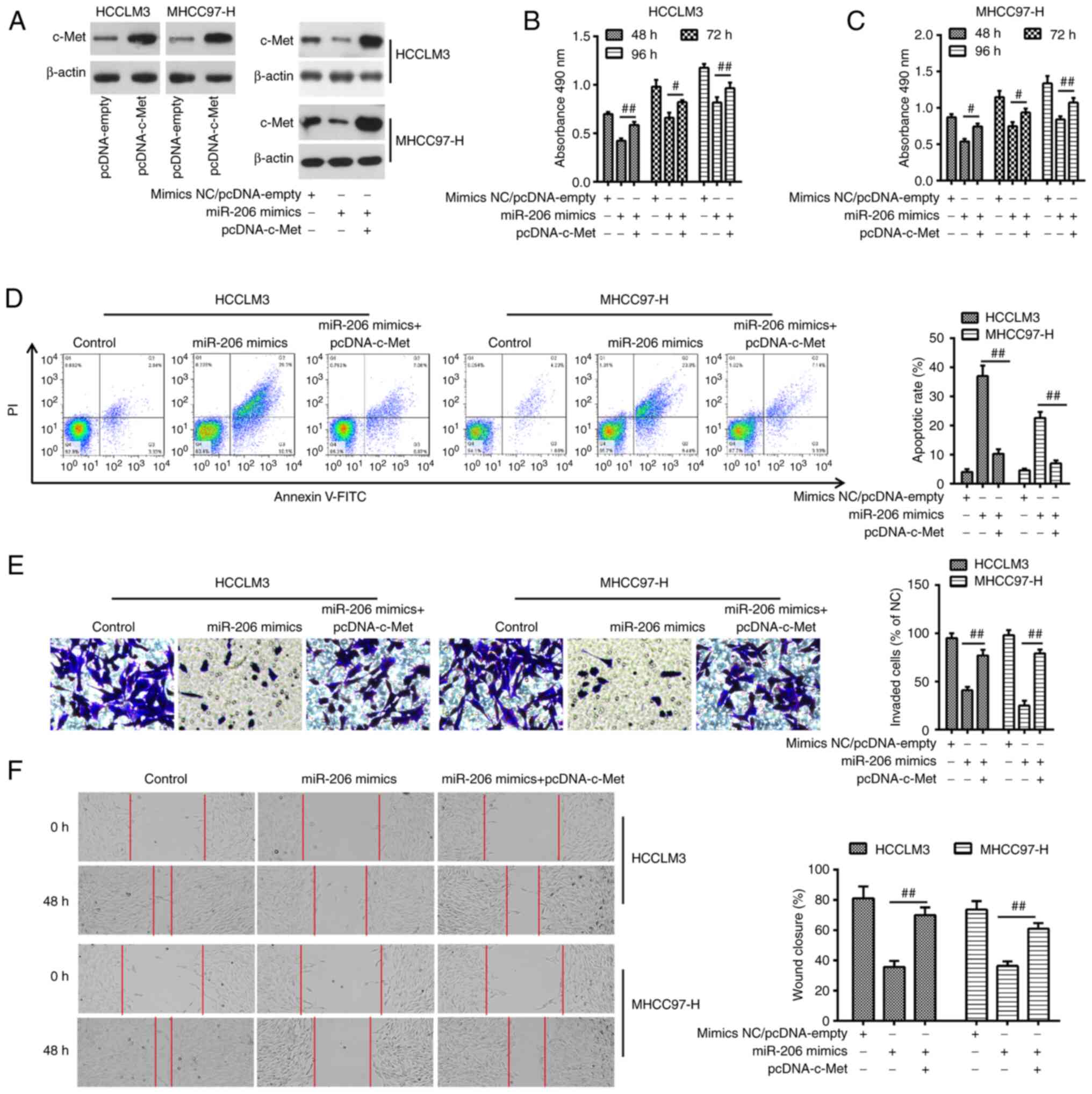
The aforementioned results led to the investigation
of whether miR-206 exerts its antitumor effect through c-Met in
HCC. To achieve this, pcDNA-c-Met overexpression vector was
co-transfected in miR-206-overexpressing MHCC97-H and HCCLM3 cells
(Fig. 5A). pcDNA-c-Met transfection
reversed the inhibitory effects of miR-206 mimics on cell viability
(Fig. 5B and C). In MHCC97-H and
HCCLM3 cells co-transfected with miR-206 mimics and pcDNA-c-Met,
the increased apoptotic rate caused by miR-206 was considerably
reduced by c-Met overexpression (Fig.
5D). c-Met overexpression also reversed the inhibitory effects
of miR-206 mimics on cell migration and invasion in MHCC97-H and
HCCLM3 cells (Fig. 5E and F). These
findings suggested a role of the target gene c-Met in the tumor
suppressor function miR-206.
miR-206 suppresses c-Met/AKT/mTOR
signaling in HCC cells
Given that the AKT/mTOR signaling pathway is one of
the most important downstream pathways of c-Met (34), and that c-Met has been shown to be a
target of miR-206, it was speculated that the c-Met/AKT/mTOR
signaling pathway may be involved in the anticancer effect of
miR-206 in HCC cells. To investigate this, western blotting was
performed to determine the changes in the expressions of the
downstream molecules of AKT/mTOR signaling, including p-Met, total
c-Met, p-AKT, total AKT, p-mTOR and total mTOR. The results showed
that miR-206 mimics transfection lowered the protein expression
levels p-c-Met, p-AKT and p-mTOR, but did not significantly alter
the levels of total c-Met, AKT and mTOR (Fig. 6). These data suggested the role of
miR-206 in suppressing the activity of c-Met/AKT/mTOR signaling in
HCC cells.
