Introduction
Ovarian high-grade serous carcinoma (OHGSC) is the
most common type of epithelial ovarian cancer, accounting for 60%
of all ovarian malignancies and 70% of ovarian cancer-related
deaths in the United States (1,2).
Survival in OHGSC is influenced by numerous factors, such as age,
cancer stage and the size of residual tumor after cytoreductive
surgery (3,4). Epigenetic dysregulation has been
recognized as a significant factor in cancer development,
progression and chemoresistance (5). These alterations involve abnormal DNA
methylation patterns, disrupted histone posttranslational
modifications, and changes in chromatin composition and/or
organization (6). Chromatin
remodeling factors (CRFs) serve a vital role in modifying chromatin
structure, regulating the accessibility of DNA to transcription
factors and machinery and thereby dynamically influencing gene
expression (7). Genome sequencing
studies have revealed a high prevalence of CRF mutations across
numerous cancer types (8). In the
context of gynecological cancer, AT-rich interaction domain 1A
(ARID1A) mutations have been identified in 35–46% of ovarian
clear cell carcinoma, 30–63% of ovarian endometrioid carcinoma, 6%
of endometrial serous carcinoma and 14% of carcinosarcoma cases
(9–11). Chromodomain-helicase-DNA-binding
protein 4 (CHD4) somatic mutations have been detected in 17%
of endometrial serous carcinoma (10), while switch/sucrose non-fermentable
(SWI/SNF) related, matrix associated, actin dependent regulator of
chromatin (SMARC) subfamily a member 4 (SMARCA4) germline
and somatic mutations have been found in 69% of cases of small cell
carcinoma of the ovary, hypercalcemic type (12). A previous study reported that
CHD4 mRNA expression is significantly higher in
platinum-resistant cases compared with in platinum-sensitive cases
of OHGSC and ovarian clear cell carcinoma (13). Despite these findings, to the best
of our knowledge, there has not been a comprehensive and
large-scale investigation exploring the association between CRFs
and OHGSC.
In the present study, a comprehensive analysis of
OHGSC cases from histological, immunohistochemical and genetic
perspectives was conducted to elucidate the role of CRF dysfunction
in OHGSC.
Materials and methods
Public data analysis
The cBioPortal (http://www.cbioportal.org/) (14,15)
was used to retrieve public whole exome sequencing data and mRNA
sequencing data for ovarian serous carcinoma. Initially,
‘Ovary/Fallopian Tube’ was selected in the ‘Select Studies for
Visualization & Analysis’ and the Ovarian Serous
Cystadenocarcinoma dataset [The Cancer Genome Atlas (TCGA),
PanCancer Atlas; https://www.cancer.gov/tcga] was chosen. This dataset
from TCGA contains whole exome sequencing data from 585 cases, and
mRNA sequencing data from 300 cases of ovarian serous carcinoma.
Genomic alteration, mRNA expression and survival data were analyzed
via the cBioPortal website by submitting a query regarding CRFs,
including ARID1A, AT-rich interaction domain 1B, actin-like
protein 6A (ACTL6A), SMARCA1, SMARCA2, SMARCA4, SMARCA5,
SMARC subfamily b member 1 (SMARCB1), SMARC
subfamily c member 1 (SMARCC1), SMARCC2, SMARC
subfamily d member 1, SMARC subfamily e member 1,
helicase-like transcription factor,
chromodomain-helicase-DNA-binding protein 1 (CHD1), CHD2,
CHD3, CHD4, CHD5, inositol-requiring 80 and
bromodomain-containing protein 9. The genetic alterations in the
‘OncoPrint’ module were analyzed and mRNA expression levels in the
‘mRNA’ module of ‘Comparison/Survival’ were displayed.
Moreover, the association of individual CRF genes
with prognosis was compared, and patients within the dataset were
categorized into two groups: One with CRF gene amplification, and
the other without genetic alteration of CRFs. The prognostic value
of individual mRNA expression levels of CRF genes in the two groups
was compared using median normalized RNA-seq by expectation
maximization values (cut off values: ACTL6A, 2,117;
SMARCC2, 3,607; CHD4, 6,891). Survival curves were
constructed using the Kaplan-Meier method, and the log-rank test
was performed using the ‘Survival’ module of ‘Comparison/Survival’
on cBioportal to analyze overall survival (OS).
Case selection and clinicopathological
characteristics
The present retrospective study adhered to the
principles outlined in The Declaration of Helsinki. This study was
approved by the Ethics Committee of Kyushu University (Fukuoka,
Japan; approval nos. 21120-01, 21037-02 and 23005-00). The case
records of the Department of Anatomic Pathology, Kyushu University
from 1988-2020 were accessed to identify cases of ovarian serous
carcinoma with available clinical data and formalin-fixed and
paraffin-embedded (FFPE) blocks of ovarian tissue. This search
yielded 318 cases of surgically resected ovarian serous carcinoma.
Cases other than primary ovarian cancer (12 cases involving
fallopian tube and 9 cases involving peritoneal cancer) were
excluded, as were cases where neoadjuvant chemotherapy had been
administered (91 cases). All cases were independently reviewed by
two pathologists (NM and TI). The typical histological structure of
OHGSC was confirmed, including papillary or solid proliferation,
severe nuclear atypia and frequent mitotic figures, in accordance
with the World Health Organization Classification of Female Genital
Tumors (16). Additionally, three
cases diagnosed with low-grade serous carcinoma were excluded.
Ultimately, this analysis included 203 cases of OHGSC. All tumor
samples were FFPE. For both immunohistochemistry (IHC) and copy
number assays, one representative FFPE block was used for each
case. Clinical data, including age, International Federation of
Gynecology and Obstetrics (FIGO) 2014 stage (17), presence of metastases, adjuvant
therapy and prognosis, were obtained from medical records.
Furthermore, one normal skeletal muscle tissue was collected from
the Department of Anatomic Pathology, Kyushu University as the
control for copy number assays.
Immunohistochemical staining
The primary antibodies used for IHC staining are
listed in Table SI. Staining
conditions were optimized by testing numerous approaches. FFPE
tissue was cut into 3 µm sections for further processing. The
paraffin-embedded sections were deparaffinized in xylene and
rehydrated in ethanol series (99, 90 and 80%). Antigen retrieval
was performed by boiling the slides at 98 or 110°C in Target
Retrieval Solution (pH 9.0; Dako; Agilent Technologies, Inc) for
ARID1A, SMARCA4, SMARCB1, SMARCC2, tri-methylation of lysine 27 of
histone H3 (H3K27me3) and p53 staining, while 10 mM sodium citrate
(pH 6.0) was used for SMARCA2, CHD4 and ACTL6A. Subsequently, 3%
hydrogen peroxide was used for blocking endogenous peroxidase
activity at room temperature for 5 min and PBS was used for
washing. Sections were then incubated with the primary antibodies
at room temperature for 90 min (CHD4, ACTL6A, H3K27me3 and p53) or
at 4°C overnight (ARID1A, SMARCA2, SMARCA4, SMARCB1 and SMARCC2).
