Introduction
According to the World Health Organization, renal cell carcinoma (RCC) is the 13th most common cause of cancer-related death worldwide (1). Globally, 2–3% of malignant tumors in adults are RCC and 90% of all malignant kidney tumors are RCC (2). On average, clear cell RCC (ccRCC) is the most common histological subtype of RCC (70–80%), followed by papillary (10–20%) and chromophobe (5%) RCC (3). Clinically, ccRCC is divided into localized ccRCC, locally advanced ccRCC and metastatic ccRCC. The treatment of ccRCC is challenging, particularly for patients with metastatic ccRCC. At present, early resection is the main type of treatment for patients with ccRCC, but it has a marginal effect on the survival and prognosis of terminally ill patients with ccRCC (4). Furthermore, renal cancer does not respond well to radiotherapy or chemotherapy; therefore, the prognosis is unfavorable for patients with this type of cancer and numerous patients die from renal cancer (5). In most cases, there are limited curative treatment options available to patients at an advanced stage of the disease. However, despite the fact that immunotherapy and molecular targeted drugs for the treatment of RCC may have a certain therapeutic effect on advanced RCC, they remain unsatisfactory due to drug resistance (6). To date, the molecular pathological mechanism of renal cancer remains unclear. Therefore, it is of great clinical value to explore the molecular mechanisms underlying ccRCC development and progression, and to identify novel diagnostic and prognostic biomarkers and therapeutic targets for ccRCC.
The gasdermin (GSDM) family consists of pore-forming proteins that serve pivotal roles in programmed cell death and inflammation (7). The GSDM protein superfamily is classified according to the GSDM-N domain, which is responsible for executing the process of pyroptosis, and comprises six members [GSDMA, GSDMB, GSDMC, GSDMD, GSDME (also known as DFNA5) and DFNB59 (also known as Pejvakin, PJVK)] (8,9). Each member of the GSDM family has unique functions and is implicated in various cellular processes. There is a structural similarity between the GSDM superfamily members (GSDMA, GSDMB, GSDMC, GSDMD, DFNA5 and DFNB59) that is essential for the induction of pyroptosis (7,10,11). Inflammasomes are involved in pyroptosis, which is a type of inflammation-dependent programmed cell death (12). With the discovery of the GSDM family, the research scope of pyroptosis has expanded. In recent years, there has been an increasing interest in pyroptosis since it has a vital role in activating the immune system and regulating tumor behavior (13). There has been evidence to suggest that the GSDM family is dysfunctional and upregulated in several types of cancer in humans, suggesting that these proteins may serve a role in tumor development (14). For example, the upregulation of GSDMA in gastric cancer cells has been shown to promote apoptosis signaling that leads to reduced cell proliferation (15). The variation in GSDM expression across different tumors, such as gastric cancer and RCC, may be due to tumor-specific factors, epigenetic changes or tissue-specific regulatory mechanisms (16). Further research is required to understand the context-specific roles of GSDM in different types of cancer. According to Lutkowska et al (17), high-resolution melting curve analysis showed that a single nucleotide polymorphism, rs8067378, increased the expression of GSDMB, further contributing to cervical carcinogenesis. Another study reported that, based on the observation that GSDMC silencing significantly reduces colorectal cancer cell proliferation, GSDMC may act as an oncogene in colorectal cancer (18). Furthermore, GSDME overexpression has been reported to promote the migration and invasion of SW480 and HCT116 colorectal cancer cells (19). Lin et al (20) demonstrated that the overexpression of GSDMD could be associated with adverse clinical outcomes in osteosarcoma, and it may act as a therapeutic target for the disease. However, to the best of our knowledge, the detailed functions and underlying mechanisms of the six GSDM family members in ccRCC development have yet to be fully elucidated and remain unknown.
A number of public databases were used in the present study to examine the expression and mutation status of GDSM genes in ccRCC. In addition, the association between the expression levels of GDSM genes and clinicopathological features, prognosis, tumor immune cell infiltration and drug sensitivity in patients with ccRCC was investigated. Consequently, the present study may improve the understanding of the role of GSDM genes in ccRCC progression, and may indicate that GSDM genes have complex and distinct functions.
Materials and methods
Ethics statement
The present study obtained ethics approval from The First Affiliated Hospital of Nanchang University (Nanchang, China) (approval no. 202012-110). Tissue samples were collected from patients who had provided written informed consent following admittance to the Department of Urology at The First Affiliated Hospital of Nanchang University.
Patients and tumor samples
In the present study, two independent pathologists confirmed 23 paired cases of ccRCC and normal tissue (2 cm away from cancer tissue). Of the 23 patients, 12 were female and 11 were male; their ages ranged between 35 and 67 years, with an mean age of 53 years. In addition, all patients underwent radical nephrectomy. Following resection, 23 pairs of matched ccRCC tissues and adjacent normal kidney tissues were stored in liquid nitrogen until they could be used at The First Affiliated Hospital of Nanchang University from August 2020 to August 2021.
Cell lines and cell culture
The following cell lines were purchased from the American Type Cell Collection: Human renal cancer cell lines (786-O, Caki-1 and ACHN) and normal kidney HK-2 cells. Cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) and 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere containing 5% CO2.
Reverse transcription (RT)-qPCR
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to isolate total RNA from 23 matched tissue samples and cell lines. According to the manufacturer's protocol, 1 µg RNA from each sample was used for cDNA synthesis (Tiangen Biotech Co., Ltd.) using the PrimeScript™ RT reagent kit (Takara Bio, Inc.) For qPCR, which was performed using a SYBR Green qPCR kit (Takara Bio, Inc.), the following conditions were applied: 95°C for 2 min, followed by 40 cycles at 95°C for 5 sec and 60°C for 10 sec. To evaluate the relative gene expression, the 2−ΔΔCq method was used (21). The primer sequences used were as follows: GAPDH, forward 5′-GCCACATCGCTCAGACACCAT-3′, reverse 5′-CCCATACGACTGCAAAGACCC; GSDMA, forward 5′-GCTAAAGCTGGTGGAGAGCA-3′ reverse 5′-GAGGAGAGCAGCTCAGGTTG-3′; GSDMB, forward 5′-CTGGATTCTGGGCTCCAAGG-3′, reverse 5′-GATGGTGGAAGCCCTGGAAA-3′; GSDMC, forward 5′-GTGGTGACAGAGGCTGTTGA-3′, reverse 5′-CTCTCTCCTTGGCCTTGACC-3′; GSDMD, forward 5′-GACCCTAACACCTGGCAGAC-3′, reverse 5′-TTCTGTGTCTGCAGCACCTC-3′; DFNA5, forward 5′-TGCTGCGCATGGGATATCTT-3′, reverse 5′-AGCTCCGCAAATGGATGGAA-3′; and PJVK, forward 5′-GGAGCAGCAGAAAGGCAGTA-3′, reverse, 5′-TTCCAGCATGCACAGACAGT-3′. All RT-qPCR assays were performed using SYBR Real-Time PCR kit (Qiagen, Inc.) at 95°C for 2 min, followed by 40 cycles at 95°C for 5 sec and 60°C for 10 sec. In order to assess relative gene expression, the 2−ΔΔCq method was used. Additionally, the GAPDH gene was used as a reference gene and each of the analyses was performed in triplicate (22).
Statistical analysis
The statistical analyses were conducted using the R package (v3.6.2) (http://www.R-project.org/). Univariate Cox regression analyses were used to evaluate the effect of clinical parameters and the mRNA expression levels of GSDM genes on survival and other clinical characteristics of patients with ccRCC. Subsequently, parameters with P<0.1 were retained for further analysis. Additionally, multivariate Cox regression analysis was performed to examine the relationship between clinical characteristics and the expression of GSDM genes. Paired Student's t-test was used to compare differences in GSDM gene expression between RCC tissues and adjacent tissues. Kruskal-Wallis test with Dunn post hoc test was used to compare more than two groups. P<0.05 was considered to indicate a statistically significant difference.
UALCAN analysis
The UALCAN database (http://ualcan.path.uab.edu) contains data on 31 types of cancer patients, including clinical and RNA-sequencing information (23). UALCAN can be used to analyze the relationship between the expression levels of target genes and the clinical characteristics of patients from TCGA dataset (https://portal.gdc.cancer.gov/). Using the online bioinformatics tool based on TCGA_kidney renal clear cell carcinoma (TCGA_KIRC) data, the association between GSDM expression and clinical pathological parameters was determined by the Kruskal-Wallis test. P<0.05 was considered to indicate a statistically significant difference.
Gene Expression Profiling Interactive Analysis (GEPIA)
The online service GEPIA (http://gepia.cancer-pku.cn/) provides expression data from a wide range of cancer types. Based on TCGA and the Genotype Tissue Expression data, the GEPIA dataset includes 9,736 tumor samples and 8,587 normal samples from healthy individuals (24). The present study investigated the expression levels of GSDM genes in cancerous and non-cancerous tissues, with the aim of evaluating their prognostic significance using the Kaplan-Meier method. Statistical analysis was performed, and P<0.05 was considered to indicate a statistically significant difference.
Survival analysis
Patient survival analysis and visualization were performed using R packages survival (https://CRAN.R-project.org/package=survival), survminer and ggplot2 (https://CRAN.R-project.org/package=survminer) based on TCGA-KIRC database. Median value was used as a cut-off value for prognostic analysis. The hazard ratios, corresponding 95% confidence intervals and Kaplan-Meier curves, which were analyzed by log-rank test, were computed and presented using the Xiantao platform (https://www.xiantao.love/).
cBioPortal
The cBioPortal (www.cbioportal.org) is a free online resource developed to comprehensively analyze cancer genomics data and clinical data from TCGA database (25). The present study collected genomic profiles corresponding to GSDM family tumor genomes, which contained mutations and putative copy-number changes as determined by the Genomic Identification of Significant Targets in Cancer, and also analyzed mRNA expression levels. Additionally, a Kaplan-Meier plot was used to determine the association between GSDM genetic mutations and the overall survival (OS) of patients with KIRC; a log-rank test was performed to compare the altered group with the unaltered group. Using the cBioPortal database, the mutation rate refers to the proportion of patients with genetic mutations among all of those who have undergone genetic sequencing.
Protein-protein interaction (PPI) network construction
A PPI network consisting of the 168 most frequently altered neighboring genes and the GSDM genes was constructed using data obtained from the cBioPortal database. The PPI network was constructed using an online program (https://www.xiantao.love/), which was based on R. P<0.05 was the cutoff criterion.
Functional enrichment analysis
Metascape (metascape.org/) combines multiple databases including the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), UniProt and DrugBank databases (26). Metascape analyzed the functional enrichment of the genes in the hub module in the study. P<0.01 was set as the cutoff criterion and significance was determined by enrichment ranking [-log10 (P-value)].
GSCALite
The GSCALite analysis platform (http://bioinfo.life.hust.edu.cn/web/GSCALite/) provides analysis of gene sets in cancer and drug sensitivity profiles (27). In addition to differentially expressed genes, GSCALite also provides details of single nucleotides, methylation, pathway activity, and microRNA (miRNA) regulation, normal tissue expression and drug sensitivity. Additionally, GSDM genes were analyzed for pathway activity, miRNA networks and drug sensitivity in ccRCC using this tool. By using the pathway activity module, the present study examined the relationship between members of the GSDM family and pathway activity. The drug-sensitivity module was used to examine the relationship between drug sensitivity and the expression levels of GSDM genes in cancer cell lines from the Genomics of Drug Sensitivity in Cancer (GDSC) database (www.cancerrxgene.org) (28) and Cancer Therapeutics Response Portal (https://portals.broadinstitute.org/ctrp/) (29). The Pearson correlation coefficient (or Spearman rank correlation coefficient, if appropriate) was employed to quantify the strength and direction of the correlation. This analysis aimed to identify genes that may be correlated with increased or decreased sensitivity to specific drugs. The Pathway Activity module based on the GSCALite database presents the difference in gene expression between pathway activity groups (activation and inhibition).
