Introduction
Lung cancer accounts for ~13% of annual cancer cases
worldwide and is the second most common type of cancer in both male
and female patients (1).
Investigation of proteins including proteins like Carcinoembryonic
Antigen (CEA), autoantibodies such as P53 autoantibodies, and gene
expression profiles in the blood or airway epithelium has yielded
promising biomarker candidates for the early detection of lung
cancer, such as epidermal growth factor receptor, c-ros
oncogene1(ROS1), KRAS expression (2). Interferon-γ (IFN-γ) is the only type
II IFN member that isa dimerized soluble molecule and consisting of
143 amino acids (3,4). IFN-γ performs roles with antiviral,
immunoregulatory and anti-tumor properties, via interactions with
specific cell-surface receptors such as IFN-γ receptor (IFNGR)
(5).
Researchers have long sought a rapid, safe, and
effective therapy for malignant tumors. However, traditional cancer
treatments, such as surgery, radiotherapy (RT), and chemotherapy
still fall short. For example, surgical excision often fails due to
cancer recurrence and metastasis. Radiotherapy which uses
high-energy X-rays, damages normal tissues, and traditional
chemotherapy is limited by severe multidrug resistance and side
effects such as nausea and Vomiting, anemia, and hair loss
(6). To overcome these challenges
and improve prognosis, intelligent nanoplatforms have been
developed for improved diagnostic and therapeutic outcomes
(7). These nanoplatforms use
naturally existing nanoparticles, such as bacterial viruses
(bacteriophages or phages), plant viruses, nucleic acid
nanoparticles (such as DNA origami), protein nanoparticles and
liposomes (8). These
bionanoparticles possess unique properties in terms of composition,
structure, shape and function, which make them valuable tools for
cancer imaging, diagnosis and therapy (9).
Liposomal IFN-γ [IFN-γ (L)] is a cytokine that
serves a key role in the maturation and function of certain immune
cells (10). The encapsulation of
IFN-γ in liposomes improves its pharmacokinetic profile, prolonging
its half-life and enhancing its stability in the bloodstream
(11). This enhanced formulation
offers several advantages, such as targeted delivery to specific
tissues and reduced systemic toxicity, making it a promising
candidate for various therapeutic applications (12). Studies have reported the potential
of IFN-γ (L) to stimulate antitumor immunity, control viral
infections and modulate the immune response in autoimmune diseases
(13,14). IFN-γ (L) inhibits melanoma growth
and metastasis by inducing antitumor immunity (15). The second generation murine IFN-γ
(L) has been reported to exhibit antitumor and antiangiogenic
effects (16). If liposomes can
overcome the current limitations like targeted delivery, and
reduced systemic toxicity, they could be considered next generation
protein therapeutics due to their ability to increase protein and
peptide (PPs) solubility and provide controlled sustained release
of PPs to decrease side effects of traditional therapy including
autoimmunity and non-specific inflammation (17).
Inactivation of the tumor suppressor gene, p53, by
somatic mutations has been reported to be associated with certain
malignant neoplasms, and its reactivation represents an attractive
therapeutic strategy for cancers (18,19).
p53 has also demonstrated the ability to induce DNA repair pathways
to minimize DNA damage (20). The
regulation of specific DNA repair is controlled via p53-mediated
transcriptional genes depending on the type of DNA damage. The p53
gene can induce crucial DNA repair genes including base excision
repair (BER), non-homologous end-joining and nucleotide excision
repair (21). Human 8-oxo
guanine-DNA glycosylase (OGG1) serves a crucial role in the repair
pathway of reactive oxygen species-induced damage, through stepwise
base excision repair (BER) (22).
In addition, OGG1 interacts with poly ADP-ribose polymerase 1
(PARP1), a DNA-damage sensor protein involved in DNA repair and
numerous other cellular processes (23). However, the effect mechanism of
IFN-γ on lung cancer lymphocytes is still not clear. The present
study examined the DNA protective effects of IFN-γ (L) on
lymphocytes from patients with lung cancer compared with healthy
individuals through the study of p53, OGG1 and PARP1 at the gene
and protein levels after treatment with IFN-γ (L).
Materials and methods
Reagents
All chemicals utilized in the present study were
purchased from Sigma-Aldrich (Merck KGaA) including IFN-γ (98%
purity; cat. no. 17001), fetal bovine serum (FBS; cat. no. F7524),
RPMI 1640 medium (RPMI-1640; cat. no. R8758) and
penicillin-streptomycin solution (cat. no. P4333). Before using
IFN-γ, the lyophilized powder was reconstituted in double-distilled
water to create the stock solution. It was then diluted in
RPMI-1640 medium containing 10% FBS and kept at 20°C.
Dose-response tests were performed to identify the
best naked IFN-γ [IFN-γ (N)] and IFN-γ (L) dosages. Various doses
(50, 100, 200,300 U/ml were administered at different time
intervals (24, 48, 72 h) at 37 °C to assess their effectiveness.
Based on this, 100 U/ml IFN-γ (N) and 100 U/ml IFN-γ (L) were
administered at a constant dosage. The 75 µM
H2O2 at 37°C for 24 h was used to induce the
oxidative stress and increase the DNA damage in lymphocytes from
healthy individuals and lung cancer patients to be used as the
positive control (PC).
Cell viability
Based on our previous study (17), cell viability was assessed using the
Cell Counting Kit-8 (CCK-8), Sigma-Aldrich (Shanghai, China) at
37°C for 4 h. In all tests, doses expected to produce a cell
viability of ~75% were used and incubated with the treatment for 24
h.
Sample preparation and enzyme-modified
comet assay
Healthy non-smoking volunteers and patients with
lung cancer (including non-small cell lung cancer and small cell
lung cancer) provided informed consent to participate. Ethical
approval for the present study was received from The Leeds East
Research Ethics Committee (approval no. 12/YH/0464; Leeds, UK), The
University of Bradford Research Ethics Sub-Committee on Research in
Human Subjects (approval no. 0405/8; Bradford, UK) and The Research
Support and Governance Office, Bradford Teaching Hospitals, NHS
Foundation (approval no. RE DA 1202; Bradford, UK). Whole blood
samples were collected and labeled for identification. Samples were
diluted in RPMI and mixed with 10% dimethyl sulfoxide. The diluted
blood solution was divided and transferred to −80°C storage. The
DNA repair capability of human lymphocytes from five healthy
volunteers and five patients with lung cancer was determined using
an Endonuclease III (Nth) and hOGG1 FLARETM Test kit (Trevigen;
cat. no. CA:4055-100-FK), according to the manufacturer's
instructions. Data analyesd by using Komet 6 software and Kinetic
Imaging (Andor Technology Ltd, Belfast) to determine the % DNA tail
and Olive tail moment (OTM).