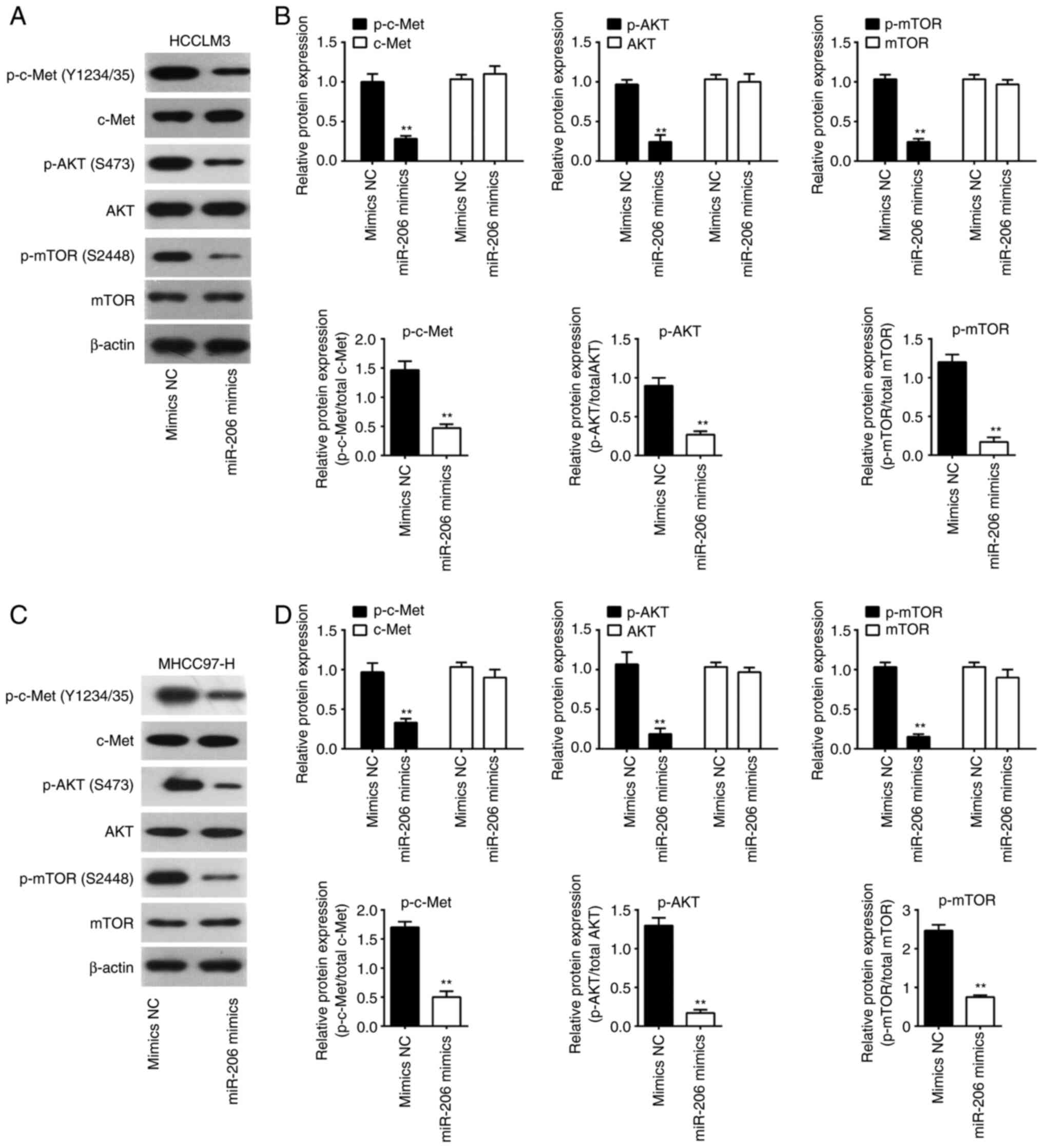 | Figure 6.Overexpression of miR-206 blocks
activation of AKT/mTOR pathway. (A and B) western blot was used to
identify the protein expression levels of c-Met, p-c-Met, AKT,
p-AKT, p-mTOR and mTOR in HCCLM3 cells after miR-206 mimics
transfection. (C and D) western blot was used to identify the
protein expression levels of c-Met, p-c-Met, AKT, p-AKT, p-mTOR and
mTOR in MHCC97-H cells after miR-206 mimics transfection. Data are
presented as the mean ± SD of three independent experiments.
**P<0.01 vs. mimics NC. HCC, hepatocellular carcinoma; miR,
microRNA; NC, negative control; p-, phosphorylated. |
lncRNA MALAT1 serves as an endogenous
sponge of miR-206 in HCC
Previous studies confirmed that lncRNAs may function
as ceRNAs by interacting with miRNAs and functionally freeing the
target genes of bound miRNAs (35,36).
Previous studies have shown that MALAT1 regulates miR-206
expression in a number of different cancers. For example, MALAT1
regulate osteosarcoma progress by modulating CDK9 expression via
sponging miR-206 (37). In
addition, MALAT1 was also reported to promote gallbladder cancer
development by acting as a molecular sponge to miR-206 (38). Thus, it was investigated whether
expression of miR-206 is also regulated by lncRNAs MALAT1 in HCC.