Tumor Immune Estimation Resource (TIMER)
The comprehensive database TIMER (http://cistrome.dfci.harvard.edu/TIMER/) allows systematic analysis of immune infiltrates associated with various types of cancer (30). In the gene module, the association between members of the GSDM family and immune cell infiltration in ccRCC was examined. The somatic copy number alteration (SCNA) module was used to compare tumor-infiltration levels among tumors with different somatic copy number changes of GSDM genes.
Results
Expression levels of different GSDM genes in patients with ccRCC
The expression levels of GSDM genes were higher in 72 paired ccRCC tissue samples compared with those in 72 paired normal renal tissue samples according to TCGA dataset (https://portal.gdc.cancer.gov/). As shown in Fig. 1A, the transcriptional levels of GSDMA, GSDMB, GSDMC and GSDMD were significantly higher in ccRCC tissues compared with those in normal kidney tissue samples, but there was no significant difference in the expression of GSDME in ccRCC tissues compared with in non-cancerous renal tissue. By contrast, the mRNA expression levels of PJVK in cancerous tissue were lower than those in normal tissue. Subsequently, based on the UALCAN database, the mRNA expression levels of GSDM genes were detected in patients with ccRCC. As shown in Fig. 1B, C, E and G, the mRNA expression levels of GSDMA, GSDMB and GSDMD were significantly elevated in ccRCC tissues compared with those in the normal kidney tissues, whereas the mRNA expression levels of DFNB59 were lower in cancer tissues than those in normal tissues in Fig. 1G. There was no difference in the expression of GSDMC and DFNA5 between cancer and non-cancer tissues (Fig. 1D and F).
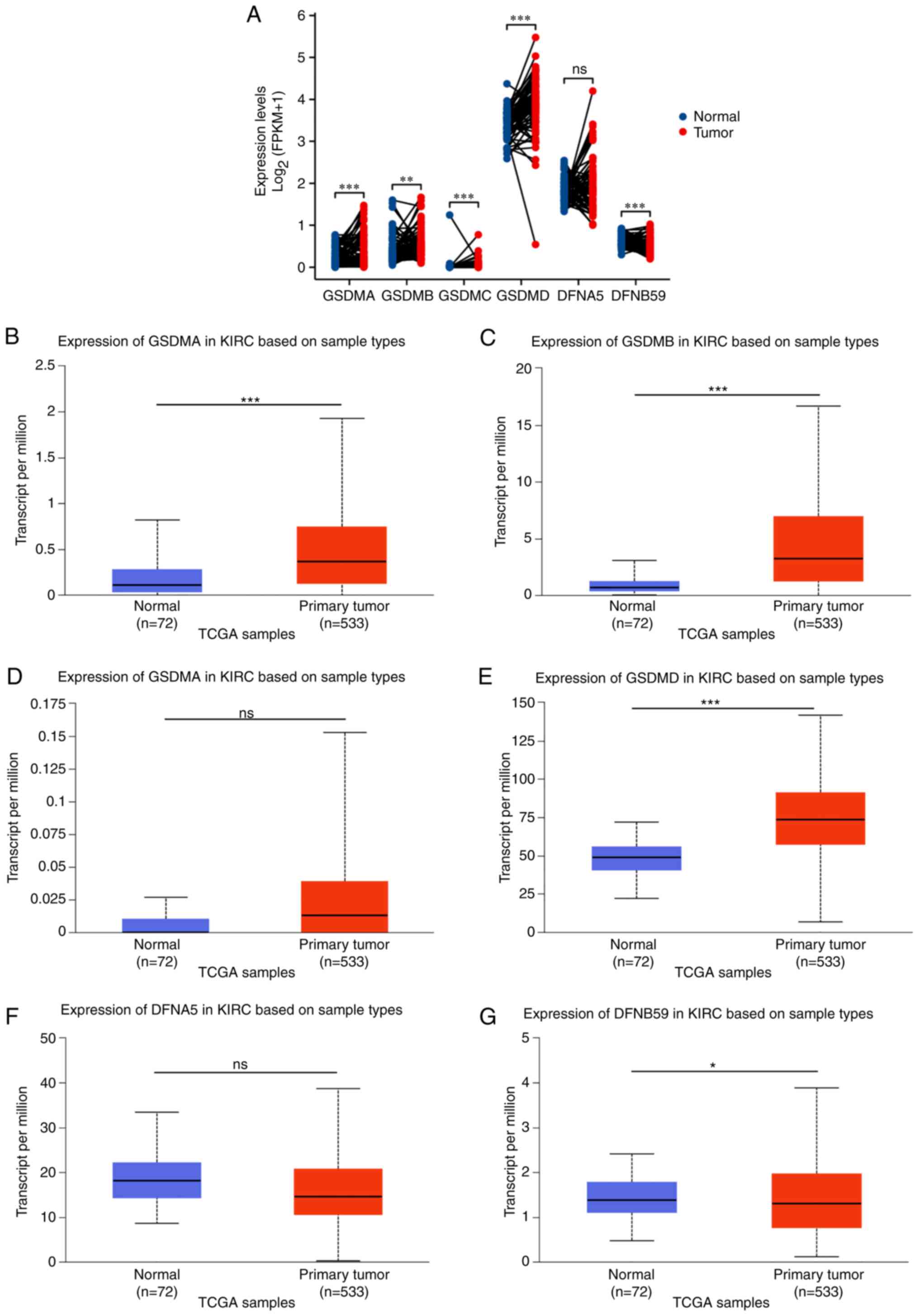 |
Figure 1.
Expression of GSDMA, GSDMB, GSDMC, GSDMD, DFNA5 and DFNB59 in KIRC based on sample types. (A) In 72 pairs of KIRC tissues and adjacent normal kidney tissues from TCGA and Genotype Tissue Expression databases, transcriptional expression of GSDM genes was evaluated. Expression levels of (B) GSDMA, (C) GSDMB, (D) GSDMC, (E) GSDMD, (F) DFNA5 and (G) DFNB59 in clear cell renal cell carcinoma and normal kidney tissues based on the UALCAN database. *P<0.05, **P<0.01, ***P<0.001. GSDM, gasdermin; KIRC, kidney renal clear cell carcinoma; ns, not significant; TCGA, The Cancer Genome Atlas.
|
To validate this conclusion, RT-qPCR analysis was performed using 23 pairs of cancer and adjacent normal tissues obtained from patients with ccRCC. The results showed that the mRNA expression levels of GSDMA, GSDMB, GSDMC, GSDMD and GSDME were decreased in normal tissues compared with those in cancer tissues, whereas the expression levels of PJVK were decreased in cancerous tissues (Fig. 2A-F). To determine the mRNA expression levels of GSDMA, GSDMB, GSDMC, GSDMD, GSDME and PJVK in kidney cancer and healthy cell lines, RT-qPCR was also conducted. The results showed that the mRNA expression levels of GSDMA, GSDMB, GSDMC and GSDMD were decreased in the normal kidney cell line HK-2, relative to those in the kidney cancer cell lines 786-O, Caki-1, and ACHN, whereas PJVK had the opposite trend (Fig. 2G).
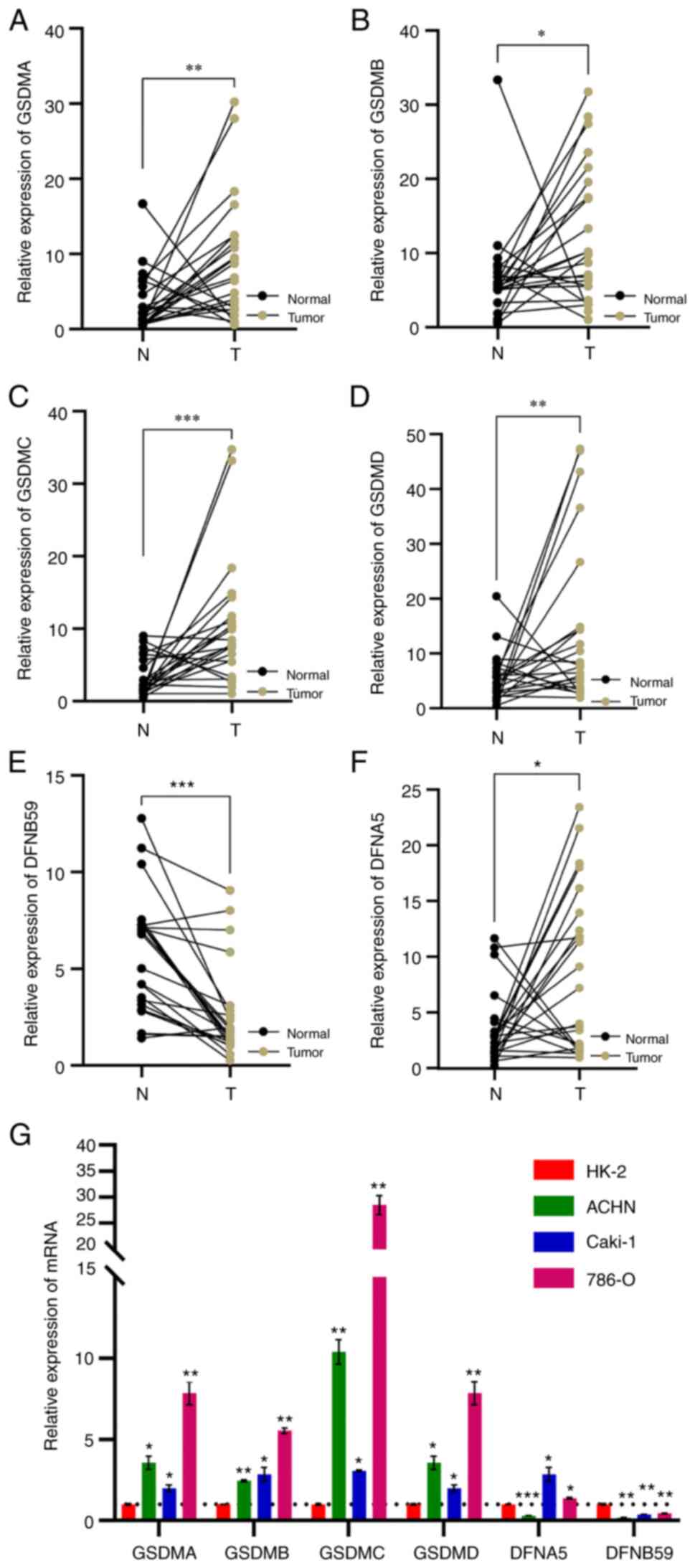 |
Figure 2.