Lymphocyte isolation
A total of 3 ml whole blood was diluted 1:1 with
0.9% saline and layered on top of 3 ml Lymphoprep™ (Axis-Shield
Diagnostics, Ltd.) in 15 ml falcon tubes. The tubes were
centrifuged for 20 min at 800 × g at 4°C. Lymphocytes were
harvested and washed with saline. Cells were re-suspended in RPMI
and used for in vitro experiments.
Preparation and characterization of
liposomes
Liposomes were prepared using the thin film
rehydration method (24) and all
measurements were performed in triplicate.
Determination of IFN-γ encapsulation
efficiency
The IFN-γ encapsulation efficiency of liposomes was
determined by an indirect procedure based on the determination of
uncoated free IFN-γ in the supernatant utilizing reversed-phase
high-performance liquid chromatography, as previously described
(25,26). Each sample was assessed in
triplicate and the loading of IFN-γ was expressed as percentage
encapsulation efficiency.
Reverse transcription-quantitative
(RT-q)PCR
Isolated lymphocytes were seeded in 6-well plates
(1×106 cells/well) and treated with 100 U/ml IFN-γ (N)
and IFN-γ (L) for 24 h at 37°C. A total of 2 mg total isolated RNA
was subjected to RT using an iScript™ cDNA synthesis kit (Bio-Rad
Laboratories, Inc.) according to the manufacturer's protocol. Each
RT-qPCR experiment was performed three times in a 10 ml reaction
mixture. The primers (MilliporeSigma; Merck KGaA) were verified
using Primer-BLAST, NCBI database
(ncbi.nlm.nih.gov/tools/primer-blast/) and presented in Table I. Data were analyzed using the
2−ΔΔCq method (16) and
normalized against the internal reference gene GAPDH in each
sample.
 | Table I.Primers for RT-qPCR analysis. |
Table I.
Primers for RT-qPCR analysis.
| Gene | Primer sequence
(5′-3′) | (Refs.) |
|---|
| p53 | F:
GGATCCTAATACGACTCACTA | (27) |
|
| R:
GGCAGTGACCCGGAAGGCA |
|
| PARP1 | F:
CCTGATCCCCCACGACTTT | (28) |
|
| R:
GCAGGTTGTCAAGCATTTC |
|
| OGG1 | F:
GGTGGCCCTAAAGGACTCTC | (29) |
|
| R:
AAGGTGCTTGGGGAATTTCT |
|
| GAPDH | F:
GGAGCGAGATCCCTCCAAAAT | (28) |
|
| R:
GGCTGTTGTCATACTTCTCATGG |
|
Western blotting analysis
Lymphocytes were seeded in 6-well plates at a
density of 1×106 cells/well. The treated cells with
IFN-γ (N) and IFN-γ (L) were incubated overnight at 37°C in the
presence of 5% CO2, washed with cold PBS and lysed by
adding 150 µl lysis buffer with 15 µl fresh protease inhibitor
cocktail (Thermo Fisher Scientific). Total protein levels were
determined using the Bio-Rad Bradford assay kit (Bio-Rad
Laboratories, Inc.) with each experiment repeated three times. Tris
buffers (pH 6.8 and 8.8) were prepared for resolving and stacking
gels. The catalysts APS and TEMED and a final concentration of
10.4% SDS were added for polyacrylamide gel polymerization with 30
µg protein/well. The blotting membranes were incubated overnight at
4°C with the primary antibody. GAPDH rabbit monoclonal primary
antibody (cat. no. ab8245) was used as a loading control. The
primary antibodies [GAPDH (1:10,000), p53 (1:1,000; cat. no. ab26);
P21 (1:1,000); cat.no. ab109520); BCL-2 (1:1000); cat. no.
ab182858] (Abcam) were diluted with TBS-T containing 1% (w/v) BSA
(Sigma fraction V; Sigma Chemicals). Proteins were transferred to a
blotting nitrocellulose membrane using the iBlot® Gel
Transfer Device (Invitrogen) for 7 min at a constant voltage of
25V. After transfer, the nitrocellulose membranes were incubated
with the blocking solution contained 1% (w/v) BSA in Tris-buffered
saline containing 0.1% Tween 20 and incubated for 1 hour at room
temperature with HRP-Donkey Anti-Rabbit IgG (CAT: ab7083, Abcam,
UK). The blots were rinsed and visulaized by enhanced
chemiluminescence substrate detection reagent [ECL substrate kit;
cat. no. ab133406 (Abcam, UK). Relative expression of the protein
was determined using image j software (version 1.54f; National
Institutes of Health).
Statistical analysis
GraphPad Prism 8 (Dotmatics) was used for
statistical analysis and One-way ANOVA followed by Dunnett's post
hoc test were conducted. Data are presented as the mean and SEM).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Encapsulation efficiency of IFN-γ
liposome
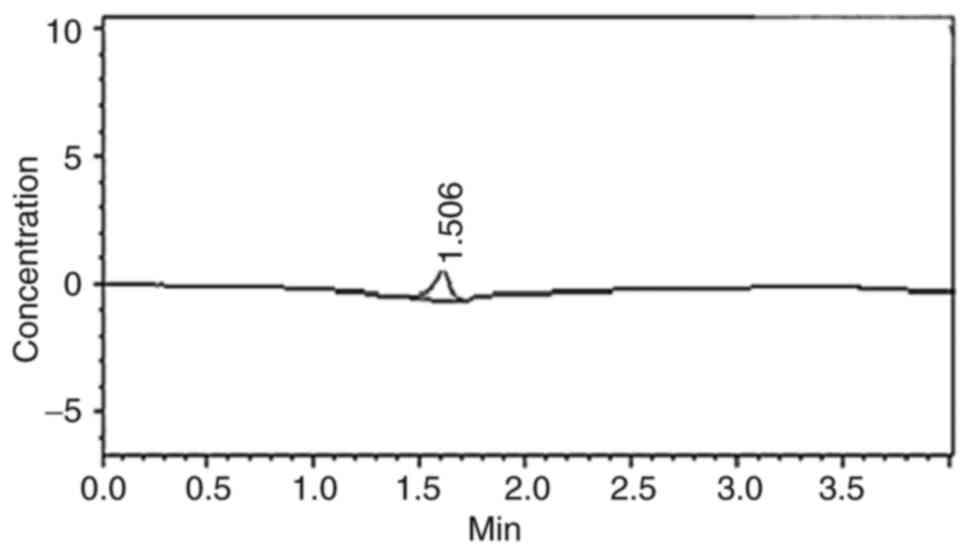
The encapsulation efficiency of IFN-γ was calculated
as follows: Encapsulation efficiency (%)=[(Total IFN-γ)-(Free
IFN-γ)]/total IFN-γ) × 100. As the initial concentration of IFN-γ
was 1.38 µg/ml and the concentration of free IFN-γ was 0.38 µg/ml,
Therefore, the concentration of encapsulated IFN-γ was 1 µg/ml and
the encapsulation efficiency was calculated to be 72%. (Fig. 1).