The EnVision-kit (Dako; Agilent Technologies, Inc) was used for
ARID1A, SMARCA2, SMARCB1, SMARCC2, H3K27me3, CHD4, ACTL6A and p53
staining, and the EnVision Flex-kit (Dako; Agilent Technologies,
Inc) was used for SMARCA4 staining to achieve better specificity,
in accordance with the manufacturer's instructions. Sections were
dehydrated in an ethanol series (95, 99 and 100%) and cleared in
xylene. Nuclear staining of stromal cells and vascular endothelium
was used as a positive internal control and stroma was used as a
negative internal control to evaluate the staining. In several
cases, p53 immunostaining was performed to support histological
diagnosis. The evaluation of IHC slides was conducted using a light
microscope by two pathologists (NM and TI) who were blinded to the
details of the patients.
Immunohistochemical scoring
The expression of ARID1A, SMARCA2, SMARCA4, SMARCB1,
SMARCC2 and H3K27me3 was considered ‘lost’ when there was a
complete absence of nuclear staining in tumor cells, while the
surrounding normal cells exhibited consistently preserved nuclear
staining. By contrast, ACTL6A, SMARCC2 and CHD4 expression were
evaluated using an H-score, calculated by multiplying the
proportion and intensity of tumor cells displaying nuclear
staining. The proportion score was determined by assessing the
percentage of tumor cells with positive nuclear staining relative
to all tumor cells on the slide (0–100%). The intensity score was
assessed using the intensity of the nuclear staining, categorized
as follows: 0, not stained; 1, weak; 2, moderate; and 3, strong.
The resulting H-score ranged from 0–300. The threshold value
between the high and low protein expression groups was determined
using the median H-score. Notably, stromal cells and vascular
endothelium exhibited positivity and served as internal positive
controls across all cases.
Copy number assay
Tumor DNA from each of the 203 cases was extracted
from FFPE blocks using the DNAstorm FFPE Kit (Biotium, Inc.),
according to the manufacturer's protocol. However, the copy number
assay could not be conducted in some cases due to limited sample
volume. Tumor DNA was successfully extracted in 154 cases for
ACTL6A, 143 cases for SMARCC2 and 140 cases for
CHD4. Normal DNA extracted from a normal skeletal muscle
tissue was used as the control. TaqMan™ Copy Number Assay (cat. no.
4400291; Thermo Fisher Scientific Inc.) was used to conduct DNA
copy number analysis. A predesigned primer and probe mix specific
for ACTL6A (Assay ID. Hs02294862_cn), SMARCC2 (Assay
ID. Hs02933711_cn) and CHD4 (Assay ID. Hs02108296_cn) was
used according to the manufacturer's instructions (Thermo Fisher
Scientific Inc.). The Rnase P gene (cat no. 4316844; Thermo
Fisher Scientific Inc.) was used as the endogenous control.
Quantitative polymerase chain reaction (qPCR) was performed with
THUNDERBIRD™ Probe qPCR Mix (Toyobo Life Science) using the ∆Cq
method (18) on a QuantStudio 3
Real Time PCR System (Thermo Fisher Scientific Inc.). The
thermocycling conditions were as follows: Hot start at 50°C for 2
min and 95°C for 1 min; followed by 50 cycles of denaturation at
95°C for 15 sec, annealing at 60°C for 60 sec and extension at 68°C
for 30 sec. The relative quantities of DNA obtained from the PCR
were analyzed using CopyCaller (Thermo Fisher Scientific Inc.) to
determine the copy numbers. Cases with negative results for
Rnase P were excluded from the analysis. Copy number
alterations (CNAs) were categorized as follows: Copy number <1,
deep deletion; 1, shallow deletion; 2, diploid; 3–4, gain and
>4, amplification.
Statistical analysis
All statistical analyses were performed using the
JMP statistical software version 17 (SAS Institute, Inc.). The data
were analyzed using Wilcoxon's rank-sum test. Multiple comparisons
of DNA copy number data and immunohistochemical expression data
were analyzed using the Wilcoxon rank-sum test with Bonferroni
correction. For survival analysis, Kaplan-Meier analysis and the
log-rank test were used. P<0.05 was considered to indicate a
statistically significant difference.
Results
Public data analysis
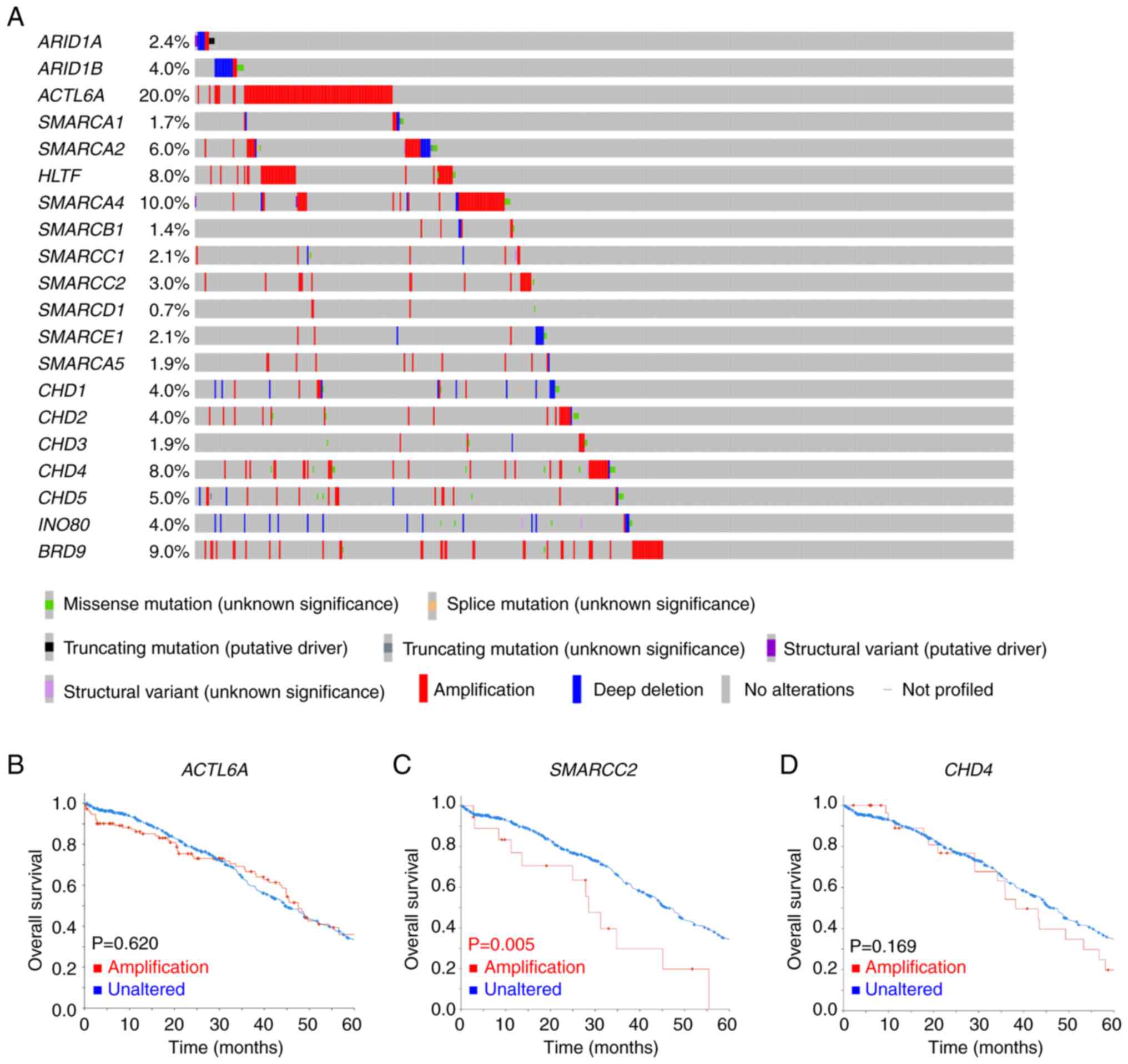
The gene alterations of CRFs in ovarian serous
carcinoma are summarized in Fig.