Reverse transcription-quantitative PCR was used to detect the mRNA expression levels of (A) GSDMA, (B) GSDMB, (C) GSDMC, (D) GSDMD, (E) GSDME and (F) PJVK in clear cell renal cell carcinoma tissues and paired adjacent normal kidney tissues. (G) Transcriptional levels of GSDMA, GSDMB, GSDMC, GSDMD, DFNA5 and DFNB59 in a normal kidney cell line and kidney cancer cell lines. ns, no significance; *P<0.05, **P<0.01, ***P<0.001 vs. HK-2 or as indicated. GSDM, gasdermin.
|
Association between GSDM genes and the clinicopathological parameters of patients with ccRCC
The association between the mRNA expression levels of GSDM genes and clinicopathological characteristics, including tumor grade, tumor stage and molecular subtype, was analyzed using the UALCAN and GEPIA databases. From grade 1 to grade 4, DFNA5 expression increased gradually and significantly (Fig. 3E). The mRNA expression levels of GSDMA and GSDMB were significantly associated with tumor grade, with mRNA expression increasing alongside tumor grade, while the mRNA expression levels of DFNB59 exhibited the opposite result, with a significant reduction in expression detected between normal and grade 1 tissues (Fig. 3A, B and F). In the case of GSDMD and GSDMC, there was a significant difference in GSDMC expression between grades 1 and 2–4 tissue, and in GSDMD expression between normal and grades 1–4 tissue (Fig. 3C and D). As for tumor stage, the mRNA expression levels of GSDMB, GSDMD and DFNA5 were significantly related to patient tumor stage, whereas the mRNA expression levels of GSDMA, GSDMC and DFNB59 were not associated with patient tumor stage (Fig. 3G-L). The lowest mRNA expression levels of GSDMB, GSDMD and DFNA5 were found in patients with stage I, and patients with more advanced tumor stages tended to exhibit higher mRNA expression levels of GSDMB, GSDMD and DFNA5. These data indicated that the differentiation of tumor cells and the staging of tumors are, to some extent, attributed to changes in GSDM expression.
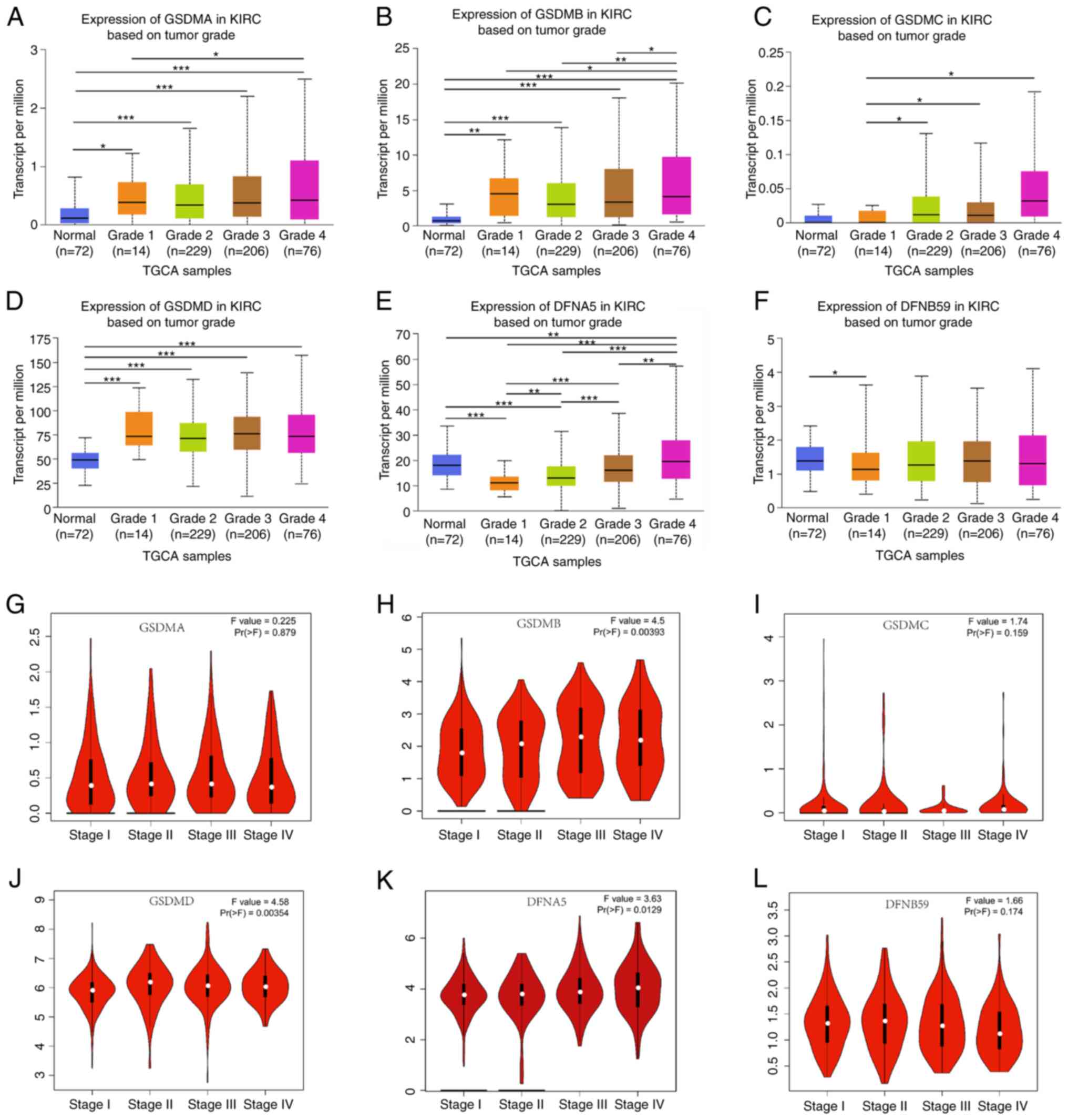 |
Figure 3.
Clinicopathological parameters associated with GSDM mRNA expression levels in patients with ccRCC. (A-F) Association of GSDM gene expression with ccRCC tumor grade. The mRNA expression levels of (A) GSDMA and (B) GSDMB were significantly associated with tumor grade, with mRNA expression increasing alongside tumor grade, while the mRNA expression levels of (F) DFNB59 exhibited the opposite result, with a significant reduction in expression detected between normal and grade 1 tissues. There was a significant difference in (C) GSDMC expression between grades 1 and 2–4 tissue, and in (D) GSDMD expression between normal and grade 1–4 tissue. (E) DFNA5 expression increased gradually and significantly from grade 1 to grade 4. (G-L) Violin plots of the association between the mRNA expression levels of GSDM genes and clinical stage in patients with ccRCC using data from the Gene Expression Profiling Interactive Analysis database. The mRNA expression levels of (H) GSDMB, (J) GSDMD and (K) DFNA5 were significantly related to pathological stage, whereas (G) GSDMA, (I) GSDMC and (L) DFNB59 were not. *P<0.05, **P<0.01, ***P<0.001. GSDM, gasdermin; KIRC, kidney renal clear cell carcinoma; TCGA, The Cancer Genome Atlas.
|
According to a retrospective study, two subtypes of ccRCC, ccA and ccB, appear to offer prognostic information. Tumors classified as ccA are associated with significantly better survival compared with those classified as ccB (31). As shown in Fig. 4A-F, the mRNA expression levels of GSDMA, GSDMB, GSDMC, DFNA5 and DFNB59 differed between ccA and ccB subtypes, whereas there was no significant difference in GSDMD expression between the ccA and ccB subtypes. In the ccB subtype of ccRCC, the expression levels of GSDMA, GSDMB, GSDMC, DFNA5 and DFNB59 were significantly higher than in the ccA subtype, whereas GSDMD expression had no significant difference.
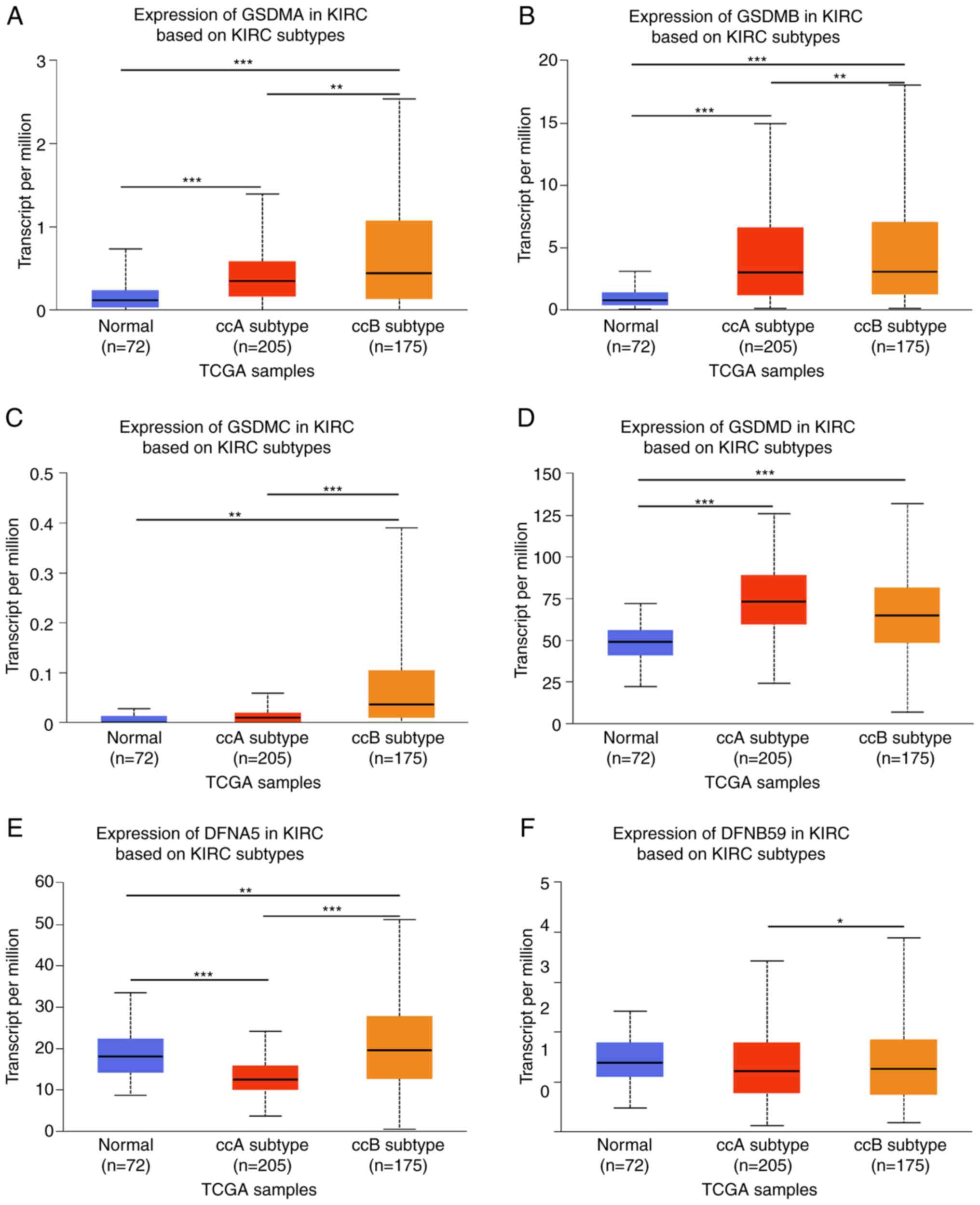 |
Figure 4.