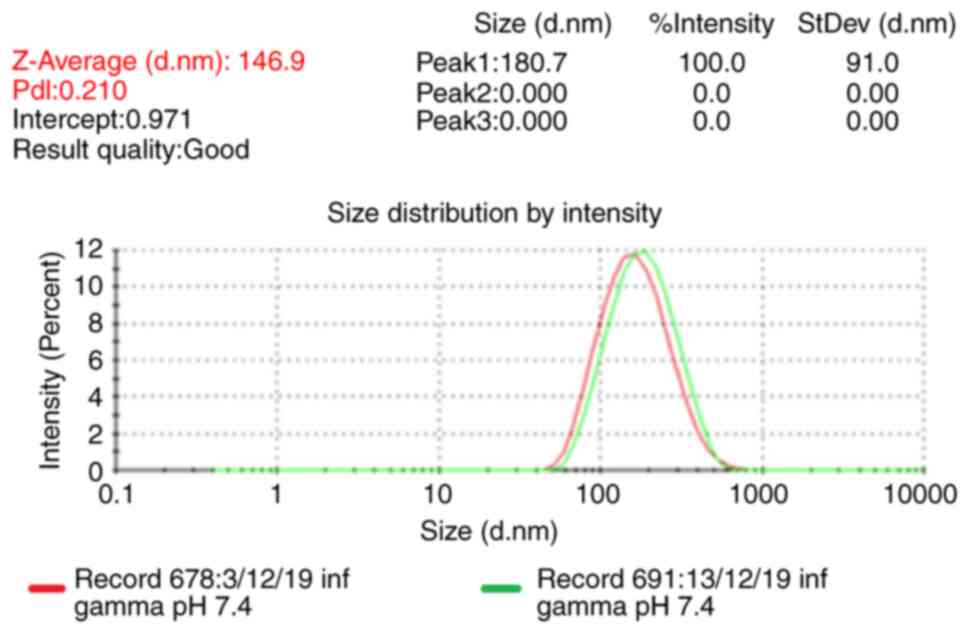
Particle size of IFN-γ liposome
The Z-Average particle size represents the
intensity-weighted mean hydrodynamic size of the entire ensemble of
particles, as measured using dynamic light scattering (DLS) was
146.9 nm with polydispersity index=0.210. To ensure liposome
stability, samples were stored at 4°C and the particle size was
measured three times, the results showed no notable increases in
the particle size over 10 days (Fig.
2).
IFN-γ has a low cytotoxicity effect
against lymphocytes
CCK-8 assay (Fig. 3)
indicated that the viability of lymphocytes from three healthy
individuals and three patients with lung cancer in different
treatment groups was >75% after 24 h treatment. Dose-response
experiments were performed to determine the optimal doses of IFN-γ
(N) and IFN-γ (L) used throughout the study with a fixed dose of
100 U/ml IFN-γ (N) and 100 U/ml of IFN-γ (L) determined to be the
optimal dose and used during the present study.
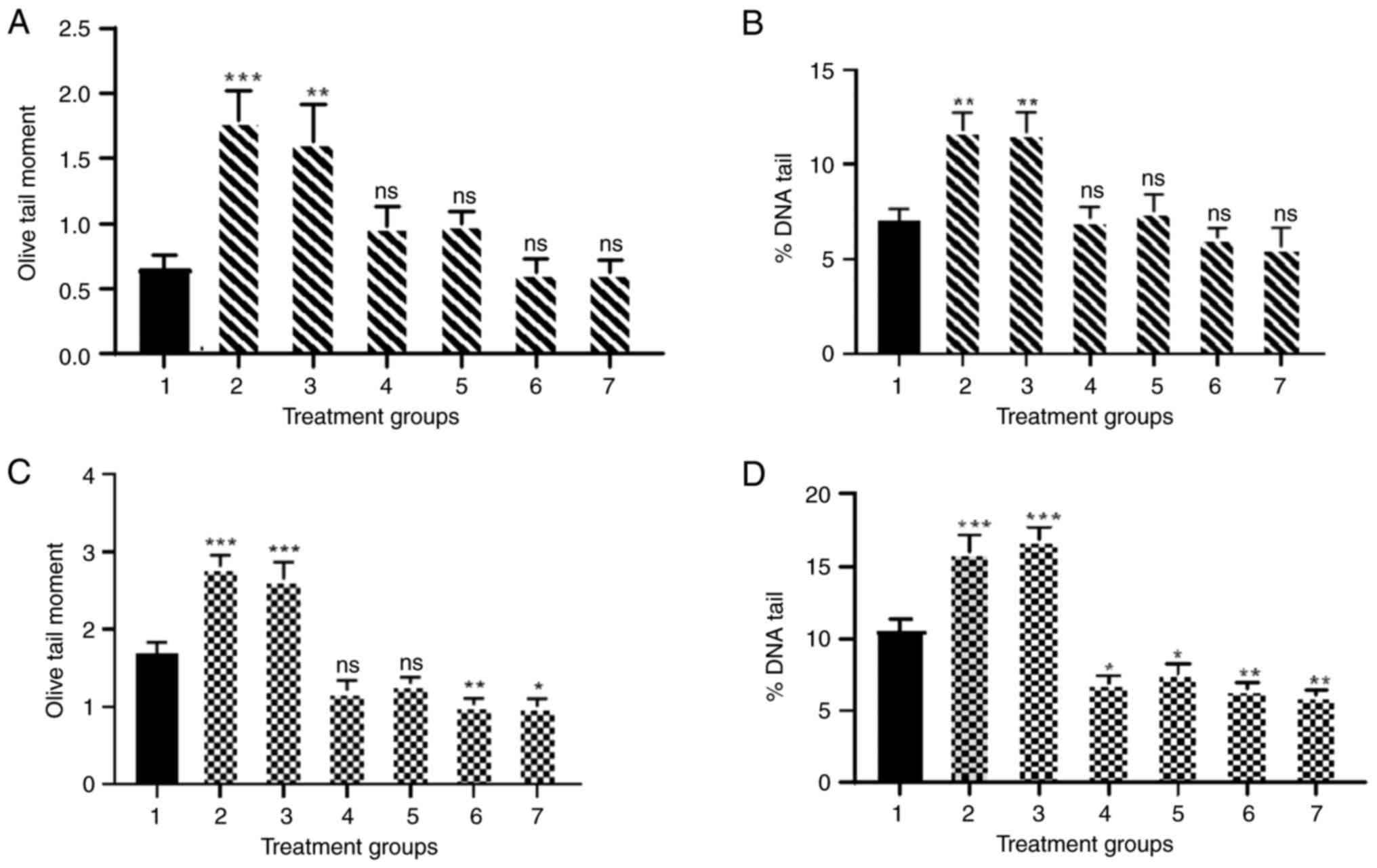 | Figure 3.DNA damage in samples from healthy
individuals and patients with lung cancer. (A) Olive tail moment
and (B) percentage DNA tail of samples from healthy individuals. 1,
NC; 2, PC + FPG; 3, PC + hOGG1; 4, 100 U/ml IFN-y (N)
+ endonuclease III; 5, 100 U/ml IFN-γ (N) + hOGG1; 6,
100 U/ml IFN-γ (L) + endonuclease III; and 7, 100 U/ml IFN-γ
(L) + hOGG1. (C) Olive tail moment and (D) percentage DNA
tail of samples from patients with lung cancer. 1, NC; 2, PC
+ endonuclease III; 3, PC + hOGG1; 4, 100 U/ml IFN-γ
(N) + endonuclease III; 5, 100 U/ml IFN-γ (N) +
hOGG1; 6, 100 U/ml IFN-γ (L) + endonuclease III; and 7, 100
U/ml IFN-γ (L) + hOGG1. Experiments were repeated at least
three times. Data are presented as the mean ± SEM. *P<0.05,
**P<0.01 and ***P<0.001 compared with NC. NC, untreated
cells; IFN-γ (N), naked IFN-γ; IFN-γ (L), liposomal IFN-γ; hOGG1,
