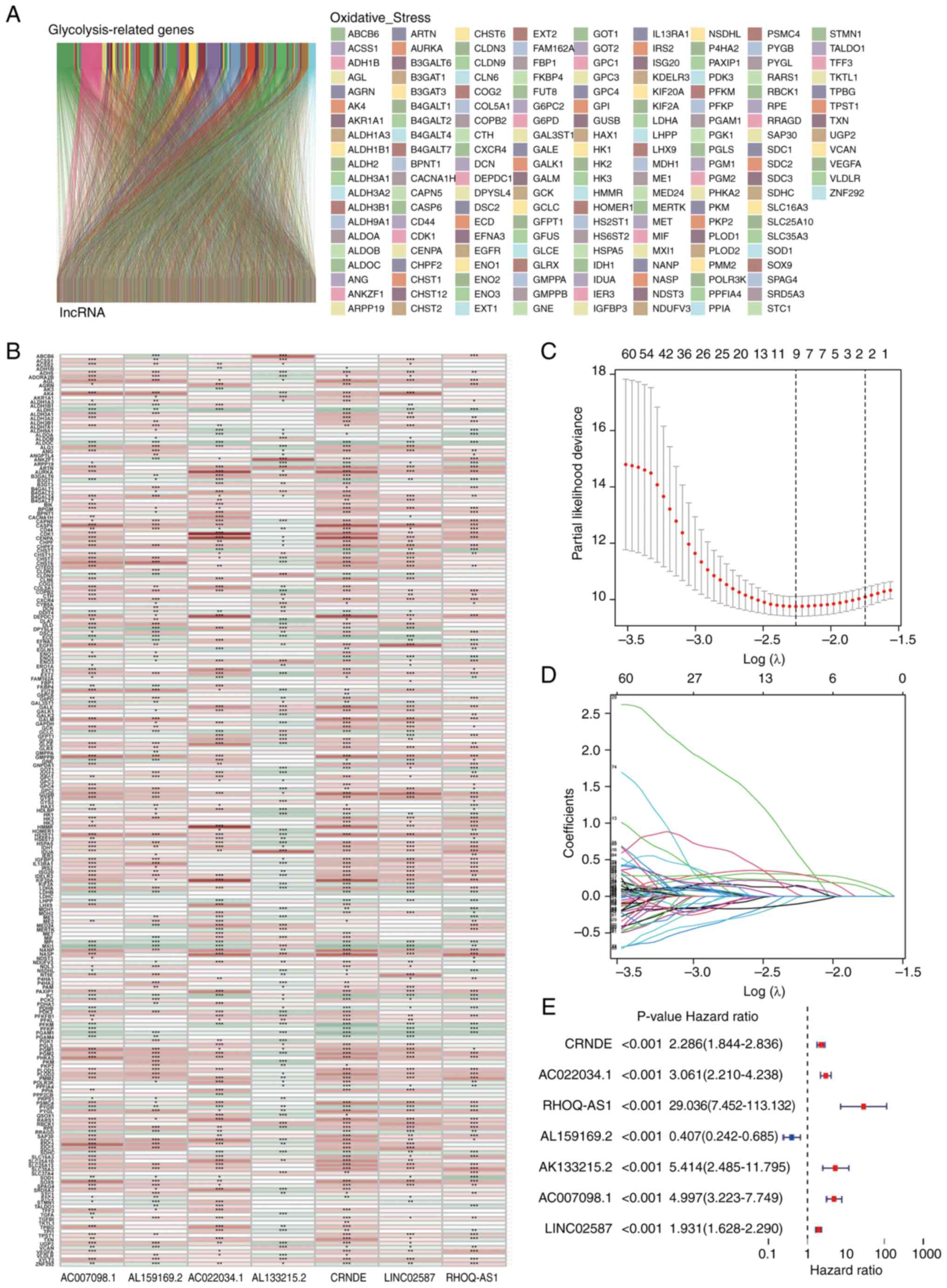