1A. Among 585 cases, 57% demonstrated CRF gene alterations,
primarily in the form of gene amplification, which was observed in
208 (35.6%) cases. The most prevalent genetic alteration among CRFs
in ovarian serous carcinoma was ACTL6A amplification
(19.5%). However, this genetic alteration did not significantly
affect OS (P=0.620; Fig. 1B).
Patients with SMARCC2 amplification (3.1%) had a
significantly shorter median OS compared with patients with
unaltered SMARCC2 (P=0.005; Fig.
1C). Patients with CHD4 amplification (5.7%) had a
notably shorter OS compared with patients with unaltered
CHD4; however, this association was not statistically
significant (P=0.169; Fig. 1D).
Furthermore, there was no significant association between
ACTL6A, SMARCC2 and CHD4 mRNA expression levels and
OS (Fig. S1A-C). However,
ACTL6A mRNA expression was significantly higher in patients
with ACTL6A amplification compared with in patients without
alterations in the ACTL6A gene (P<0.001; Fig. S1D).
Clinicopathological data analysis
Table I presents the
clinical characteristics of patients with OHGSC included in the
present histological study. The age of the patients ranged from
28–87 years (mean, 58.3 years; median, 58 years). Out of the 203
cases, 148 (72.9%) had available 5-year clinical follow-up data,
with a mean follow-up duration of 61.9 months (1–233 months). The
majority of patients (78.4%) presented with advanced stage disease
(III–IV). In total, there were 124 cases (61.1%) with recurrence,
and 81 cases (39.9%) resulted in disease-related death.
 | Table I.Clinicopathological features of
patients with OHGSC (n=203). |
Table I.
Clinicopathological features of
patients with OHGSC (n=203).
| Characteristic | No. (%) |
|---|
| FIGO stage |
|
| I | 21 (10.3) |
| II | 23 (11.3) |
|
III | 114 (56.2) |
| IV | 45 (22.2) |
| T stage |
|
| 1 | 30 (14.8) |
| 2 | 37 (18.2) |
| 3 | 136 (67.0) |
| N stage |
|
| 0 | 68 (33.5) |
| 1 | 104 (51.2) |
| X | 31 (15.3) |
| M stage |
|
| 0 | 158 (77.8) |
| 1 | 45 (22.2) |
| Recurrence |
|
| - | 79 (38.9) |
| + | 124 (61.1) |
| Adjuvant
chemotherapy |
|
| - | 8 (3.9) |
| + | 195 (96.1) |
| Tumor-related
mortality |
|
|
NED | 52 (25.6) |
|
AWD | 15 (7.4) |
|
DOD | 81 (39.9) |
| NA | 55 (27.1) |
Regarding treatment, all cases underwent surgical
resection. Additionally, 195 cases (96.1%) received adjuvant
chemotherapy, consisting of various regimens, such as paclitaxel
and carboplatin, docetaxel and carboplatin, cyclophosphamide,
doxorubicin and cisplatin, cyclophosphamide, epirubicin and
cisplatin, or paclitaxel and cisplatin. Furthermore, 9 cases
received maintenance treatment, which supplemented chemotherapy
with bevacizumab treatment.
Immunohistochemical results
Figs. 2, S2 and S3
display representative images of IHC staining, while the summarized
IHC results are presented in Table
II. ARID1A, SMARCA2, SMARCA4, SMARCB1, SMARCC2 and H3K27me3
were predominantly expressed in the nucleus of the tumor cells
(Fig. S2). Nuclear staining of
CRFs was lost in 8.9% of cases. Specifically, loss of nuclear
ARID1A staining occurred in 2.5% of cases, SMARCA2 in 2.5% of cases
and SMARCA4 in 3.9% of cases (Table
II; Fig. 2A-C). Additionally,
2.5% of cases exhibited loss of H3K27me3 expression (Table II; Fig.
2D). The nuclear staining of these markers in stromal cells and
vascular endothelium served as a positive internal control. Loss of
ARID1A, SMARCA2 and SMARCA4 protein expression was mutually
exclusive, and the loss of H3K27me3 was not related to loss of the
protein expression of these CRFs. However, SMARCC2 and SMARCB1
protein expression was retained in all cases (Table II). The intensity of ACTL6A,
SMARCC2 and CHD4 protein expression was higher in tumor cells
compared with in stromal cells or lymphocytes (Fig. 2E-G). In addition, the nucleus of
OHGSC tumor cells was strongly and diffusely positive for p53
(Fig. 2H).