human 8-oxo guanine-DNA glycosylase; PC, positive control of 75 µm
H2O2; ns, not significant; FPG,
formamidopyrimidine (fapy)-DNA glycosylase) repair enzyme. |
IFN-γ (L) reduced DNA damage in the
lymphocytes of patients with lung cancer
The results demonstrated the concentration-response
for 100 U/ml IFN-γ (N) and IFN-γ (L) in the presence of
endonuclease III and hOGG1 enzymes using % DNA tail and OTM, which
indicated the extent of DNA damage in lymphocytes. The DNA of
lymphocytes from healthy volunteers treated with 100 U/ml IFN-γ (N)
and IFN-γ (L) demonstrated no significant change in DNA damage
compared with untreated cells (Fig. 3A
and B). However, lymphocyte DNA from patients with lung cancer
(Fig. 3C and D) showed a
significant decrease in % DNA tail for IFN-γ (N) (P<0.05) and
(P<0.01) for IFN-γ (L) in lymphocytes from patients with lung
cancer. Moreover, the IFN-γ (L) with endonuclease III showed a
significant reduction in OTM compared with untreated cells
(P<0.01). hOGG1 enzymes with IFN-γ (L) also demonstrated a
significant decrease in DNA damage in lymphocytes from patients
with lung cancer (P<0.05).
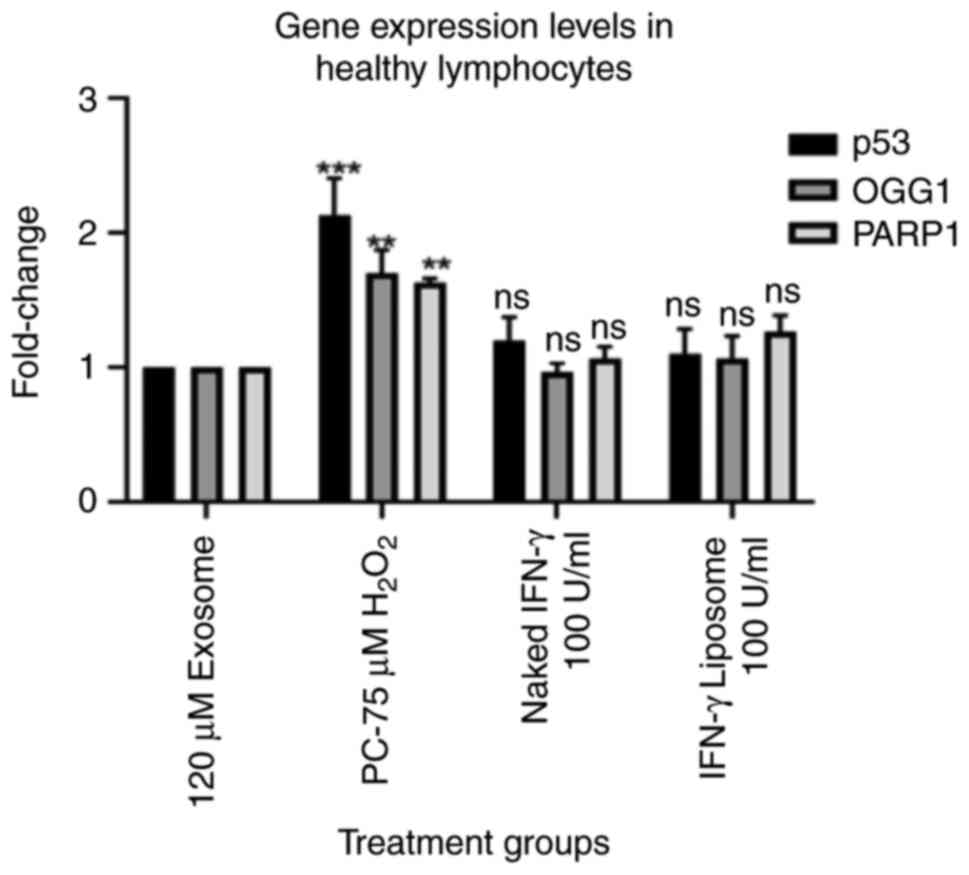
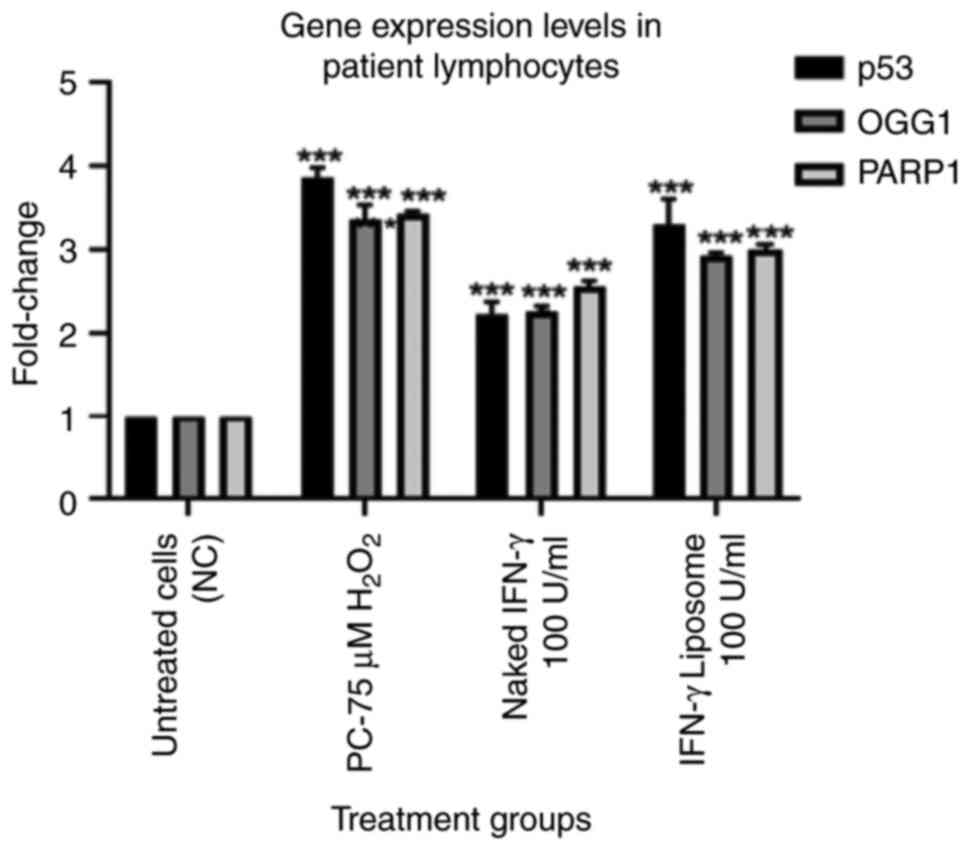
IFN-γ upregulates the gene expression
of p53, PARP1 and OGG1 genes in lymphocytes from patients with lung
cancer
The gene expression levels of p53, PARP1 and OGG1
were evaluated using RT-qPCR. The results indicated that 100 U/ml
IFN-γ (N) and 100 U/ml IFN-γ (L) treatments had no detectable
effects on the mRNA expression levels of p53, PARP1 and OGG1 in
lymphocytes from healthy individuals (Fig. 4). IFN-γ treatment significantly
increased the mRNA expression levels of p53, PARP1 and OGG1 in
lymphocytes from patients with lung cancer (P<0.001). However,
IFN-γ (L) upregulated the targeted genes markedly more than the
naked form (Fig. 5).