To determine whether miR-206 interacted with MALAT1 in HCC, MALAT1
expression levels were first measured in HCC tissues. The results
of RT-qPCR indicated that the expression of MALAT1 was
significantly higher in HCC tissues compared with that in
tumor-adjacent tissues (Fig. 7A),
which is consistent with previous studies (39–41).
Furthermore, a negative correlation was observed between MALAT1
expression and miR-206 levels in HCC tissues (Fig. 7B). Taken a step further, we examined
the expression levels of MALAT1 in HCC cells. The data showed that
MALAT1 was remarkably increased in MHCC97-H, Huh-7, Hep3B and
HCCLM3 compared with that in the immortalized non-tumorigenic human
hepatocyte cell line, MIHA, especially in MHCC97-H and HCCLM3 cells
(Fig. 7C). Subsequently, we found
that lncRNA MALAT1 had a complementary sequence of miR-206 using
Starbase v.2.0 (Fig. 7D). Then,
dual-luciferase reporter assay was performed to further investigate
the interaction of miR-206 with MALAT1. Luciferase reporter gene
assay showed that overexpression of miR-206 could significantly
inhibit the reporter activity of wt-MALAT1, whereas inhibition of
miR-206 promoted the reporter activity. Similarly, we observed that
the luciferase activity did not change significantly when the
targeted sequence of MALAT1 was mutated in the miR-206-binding site
(Fig. 7E). To further confirm the
relationship between miR-206 and MALAT1, we also detected the
expression of MALAT1 in miR-206 mimics transfected MHCC97-H cells
and miR-206 inhibitor transfected Hun7 cells. As shown in Fig. 7F, overexpression of miR-206 markedly
decreased the expression of MALAT1, whilst knockdown of miR-206
promoted the expression of MALAT1. Furthermore, MHCC97-H cells
which had high original MALAT1 expression levels were selected to
silence MALAT1 expression by siRNA, while Huh7 cells which had low
original MALAT1 expression levels were selected to enhance the
expression of MALAT1 by pcDNA-MALAT1. The interfering and
overexpressing efficiencies were confirmed by RT-PCR (Fig. 7G). We subsequently tested whether
MALAT1 influenced the miR-206 expression. miR-206 was significantly
increased after knockdown of MALAT1, whereas the miR-206 expression
level could also be repressed by MALAT1 overexpression (Fig. 7H). These findings may explain why
miR-206 was expressed at low levels in HCC.
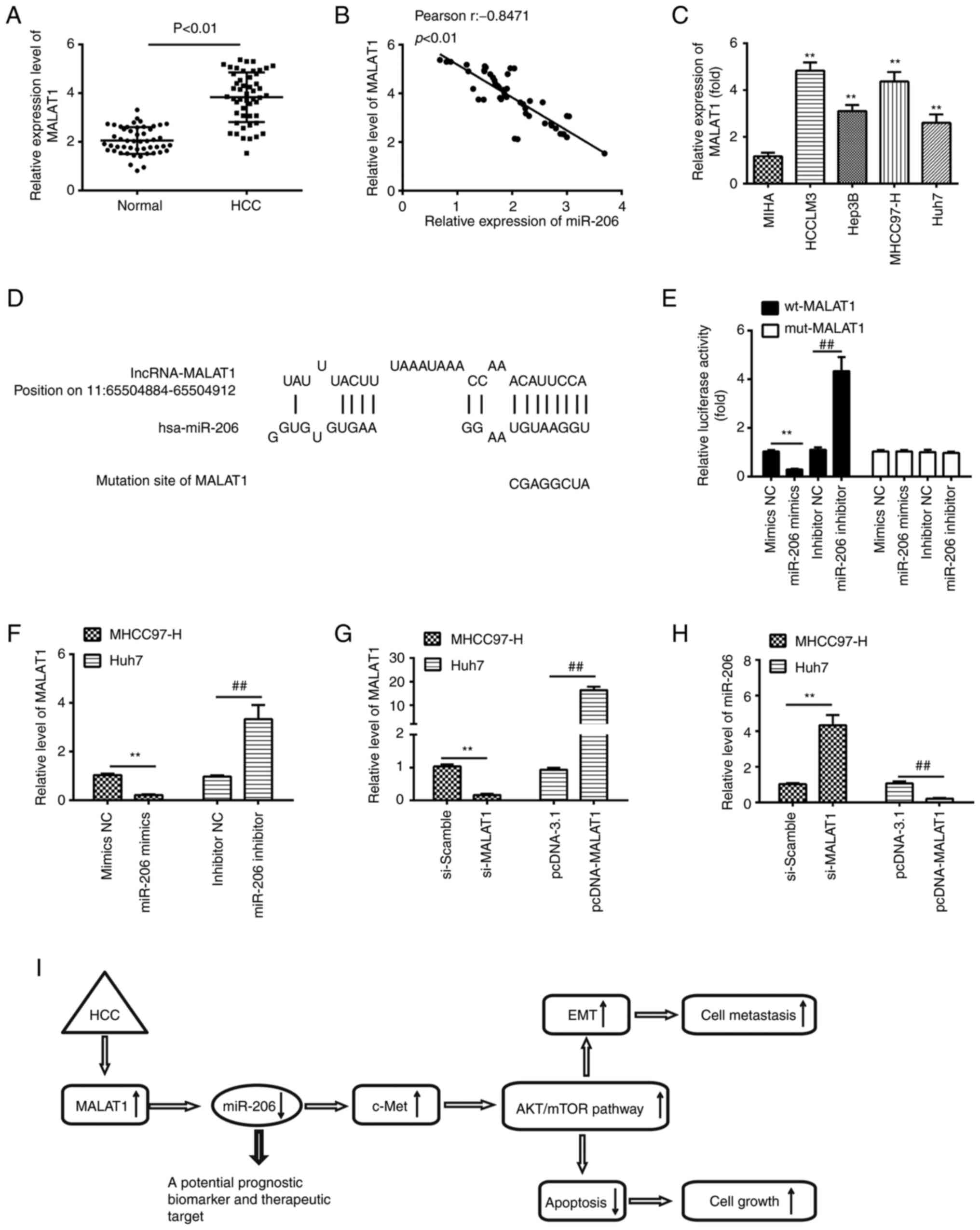 | Figure 7.LncRNA MALAT1 negatively regulates
miR-206 expression in HCC. (A) Expression of MALAT1 in HCC and
matched tumor-adjacent tissues (n=50). (B) A negative correlation
was identified between MALAT1 and miR-206 in a cohort with 50
patients with HCC. (C) The expression of MALAT1 in HCC cell lines
MHCC97-H, HCCLM3, Huh-7 and Hep3B; MIHA used as NC. Data are
presented as the mean ± SD of three independent experiments.