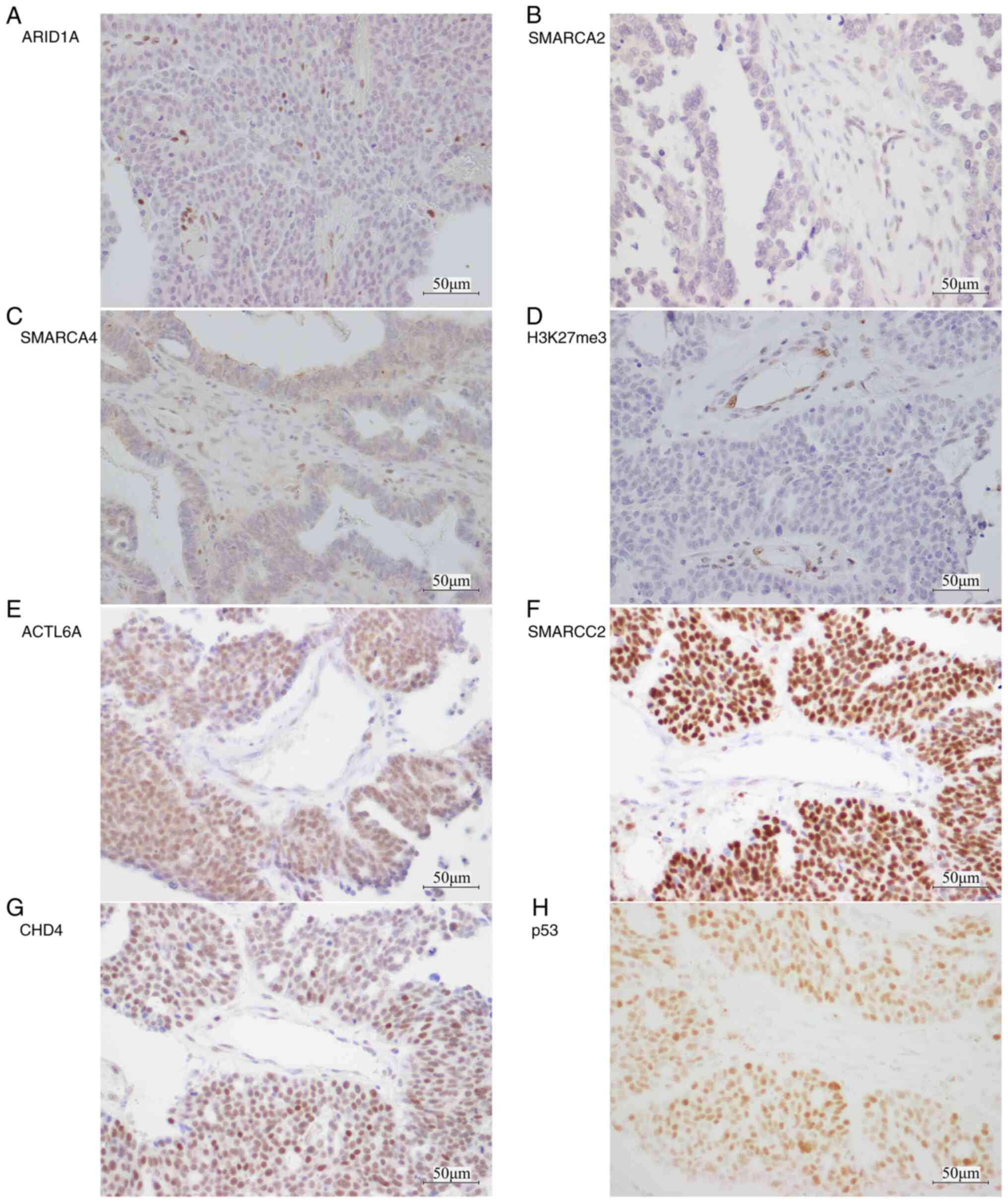 | Figure 2.Representative immunohistochemistry
images for chromatin remodeling factors and H3K27me3 in ovarian
high-grade serous carcinoma. Loss of nuclear staining in tumor
cells for (A) ARID1A, (B) SMARCA2, (C) SMARCA4 and (D) H3K27me3.
(E) ACTL6A, (F) SMARCC2 and (G) CHD4 exhibited stronger nuclear
expression in tumor cells compared with stromal cells and
lymphocytes. Stromal cells and vascular endothelium demonstrated
consistently preserved nuclear staining, serving as a positive
internal control. (H) Aberrant expression of p53 in the nucleus of
tumor cells. Scale bar, 50 µm. ARID1A, AT-rich interaction domain
1A; SMARCA, switch/sucrose non-fermentable related, matrix
associated, actin dependent regulator of chromatin subfamily a;
H3K27me3, tri-methylation of lysine 27 of histone H3;
ACTL6A, actin-like protein 6A; SMARCC2,
switch/sucrose non-fermentable related, matrix associated, actin
dependent regulator of chromatin subfamily c member 2; CHD4,
chromodomain-helicase-DNA-binding protein 4. |
 | Table II.Immunohistochemistry results. |
Table II.
Immunohistochemistry results.
| Antibody | Positivity | No. (%) |
|---|
| ARID1A | Lost | 5/203 (2.5) |
|
| Retained | 198/203 (97.5) |
| SMARCA2 | Lost | 5/203 (2.5) |
|
| Retained | 198/203 (97.5) |
| SMARCA4 | Lost | 8/203 (3.9) |
|
| Retained | 195/203 (96.1) |
| SMARCB1 | Lost | 0/203 (0) |
|
| Retained | 203/203 (100) |
| SMARCC2 | Lost | 0/203 (0) |
|
| Retained | 203/203 (100) |
| H3K27me3 | Lost | 5/203 (2.5) |
|
| Retained | 198/203 (97.5) |
The H-score of ACTL6A, SMARCC2 and CHD4 ranged from
140–220 (mean 169.66), 140–210 (mean 180.39), and 60–210 (mean
168.82), respectively. The median H-scores: 170 for ACTL6A, 180 for
SMARCC2 and 170 for CHD4, were used as the cutoff values for
distinguishing low and high CRF expression; representative images
of high and low ACTL6A, SMARCC2 and CHD4 staining are shown
(Fig. S3A-F).
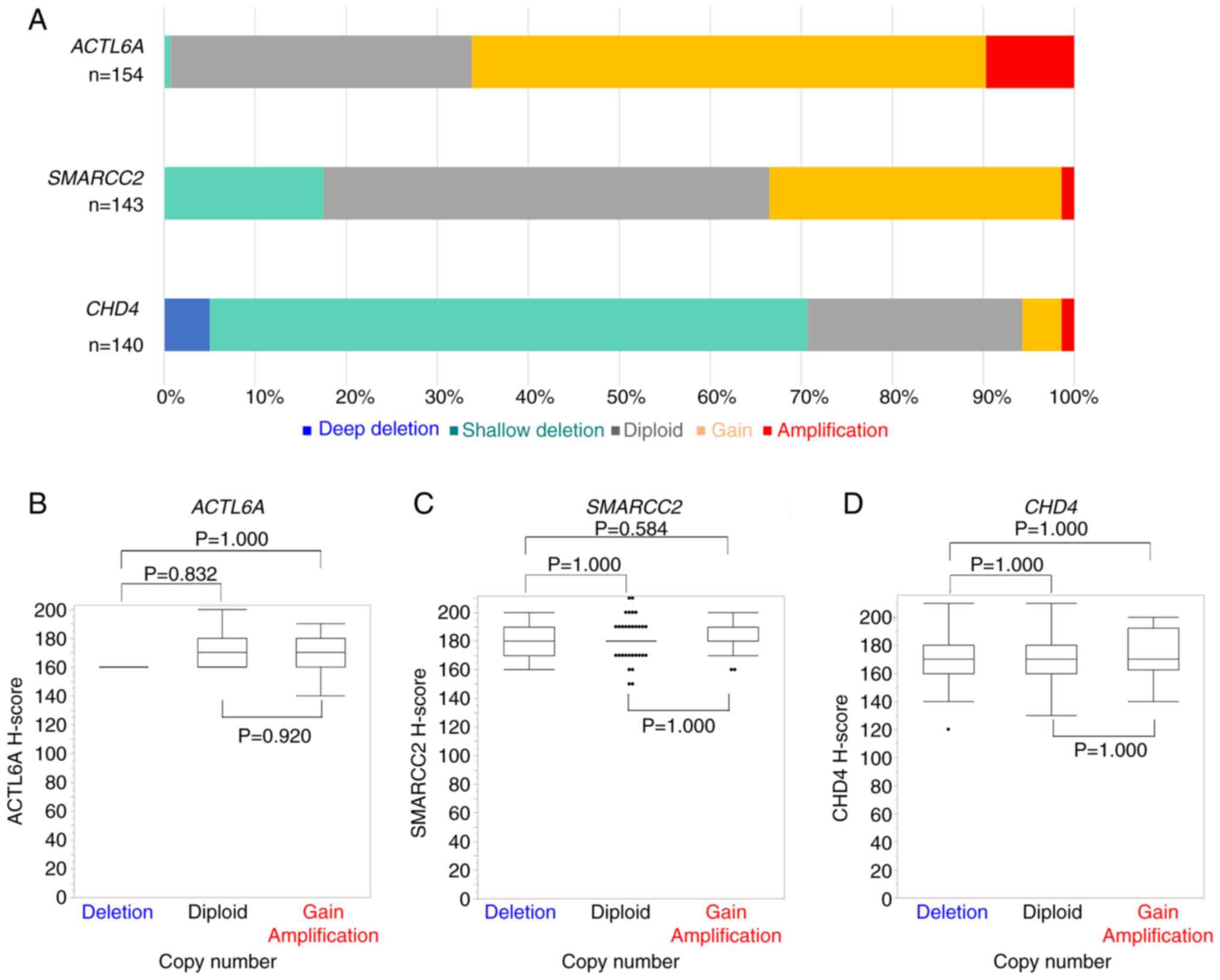
Copy number analysis
Fig. 3A presents a
summary of the DNA copy numbers of ACTL6A, SMARCC2 and
CHD4 in OHGSC. Results were obtained for 154 cases for
ACTL6A, 143 cases for SMARCC2 and 140 cases for
CHD4, out of a total of 203 cases. For comparison, normal
skeletal muscle tissue was used as the control and the copy numbers
were normalized to Rnase P, which served as the internal
control. The copy numbers of ACTL6A, SMARCC2 and CHD4
ranged from 1–8 (mean 3.09), 1–7 (mean 2.27), and 0–5 (mean 1.34),
respectively.