IFN-γ increases protein expression
levels of p53, OGG1 and PARP1 in lymphocytes from patients with
lung cancer
The results of the present study demonstrated that
p53, OGG1 and PARP1 protein expression levels in the lymphocytes of
healthy individuals were not significantly affected by 100 U/ml
IFN-γ (N) or 100 U/ml IFN-γ (L) (Fig.
6). However, p53, OGG1 and PARP1 protein expression levels in
lymphocytes from patients with lung cancer showed a statistically
significant increase when compared with the untreated cells
(Fig. 7). Treatment with 100 U/ml
IFN-γ (N) and 100 U/ml IFN-γ (L) significantly increased p53 levels
in lymphocytes from patients with lung cancer by 1.8 and 1.9-fold,
respectively. Moreover, compared with the control group, the OGG1
levels for both the naked and liposomal forms of 100 U/ml IFN-γ
increased by 1.9-fold. Furthermore, 100 U/ml IFN-γ (N) increased
the protein expression levels of PARP1 by 1.2-fold, whereas 100
U/ml IFN-γ (L) increased it by ~1.8-fold. IFN-γ (L) had a greater
impact on the protein levels of targeted proteins compared with the
naked form.
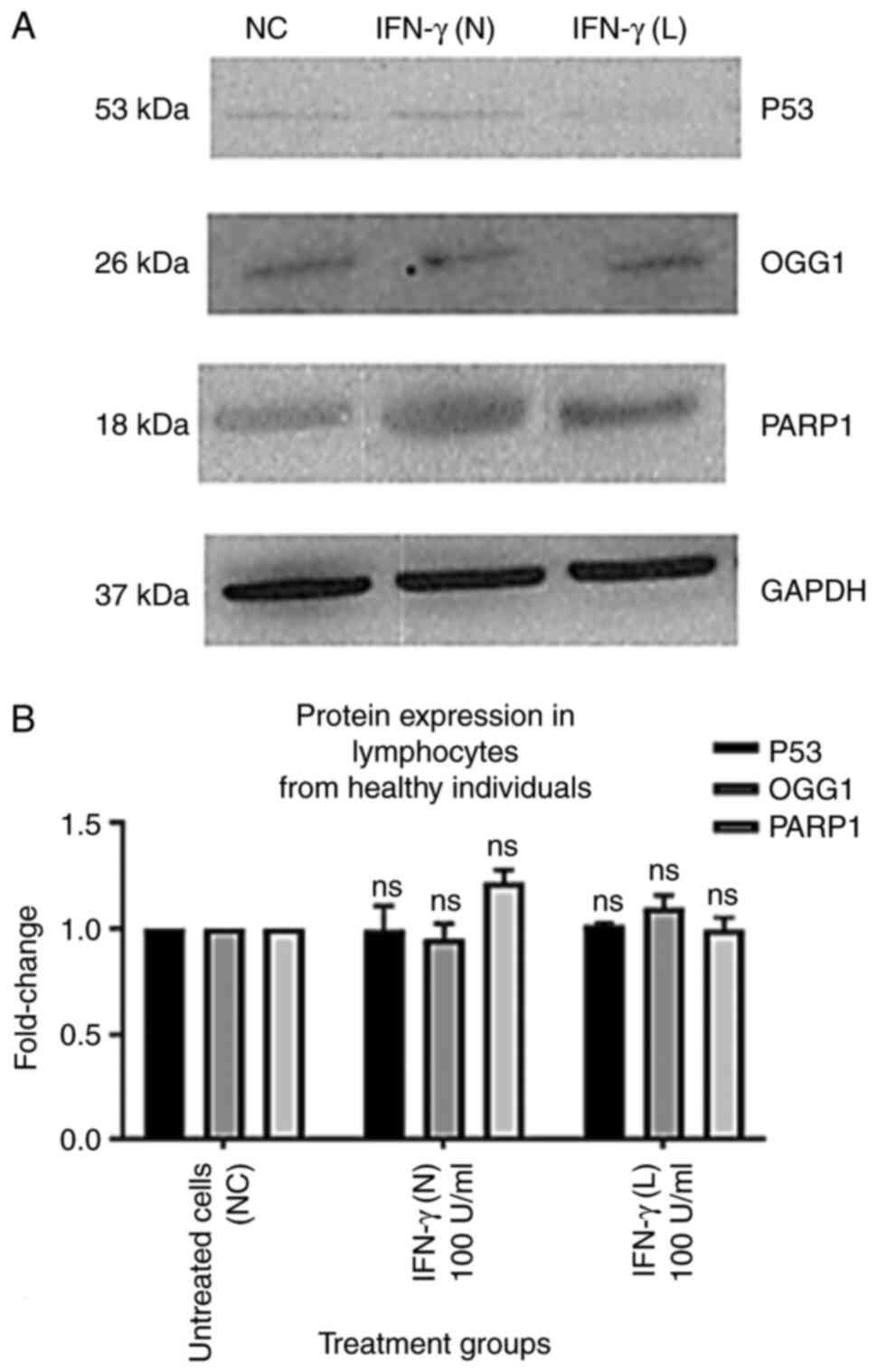 | Figure 6.Effect of 100 U/ml IFN-γ (N) and 100
U/ml IFN-γ (L) on the protein expression levels of p53, OGG1 and
PARP1 in lymphocytes from healthy individuals. All data from the
treatment groups were compared with the NC and normalized against
the internal reference protein, GAPDH. The experiment was repeated
three times in three different individuals. The treatment groups
included NC, 100 U/ml IFN-γ (N) and 100 U/ml IFN-γ (L). IFN-γ in
both forms did not demonstrate any significant effect on p53, OGG1
and PARP1 protein levels compared with the NC. (A) Immunoblot
analysis of p53, OGG1 and PARP1 proteins in lymphocytes from
healthy individuals treated with 100 U/ml IFN-γ (N) and 100 U/ml
IFN-γ (L). (B) Bar graphs presented fold changes in protein
expression levels. Data are presented as the mean ± SEM of three
experiments. NC, untreated cells; ns, not significant; OGG1, 8-oxo
guanine-DNA glycosylase; IFN-γ (N), naked IFN-γ; IFN-γ (L),
liposomal IFN-γ; PARP1, poly ADP-ribose polymerase 1. |
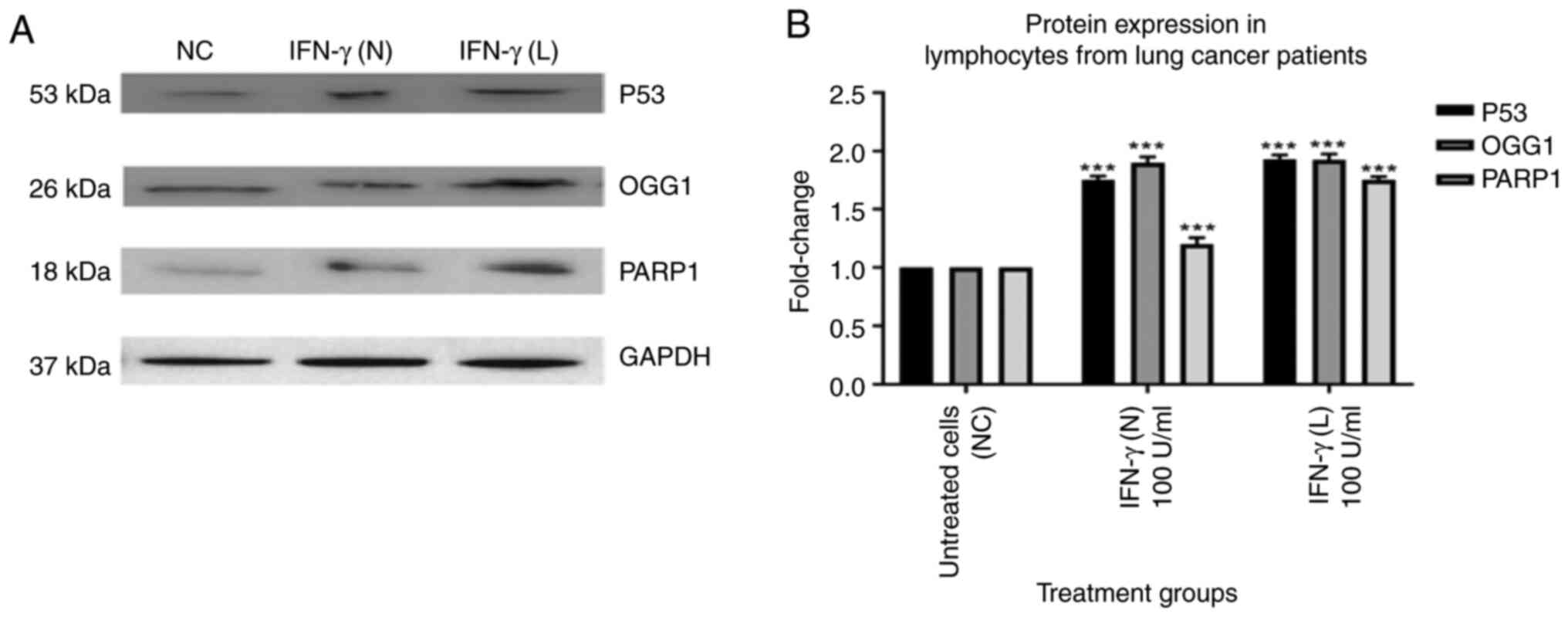 | Figure 7.Effect of 100 U/ml IFN-γ (N) and 100
U/ml IFN-γ (L) on the protein expression levels of p53, OGG1 and