GSDM family mRNA expression levels in KIRC ccA and ccB subtypes. (A-F) mRNA expression levels of GSDM family members in ccA and ccB subtypes of KIRC. The mRNA expression levels of (A) GSDMA, (B) GSDMB, (C) GSDMC, (E) DFNA5 and (F) DFNB59 differed between ccA and ccB subtypes, whereas there was no significant difference in (D) GSDMD expression between the ccA and ccB subtypes. *P<0.05, **P<0.01, ***P<0.001. GSDM, gasdermin; KIRC, kidney renal clear cell carcinoma; TCGA, The Cancer Genome Atlas.
|
Prognostic value of mRNA expression of GSDM genes in patients with ccRCC
The prognostic values of the mRNA expression levels of GSDM genes in patients with ccRCC were assessed based on TCGA dataset. As shown in Fig. 5A-F, the majority of GSDM genes were significantly associated with the prognosis of patients with ccRCC. Compared with low expression, high GSDMB (HR=2.49, 95% CI: 1.84–3.37, P<0.001), GSDMC (HR=1.74, 95% CI: 1.26–2.41, P=0.001), GSDMD (HR=1.63, 95% CI: 1.19–2.25, P=0.003), GSDME (HR=1.80, 95% CI: 1.34–2.43, P<0.001) and PJVK (HR=2.30, 95% CI: 1.69–3.12, P<0.001) expression was significantly associated with poor OS in patients with ccRCC. However, GSDMA was not associated with the OS of patients with ccRCC. As shown in Fig. 5G-L, the relationship between the mRNA expression levels of distinct GSDM genes and progression-free interval (PFI) of patients with ccRCC was further analyzed. Higher mRNA expression levels of GSDMB (HR=1.77, 95% CI: 1.28–2.46, P=0.001), GSDMC (HR=1.78, 95% CI: 1.27–2.51, P=0.001), GSDMD (HR=1.53, 95% CI: 1.10–2.13, P=0.013), GSDME (HR=2.20, 95% CI: 1.61–3.01, P<0.001) and PJVK (HR=1.51, 95% CI: 1.09–2.09, P=0.014) were significantly associated with a shorter PFI in patients with ccRCC, whereas the mRNA expression levels of GSDMA had no effect on the PFI of patients with ccRCC. These data indicated that elevated mRNA expression levels of GSDMB, GSDMC, GSDMD, GSDME and PJVK are significantly associated with the prognosis of patients with ccRCC, and they may serve as prognostic biomarkers for predicting patient survival and disease progression.
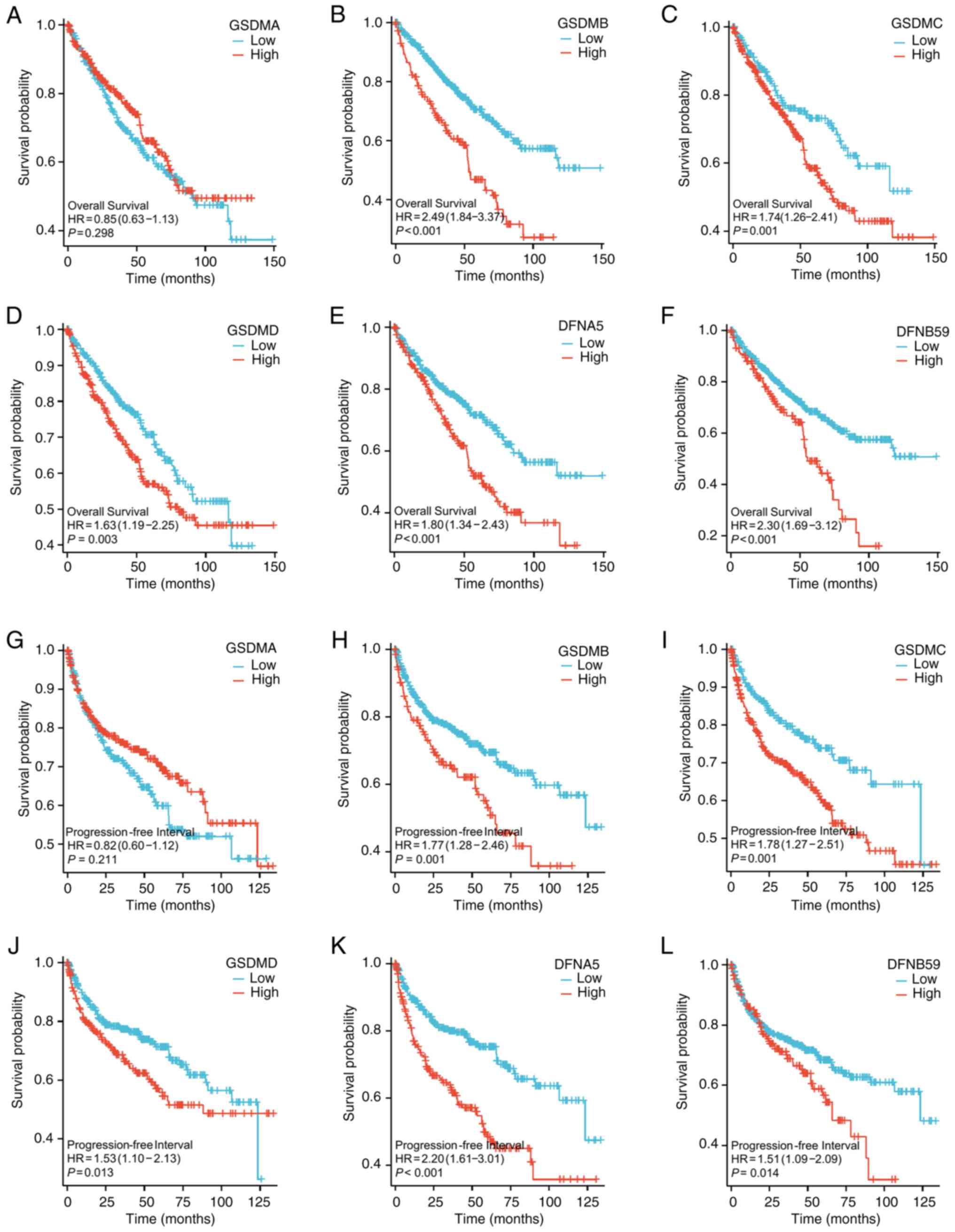 |
Figure 5.
Prognostic value of GSDM-encoding mRNA expression levels in patients with ccRCC. (A-F) Overall survival of patients with clear cell renal cell carcinoma with high or low GSDMA, GSDMB, GSDMC, GSDMD, DFNA5 and DFNB59 expression. (B) GSDMB, (C) GSDMC (D) GSDMD, (E) DFNA5 and (F) DFNB59 mRNA expression levels were significantly associated with poor OS in patients with ccRCC, while (A) GSDMA was not associated with the OS of patients with ccRCC. (G-L) Progression-free interval of patients with clear cell renal cell carcinoma with high or low GSDMA, GSDMB, GSDMC, GSDMD, DFNA5 and DFNB59 expression. (H) GSDMB, (I) GSDMC, (J) GSDMD, (K) DFNA5 and (L) DFNB59 were significantly associated with a shorter PFI in patients with ccRCC, whereas the mRNA expression levels of (G) GSDMA had no effect on the PFI of patients with ccRCC. GSDM, gasdermin.
|
Independent prognostic value of GSDM mRNA expression levels in terms of OS in patients with ccRCC
Since the mRNA expression levels of GSDMB, GSDMC, GSDMD, GSDME and PJVK were revealed to be significantly associated with the prognosis of patients with ccRCC, the present study further assessed the independent prognostic value of the mRNA expression levels of GSDMs in terms of OS in patients with ccRCC based on TCGA database via Cox survival regression analysis (32). As determined by univariate analysis, high mRNA expression levels of GSDMB (HR=1.977, 95% CI: 1.613–2.423, P<0.001), GSDMD (HR=1.523, 95% CI: 1.154–2.010, P=0.003), GSDME (HR=1.532, 95% CI: 1.225–1.916, P<0.001) and PJVK (HR=2.317, 95% CI: 1.624–3.305, P<0.001) were associated with the poor OS of patients with ccRCC, whereas GSDMA (HR=0.972, 95% CI: 0.675–1.398, P=0.877) and GSDMC (HR=0.926, 95% CI: 0.532–1.611, P=0.785) were not significantly associated with poor OS (Table SI). Multivariate analysis showed that high mRNA expression levels of GSDMB (HR=1.698, 95% CI: 1.249–2.307, P<0.001) were independently associated with a significantly shorter OS for patients with ccRCC. These results indicated that the expression of GSDMB may be an independent prognostic factor of OS in patients with ccRCC.
Genetic mutations in GSDM genes and their association with the OS of patients with ccRCC
Using the cBioPortal online tool, the genetic alterations in GSDM genes and their association with the OS of patients with ccRCC was analyzed to explore the potential expression pattern of GSDM genes. The genetic alterations of GSDM genes and their alteration frequency are shown in Fig. 6A and B. Among the 446 sequenced patients with ccRCC, genetic alteration was observed in 110 patients, with a 33% mutation rate. It was revealed that GSDMB, GSDMC and PJVK exhibited the lowest mutation rate (4%). GSDME, GSDMD and GSDMA were the top three genes for genetic alterations, and their mutation rates were 8, 6 and 7%, respectively. Furthermore, the results were plotted using a Kaplan-Meier plot and analyzed by log-rank test, which indicated that the group with GSDM gene mutations was related to a shorter OS in ccRCC (Fig. 6C; P=2.261×10−4). These results suggested that genetic alteration of GSDM genes could significantly affect the prognosis of patients with ccRCC. As shown in Fig. 6D, the miRNA-gene expression regulation network contains information on the potential regulatory effects of miRNAs on GSDM genes.
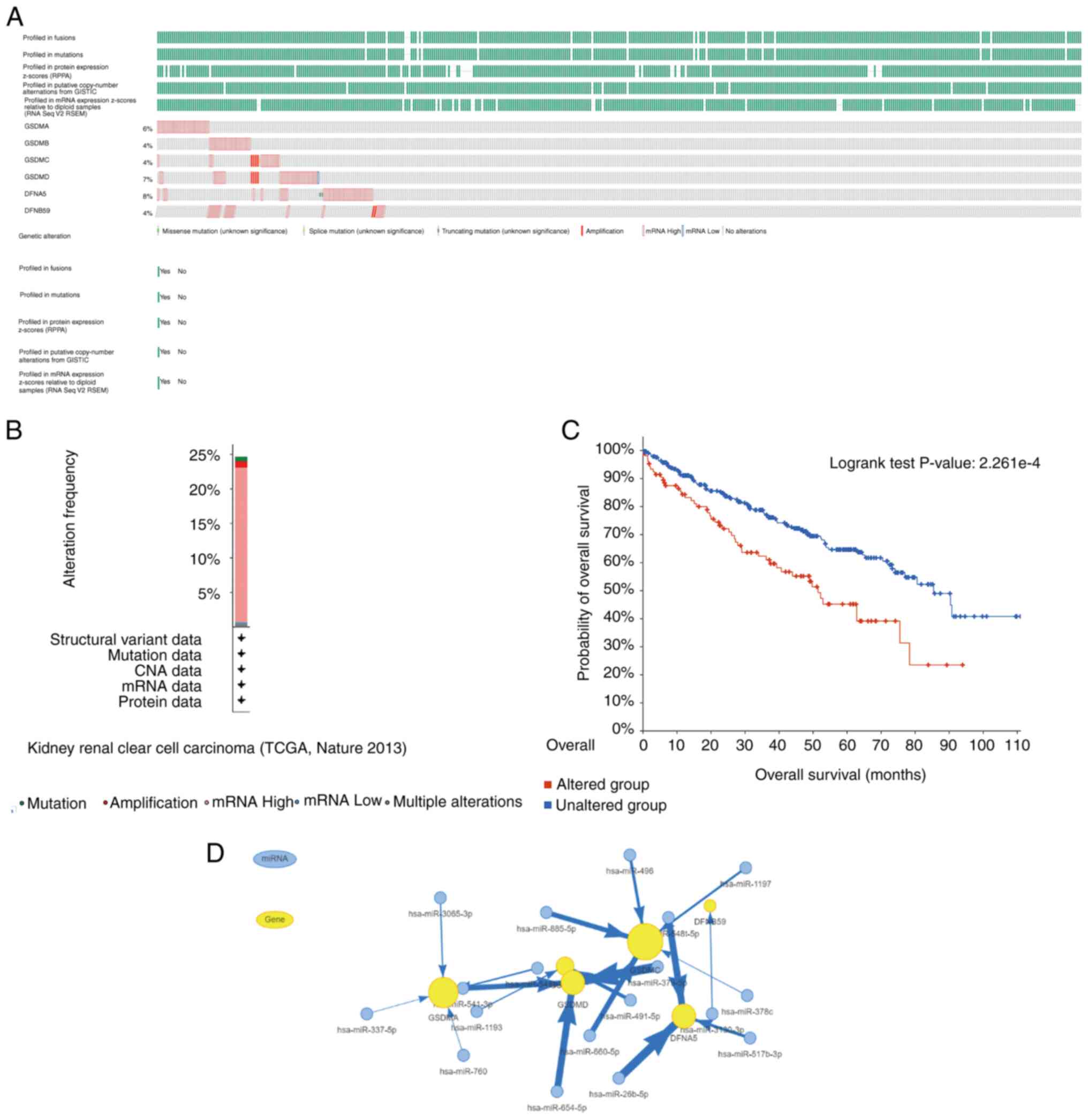 |
Figure 6.