**P<0.01 vs. MIHA. (D) Schematic representation of the predicted
binding site for miR-206 and lncRNA MALAT1. (E) Relative luciferase
activity of MALAT1 wt or mut in 293T cells following transfection
with the miR-206 mimics, inhibitor or corresponding NC. Data are
presented as the mean ± SD of three independent experiments.
**P<0.01; ##P<0.01. (F) lncRNA MALAT1 expression
levels following transfection with miR-206 mimic or miR-206
inhibitor were measured by reverse transcription-quantitative PCR.
**P<0.01; ##P<0.01. (G) Verification of successful
downregulation of MALAT1 by siRNA and overexpression of MALAT1 by
pcDNA-MALAT1 vector transfections in MHCC97-H and Huh-7 cells.
**P<0.01; ##P<0.01. (H) MALAT1 knockdown
increased, whereas overexpression of MALAT1 decreased miR-206
expression in HCCLM3 and Huh-7 cells. **P<0.01;
##P<0.01. (I) lncRNA MALAT1 increases c-Met
expression via sequestering miR-206 at post-transcription level,
leading to the activation of AKT/mTOR signaling pathway, thus
promotes HCC cell growth, migration and invasion. HCC,
hepatocellular carcinoma; hsa, Homo sapiens; lncRNA, long
non-coding RNA; miR, microRNA; NC, negative control. |
Discussion
In the present study, it was shown that miR-206
expression is downregulated in human HCC tissues and cell lines,
and significantly associated with poor prognosis. Furthermore,
miR-206 overexpression reduced cell viability, induced apoptosis
and suppressed migration and invasion through AKT/mTOR pathway by
downregulating c-Met. Data from this study also revealed that the
miR-206 expression was regulated by lncRNA MALAT1 in HCC. These
results suggested that miR-206 may function as a tumor suppressor
in HCC and may serve as a novel and promising therapeutic target
for non-small cell lung cancer (NSCLC).
Increasing evidence indicates that miRNAs may be
valuable as a target for diagnostic and therapeutic purposes in
various malignancies, including HCC (42,43).
In particular, previous studies have revealed that a number of
miRNAs are dysregulated and exert essential roles in HCC
progression; these miRNAs include miR-342-3p, miR-125b and miR-9-5p
(16,44,45).
Li et al (46) found that
miR-20b-5p promoted HCC cell viability, migration and invasion by
downregulating cytoplasmic polyadenylation element-binding protein
3. Cao et al (47) reported
that miR-23b was a tumor suppressor which may regulate HCC
migration and invasion by targeting Pyk2 regulation of EMT. More
significantly, in non-human primates with chronic HCV infection,
the administration of locked nucleic acids specific for miR-122
inhibited long-term viral viability, supporting its use as a
therapeutic agent for HCC (48).
Additionally, a multi-center phase IIA trial involving patients
with HCC and the miR-122 antagonist miravirsen showed a decreased
viral burden in patients with HCC following miravirsen treatment
(49). Morpholino-anti-miR-487a
oligomers were also used to effectively silence miR-487a in mouse
models, which prevented the progression of HCC tumors without
causing any harm to the mice in terms of weight loss, obvious
abnormalities or animal mortality (50). The present study validated that
miR-206 was lowly expressed in HCC tissues and cell lines, and its
expression was significantly associated with advanced tumor
nodularity, TNM stage and venous infiltration. Notably, our data
provided evidence that miR-206 may have great potential as a
prognostic biomarker for HCC.
A number of previous studies have reported that
miR-206 acts as a tumor suppressor in various types of human
tumors. For instance, Liu et al (51) demonstrated that miR-206 suppressed
the viability of head and neck squamous cell carcinoma cells by
targeting HDAC6. Similarly, Shao et al (19) discovered that miR-206 had an
inhibitory effect on gastric cancer cell proliferation. Xiao et
al (52) demonstrated that
miR-206 inhibited clear-cell renal cell carcinoma proliferation
through inducing cell cycle arrest by targeting the cell cycle
related genes CDK4, CDK9 and CCND1. In addition, the function of
miR-206 was explored in HCC. For example, Liu et al
(53) reported miR-206 could
suppress HCC cell dedifferentiation and liver cancer stem cells
expansion by targeting EGFR signaling. Another study performed by
Liu et al showed that miR-206 prevented HCC by attenuating
TGFβ1 overproduction, disrupted the communication of malignant
hepatocytes with CTLs (cytotoxic T lymphocytes) and Tregs
(regulatory T cells) (54). In the
present study, overexpression of miR-206 suppressed cell viability
and invasion, and induced apoptosis in HCC cells. Previous studies
have highlighted the notable effects of miR-206 during the process
of EMT and metastasis (32,55). The current study results
demonstrated that overexpression of miR-206 resulted in a
significant upregulation of E-cadherin and a significant
downregulation of N-cadherin, Vimentin and Fibronectin in MHCC97-H
and HCCLM3 cells. These data suggested that miR-206 may suppress
the proliferative and invasive abilities of HCC cells and may serve
as a tumor suppressor in HCC.
c-Met is a well-known oncogene linked to development
of numerous malignancies (56,57).