PARP1 in lymphocytes from patients with lung cancer. All data from
the treatment groups were compared with the NC and normalized
against the internal reference protein, GAPDH. The experiment was
repeated three times in three different individuals. The treatment
groups included NC, 100 U/ml IFN-γ (N) and 100 U/ml IFN-γ (L).
IFN-γ in both forms significantly increased the protein expression
levels of p53, OGG1 and PARP1. The protein expression levels of
p53, OGG1 and PARP1 in lymphocytes from patients with lung cancer
showed a significant increase after treatment with IFN-γ compared
with the NC. (A) Immunoblot analysis of the p53, OGG1 and PARP1
proteins in lymphocytes from patients with lung cancer treated with
100 U/ml IFN-γ (N) and 100 U/ml IFN-γ (L). (B) Bar graphs
presenting fold changes in protein expression levels. Data are
presented as the mean ± SEM of three experiments. ***P<0.001
compared with the NC. ns, not significant; NC, untreated cells;
OGG1, 8-oxo guanine-DNA glycosylase; IFN-γ (N), naked IFN-γ; IFN-γ
(L), liposomal IFN-γ; PARP1, poly ADP-ribose polymerase 1. |
Discussion
The purpose of the present study was to analyze the
effect of IFN-γ on the peripheral lymphocytes of patients with lung
cancer and the ability of IFN-γ to protect against oxidative
stress.
Lymphocytes were selected as the model cells for the
present investigation. High levels of DNA damage in lymphocytes may
be caused by genetically impaired DNA repair mechanisms (27). Peripheral lymphocytes are an
excellent model for evaluating the sensitivity of the genome to
mutagens, which is determined by measuring genotoxic events
triggered by chemical or physical agents (28).
The results of the hOGG-1 and endonuclease III comet
modified test showed that IFN-γ (N) and IFN-γ (L) were able to
repair DNA damage in human lymphocytes derived from patients with
lung cancer and healthy persons compared with untreated cells.
IFN-γ (L) decreased DNA damage more effectively than IFN-γ (N)
compared with untreated cells. These findings were consistent with
previous reports in which lymphocytes from patients with lung
cancer were treated with IFN-γ in both forms and which revealed
that DNA damage was reduced compared with untreated cells (29,30).