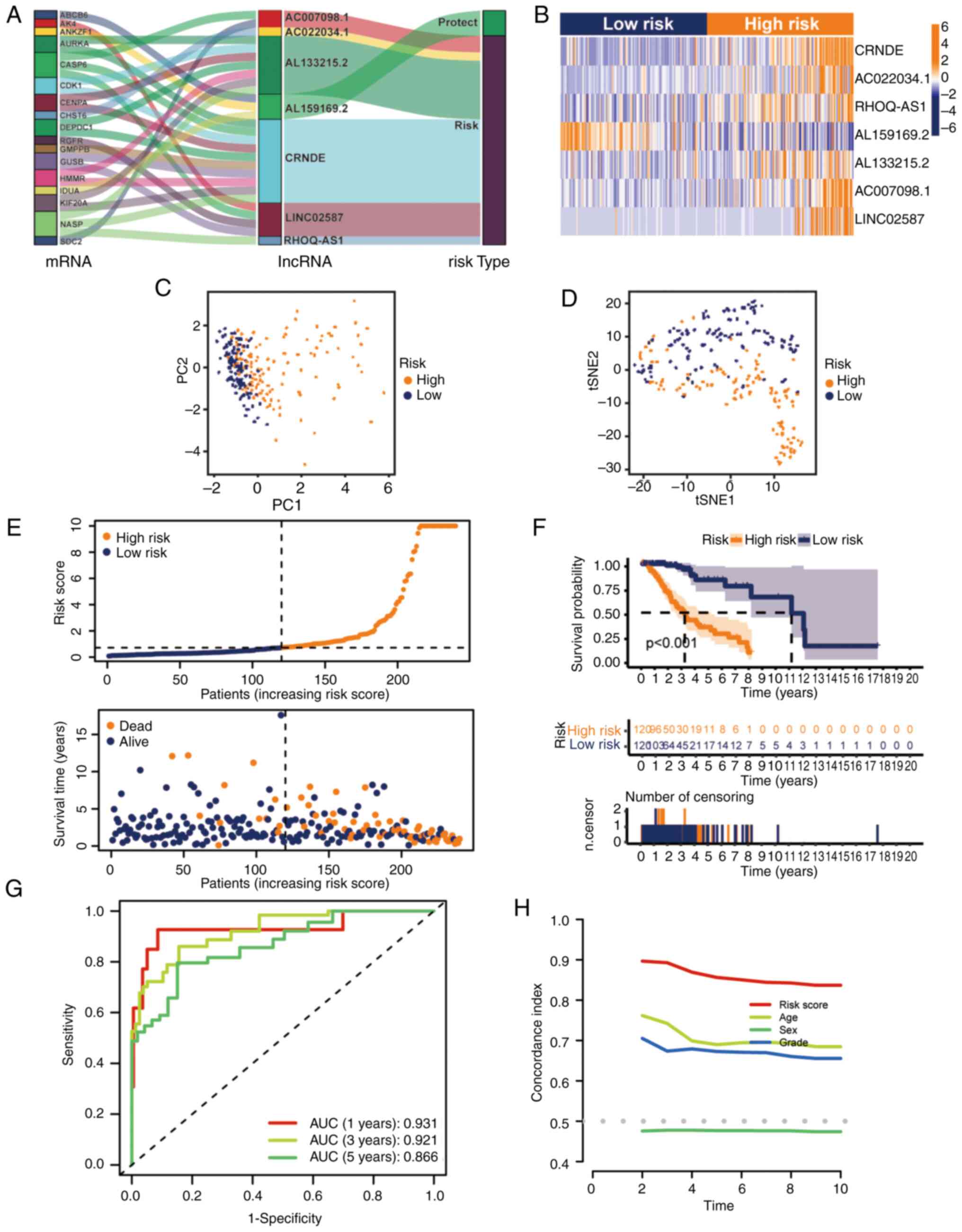