For example, upregulation of c-Met promotes the progression and
development of multiple myeloma (58). Notably, it has also been shown in
several types of cancers including NSCLC and colorectal cancer that
miR-206 exerted its tumor-suppressive effects by targeting c-Met
(32,59). For example, miR-206 targets c-Met to
inhibit tumor cell proliferation, migration and colony formation in
NSCLC (60). Given the relationship
between miR-206 and c-Met (31–33),
it was hypothesized that, in HCC, the anti-oncogenic function of
miR-206 was achieved through suppressing c-Met expression. In the
present study, c-Met was identified to be a target of miR-206 using
online informatics tools. It was also shown that there was an
inverse correlation between miR-206 and c-Met in tumor tissues.
Notably, c-Met overexpression reversed the suppressive effects
induced by miR-206 mimics on HCC cell invasion and viability.
Interestingly, a previous study reported the similar results that
miR-206 inhibited HCC cell proliferation and migration by
modulating c-MET expression (61).
In this study, we further determined that this targeting
relationship between miR-206 and c-Met affected the AKT/mTOR
pathway signaling pathway, thus affecting cell proliferation,
invasion and migration. The protein expression levels of AKT/mTOR
pathway, one of downstream signaling pathways regulated by c-Met
(62,63) were assessed, and it was revealed
that miR-206 suppresses the activity of c-Met/AKT/mTOR signaling
pathway. These results suggested that miR-206 suppressed c-Met to
inhibit AKT/mTOR signaling pathway and therefore inhibit the
malignancy of HCC cells.
Although the present study data showed that miR-206
was expressed at a low level in HCC tissues and cell lines, it was
still unclear how miR-206 is altered. lncRNAs regulate the
expression and activity of miRNAs by acting as miRNA sponges
(64). Thus, whether there are
lncRNAs that regulate the expression of miR-206 in HCC is a
critical issue. In the present study, lncRNA MALAT1 was speculated
to be a potential regulator of miR-206 according to previous
studies. For example, Sun et al (65) showed that MALAT1 promoted bone
marrow-derived mesenchymal stem cell differentiation into
endothelial cells by sponging miR-206. In addition, MALAT1
regulates osteosarcoma progress by modulating CDK9 expression by
sponging miR-206 (37). MALAT1
silencing suppressed cell viability, migration, invasion and
vasoformation of hemangioma endothelial cells through modulating
miR-206 (66). However, it is not
known whether MALAT1 and miR-206 interact with each other directly
or indirectly in HCC cells. Expression of MALAT1 was upregulated in
HCC tissues, and there was an inverse correlation between miR-206
and MALAT1 expression in HCC tissues. Furthermore, silencing MALAT1
by siRNA increased, whereas overexpression of MALAT1 by
pcDNA-MALAT1 repressed the expression of miR-206. All data suggest
that downregulation of miR-206 is regulated by MALAT1 in HCC.
However, this study has limitations. For example,
the sample size was small, thus the relationship between miR-206
and clinicopathological features of HCC should be further explored
in a large number of samples. Previous studies have reported that
miR-206 serves an important role in various tumor cells by
targeting c-Met, such as in NSCLC (32) and gastric cancer (67). Thus, the relationship between
miR-206 and c-Met was investigated, attempting to explain how the
lowly expressed miR-206 may serve a role in the development of HCC.
Results from the present study demonstrated that miR-206 may
function as a tumor suppressor by targeting c-Met. However, whether
there are additional functional targets of miR-206 in HCC needs to
be further investigated.
In conclusion, miR-206 is downregulated in HCC
tissues and cell lines. We also find that lncRNA MALAT1 increases
c-Met expression via sequestering miR-206 at post-transcription
level, leading to the activation of AKT/mTOR signaling pathway,
thus promotes HCC cell growth, migration and invasion. miR-206 is
likely controlled by the lncRNA MALAT1 and may function as a tumor
suppressor by blocking the c-Met/AKT/mTOR signaling pathway to
induce apoptosis and inhibit HCC cell viability and invasion
(Fig. 7I). Our findings takes a
further step into the mechanism of miR-206 in HCC, and it is
suggested that developing targets to miR-206 may lead to new
approaches to treat HCC.
Acknowledgements
Not applicable.
Funding
This study was supported by The Talent Project of Henan
University of Science and Technology (grant no. 2020HAUST052).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JW, GY and BZ performed the experiments, contributed
to data analysis and wrote the paper. JW, GY, BZ and ZZ analyzed
the data. YF conceptualized the study design, contributed to data
analysis and experimental materials. JW and YF confirm the
authenticity of all the raw data. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
This research was approved by the ethics committee
of the First Affiliated Hospital of Henan University of Science and
Technology (Luoyang, China) (Ethics approval number: 2019-006). All
patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang J, Zhou ZG, Huang ZX, Yang KL, Chen
JC, Chen JB, Xu L, Chen MS and Zhang YJ: Prospective, single-center
cohort study analyzing the efficacy of complete laparoscopic
resection on recurrent hepatocellular carcinoma. Chin J Cancer.