Notably, CNAs with increased copy numbers were
predominant in ACTL6A and SMARCC2, while CHD4
CNAs primarily exhibited decreased copy numbers. Among the cases
assessed, 102 out of 154 (66.2%) demonstrated ACTL6A copy
number gain or gene amplification, 48 out of 143 (33.5%) showed
SMARCC2 copy number gain or gene amplification, and 99 out
of 140 (70.7%) demonstrated CHD4 shallow deletion or deep
deletion. The relationship between ACTL6A, SMARCC2 and
CHD4 copy numbers and the immunohistochemical expression of
these proteins was assessed. However, no statistically significant
association was observed between the copy numbers and the protein
expression of ACTL6A (Fig. 3B),
SMARCC2 (Fig. 3C) and CHD4
(Fig. 3D).
Relationship between prognosis and
CRFs
The survival analyses for copy numbers of CRFs are
summarized in Fig. S4, while the
association of protein expression with OS is presented in Fig. 4.
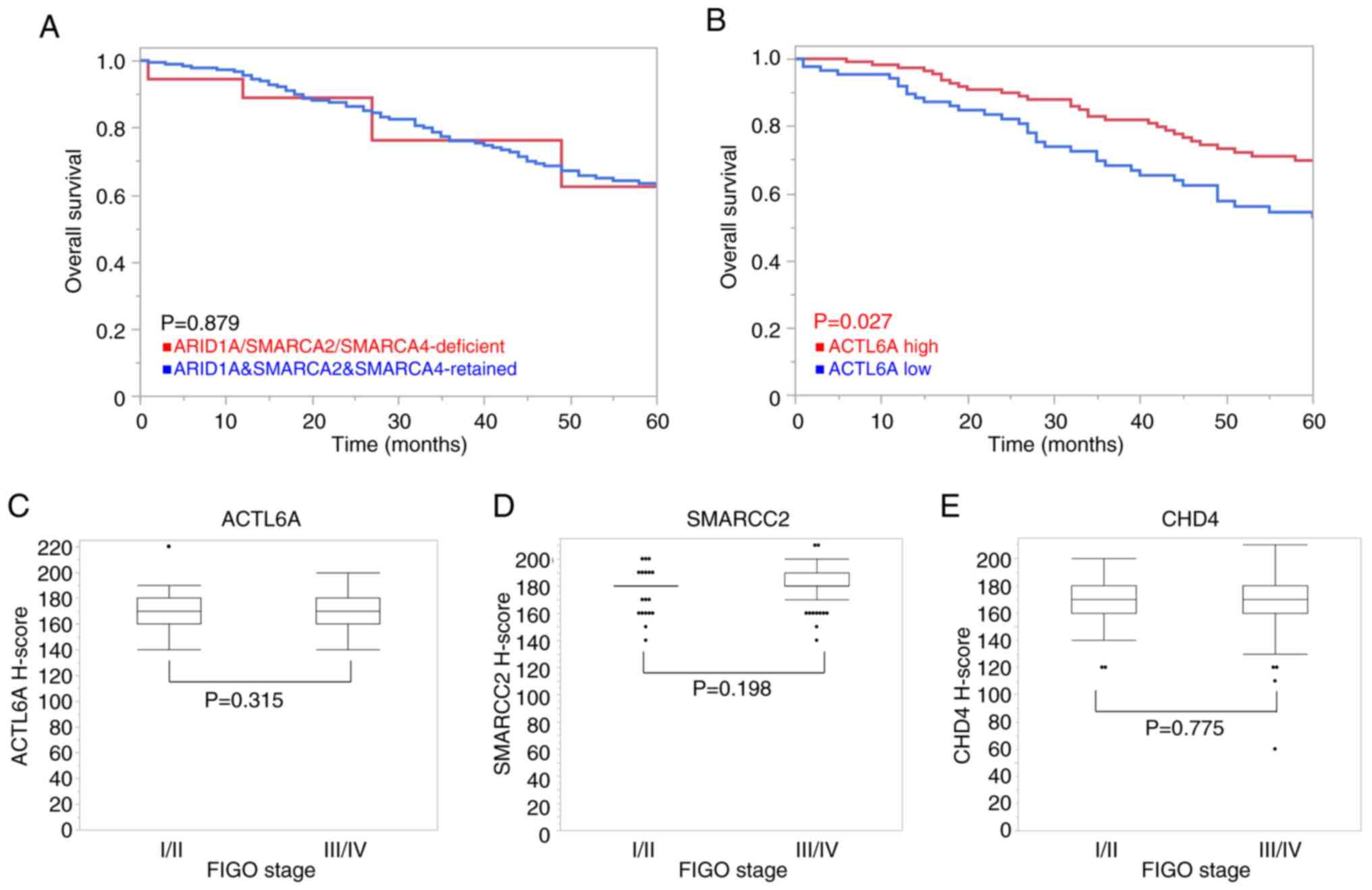 | Figure 4.Relationship among CRF protein
expression, overall survival and clinical stages in patients with
OHGSC. (A) Kaplan-Meier survival analysis was performed for
patients with a deficiency of any of the ARID1A, SMARCA2 and
SMARCA4 protein levels or with retained protein levels. Cases with
a deficiency of any of the ARID1A, SMARCA2 or SMARCA4 proteins
demonstrated similar outcome trends compared with those with
retained expression, and there was no statistically significant
difference. (B) Kaplan-Meier analysis demonstrated the prognostic
differences based on ACTL6A protein expression. Patients with high
ACTL6A protein expression exhibited a more favorable outcome
compared with those with low ACTL6A expression. The threshold value
between the high and low groups was determined by the median
H-score, 170. The relationship between (C) ACTL6A, (D) SMARCC2 and
(E) CHD4 protein expression levels and the FIGO stage. (D) Patients
with higher FIGO stage tended to have higher SMARCC2 expression,
but this was not statistically significant. (C) ACTL6A and (E) CHD4
protein levels were unchanged between the FIGO stages. CRF,
chromatin remodeling factor; OHGSC, ovarian high-grade serous
carcinoma; FIGO, International Federation of Gynecology and
Obstetrics; ARID1A, AT-rich interaction domain 1A; SMARCA,
switch/sucrose non-fermentable related, matrix associated, actin
dependent regulator of chromatin subfamily a; ACTL6A, actin-like
protein 6A; SMARCC2, switch/sucrose non-fermentable related, matrix
associated, actin dependent regulator of chromatin subfamily c
member 2; CHD4, chromodomain-helicase-DNA-binding protein 4. |
Regarding the CNAs of ACTL6A, SMARCC2 and
CHD4, changes in copy number of these genes demonstrated no
statistically significant association with the OS of patients
(P=0.434, P=0.629 and P=0.578, respectively; Fig. S4A-C). Similarly, there was no
significant association between the copy numbers of these CRFs and
the FIGO stage (P=0.506, P=0.862 and P=0.974, respectively;
Fig. S4D-F). However, although not
significant (P=0.094), in patients with FIGO stage III/IV OHGSC,
copy number gain or amplification in either ACTL6A, SMARCC2
or CHD4 demonstrated unfavorable outcome trends compared
with patients with diploid ACTL6A, SMARCC2 and CHD4
(Fig. S4G).