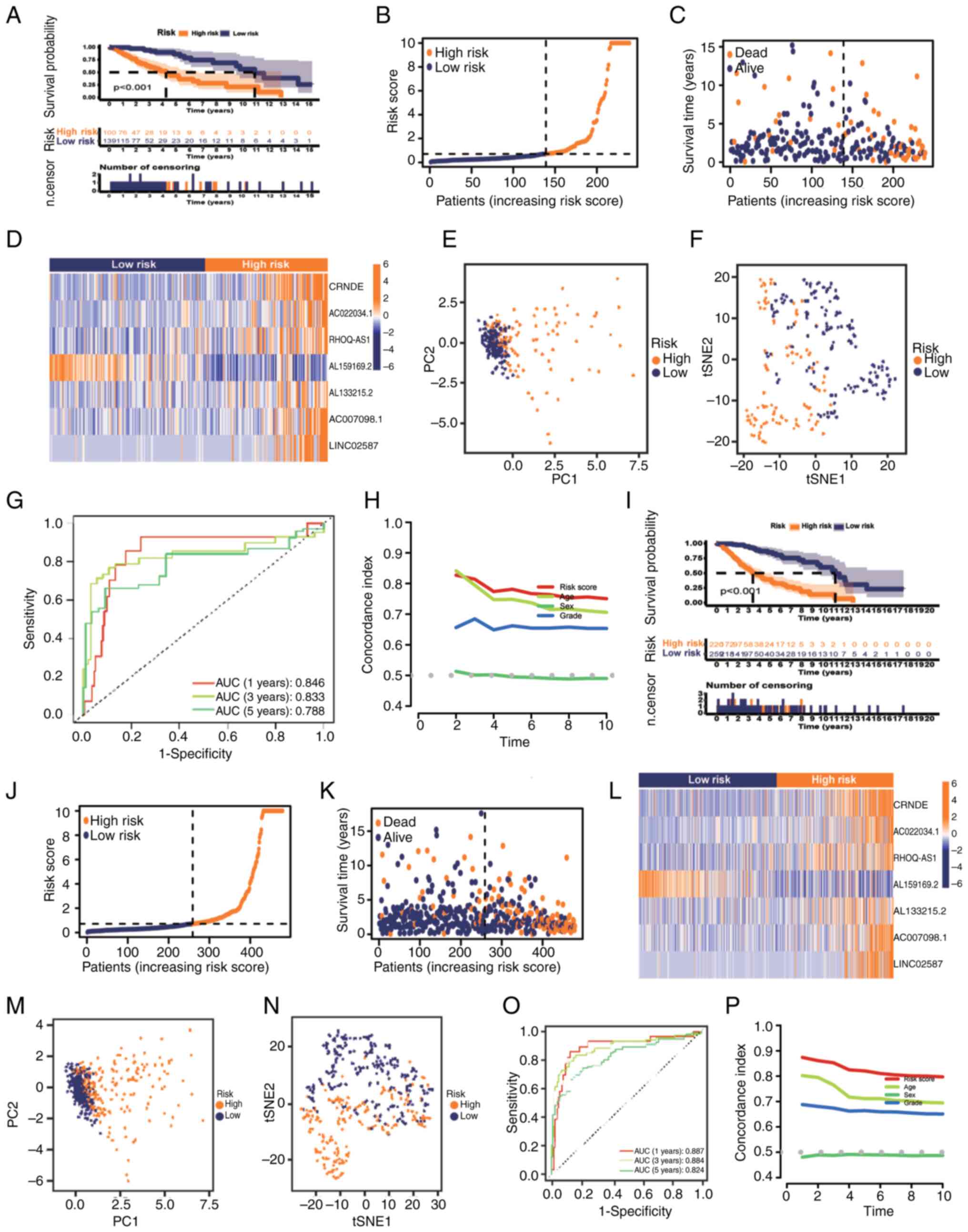