Nonetheless, the reduction in DNA damage caused by IFN-γ (L) was
greater than the reduction caused by IFN-γ (N). This may be a
consequence of the increased biocompatibility and cellular
reactivity of liposomes as compared with compounds larger particles
(24).
The tumor-suppressor, p53, provides a protective
effect against the development of cancer by serving a crucial part
in genomic stability-maintaining homeostasis and repairing
processes (31). In addition,
certain DNA repair mechanisms, including the BER pathway which
depends on the activity of the OGG1 and PARP1 proteins, are
important to protect genetic integrity and prevent mutations that
can cause disease or cell death (32). Therefore, it is important to unravel
the effect of IFN-γ on the gene expression and protein expression
levels of p53, OGG1 and PARP1.
In the present study, there was a significant
upregulation of the p53 gene and increase in the protein expression
level of p53 in lymphocytes from patients with lung cancer after 24
h of treatment with both the liposome and naked forms of IFN-γ.
These findings suggested that IFN-γ may encourage p53-mediated cell
cycle arrest and DNA repair in patients with lung cancer and that
the protective effects caused by IFN-γ might be dependent on the
tumor suppressor activity of the p53 gene. This is similar to a
previous study which reported that IFN-γ activated p53 expression
in melanoma cancer, resulted in the triggering of certain cellular
stressors, such as those brought on by DNA damage and replication
stress caused by misregulated oncogenes (33).
Previous research has shown that p53 may also
influence the transcriptional expression of BER genes, including
OGG1 and PARP1 (34). Similarly,
the findings of the present study demonstrated that protein and
mRNA expression levels of OGG1 were increased in lymphocytes from
patients with lung cancer after treatment with IFN-γ (L) and IFN-γ
(N). Moreover, PARP1 levels were significantly affected by both
forms of IFN-γ in lymphocytes from patients with lung cancer and
healthy individuals.
However, the p53, OGG1 and PARP1 protein and mRNA
expression levels in lymphocytes from healthy individuals after
treatment with IFN-γ liposome and naked forms were barely
detectable. The findings were supported by previously reported
research in which the MDM2 proto-oncogene maintains p53 at a low
level in normal cells (35,36).
Furthermore, the present study demonstrated that
expression of p53, OGG1 and PARP1 genes in lymphocytes from healthy
individuals and patients with lung cancer was up-regulated by
stimulation with H2O2 which used as a
positive control, these results were consistent with a previous
study, which reported that H2O2 induced
apoptosis in H9C2 cells by an increase in p53
expression (30,37). Taken together, the findings of the
present study suggested that IFN-γ may prevent lung cancer by
stopping tumor cell cycles via induction of the expression of p53,
OGG1 and PARP1 genes and increasing protein levels, and that
liposomes may be a more effective alternative drug delivery
strategy in certain conditions such as severe side effects and
chemotherapy resistance.
The potential mechanism of the effect of IFN-γ on
lymphocytes is complicated, and further work is needed to evaluate
the mechanism in vitro and in vivo. The mechanism of
IFN-γ effect on lymphocytes is intricate and not fully understood.
The study may not have comprehensively explained all facets of this
complex mechanism, indicating the need for further research in this
area. The study primarily relied on in vitro (cell culture)
models, which may not completely represent the intricacies of
immune responses that occur in living organisms (in vivo).
Findings from in vitro experiments might not always directly
apply in vivo situations.
Acknowledgements
The authors would like to thank University of
Bradford in the UK, Mutah University, and Philadelphia University
in Jordan for use of laboratories and machines.
Funding
The present study was funded by the University of Bradford
(grant no. 43091/205100/DB071).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DA, MA and HANAJ conceptualized the study; MA
performed the study methodology; WH, YAH, MO, HKMS and IAT
performed the formal analysis; DA investigated the present study;
ASAW supplied the resources for the present study; MA and BA wrote
the original draft preparation; HANAJ, BA, WH, NRM and ASAW
analyzed and interpreted data, reviewed and edited the manuscript;
NRM, YAH, MO and IAT performed study visualization; and DA
supervised, performed project administration and acquired funding
for the present study. All authors have read and approved the final
version of the manuscript. MA, BA and HANAJ confirmed the
authenticity of all the raw data.
Ethics approval and consent to
participate
Ethical approval was obtained to perform the Comet
repair assay, RT-qPCR and western blotting for the study of IFN-γ
(N) and IFN-γ (L). The present study received ethical approval from
The Leeds East Research Ethics Committee (approval no. 12/YH/0464;
Leeds, UK), The University of Bradford Research Ethics
Sub-Committee on Research in Human Subjects (approval no. 0405/8;
Bradford, UK) and The Research Support and Governance Office,
Bradford Teaching Hospitals, NHS Foundation (approval no. RE DA
1202; Bradford, UK). Informed consent was obtained from all
participants prior to participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Oo AM, Mohd Adnan LH, Nor NM, Simbak N,
Ahmad NZ and Lwin OM: Immunomodulatory effects of flavonoids: An
experimental study on natural-killer-cell-mediated cytotoxicity
against lung cancer and cytotoxic granule secretion profile.
Proceedings Singapore Healthcare. 30:279–285. 2020. View Article : Google Scholar
|
|
2
|
Daud NNNNM, Septama AW, Simbak N, Bakar
NHA and Rahmi EP: Synergistic effect of flavonoids from artocarpus
heterophyllus heartwoods on anticancer activity of cisplatin
against H460 and MCF-7 cell lines. Nat Product Sci. 25:311–316.
2019. View Article : Google Scholar
|
|
3
|
Miller CH, Maher SG and Young HA: Clinical
use of interferon-γ. Ann N Y Acad Sci. 1182:69–79. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferreira VL, Borba HHL, Bonetti AdF,
Leonart LP and Pontarolo R: Cytokines and interferons: Types and
functions. Autoantibodies Cytokines. 13:2018.
|
|
5
|
Castro F, Cardoso AP, Gonçalves RM, Serre
K and Oliveira MJ: Interferon-gamma at the crossroads of tumor
immune surveillance or evasion. Front Immunol. 9:8472018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hishinuma S, Ogata Y, Tomikawa M, Ozawa I,
Hirabayashi K and Igarashi S: Patterns of recurrence after curative
resection of pancreatic cancer, based on autopsy findings. J
Gastrointest Surg. 10:511–518. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Y, Bao Q, Yang S, Yang M and Mao C:
Bionanoparticles in cancer imaging, diagnosis, and treatment. View.