Moreover, although not significant (P=0.128),
patients with FIGO stage III/IV with shallow or deep deletions in
either ACTL6A, SMARCC2 or CHD4 demonstrated
unfavorable outcome trends compared with those with ACTL6A,
SMARCC2 and CHD4 diploids (Fig. S4H). However, due to the small
number of cases, the effect of CRF CNAs on FIGO stage I/II cases
could not be analyzed.
In cases with decreased protein levels of ARID1A,
SMARCA2 or SMARCA4, similar outcome trends were demonstrated in OS
compared with those with retained expression levels, and there was
no statistically significant difference (P=0.879; Fig. 4A). Notably, patients with high
ACTL6A protein levels demonstrated a statistically longer OS
compared with patients with low ACTL6A protein levels (P=0.027;
Fig. 4B).
Regarding the association with FIGO stage, higher
SMARCC2 protein expression was detected in patients with a higher
FIGO stage, but this relationship was not statistically significant
(P=0.198; Fig. 4D). Likewise, the
difference between ACTL6A and CHD4 protein levels and FIGO stage
was not found to be significantly different (P=0.315 and P=0.775,
respectively; Fig. 4C and E).
Discussion
The present study conducted a comprehensive analysis
of the relationship between CRF alterations and the
clinicopathological features of OHGSC. The findings revealed CNAs
in ACTL6A, SMARCC2 and CHD4 in OHGSC, as well as
protein loss of ARID1A (2.5%), SMARCA2 (2.5%) and SMARCA4 (3.9%),
indicating possible gene alterations. Notably, low protein
expression levels of ACTL6A were identified as a positive indicator
of shortened OS in patients with OHGSC.
Adenosine triphosphate (ATP)-dependent chromatin
remodeling complexes regulate the chromatin packing state by
sliding, ejecting and restructuring the nucleosome for
transcriptional regulation (19).
The Brg1-associated factor (BAF) complex is composed of a central
ATPase (SMARCA2 or SMARCA4) and multiple BAFs, including ARID1A,
ACTL6A and SMARCC2, which are assembled in a combinatorial fashion
to dictate functional specificity (20). Overall, complex stoichiometry is
influenced by individual BAFs that can regulate the expression of
other subunits (21).
CHD4 is a core component of the nucleosome
remodeling and deacetylase complex that combines chromatin
remodeling activity with histone deacetylase and demethylase
functions, which are involved in transcriptional repression
(22). CHD4 comprises a core
ATPase/helicase domain flanked by two plant homeodomain motifs that
recognize modifications of histone tails, tandem chromodomains and
carboxyl-terminal domains (23).
Previous studies have highlighted the amplification
and upregulation of ACTL6A in numerous types of cancer, such
as ovarian cancer, glioma, squamous cell carcinoma, osteosarcoma
and hepatocellular carcinoma (24–28).
ACTL6A has been implicated in promoting metastasis and
epithelial-mesenchymal transition in hepatocellular carcinoma
(28) and colon cancer (29). Additionally, it has been reported to
serve a role in tumorigenesis in head and neck squamous cell
carcinoma (26) and glioma
(25) by activating the Hippo/YAP
pathway. In the context of ovarian cancer, high mRNA expression of
ACTL6A has been reported to be associated with shortened OS
(24) and platinum resistance
(30). In the present study, public
data demonstrated no significant association between ACTL6A
mRNA expression and prognosis in patients with ovarian serous
carcinoma; however, low ACTL6A protein expression levels, as
detected by IHC, were associated with decreased OS. Thus, the
protein levels and mRNA levels may not comparable. Differences
between mRNA and protein levels may arise due to technical or
biological reasons, such as post-transcriptional regulation
(31). The process of mRNA
stabilization needs to be elucidated to prove these
divergences.
Additionally, despite a ACTL6A copy number
gain or gene amplification in 66.2% of the cases tested, a
statistically significant association between the copy number and
protein levels of ACTL6A was not found. The different steps in the
gene expression pathway each involve a complex process that confers
regulatory control. Likewise, other epigenetic factors, such as
microRNAs and ubiquitination, may have contributed to ACTL6A
protein expression. Moreover, ACTL6A may affect the expression of
oncogenes although ACTL6A mRNA expression was not related to
prognosis. According to the present results, poor prognosis may be
caused by the dysregulated transcription of other oncogenes or
tumor suppressor genes following the decrease in ACTL6A protein
expression. In vitro or in vivo analysis using
protein knockdown or gene knockout of ACTL6A may be required
to evaluate these hypotheses.
In the present study, it was demonstrated that
patients with a higher FIGO stage tended to exhibit higher SMARCC2
protein expression. Furthermore, the analysis of TCGA data
demonstrated that patients with SMARCC2 amplification had a
shorter median OS compared with patients with wild-type
SMARCC2 in ovarian serous carcinoma. Several studies have
indicated that SMARCC2 is deficiently expressed in cancer (32,33).
SMARCC2 has been reported to inhibit tumor development by mediating
the expression of the transcription factor early growth response 1
via chromatin remodeling, and by inhibiting activation of the
phosphoinositide 3-kinase-AKT pathway in glioblastoma (33). However, several studies have
reported SMARCC2 gene amplification in cancer, such as
follicular lymphoma (34) and
hepatocellular carcinoma (35), in
line with the results of the present study. These results
collectively suggested that SMARCC2 function varies according to
tumor type, and that aberrant SMARCC2 expression could be involved
in the regulation of numerous cellular functions, such as cell
proliferation and the cell cycle of the tumor.