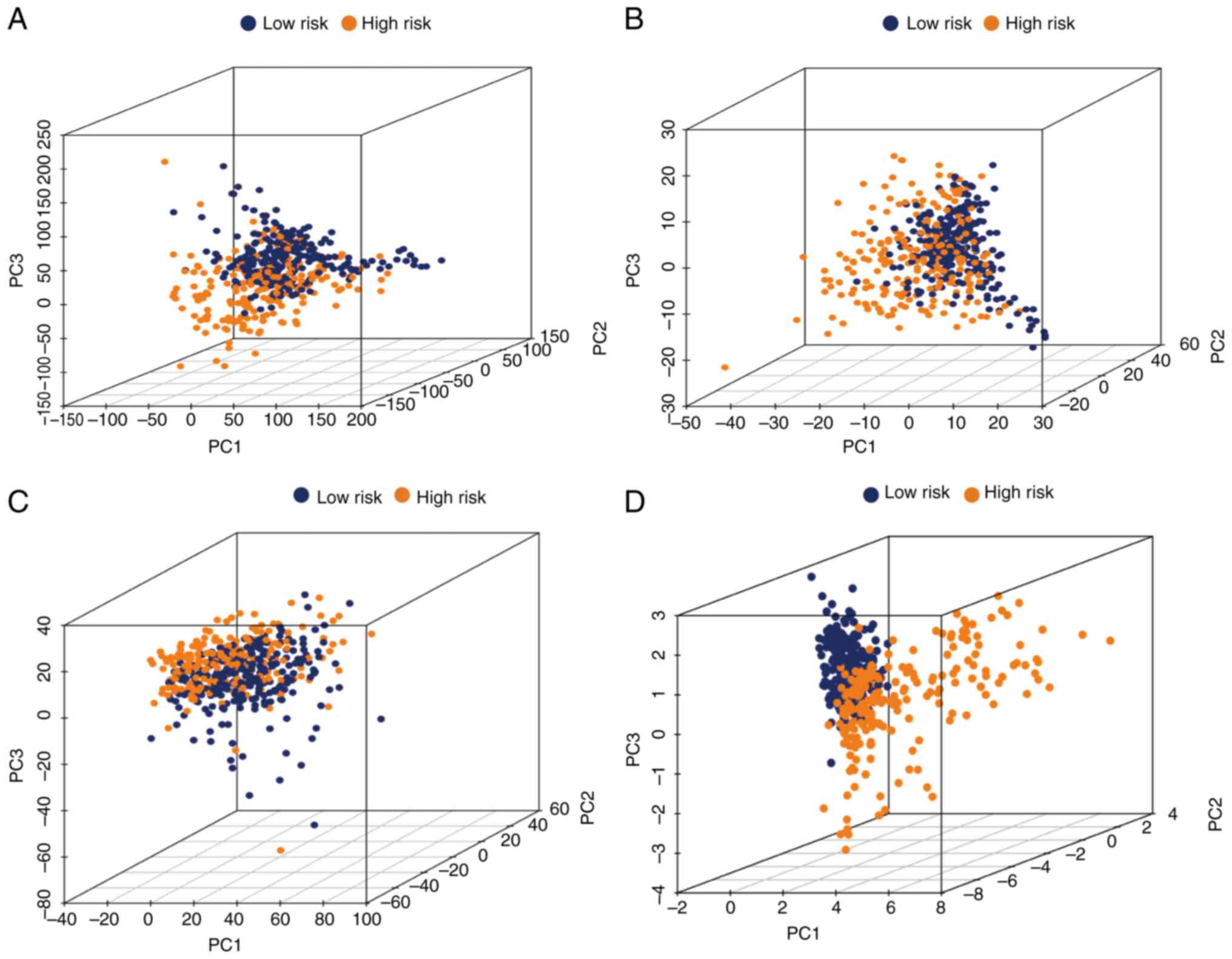