GSDM gene expression and mutation analysis in ccRCC, and interaction network between GSDM genes and miRNAs. (A) Genetic changes in the GSDM genes in ccRCC. (B) Based on the cBioPortal database, frequency of GSDM gene alterations. (C) Kaplan-Meier plot of the overall survival of patients with ccRCC with or without GSDM gene alterations. (D) Network of GSDM genes and miRNAs. ccRCC, clear cell renal cell carcinoma; GSDM, gasdermin; miR/miRNA, microRNA.
|
Predicted functions and pathways associated with the changes in GSDM genes and the 168 most frequently altered neighbor genes in patients with ccRCC
After analyzing the genetic alterations of GSDM genes and their prognostic values in ccRCC, the 168 neighboring genes related to the GSDM gene mutants were analyzed, and an integrated network was generated. cBioPortal was used to screen out the top 168 genes, which were co-expressed and associated with the GSDM genes, and the PPI network was created using R software. FBF1, MICAL1, CHFR, FCHSD1 and AIFM3 genes were found to be most closely associated with mutations in GSDM genes, and thus may contribute to the initiation and progression of ccRCC (Fig. 7A). To gain a better understanding of the associated functions of GSDM genes in ccRCC, a functional GO and KEGG enrichment analysis of GSDM genes and the 168 neighbor genes was performed in Metascape. As shown in Fig. 7B-D, biological processes, such as GO: 0070269 (‘pyroptosis’), GO: 1901222 (‘regulation of NIK/NF-kappaB signaling’), GO:0098552 (‘side of membrane’), GO:0002252 (‘immune effector process’), GO:0045321 (‘leukocyte activation’) and GO:0002577 (‘regulation of antigen processing and presentation’) were significantly associated with GSDM gene mutations in ccRCC. Cellular components, including GO: 0030139 (‘endocytic vesicle’), GO: 0030029 (‘actin filament-based process’), GO: 0072559 (‘NLRP3 inflammasome complex’), and GO: 0007163 (‘establishment or maintenance of cell polarity’) were also significantly associated with GSDM gene alterations. In addition, GSDM gene mutations were associated with molecular functions, such as GO: 0071417 (‘cellular response to organonitrogen compound’), GO: 0070820 (‘tertiary granule’), GO: 0033218 (‘amide binding’), GO: 0061462 (‘protein localization to lysosome’), GO: 0010942 (‘positive regulation of cell death’) and GO: 0043086 (‘negative regulation of catalytic activity’). KEGG pathway analysis demonstrated the involvement of the GSDM gene family with hsa04613 (‘Neutrophil extracellular trap formation’), hsa05150 (‘Staphylococcus aureus infection’) and hsa04072 (‘phospholipase D signaling pathway’).
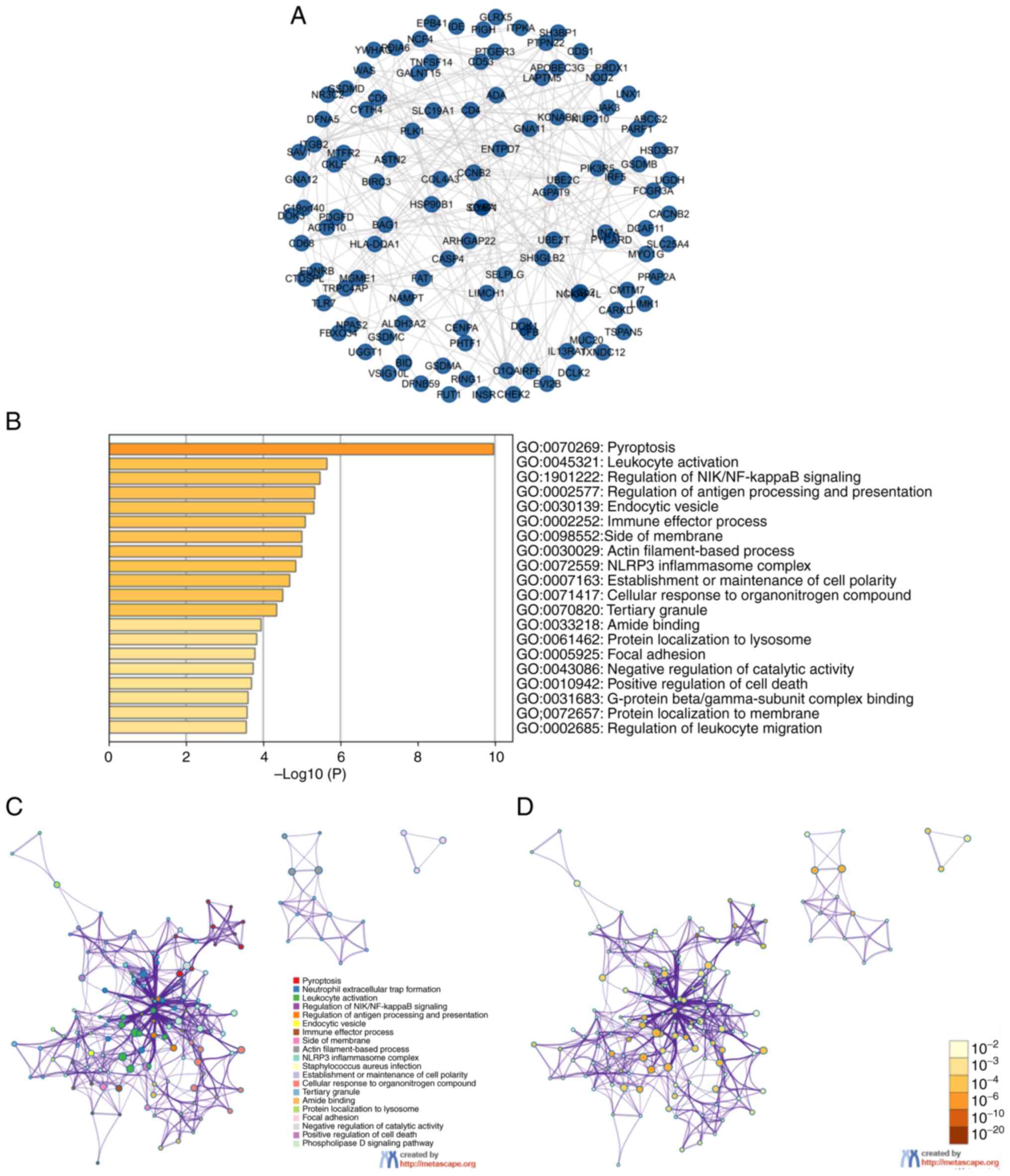 |
Figure 7.
(A) Protein-protein interaction network and (B) enrichment analysis of GSDM genes and 168 neighboring genes related to the mutations of GSDM genes in clear cell renal cell carcinoma. (C) GO and Kyoto Encyclopedia of Genes and Genomes functional enrichment analysis of GSDM genes and the 168 neighboring genes generated by Metascape. (D) GO and Kyoto Encyclopedia of Genes and Genomes functional enrichment analysis of GSDM genes and the 168 neighboring genes generated by Metascape. GO, Gene Ontology; GSDM, gasdermin.
|
Correlation of immune cell infiltration with GSDM gene expression in ccRCC
A correlation analysis was conducted using immune cell infiltration data to examine the relationship between GSDM expression and immune cell infiltration based on the TIMER database. It has been suggested that immune cells infiltrate around tumor tissue and form the foundation of the tumor microenvironment (TME) (33). Different types of immune cells often invade tumors along with the stroma, forming the TME, which can either support or impede cancer growth (34). T cells, not macrophages, are considered the dominant immune subset in ccRCC, which is one of the most heavily immune infiltrated solid tumor types (35). Therefore, the present study explored the correlations between GSDM genes and immune infiltration using the TIMER database. As shown in Fig. 8A-D, positive correlations existed between the abundance of CD4+ T cells and the expression levels of all GSDM genes, except for DFNA5. GSDMA was also positively correlated with infiltration of macrophages, neutrophils, B cells and dendritic cells, whereas GSDMB was only positively correlated with infiltration of neutrophils. GSDMC was positively correlated with infiltration of CD4+ T cells and macrophages. GSDMD was only positively correlated with infiltration of CD8+ T cells, CD4+ T cells, neutrophils and dendritic cells. DFNA5 was found to have no correlation with infiltration of CD8+ T cells and CD4+ T cells, whereas PJVK showed no association with neutrophils and macrophages. The association between the SCNAs of GSDM genes and immune infiltration was performed according to the SCNA module of the TIMER database. As shown in Fig. 9A-F, the SCNAs of GSDMA, DFNB59 and GSDMB were significantly associated with the infiltrating levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells, whereas those of GSDMC and GSDMD were associated with the infiltrating levels of CD8+ T cells and macrophages. The SCNA of DFNA5 was significantly associated with B cells and CD4+ T cells. Overall, immune infiltration may be closely associated with GSDM genes in patients with ccRCC.