3:202000272022. View Article : Google Scholar
|
|
8
|
de Ruiter MV, Klem R, Luque D, Cornelissen
JJ and Castón JR: Structural nanotechnology: Three-dimensional
cryo-EM and its use in the development of nanoplatforms for in
vitro catalysis. Nanoscale. 11:4130–4146. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Z and Chen H: The recent progress of
inorganic-based intelligent responsive nanoplatform for tumor
theranostics. View. 3:202200092022. View Article : Google Scholar
|
|
10
|
Koizumi S-i, Wakita D, Sato T, Mitamura R,
Izumo T, Shibata H, Kiso Y, Chamoto K, Togashi Y, Kitamura H and
Nishimura T: Essential role of Toll-like receptors for dendritic
cell and NK1. 1+ cell-dependent activation of type 1 immunity by
Lactobacillus pentosus strain S-PT84. Immunol Lett. 120:14–19.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ramos TI, Villacis-Aguirre CA, Santiago
Vispo N, Santiago Padilla L, Pedroso Santana S, Parra NC and Alonso
JRT: Forms and methods for interferon's encapsulation.
Pharmaceutics. 13:15332021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jailani MTM, Totten J, Nevin J and Halbert
G: ID 180. Fabrication, characterisation and in vitro study of
dual-loaded irinotecan/cisplatin liposomes. J Pharmacy Bioallied
Sci. 12:2020.
|
|
13
|
Hirai M, Kimura R, Takeuchi K, Hagiwara Y,
Kawai-Hirai R, Ohta N, Igarashi N and Shimuzu N: Structure of
liposome encapsulating proteins characterized by X-ray scattering
and shell-modeling. J Synchrotron Radiat. 20:869–874. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao H, Wu L, Yan G, Chen Y, Zhou M, Wu Y
and Li Y: Inflammation and tumor progression: Signaling pathways
and targeted intervention. Signal Transduct Target Ther. 6:2632021.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kalm M, Andreasson U, Björk-Eriksson T,
Zetterberg H, Pekny M, Blennow K, Pekna M and Blomgren K: C3
deficiency ameliorates the negative effects of irradiation of the
young brain on hippocampal development and learning. Oncotarget.
7:193822016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wirth M, Plattner V and Gabor F:
Strategies to improve drug delivery in bladder cancer therapy.
Expert Opin Drug Deliv. 6:727–744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pisal DS, Kosloski MP and Balu-Iyer SV:
Delivery of therapeutic proteins. J Pharm Sci. 99:2557–2575. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu H, Gao HY, Guo H, Wang GZ, Yang YQ, Hu
Q, Liang LJ, Zhao Q, Xie DW, Rao Y and Zhou GB: Upregulation of
wild-type p53 by small molecule-induced elevation of NQO1 in
non-small cell lung cancer cells. Acta Pharmacol Sinica.
43:692–702. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Almajali B, Johan MF, Al-Wajeeh AS, Wan
Taib WR, Ismail I, Alhawamdeh M, Al-Tawarah NM, Ibrahim WN,
Al-Rawashde FA and Al-Jamal HAN: Gene expression profiling and
protein analysis reveal suppression of the C-Myc oncogene and
inhibition JAK/STAT and PI3K/AKT/mTOR signaling by thymoquinone in
acute myeloid leukemia cells. Pharmaceuticals. 15:3072022.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Williams AB and Schumacher B: p53 in the
DNA-damage-repair process. Cold Spring Harb Perspect Med.
6:a0260702016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu X, Jin S, Ma Y, Fan Z, Yan Z, Li W,
Song Q, You W, Lyu Z, Song Y, et al: miR-30a-5p enhances paclitaxel
sensitivity in non-small cell lung cancer through targeting BCL-2
expression. J Mol Med (Berl). 95:861–871. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Slupphaug G, Kavli B and Krokan HE: The
interacting pathways for prevention and repair of oxidative DNA
damage. Mutat Res. 531:231–251. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Noren Hooten N, Kompaniez K, Barnes J,
Lohani A and Evans MK: Poly(ADP-ribose) polymerase 1 (PARP-1) binds
to 8-oxoguanine-DNA glycosylase (OGG1). J Biol Chem.
286:44679–44690. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Alhawamdeh M, Isreb M, Aziz A, Jacob BK,
Anderson D and Najafzadeh M: Interferon-γ liposome: A new system to
improve drug delivery in the treatment of lung cancer. ERJ Open
Res. 7:00555–2020. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Van Slooten M, Boerman O, Romøren K, Kedar
E, Crommelin D and Storm G: Liposomes as sustained release system
for human interferon-γ: Biopharmaceutical aspects. Biochim Biophys
Acta. 1530:134–145. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Haggag YA, Matchett KB, Falconer RA, Isreb
M, Jones J, Faheem A, McCarron P and El-Tanani M: Novel ran-RCC1
inhibitory peptide-loaded nanoparticles have anti-cancer efficacy
in vitro and in vivo. Cancers. 11:2222019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fouad YA and Aanei C: Revisiting the
hallmarks of cancer. Am J Cancer Res. 7:10162017.PubMed/NCBI
|
|
28
|
Collins AR: The comet assay for DNA damage
and repair: principles, applications, and limitations. Mol
Biotechnol. 26:249–261. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alhawamdeh MF: Applying a new technique,
the interferon gamma liposomal delivery system to improve drug
delivery in the treatment of lung cancer. University of Bradford;
2021, PubMed/NCBI
|
|
30
|
Rivlin N, Brosh R, Oren M and Rotter V:
Mutations in the p53 tumor suppressor gene: important milestones at
the various steps of tumorigenesis. Genes Cancer. 2:466–474. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yin T, Wang P, Li J, Wang Y, Zheng B,
Zheng R, Cheng D and Shuai X: Tumor-penetrating codelivery of siRNA
and paclitaxel with ultrasound-responsive nanobubbles
hetero-assembled from polymeric micelles and liposomes.
Biomaterials. 35:5932–5943. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lieschke E, Wang Z, Kelly GL and Strasser
A: Discussion of some ‘knowns’ and some ‘unknowns’ about the tumour
suppressor p53. J Mol Cell Biol. 11:212–223. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chatterjee N and Walker GC: Mechanisms of
DNA damage, repair, and mutagenesis. Environ Mol Mutagen.
58:235–263. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fukui T, Matsui K, Kato H, Takao H,
Sugiyama Y, Kunieda K and Saji S: Significance of apoptosis induced
by tumor necrosis factor-α and/or interferon-γ against human
gastric cancer cell lines and the role of the p53 gene. Surg Today.
33:847–853. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thiem A, Hesbacher S, Kneitz H, di Primio
T, Heppt MV, Hermanns HM, Goebeler M, Meierjohann S, Houben R and
Schrama D: IFN-gamma-induced PD-L1 expression in melanoma depends
on p53 expression. J Exp Clin Cancer Res. 38:3972019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Midgley CA and Lane DP: p53 protein
stability in tumour cells is not determined by mutation but is
dependent on Mdm2 binding. Oncogene. 15:1179–1189. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang M, Zhang Y, Xu E, Mohibi S, de Anda
DM, Jiang Y, Zhang J and Chen X: Rbm24, a target of p53, is
necessary for proper expression of p53 and heart development. Cell
Death Differ. 25:1118–1130. 2018. View Article : Google Scholar : PubMed/NCBI
|