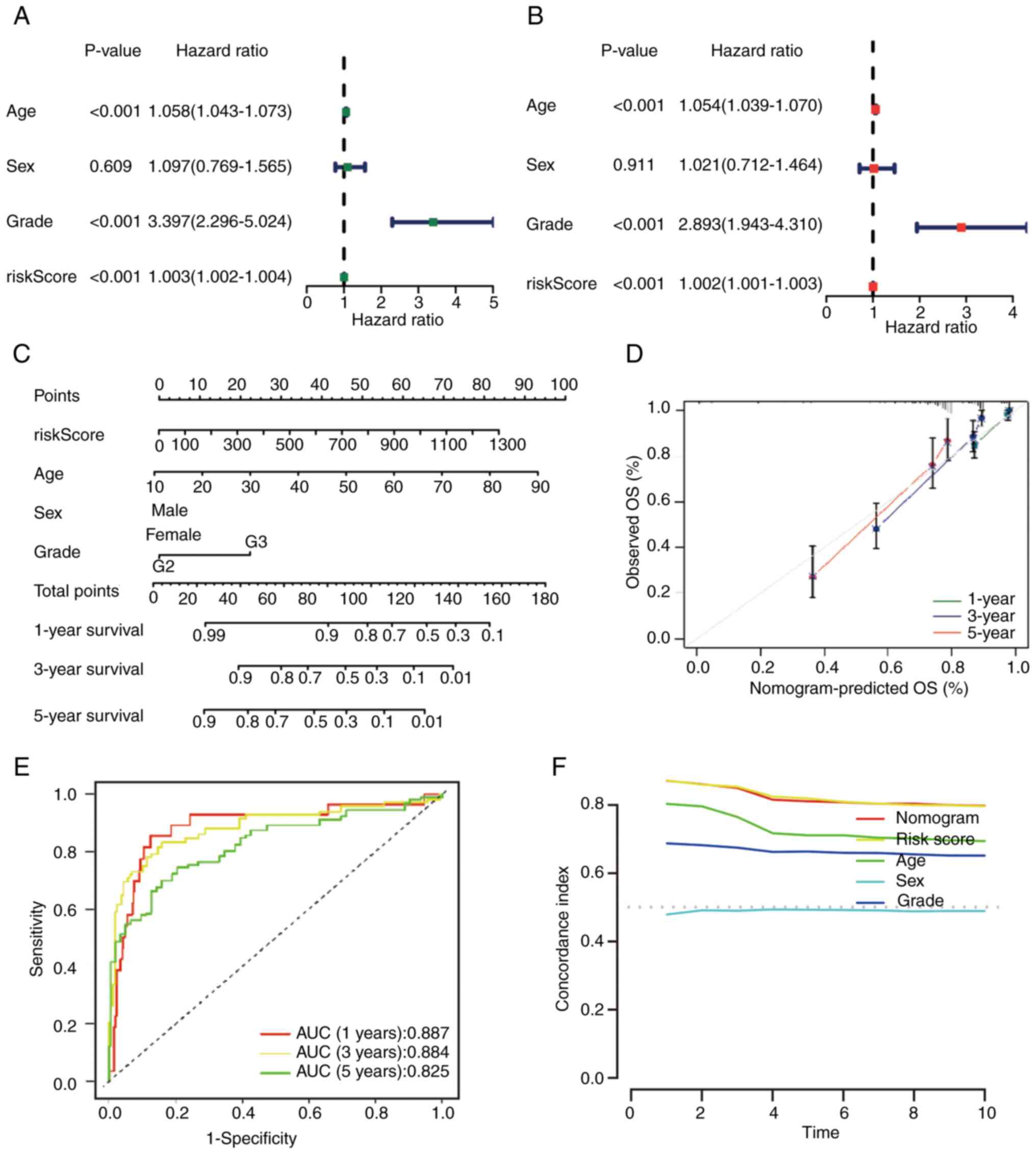