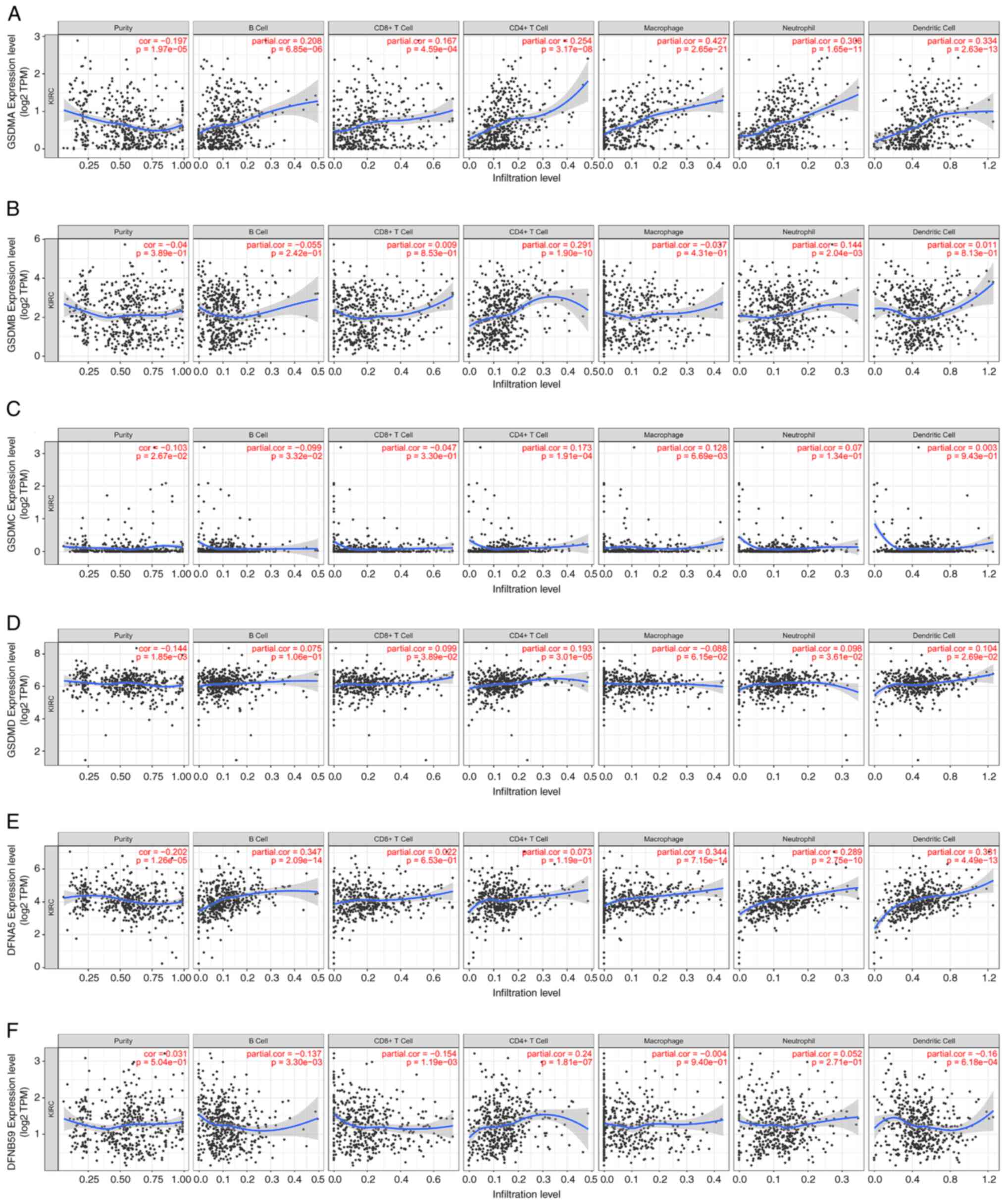 |
Figure 8.
Correlation between GSDM genes, including (A) GSDMA, (B) GSDMB, (C) GSDMC, (D) GSDMD, (E) GSDME and (F) PJVK, and immune cell infiltration based on TIMER database. TIMER, Tumor Immune Estimation Resource. GSDM, gasdermin.
|
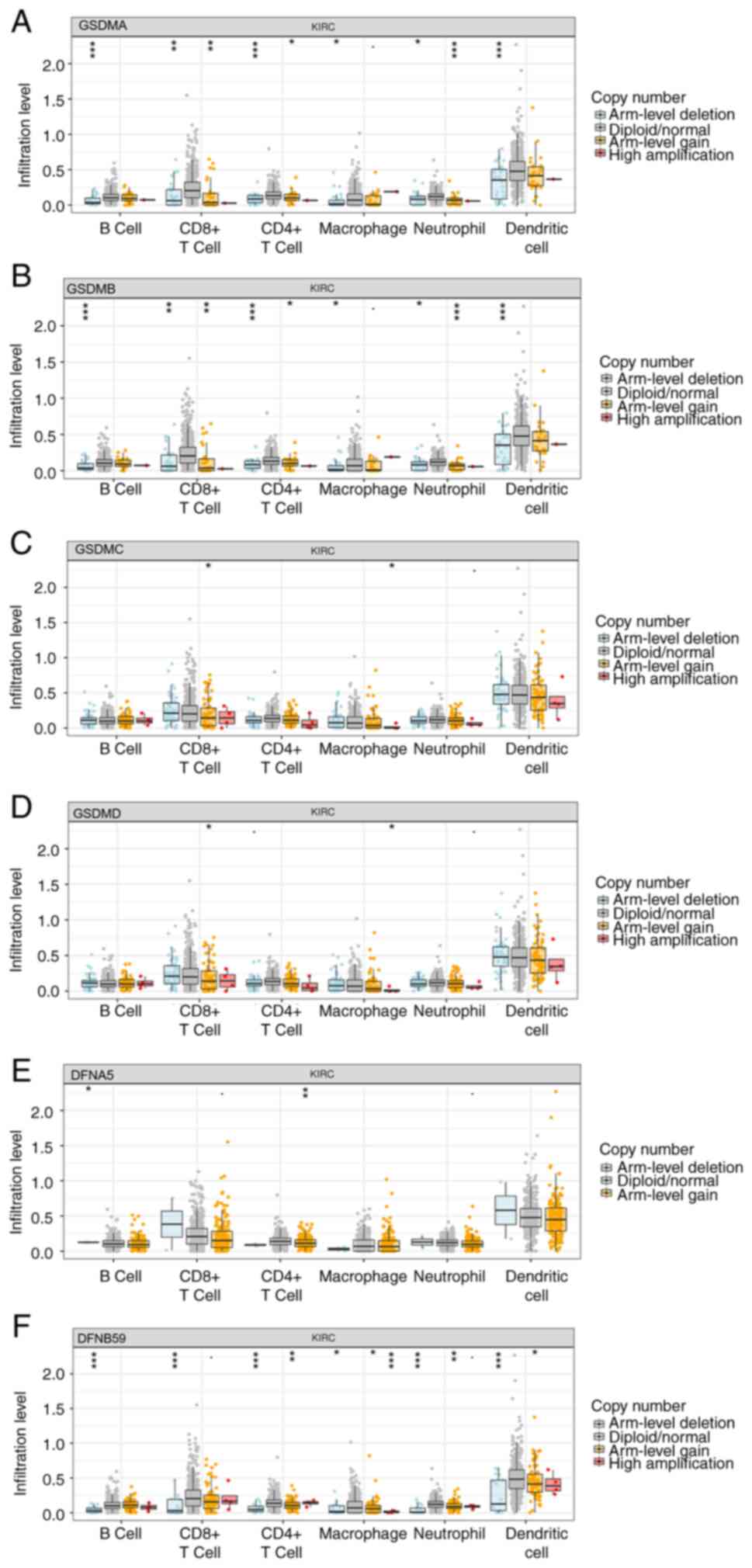 |
Figure 9.
Analysis of the correlation between GSDMs and immune cells. Comparison of tumor infiltration levels in clear cell renal cell carcinoma with different somatic copy number alterations for (A) GSDMA, (B) GSDMB, (C) GSDMC, (D) GSDMD, (E) GSDME and (F) PJVK. *P<0.05, **P<0.01, ***P<0.001. GSDM, gasdermin; KIRC, kidney renal clear cell carcinoma.
|
Drug sensitivity analysis of GSDM genes
To identify acquired drug sensitivities, the present study assessed the relationship between GSDM gene expression and IC50 values of molecules from the GDSC database and Cancer Therapeutics Response Portal. The results showed that the expression of DFNA5, GSDMC and GSDMD showed a weak negative correlation with drugs or small molecules based on the GDSC database, whereas there was a positive correlation between DFNA5 expression and drug or small molecule activity, and the expression of GSDMB exhibited negative correlations with small molecules based on the Cancer Therapeutics Response Portal, indicating they could potentially be used as new markers for drug sensitivity screening (Fig. 10A and B).
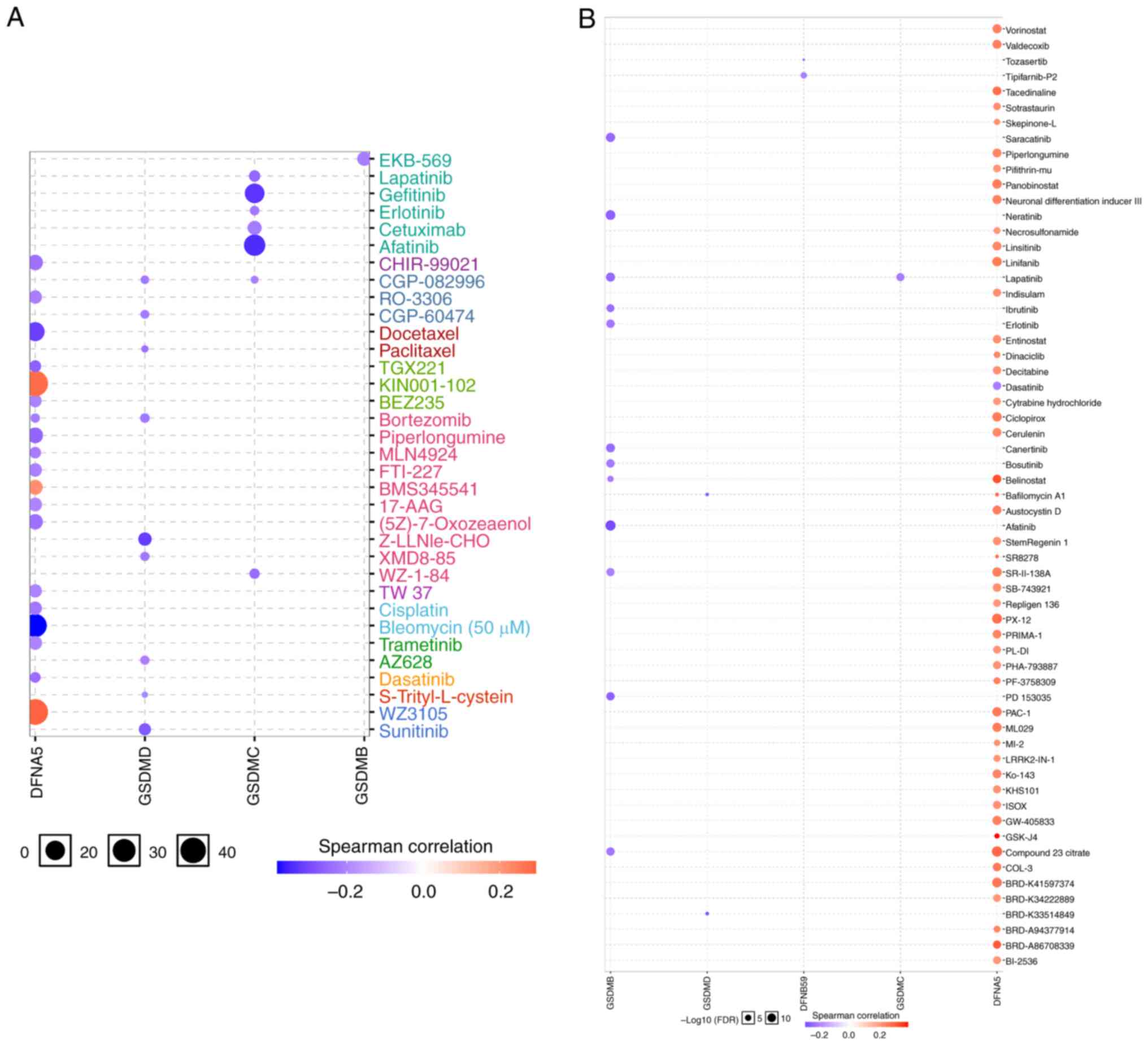 |
Figure 10.