Additionally, patients in FIGO stage III/IV who have
copy number gain or amplification in either ACTL6A, SMARCC2
or CHD4 had a poor prognosis compared with those of
ACTL6A, SMARCC2 and CHD4 diploids.
In the present study, 8.9% of cases exhibited a
deficiency in either ARID1A, SMARCA2 or SMARCA4 protein levels.
Notably, the deficiency in ARID1A, SMARCA2 and SMARCA4 was found to
be mutually exclusive in this analysis. This exclusivity is
attributed to the biochemical and functional heterogeneity of BAF
complexes (36), and numerous
epigenetic mechanisms are involved in the instability and silencing
of SMARCA2, SMARCA4 and other subunits of the BAF complex (37). The BAF-chromatin remodeling complex,
with its mutually exclusive ATPases SMARCA2 and SMARCA4, is
essential for the transcriptional activation of numerous genes
(38).
Furthermore, changes in nucleosome distribution
pattern and density have been linked to reduced levels of H3K27me3
in chromatin remodeling enzyme mutants (39). However, in the present study, the
loss of H3K27me3 was not found to be associated with the loss of
CRFs expression.
The molecular biology of OHGSC is characterized by
genomic complexity, often lacking targetable oncogenic alterations
(40). Nonetheless, the present
study revealed aberrant protein expression and CNAs of CRFs in
OHGSC, which supports their potential use as therapeutic
targets.
One limitation of the present study is the lack of
in vivo confirmation of CRF expression in OHGSC. Thus,
future studies using animal models are required to validate these
findings.
There are ongoing developments in drugs that target
genomic abnormalities of CRFs and combination therapies aimed at
enhancing the therapeutic effects of anticancer drugs (41). In a previous study, the histone
deacetylase inhibitor romidepsin, which targets CHD4, was
demonstrated to suppress the progression of metastases in ovarian
cancer both in vitro and in vivo (42). Additionally, panobinostat was
reported to counteract ACTL6A-induced cisplatin resistance by
inhibiting the repair of cisplatin-DNA adducts in vivo
(30).
In conclusion, the present study demonstrated copy
number and protein expression alterations of CRFs in OHGSC.
Notably, the protein expression levels of ACTL6A were found to be
associated with poor prognosis. These findings suggested that CRFs
could be prognostic markers for OHGSC. However, further research is
required to fully understand the mechanisms through which CRFs
contribute to transcriptional aberration of oncogenes, particularly
in the context of other epigenetic factors in OHGSC, and to
investigate whether they are potential therapeutic targets for
OHGSC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Ms. Haruka Toki, Ms.
Nahoko Ieiri, Ms. Jumi Yahiro, Ms. Motoko Tomita and Ms. Mami
Nakamizo (Department of Anatomic Pathology, Kyushu University), for
their technical support for this study.
Funding
This study received financial support from the ‘FUKUOKA’ OBGYN
Researcher's Charity Foundation Fund, Japan, through
grants-in-aid.
Availability of data and materials
The data generated in the present study may be found
in The Cancer Genome Atlas or at the following URL: https://www.cancer.gov/tcga. The other data generated
in the present study may be requested from the corresponding
author.
Authors' contributions
NM and TI conducted the research and wrote the
article. KK, YK, TT, MN and FN contributed to the sample collection
and research design. YO designed the research and gave final
approval of the article. NM, TI and YO confirm the authenticity of
all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was conducted in accordance with the
principles of The Declaration of Helsinki. The study protocol was
approved by the Ethics Committee of Kyushu University (Fukuoka,
Japan; approval nos. 21120-01, 21037-02 and 23005-00). There was an
opt-out approach for consent where participants were informed of
the trial on the homepage and were invited to opt-out if
preferred.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ACTL6A
|
actin-like protein 6A
|
|
ARID1A
|
AT-rich interaction domain 1A
|
|
ATP
|
adenosine triphosphate
|
|
CHD
|
chromodomain-helicase-DNA-binding
protein
|
|
CNA
|
copy number alteration
|
|
CRF
|
chromatin remodeling factor
|
|
FFPE
|
formalin-fixed and
paraffin-embedded
|
|
FIGO
|
International Federation of Gynecology
and Obstetrics
|
|
H3K27me3
|
tri-methylation of lysine 27 of
histone H3
|
|
IHC
|
immunohistochemistry
|
|
NGS
|
next-generation sequencing
|
|
OHGSC
|
ovarian high-grade serous
carcinoma
|
|
OS
|
overall survival
|
|
PCR
|
polymerase chain reaction
|
|
SWI/SNF
|
switch/sucrose non-fermentable
|
|
SMARCA
|
SWI/SNF related, matrix associated,
actin dependent regulator of chromatin subfamily a
|
|
SMARCB
|
SWI/SNF related, matrix associated,
actin dependent regulator of chromatin subfamily b
|
|
SMARCC
|
SWI/SNF related, matrix associated,
actin dependent regulator of chromatin subfamily c
|
|
TCGA
|
The Cancer Genome Atlas
|
References
|
1
|
Kim J, Park EY, Kim O, Schilder JM, Coffey
DM, Cho CH and Bast RC Jr: Cell origins of high-grade serous
ovarian cancer. Cancers (Basel). 10:4332018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peres LC, Cushing-Haugen KL, Köbel M,
Harris HR, Berchuck A, Rossing MA, Schidkraut JM and Doherty JA:
Invasive epithelial ovarian cancer survival by histotype and
disease stage. J Natl Cancer Inst. 111:60–68. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barlin JN, Long KC, Tanner EJ, Gardner GJ,
Leitao MM Jr, Levine DA, Sonoda Y, Abu-Rustum NR, Barakat RR and
Chi DS: Optimal (≤1 cm) but visible residual disease: Is extensive
debulking warranted? Gynecol Oncol. 130:284–288. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dao F, Schlappe BA, Tseng J, Lester J,
Nick AM, Lutgendorf SK, McMeekin S, Coleman RL, Moore KN, Karlan
BY, et al: Characteristics of 10-year survivors of high-grade
serous ovarian carcinoma. Gynecol Oncol. 141:260–263. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iacobuzio-Donahue CA: Epigenetic changes
in cancer. Annu Rev Pathol. 4:229–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baylin SB and Jones PA: Epigenetic
determinants of cancer. Cold Spring Harb Perspect Biol.
8:a0195052016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hasan N and Ahuja N: The emerging roles of
ATP-dependent chromatin remodeling complexes in pancreatic cancer.
Cancers (Basel). 11:18592019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kadoch C and Crabtree GR: Mammalian
SWI/SNF chromatin remodeling complexes and cancer: Mechanistic
insights gained from human genomics. Sci Adv. 1:e15004472015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khalique S, Naidoo K, Attygalle AD,
Kriplani D, Daley F, Lowe A, Campbell J, Jones T, Hubank M, Fenwick
K, et al: Optimised ARID1A immunohistochemistry is an accurate
predictor of ARID1A mutational status in gynaecological cancers.