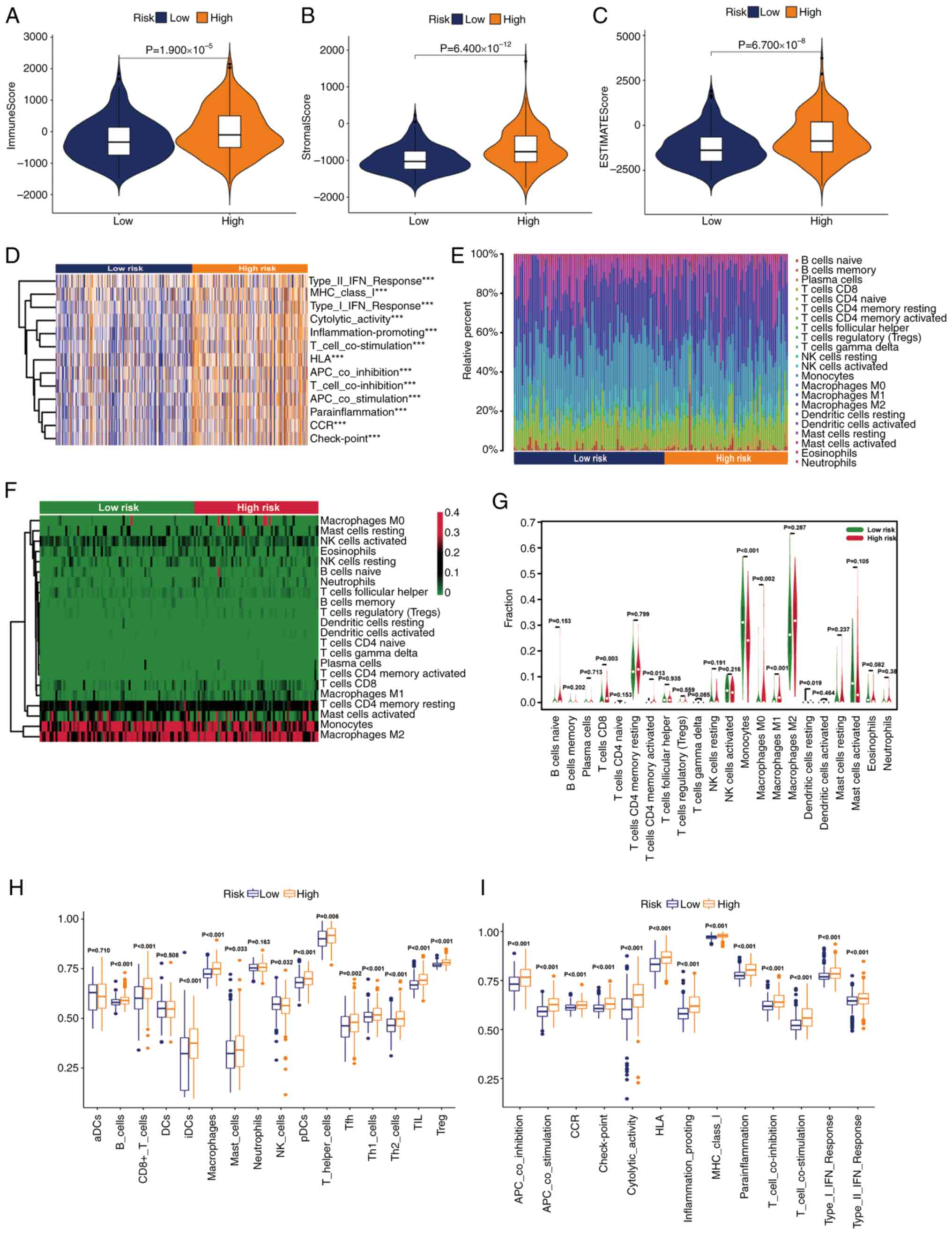