Analysis of the Spearman correlation between GSDMs and drug sensitivity. (A) Using GSCALite, the correlation between drug sensitivity and GSDM genes in the Genomics of Drug Sensitivity in Cancer was determined. (B) Using GSCALite, the correlation between drug sensitivity and GSDM genes in the Cancer Therapeutics Response Portal. GSDM, gasdermin.
|
Pathway enrichment of GSDM genes in ccRCC
The cancer-related pathways associated with GSDM genes were determined using the GSCALite database. There were 10 pathways associated with the function of GSDM genes in ccRCC (Fig. 11). The expression levels of most GSDM genes were inhibited in DNA damage response and hormone androgen receptor-related pathways. In addition, epithelial-mesenchymal transition (EMT) was associated with the activation of most GSDM genes, and the expression levels of GSDMA, GSDMB, GSDMC, GSDMD and DFNA5 were also related to apoptosis. Taken together, these findings indicated that changes in the target genes could lead to changes in multiple signaling pathways.
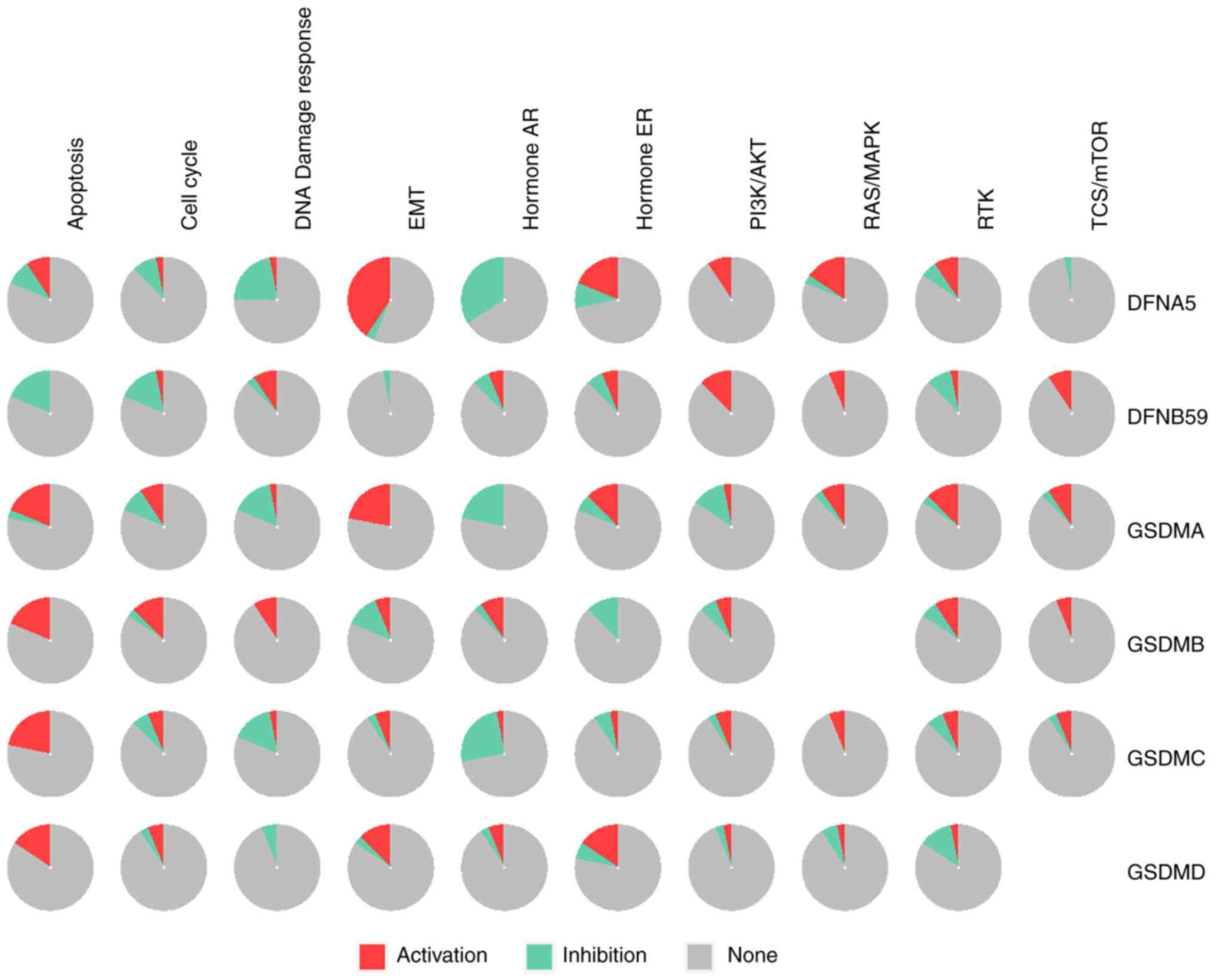 |
Figure 11.
Pathway enrichment of the six GSDM genes were analyzed using GSCALite. AR, androgen receptor; EMT, epithelial-mesenchymal transition; ER, estrogen receptor; GSDM, gasdermin; RTK, receptor tyrosine kinase.
|
Discussion
Pyroptosis, which is characterized by the release of proinflammatory signals and cytokines, promoting an immune response against tumors, is a programmed cell death process initiated by members of the GSDM family, which has been shown to be associated with a tumor-forming mechanism and also as a therapeutic component in various antitumor strategies (36). Numerous studies have detected dysfunctional and abnormal expression of the GSDM family in cancer (17,18), implying that GSDM genes may serve a role in tumorigenesis. To date, however, it remains unclear whether mRNA expression of specific GSDM family members is strongly associated with outcome in patients with ccRCC. To the best of our knowledge, there has been no systematic assessment of the involvement of GSDM family members in ccRCC. The present study quantitatively assessed the mRNA expression levels, genetic changes, functional enrichment, immune infiltration, drug sensitivity and prognostic value of GSDM genes in patients with ccRCC. When discussing the correlation between gene expression level and infiltrated immune cells, it was found that some of the correlations were weak, but there was a certain trend toward positive or negative correlation. The weak correlation between gene expression levels and immune molecules in the TIMER database may be due to a biological variability among different cancer types, sample heterogeneity from diverse studies and tumor immune evasion mechanisms. Variations in cancer biology, genetic backgrounds and tumor environments contribute to this variability, while differences in sample collection and processing across studies add to the heterogeneity. Additionally, tumors often adapt by altering surface molecules to evade immune detection, further weakening the correlation between gene expression and immune responses. By elucidating the function and mechanism of the GSDM family, more effective novel therapeutics could be developed that target GSDM genes.
The GSDMA gene is expressed in epithelial cells and has been reported to be associated with autoimmune conditions and cancer, participating in various cellular processes, including pyroptosis, cell cycle, DNA damage response, EMT, androgen receptor signaling pathway and apoptosis, which are highly related to the occurrence, development, metastasis and prognosis of tumors (37). The GSDMA gene is frequently downregulated in gastric cancer (15), whereas a higher expression of GSDMA was observed in ccRCC tissues compared with in normal tissues in the present study. However, patients with ccRCC and a high expression of GSDMA showed no statistically significant difference in OS or PFI compared with patients with low GSDMA expression. This suggests that GSDMA may not play a significant role in the prognosis of ccRCC. The possible reason for the aforementioned phenomenon may be attributed to the regulation of GSDMA by non-coding RNAs, such as miRNA-3065-3p and miRNA-760.
Several studies have indicated that GSDMB upregulation occurs in various types of cancer, including human epidermal growth factor 2-positive breast cancer and gastric cancer, where it may contribute to cancer progression and metastasis (38,39). In addition, GSDMB upregulation has been shown to enhance bladder cancer cell invasion and proliferation by binding to STAT3 and activating STAT3 signaling (40). The upregulation of GSDMB may also have a role in the development and progression of cervical squamous cell carcinoma (14). In the present study, an elevated expression of GSDMB was observed in kidney cancer tissues compared with that in normal tissues, and the expression of GSDMB was associated with tumor grade and pathological stage. In addition, there was a significant association between higher expression of GSDMB in patients with ccRCC and poor OS, thus suggesting that GSDMB may function as an oncogene. Moreover, a previous study revealed that the expression of GSDMB is associated with CD4+ T cells and neutrophils in ccRCC, indicating that GSDMB shows potential in controlling immune infiltrates (41). These findings suggested that elevated expression levels of GSDMB may independently affect the prognosis of ccRCC.
GSDMC acts as a potential tumor suppressor gene, inhibiting cancer cells from proliferating when GSDMC is overexpressed in gastric cancer (42). By contrast, several studies have shown that the expression of GSDMC is elevated in various types of cancer, such as colorectal cancer, lung adenocarcinoma and melanoma, and that it may promote metastasis in patients with melanoma (43,44). In the present study, the expression of GSDMC in the qPCR experimental results was not consistent with the results from TCGA. It was hypothesized that this difference may be because TCGA database contained ethnic differences, which could contribute to tumor heterogeneity. Furthermore, there was no relationship identified between the expression levels of GSDMC and tumor stage and grade. These findings suggested that GSDMC may not serve as a suitable prognostic indicator for ccRCC.
In the GSDM family, most research has focused on GSDMD (45). GSDMD plays a significant role in the regulation of pyroptosis and the sensitivity of cancer therapies (46). As a result of GSDMD downregulation, the risk of developing gastric cancer is elevated (16), whereas in non-small-cell lung cancer, its upregulation is associated with a poor prognosis (47). In the present study, compared with in adjacent normal tissues, it was revealed that GSDMD expression was highly expressed in ccRCC tissues. In addition, there was a significant association between GSDMD expression and poor OS in patients with ccRCC, which supports its function as an oncogene.
DFNA5 (GSDME) is associated with numerous types of cancer and causes pyroptosis in cells after chemical treatment (48). GSDME, which acts as a candidate tumor suppressor, is a transcriptional target of p53 and is silenced in gastric cancer and colorectal carcinoma (49,50). Several chemotherapy agents, including etoposide, topotecan, CPT-11 and cisplatin, induce pyroptosis in cancer cells by triggering the GSDME pathway, whereas they promote apoptosis in cells lacking the GSDME pathway (51). The present study detected reduced expression levels of GSDME in normal tissues compared with those in paired kidney cancer tissues; however, based on the data obtained from TCGA database, no discernible variation in mRNA expression was observed. The reason for this phenomenon may be due to genetic heterogeneity among different ethnicities based on TCGA database. The present results revealed that the expression of GSDME was strongly positively correlated with drugs and small molecules. A previous study suggested that a loss of GSDME can reduce the effectiveness of some chemotherapy drugs (52). Furthermore, in the present study, a high level of GSDME expression was revealed to be strongly associated with poor OS, PFI and cancer stage, suggesting an oncogenic role of GSDME; these aforementioned results are similar to those of Yao et al (53). Differences in GSDME in cancer research could be due to variations in the experiments, tissue types and cancer stages. The exact manner by which GSDME functions is still being studied, with ongoing research focusing on its complex roles in different types of cancer.
Presently, there are few specific studies available on the role of PJVK in human cancer. According to the present study, PJVK was increased in normal tissues compared with that in kidney cancer tissues. This finding suggested that PJVK may play an inhibitory role in ccRCC; however, high PJVK expression was significantly associated with poor outcomes. Subsequently, it was revealed that PJVK was regulated by miRNA-378c. The involvement of PJVK in cancer tumorigenicity has yet to be fully elucidated.
The present study explored the detailed function and molecular mechanism of the GSDM family in ccRCC biology. Using data from several bioinformatics platforms, the expression and prognostic value of GSDM genes were evaluated in ccRCC. Furthermore, qPCR was performed to detect the expression levels of GSDM genes in ccRCC tissues. However, the present study has some limitations. First, although higher mRNA expression levels of GSDMB, GSDMD and DFNA5 were revealed to be significantly related to patient tumor stage and grade, and GSDMB emerged as an independent prognostic factor for shorter OS of patients with ccRCC, further investigation using a substantial number of samples is needed to validate these findings and to explore the clinical application of GSDM members in ccRCC treatment. Additionally, the results of the present study are promising, but they are based on the analysis of bioinformatics data; therefore, a large number of functional and mechanistic experiments are required to confirm the conclusions.