35:252016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li X, Li C, Zhang L, Wu M, Cao K, Jiang F,
Chen D, Li N and Li W: The significance of exosomes in the
development and treatment of hepatocellular carcinoma. Mol Cancer.
19:12020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rizzo A, Ricci AD, Gadaleta-Caldarola G
and Brandi G: First-line immune checkpoint inhibitor-based
combinations in unresectable hepatocellular carcinoma: Current
management and future challenges. Expert Rev Gastroenterol Hepatol.
15:1245–1251. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rizzo A, Nannini M, Novelli M, Dalia Ricci
A, Scioscio VD and Pantaleo MA: Dose reduction and discontinuation
of standard-dose regorafenib associated with adverse drug events in
cancer patients: A systematic review and meta-analysis. Ther Adv
Med Oncol. 12:17588359209369322020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
De Lorenzo S, Tovoli F, Barbera MA, Garuti
F, Palloni A, Frega G, Garajovà I, Rizzo A, Trevisani F and Brandi
G: Metronomic capecitabine vs best supportive care in Child-Pugh B
hepatocellular carcinoma: A proof of concept. Sci Rep. 8:99972018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rizzo A, Ricci AD, Di Federico A, Frega G,
Palloni A, Tavolari S and Brandi G: Predictive biomarkers for
checkpoint inhibitor-based immunotherapy in hepatocellular
carcinoma: Where do we stand? Front Oncol. 11:8031332021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pahlavan Y, Mohammadi Nasr M, Dalir
Abdolahinia E, Pirdel Z, Razi Soofiyani S, Siahpoush S and Nejati
K: Prominent roles of microRNA-142 in cancer. Pathol Res Pract.
216:1532202020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ali Syeda Z, Langden SSS, Munkhzul C, Lee
M and Song SJ: Regulatory mechanism of MicroRNA expression in
cancer. Int J Mol Sci. 21:17232020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hussen BM, Hidayat HJ, Salihi A, Sabir DK,
Taheri M and Ghafouri-Fard S: MicroRNA: A signature for cancer
progression. Biomed Pharmacother. 138:1115282021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fornari F, Gramantieri L, Callegari E,
Shankaraiah RC, Piscaglia F, Negrini M and Giovannini C: MicroRNAs
in animal models of HCC. Cancers (Basel). 11:19062019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yin LC, Xiao G, Zhou R, Huang XP, Li NL,
Tan CL, Xie FJ, Weng J and Liu LX: MicroRNA-361-5p inhibits
tumorigenesis and the EMT of HCC by targeting Twist1. Biomed Res
Int. 2020:88918762020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang T, Guan LY, Ye YS, Liu HY and Li R:
MiR-874 inhibits metastasis and epithelial-mesenchymal transition
in hepatocellular carcinoma by targeting SOX12. Am J Cancer Res.
7:1310–1321. 2017.PubMed/NCBI
|
|
15
|
Li DP, Fan J, Wu YJ, Xie YF, Zha JM and
Zhou XM: MiR-155 up-regulated by TGF-β promotes
epithelial-mesenchymal transition, invasion and metastasis of human
hepatocellular carcinoma cells in vitro. Am J Transl Res.
9:2956–2965. 2017.PubMed/NCBI
|
|
16
|
Komoll RM, Hu Q, Olarewaju O, von Döhlen
L, Yuan Q, Xie Y, Tsay HC, Daon J, Qin R, Manns MP, et al:
MicroRNA-342-3p is a potent tumour suppressor in hepatocellular
carcinoma. J Hepatol. 74:122–134. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Khare S, Khare T, Ramanathan R and Ibdah
JA: Hepatocellular carcinoma: The role of MicroRNAs. Biomolecules.
12:6452022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei L, Wang X, Lv L, Liu J, Xing H, Song
Y, Xie M, Lei T, Zhang N and Yang M: The emerging role of microRNAs
and long noncoding RNAs in drug resistance of hepatocellular
carcinoma. Mol Cancer. 18:1472019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X,
Yang Y, Xiao B and Guo J: LncRNA-RMRP promotes carcinogenesis by
acting as a miR-206 sponge and is used as a novel biomarker for
gastric cancer. Oncotarget. 7:37812–37824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma F, Wang SH, Cai Q, Jin LY, Zhou D, Ding
J and Quan ZW: Long non-coding RNA TUG1 promotes cell proliferation
and metastasis by negatively regulating miR-300 in gallbladder
carcinoma. Biomed Pharmacother. 88:863–869. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang CZ: Long non-coding RNA FTH1P3
facilitates oral squamous cell carcinoma progression by acting as a
molecular sponge of miR-224-5p to modulate fizzled 5 expression.