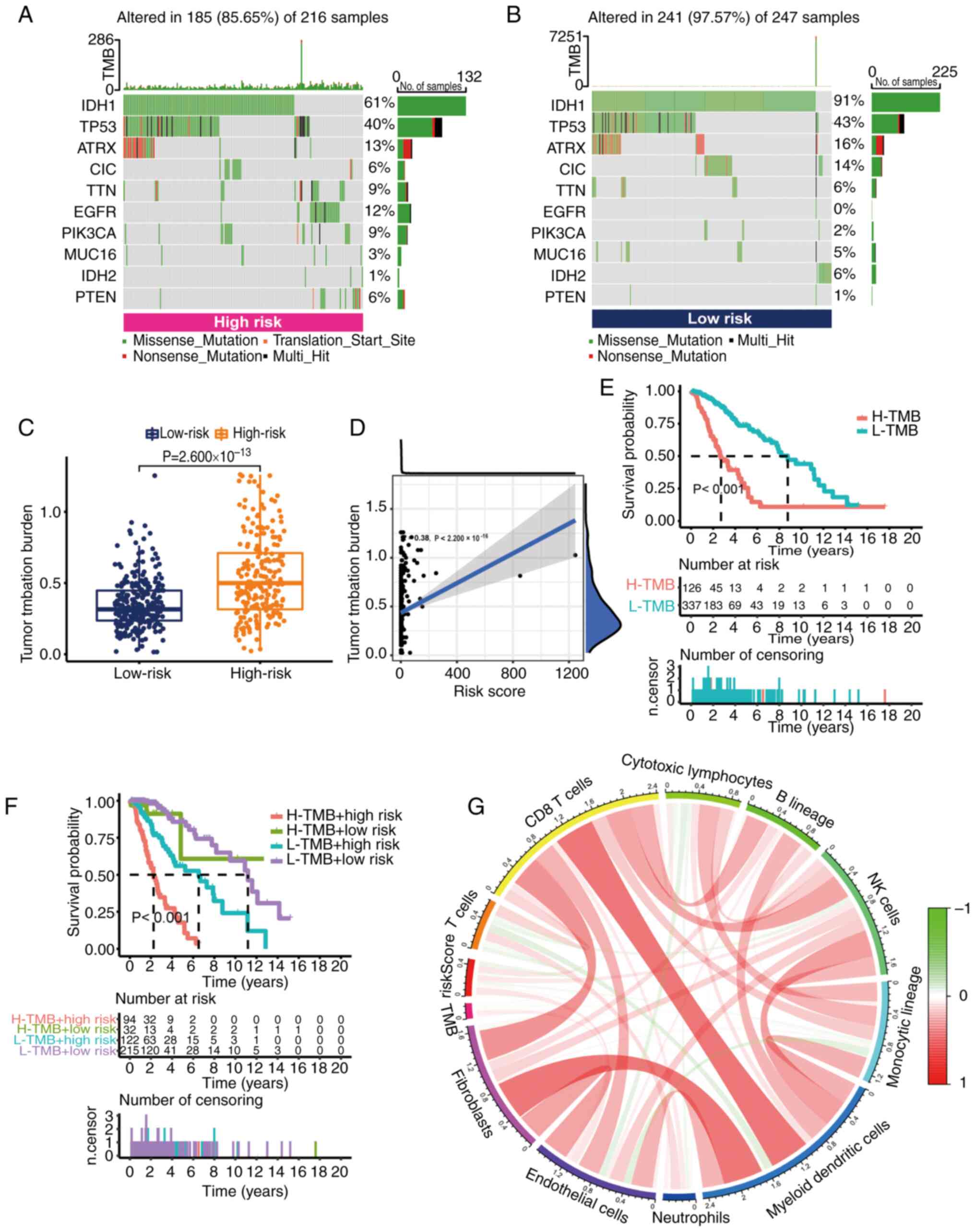