Pathol Clin Res. 4:154–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Le Gallo M, O'Hara AJ, Rudd ML, Urick ME,
Hansen NF, O'Neil NJ, Price JC, Zhang S, England BM, Godwin AK, et
al: Exome sequencing of serous endometrial tumors identifies
recurrent somatic mutations in chromatin-remodeling and ubiquitin
ligase complex genes. Nat Genet. 44:1310–1315. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wiegand KC, Shah SP, Al-Agha OM, Zhao Y,
Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et
al: ARID1A mutations in endometriosis-associated ovarian
carcinomas. N Engl J Med. 363:1532–1543. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ramos P, Karnezis AN, Craig DW, Sekulic A,
Russell ML, Hendricks WPD, Corneveaux JJ, Barrett MT, Shumansky K,
Yang Y, et al: Small cell carcinoma of the ovary, hypercalcemic
type, displays frequent inactivating germline and somatic mutations
in SMARCA4. Nat Genet. 46:427–429. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oyama Y, Shigeta S, Tokunaga H, Tsuji K,
Ishibashi M, Shibuya Y, Shimada M, Yasuda J and Yaegashi N: CHD4
regulates platinum sensitivity through MDR1 expression in ovarian
cancer: A potential role of CHD4 inhibition as a combination
therapy with platinum agents. PLoS One. 16:e02510792021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cree IA, White VA, Indave BI and Lokuhetty
D: Revising the WHO classification: Female genital tract tumours.
Histopathology. 76:151–156. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Prat J and Oncology FCoG: FIGO's staging
classification for cancer of the ovary, fallopian tube, and
peritoneum: Abridged republication. J Gynecol Oncol. 26:87–89.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Clapier CR, Iwasa J, Cairns BR and
Peterson CL: Mechanisms of action and regulation of ATP-dependent
chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 18:407–422.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He S, Wu Z, Tian Y, Yu Z, Yu J, Wang X, Li
J, Liu B and Xu Y: Structure of nucleosome-bound human BAF complex.
Science. 367:875–881. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wade SL, Langer LF, Ward JM and Archer TK:
MiRNA-mediated regulation of the SWI/SNF chromatin remodeling
complex controls pluripotency and endodermal differentiation in
human ESCs. Stem Cells. 33:2925–2935. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ramírez J and Hagman J: The Mi-2/NuRD
complex: A critical epigenetic regulator of hematopoietic
development, differentiation and cancer. Epigenetics. 4:532–536.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Shih DJH and Lin SY: The tale of
CHD4 in DNA damage response and chemotherapeutic response. J Cancer
Res Cell Ther. 3:0522019.PubMed/NCBI
|
|
24
|
Chen PM, Wong CN, Wong CN and Chu PY:
Actin-like Protein 6A expression correlates with cancer stem
cell-like features and poor prognosis in ovarian cancer. Int J Mol
Sci. 24:20162023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ji J, Xu R, Zhang X, Han M, Xu Y, Wei Y,
Ding K, Wang S, Huang B, Chen A, et al: Actin like-6A promotes
glioma progression through stabilization of transcriptional
regulators YAP/TAZ. Cell Death Dis. 9:5172018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saladi SV, Ross K, Karaayvaz M, Tata PR,
Mou H, Rajagopal J, Ramaswamy S and Ellisen LW: ACTL6A is
co-amplified with p63 in squamous cell carcinoma to drive YAP
activation, regenerative proliferation, and poor prognosis. Cancer
Cell. 31:35–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun W, Wang W, Lei J, Li H and Wu Y:
Actin-like protein 6A is a novel prognostic indicator promoting
invasion and metastasis in osteosarcoma. Oncol Rep. 37:2405–2417.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xiao S, Chang RM, Yang MY, Lei X, Liu X,
Gao WB, Xiao JL and Yang LY: Actin-like 6A predicts poor prognosis
of hepatocellular carcinoma and promotes metastasis and
epithelial-mesenchymal transition. Hepatology. 63:1256–1271. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zeng Z, Yang H and Xiao S: ACTL6A
expression promotes invasion, metastasis and epithelial mesenchymal
transition of colon cancer. BMC Cancer. 18:10202018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiao Y, Lin FT and Lin WC: ACTL6A promotes
repair of cisplatin-induced DNA damage, a new mechanism of platinum
resistance in cancer. Proc Natl Acad Sci USA. 118:e20158081182021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Buccitelli C and Selbach M: mRNAs,
proteins and the emerging principles of gene expression control.
Nat Rev Genet. 21:630–644. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yamamoto T, Kohashi K, Yamada Y, Kawata J,
Sakihama K, Matsuda R, Koga Y, Aishima S, Nakamura M and Oda Y:
Relationship between cellular morphology and abnormality of SWI/SNF
complex subunits in pancreatic undifferentiated carcinoma. J Cancer
Res Clin Oncol. 148:2945–2957. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li C, Wang T, Gu J, Qi S, Li J, Chen L, Wu
H, Shi L, Song C and Li H: SMARCC2 mediates the regulation of DKK1
by the transcription factor EGR1 through chromatin remodeling to
reduce the proliferative capacity of glioblastoma. Cell Death Dis.
13:9902022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Havranek O, Westin JR, Zhang M, Rawal S,
Kwak LW, Neelapu SS and Davis RE: Integrated analysis of genomic
and gene expression profiles in follicular lymphoma reveals subsets
and driver genes of potential microenvironmental importance. Blood.
122:2487–2490. 2013. View Article : Google Scholar
|
|
35
|
Hu B, Lin JZ, Yang XB and Sang XT: The
roles of mutated SWI/SNF complexes in the initiation and
development of hepatocellular carcinoma and its regulatory effect
on the immune system: A review. Cell Prolif. 53:e127912020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Raab JR, Runge JS, Spear CC and Magnuson
T: Co-regulation of transcription by BRG1 and BRM, two mutually
exclusive SWI/SNF ATPase subunits. Epigenetics Chromatin.
10:622017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Marquez SB, Thompson KW, Lu L and Reisman
D: Beyond mutations: Additional mechanisms and implications of
SWI/SNF complex inactivation. Front Oncol. 4:3722014.PubMed/NCBI
|
|
38
|
Wilson BG, Helming KC, Wang X, Kim Y,
Vazquez F, Jagani Z, Hahn WC and Roberts CWM: Residual complexes
containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4
(BRG1) mutation. Mol Cell Biol. 34:1136–1144. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang T, Wang D, Tian G, Sun L, Yang M, Yin
X, Xiao J, Sheng Y, Zhu D, He H and Zhou Y: Chromatin remodeling
complexes regulate genome architecture in Arabidopsis. Plant Cell.
34:2638–2651. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bowtell DD, Böhm S, Ahmed AA, Aspuria PJ,
Bast RC Jr, Beral V, Berek JS, Birrer MJ, Blagden S, Bookman MA, et
al: Rethinking ovarian cancer II: Reducing mortality from
high-grade serous ovarian cancer. Nat Rev Cancer. 15:668–679. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang FL and Li DQ: Targeting
chromatin-remodeling factors in cancer cells: Promising molecules
in cancer therapy. Int J Mol Sci. 23:2022.
|
|
42
|
Wang J, Zhong F, Li J, Yue H, Li W and Lu
X: The epigenetic factor CHD4 contributes to metastasis by
regulating the EZH2/β-catenin axis and acts as a therapeutic target
in ovarian cancer. J Transl Med. 21:382023. View Article : Google Scholar : PubMed/NCBI
|