Gene. 607:47–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song W, Wenhui Z, Ruiqiang Y, Hu X, Shi T,
Wang M and Zhang H: Long noncoding RNA PP7080 promotes
hepatocellular carcinoma development by sponging mir-601 and
targeting SIRT1. Bioengineered. 12:1599–1610. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guan MM, Rao QX, Huang ML, Wang LJ, Lin
SD, Chen Q and Liu CH: Long noncoding RNA TP73-AS1 targets
MicroRNA-329-3p to regulate expression of the SMAD2 gene in human
cervical cancer tissue and cell lines. Med Sci Monit. 25:8131–8141.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhuang LK, Yang YT, Ma X, Han B, Wang ZS,
Zhao QY, Wu LQ and Qu ZQ: MicroRNA-92b promotes hepatocellular
carcinoma progression by targeting Smad7 and is mediated by long
non-coding RNA XIST. Cell Death Dis. 7:e22032016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen X, Tong ZK and Zhou JY, Yao YK, Zhang
SM and Zhou JY: MicroRNA-206 inhibits the viability and migration
of human lung adenocarcinoma cells partly by targeting MET. Oncol
Lett. 12:1171–1177. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ren XL, He GY, Li XM, Men H, Yi LZ, Lu GF,
Xin SN, Wu PX, Li YL, Liao WT, et al: MicroRNA-206 functions as a
tumor suppressor in colorectal cancer by targeting FMNL2. J Cancer
Res Clin Oncol. 142:581–592. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liang Z, Bian X and Shim H: Downregulation
of microRNA-206 promotes invasion and angiogenesis of triple
negative breast cancer. Biochem Biophys Res Commun. 477:461–466.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yunqiao L, Vanke H, Jun X and Tangmeng G:
MicroRNA-206, down-regulated in hepatocellular carcinoma,
suppresses cell proliferation and promotes apoptosis.
Hepatogastroenterology. 61:1302–1307. 2014.PubMed/NCBI
|
|
30
|
Iwatsuki M, Mimori K, Yokobori T, Ishi H,
Beppu T, Nakamori S, Baba H and Mori M: Epithelial-mesenchymal
transition in cancer development and its clinical significance.
Cancer Sci. 101:293–299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen QY, Jiao DM, Yan L, Wu YQ, Hu HZ,
Song J, Yan J, Wu LJ, Xu LQ and Shi JG: Comprehensive gene and
microRNA expression profiling reveals miR-206 inhibits MET in lung
cancer metastasis. Mol Biosyst. 11:2290–2302. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen QY, Jiao DM, Wu YQ, Chen J, Wang J,
Tang XL, Mou H, Hu HZ, Song J, Yan J, et al: MiR-206 inhibits
HGF-induced epithelial-mesenchymal transition and angiogenesis in
non-small cell lung cancer via c-Met/PI3k/Akt/mTOR pathway.
Oncotarget. 7:18247–18261. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Fa X and Zhang Q: MicroRNA-206
exerts anti-oncogenic functions in esophageal squamous cell
carcinoma by suppressing the c-Met/AKT/mTOR pathway. Mol Med Rep.
19:1491–1500. 2019.PubMed/NCBI
|
|
34
|
Yang H, Wen L, Wen M, Liu T, Zhao L, Wu B,
Yun Y, Liu W, Wang H, Wang Y and Wen N: FoxM1 promotes
epithelial-mesenchymal transition, invasion, and migration of
tongue squamous cell carcinoma cells through a c-Met/AKT-dependent
positive feedback loop. Anticancer Drugs. 29:216–226. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dasgupta P, Kulkarni P, Majid S, Shahryari
V, Hashimoto Y, Bhat NS, Shiina M, Deng G, Saini S, Tabatabai ZL,
et al: MicroRNA-203 inhibits long noncoding RNA HOTAIR and
regulates tumorigenesis through epithelial-to-mesenchymal
transition pathway in renal cell carcinoma. Mol Cancer Ther.
17:1061–1069. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ren D, Zheng H, Fei S and Zhao JL: MALAT1
induces osteosarcoma progression by targeting miR-206/CDK9 axis. J
Cell Physiol. 234:950–957. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang SH, Zhang WJ, Wu XC, Zhang MD, Weng
MZ, Zhou D, Wang JD and Quan ZW: Long non-coding RNA Malat1
promotes gallbladder cancer development by acting as a molecular
sponge to regulate miR-206. Oncotarget. 7:37857–37867. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li C, Miao R, Liu S, Wan Y, Zhang S, Deng
Y, Bi J, Qu K, Zhang J and Liu C: Down-regulation of miR-146b-5p by
long noncoding RNA MALAT1 in hepatocellular carcinoma promotes
cancer growth and metastasis. Oncotarget. 8:28683–28695. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu D, Zhu Y, Pang J, Weng X, Feng X and
Guo Y: Knockdown of long non-coding RNA MALAT1 inhibits growth and
motility of human hepatoma cells via modulation of miR-195. J Cell
Biochem. 119:1368–1380. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen L, Yao H, Wang K and Liu X: Long
non-coding RNA MALAT1 regulates ZEB1 expression by sponging
miR-143-3p and promotes hepatocellular carcinoma progression. J
Cell Biochem. 118:4836–4843. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu H, Tao J, Li X, Zhang T, Zhao L, Wang
Y, Zhang L, Xiong J, Zeng Z, Zhan N, et al: MicroRNA-206 prevents
the pathogenesis of hepatocellular carcinoma by modulating
expression of met proto-oncogene and cyclin-dependent kinase 6 in
mice. Hepatology. 66:1952–1967. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ren J, Huang HJ, Gong Y, Yue S, Tang LM
and Cheng SY: MicroRNA-206 suppresses gastric cancer cell growth
and metastasis. Cell Biosci. 4:262014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang M, Wei S, Zhao H, Zhou D and Cui X:
The role of miRNA125b in the progression of hepatocellular
carcinoma. Clin Res Hepatol Gastroenterol. 45:1017122021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang L, Cui M, Cheng D, Qu F, Yu J, Wei Y,
Cheng L, Wu X and Liu X: miR-9-5p facilitates hepatocellular
carcinoma cell proliferation, migration and invasion by targeting
ESR1. Mol Cell Biochem. 476:575–583. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li Z, Wu L, Tan W, Zhang K, Lin Q, Zhu J,
Tu C, Lv X and Jiang C: MiR-20b-5p promotes hepatocellular
carcinoma cell proliferation, migration and invasion by
down-regulating CPEB3. Ann Hepatol. 23:1003452021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cao J, Liu J, Long J, Fu J, Huang L, Li J,
Liu C, Zhang X and Yan Y: microRNA-23b suppresses
epithelial-mesenchymal transition (EMT) and metastasis in
hepatocellular carcinoma via targeting Pyk2. Biomed Pharmacother.