In conclusion, the present study revealed that GSDMB was significantly upregulated in ccRCC tissues and that upregulated GSDMB expression could act as an independent predictor for the OS of patients with ccRCC. Moreover, the abnormal expression levels of GSDMB, GSDMD and DFNA5 were significantly associated with the tumor stage and grade of ccRCC, and DFNA5 expression exhibited a positive correlation with the activity of drugs or small molecules. These findings suggested that GSDMB could serve as a potential biomarker and may contribute to the future identification of promising therapeutic agents for patients with ccRCC.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank two independent senior pathologists, Dr Jun Zhou and Dr Zhiwei Chen for their technical assistance (Department of Pathology, The Affiliated Hospital of Putian University, Putian, China).
Funding
This work was supported by grants from the Natural Science Foundation of Fujian Province (grant no. 2019J01153), the Startup Fund for Scientific Research, Fujian Medical University (grant no. 2019QH1053) and the Training Program for Young and Middle-aged Elite Talents of Fujian Provincial Health Commission (grant no. 2021GGA014).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HH and XD conceived and directed the project. RC and JW performed bioinformatics analysis and conducted experiments. RC and HH confirm the authenticity of all the raw data. JC was responsible for data interpretation and wrote the manuscript and revised it. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All subjects provided written informed consent. In accordance with The Declaration of Helsinki, the present study was approved by the Institutional Ethics Committee of The First Affiliated Hospital of Nanchang University (approval no. 202012-110).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Capitanio U, Bensalah K, Bex A, Boorjian SA, Bray F, Coleman J, Gore JL, Sun M, Wood C and Russo P: Epidemiology of renal cell carcinoma. Eur Urol. 75:74–84. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fuchs HE and Jemal A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Courthod G, Tucci M, Di Maio M and Scagliotti GV: Papillary renal cell carcinoma: A review of the current therapeutic landscape. Crit Rev Oncol Hematol. 96:100–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bellido JA, Palou J, Hübner M, Pascual M, Sagristá R, Martínez E, Martínez J, Rosales A and Villavicencio H: Early ultrasound detection of renal tumors in patients with end stage renal disease in dialysis. Arch Esp Urol. 60:1.079–1.083. 2007.(In Spanish). PubMed/NCBI
|
|
5
|
Tunio MA, Hashmi A and Rafi M: Need for a new trial to evaluate postoperative radiotherapy in renal cell carcinoma: A meta-analysis of randomized controlled trials. Ann Oncol. 21:1839–1845. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Wu G, Xu Y, Li J, Ruan N, Chen Y, Zhang Q and Xia Q: Porcupine inhibitor LGK974 downregulates the Wnt signaling pathway and inhibits clear cell renal cell carcinoma. Biomed Res Int. 2020:25276432020.PubMed/NCBI
|
|
7
|
Kovacs SB and Miao EA: Gasdermins: Effectors of pyroptosis. Trends Cell Biol. 27:673–684. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Broz P, Pelegrín P and Shao F: The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol. 20:143–157. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang X and Tang Z: Role of gasdermin family proteins in cancers (review). Int J Oncol. 63:1002023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi J, Gao W and Shao F: Pyroptosis: Gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci. 42:245–254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuan YY, Xie KX, Wang SL and Yuan LW: Inflammatory caspase-related pyroptosis: Mechanism, regulation and therapeutic potential for inflammatory bowel disease. Gastroenterol Rep (Oxf). 6:167–176. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Liu X, Bai X, Lin Y, Li Z, Fu J, Li M, Zhao T, Yang H, Xu R, et al: Melatonin prevents endothelial cell pyroptosis via regulation of long noncoding RNA MEG3/miR-223/NLRP3 axis. J Pineal Res. 64:2018. View Article : Google Scholar
|
|
13
|
Zhao P, Wang M, Chen M, Chen Z, Peng X, Zhou F, Song J and Qu J: Programming cell pyroptosis with biomimetic nanoparticles for solid tumor immunotherapy. Biomaterials. 254:1201422020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li L, Li Y and Bai Y: Role of GSDMB in pyroptosis and cancer. Cancer Manag Res. 12:3033–3043. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saeki N, Kim DH, Usui T, Aoyagi K, Tatsuta T, Aoki K, Yanagihara K, Tamura M, Mizushima H, Sakamoto H, et al: GASDERMIN, suppressed frequently in gastric cancer, is a target of LMO1 in TGF-beta-dependent apoptotic signalling. Oncogene. 26:6488–6498. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang WJ, Chen D, Jiang MZ, Xu B, Li XW, Chu Y, Zhang YJ, Mao R, Liang J and Fan DM: Downregulation of gasdermin D promotes gastric cancer proliferation by regulating cell cycle-related proteins. J Dig Dis. 19:74–83. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lutkowska A, Roszak A, Lianeri M, Sowińska A, Sotiri E and Jagodziński PP: Analysis of rs8067378 polymorphism in the risk of uterine cervical cancer from a polish population and its impact on gasdermin B expression. Mol Diagn Ther. 21:199–207. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Miguchi M, Hinoi T, Shimomura M, Adachi T, Saito Y, Niitsu H, Kochi M, Sada H, Sotomaru Y, Ikenoue T, et al: Gasdermin C is upregulated by inactivation of transforming growth factor β receptor type II in the presence of mutated Apc, promoting colorectal cancer proliferation. PLoS One. 11:e01664222016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mu M, Yu Q, Zhang Q, Guo J, Wang X, Sun X and Yu J: A pan-cancer analysis of molecular characteristics and oncogenic role of gasdermins. Cancer Cell Int. 22:802022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li R, Wei H, Wang S, Huang Z, Chen H, Zhang S, Lin J and Zhong G: Gasdermin D expression and clinicopathologic outcome in primary osteosarcoma patients. Int J Clin Exp Pathol. 13:3149–3157. 2020.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen R, Zhang Z, Hu B, Jiang M, Zheng P, Deng W, Fu B and Sun T: Identification of the expression and clinical significance of E2F family in clear cell renal cell carcinoma. Int J Gen Med. 15:1193–1212. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and Varambally S: UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang Z, Li C, Kang B, Gao G, Li C and Zhang Z: GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45((W1)): W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al: Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu CJ, Hu FF, Xia MX, Han L, Zhang Q and Guo AY: GSCALite: A web server for gene set cancer analysis. Bioinformatics. 34:3771–3772. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et al: Genomics of drug sensitivity in cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41:(Database Issue). D955–961. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, Ebright RY, Stewart ML, Ito D, Wang S, et al: An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 154:1151–1161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brannon AR, Reddy A, Seiler M, Arreola A, Moore DT, Pruthi RS, Wallen EM, Nielsen ME, Liu H, Nathanson KL, et al: Molecular stratification of clear cell renal cell carcinoma by consensus clustering reveals distinct subtypes and survival patterns. Genes Cancer. 1:152–163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, et al: An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 173:400–416.e11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
T G S: Innate and adaptive immune cells in Tumor microenvironment. Gulf J Oncolog. 1:77–81. 2021.PubMed/NCBI
|
|
34
|
Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, et al: Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 313:1960–1964. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, Ries CH, Ailles L, Jewett MAS, Moch H, et al: An immune atlas of clear cell renal cell carcinoma. Cell. 169:736–749.e18. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fang Y, Tian S, Pan Y, Li W, Wang Q, Tang Y, Yu T, Wu X, Shi Y, Ma P and Shu Y: Pyroptosis: A new frontier in cancer. Biomed Pharmacother. 121:1095952020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yu P, Chen X and Peng C: A new perspective in pyroptosis: Lifting the veil on GSDMA activation. Front Med. 17:581–583. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hergueta-Redondo M, Sarrio D, Molina-Crespo Á, Vicario R, Bernadó-Morales C, Martínez L, Rojo-Sebastián A, Serra-Musach J, Mota A, Martínez-Ramírez Á, et al: Gasdermin B expression predicts poor clinical outcome in HER2-positive breast cancer. Oncotarget. 7:56295–56308. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Komiyama H, Aoki A, Tanaka S, Maekawa H, Kato Y, Wada R, Maekawa T, Tamura M and Shiroishi T: Alu-derived cis-element regulates tumorigenesis-dependent gastric expression of GASDERMIN B (GSDMB). Genes Genet Syst. 85:75–83. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
He H, Yi L, Zhang B, Yan B, Xiao M, Ren J, Zi D, Zhu L, Zhong Z, Zhao X, et al: USP24-GSDMB complex promotes bladder cancer proliferation via activation of the STAT3 pathway. Int J Biol Sci. 17:2417–2429. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cui Y, Zhou Z, Chai Y and Zhang Y: Upregulated GSDMB in clear cell renal cell carcinoma is associated with immune infiltrates and poor prognosis. J Immunol Res. 2021:77535532021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ruan J: Structural insight of gasdermin family driving pyroptotic cell death. Adv Exp Med Biol. 1172:189–205. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Feng S, Fox D and Man SM: Mechanisms of gasdermin family members in inflammasome signaling and cell death. J Mol Biol. 430:3068–3080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wei J, Xu Z, Chen X, Wang X, Zeng S, Qian L, Yang X, Ou C, Lin W, Gong Z and Yan Y: Overexpression of GSDMC is a prognostic factor for predicting a poor outcome in lung adenocarcinoma. Mol Med Rep. 21:360–370. 2020.PubMed/NCBI
|
|
45
|
Orning P, Lien E and Fitzgerald KA: Gasdermins and their role in immunity and inflammation. J Exp Med. 216:2453–2465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang YY, Liu XL and Zhao R: Induction of pyroptosis and its implications in cancer management. Front Oncol. 9:9712019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gao J, Qiu X, Xi G, Liu H, Zhang F, Lv T and Song Y: Downregulation of GSDMD attenuates tumor proliferation via the intrinsic mitochondrial apoptotic pathway and inhibition of EGFR/Akt signaling and predicts a good prognosis in non-small cell lung cancer. Oncol Rep. 40:1971–1984. 2018.PubMed/NCBI
|
|
48
|
Wang Y, Yin B, Li D, Wang G, Han X and Sun X: GSDME mediates caspase-3-dependent pyroptosis in gastric cancer. Biochem Biophys Res Commun. 495:1418–1425. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Akino K, Toyota M, Suzuki H, Imai T, Maruyama R, Kusano M, Nishikawa N, Watanabe Y, Sasaki Y, Abe T, et al: Identification of DFNA5 as a target of epigenetic inactivation in gastric cancer. Cancer Sci. 98:88–95. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kim MS, Chang X, Yamashita K, Nagpal JK, Baek JH, Wu G, Trink B, Ratovitski EA, Mori M and Sidransky D: Aberrant promoter methylation and tumor suppressive activity of the DFNA5 gene in colorectal carcinoma. Oncogene. 27:3624–3634. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang X and Zhang H: Chemotherapy drugs induce pyroptosis through caspase-3-dependent cleavage of GSDME. Sci China Life Sci. 61:739–740. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang Y, Gao W, Shi X, Ding J, Liu W, He H, Wang K and Shao F: Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 547:99–103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yao L, Li J, Xu Z, Yan Y and Hu K: GSDMs are potential therapeutic targets and prognostic biomarkers in clear cell renal cell carcinoma. Aging (Albany NY). 14:2758–2774. 2022. View Article : Google Scholar : PubMed/NCBI
|

