89:642–650. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Callegari E, Gramantieri L, Domenicali M,
D'Abundo L, Sabbioni S and Negrini M: MicroRNAs in liver cancer: A
model for investigating pathogenesis and novel therapeutic
approaches. Cell Death Differ. 22:46–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Janssen HLA, Reesink HW, Lawitz EJ, Zeuzem
S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A,
Zhou Y, et al: Treatment of HCV infection by targeting microRNA. N
Engl J Med. 368:1685–1694. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chang RM, Xiao S, Lei X, Yang H, Fang F
and Yang LY: miRNA-487a promotes proliferation and metastasis in
hepatocellular carcinoma. Clin Cancer Res. 23:2593–2604. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu F, Zhao X, Qian Y, Zhang J, Zhang Y
and Yin R: MiR-206 inhibits head and neck squamous cell carcinoma
cell progression by targeting HDAC6 via PTEN/AKT/mTOR pathway.
Biomed Pharmacother. 96:229–237. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xiao H, Xiao W, Cao J, Li H, Guan W, Guo
X, Chen K, Zheng T, Ye Z, Wang J and Xu H: miR-206 functions as a
novel cell cycle regulator and tumor suppressor in clear-cell renal
cell carcinoma. Cancer Lett. 374:107–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu C, Li J, Wang W, Zhong X, Xu F and Lu
J: miR-206 inhibits liver cancer stem cell expansion by regulating
EGFR expression. Cell Cycle. 19:1077–1088. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu N, Steer CJ and Song G: MicroRNA-206
enhances antitumor immunity by disrupting the communication between
malignant hepatocytes and regulatory T cells in c-Myc mice.
Hepatology. 76:32–47. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yin K, Yin W, Wang Y, Zhou L, Liu Y, Yang
G, Wang J and Lu J: MiR-206 suppresses epithelial mesenchymal
transition by targeting TGF-β signaling in estrogen receptor
positive breast cancer cells. Oncotarget. 7:24537–24548. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang J and Babic A: Regulation of the MET
oncogene: Molecular mechanisms. Carcinogenesis. 37:345–355. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Marano L, Chiari R, Fabozzi A, De Vita F,
Boccardi V, Roviello G, Petrioli R, Marrelli D, Roviello F and
Patriti A: c-Met targeting in advanced gastric cancer: An open
challenge. Cancer Lett. 365:30–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ho-Yen CM, Jones JL and Kermorgant S: The
clinical and functional significance of c-Met in breast cancer: A
review. Breast Cancer Res. 17:522015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lyu J, Sun Y, Li X and Ma H: MicroRNA-206
inhibits the proliferation, migration and invasion of colorectal
cancer cells by regulating the c-Met/AKT/GSK-3β pathway. Oncol
Lett. 21:1472021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dai C, Xie Y, Zhuang X and Yuan Z: MiR-206
inhibits epithelial ovarian cancer cells growth and invasion via
blocking c-Met/AKT/mTOR signaling pathway. Biomed Pharmacother.
104:763–770. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang Y, Tai Q, Zhang J, Kang J, Gao F,
Zhong F, Cai L, Fang F and Gao Y: MiRNA-206 inhibits hepatocellular
carcinoma cell proliferation and migration but promotes apoptosis
by modulating cMET expression. Acta Biochim Biophys Sin (Shanghai).
51:243–253. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sun C, Liu Z, Li S, Yang C, Xue R, Xi Y,
Wang L, Wang S, He Q, Huang J, et al: Down-regulation of c-Met and
Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell
proliferation, migration and colony formation. Oncotarget.
6:25533–25574. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Boccaccio C, Luraghi P and Comoglio PM:
MET-mediated resistance to EGFR inhibitors: An old liaison rooted
in colorectal cancer stem cells. Cancer Res. 74:3647–3651. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shen SN, Li K, Liu Y, Yang CL, He CY and
Wang HR: Down-regulation of long noncoding RNA PVT1 inhibits
esophageal carcinoma cell migration and invasion and promotes cell
apoptosis via microRNA-145-mediated inhibition of FSCN1. Mol Oncol.
13:2554–2573. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sun X, Luo L and Li J: LncRNA MALAT1
facilitates BM-MSCs differentiation into endothelial cells via
targeting miR-206/VEGFA axis. Cell Cycle. 19:3018–3028. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wang S, Ren L, Shen G, Liu M and Luo J:
The knockdown of MALAT1 inhibits the proliferation, invasion and
migration of hemangioma endothelial cells by regulating
MiR-206/VEGFA axis. Mol Cell Probes. 51:1015402020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zheng Z, Yan D, Chen X, Huang H, Chen K,
Li G, Zhou L, Zheng D, Tu L and Dong XD: MicroRNA-206: Effective
inhibition of gastric cancer progression through the c-Met pathway.
PLoS One. 10:e01287512015. View Article : Google Scholar : PubMed/NCBI
|