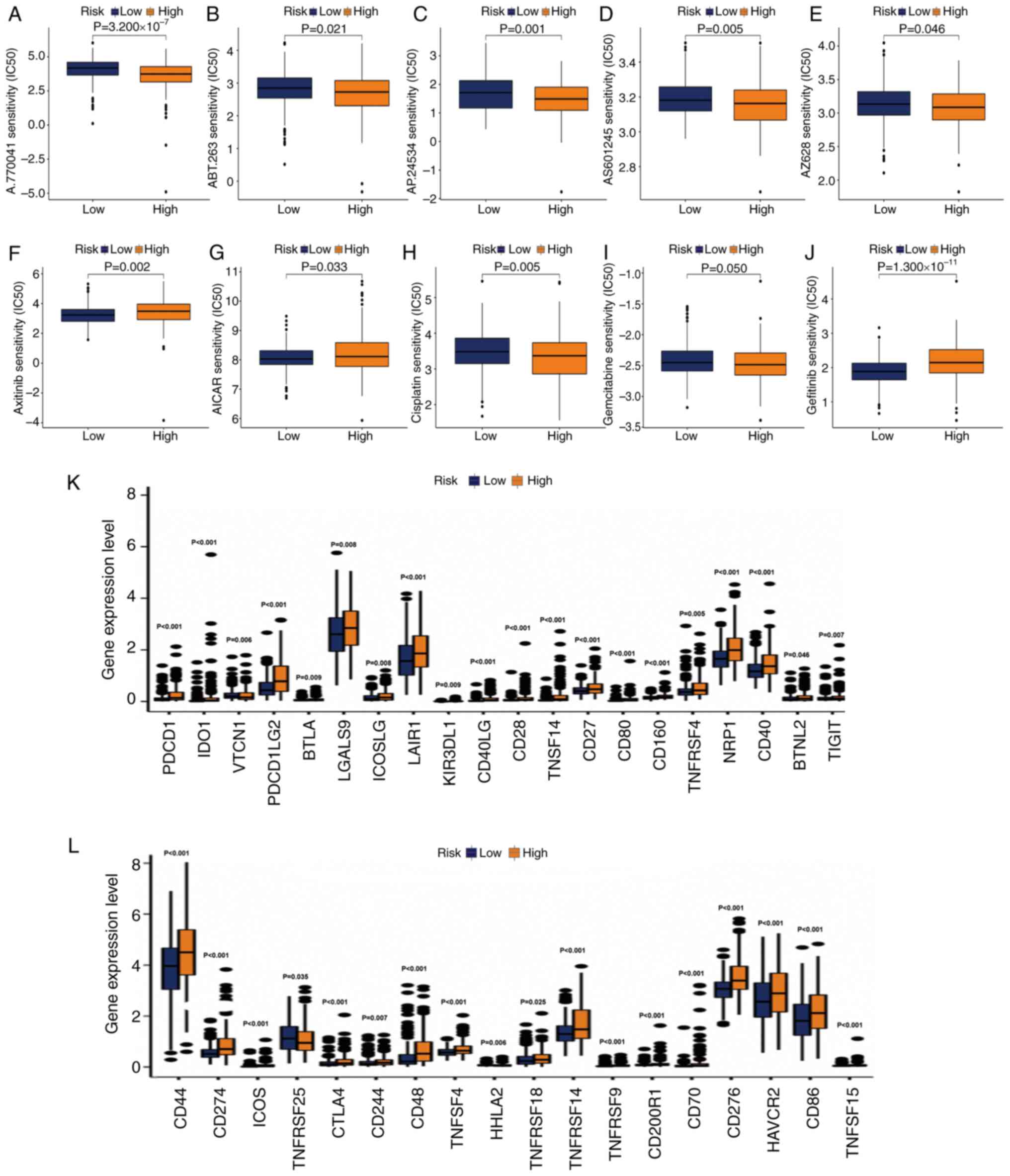
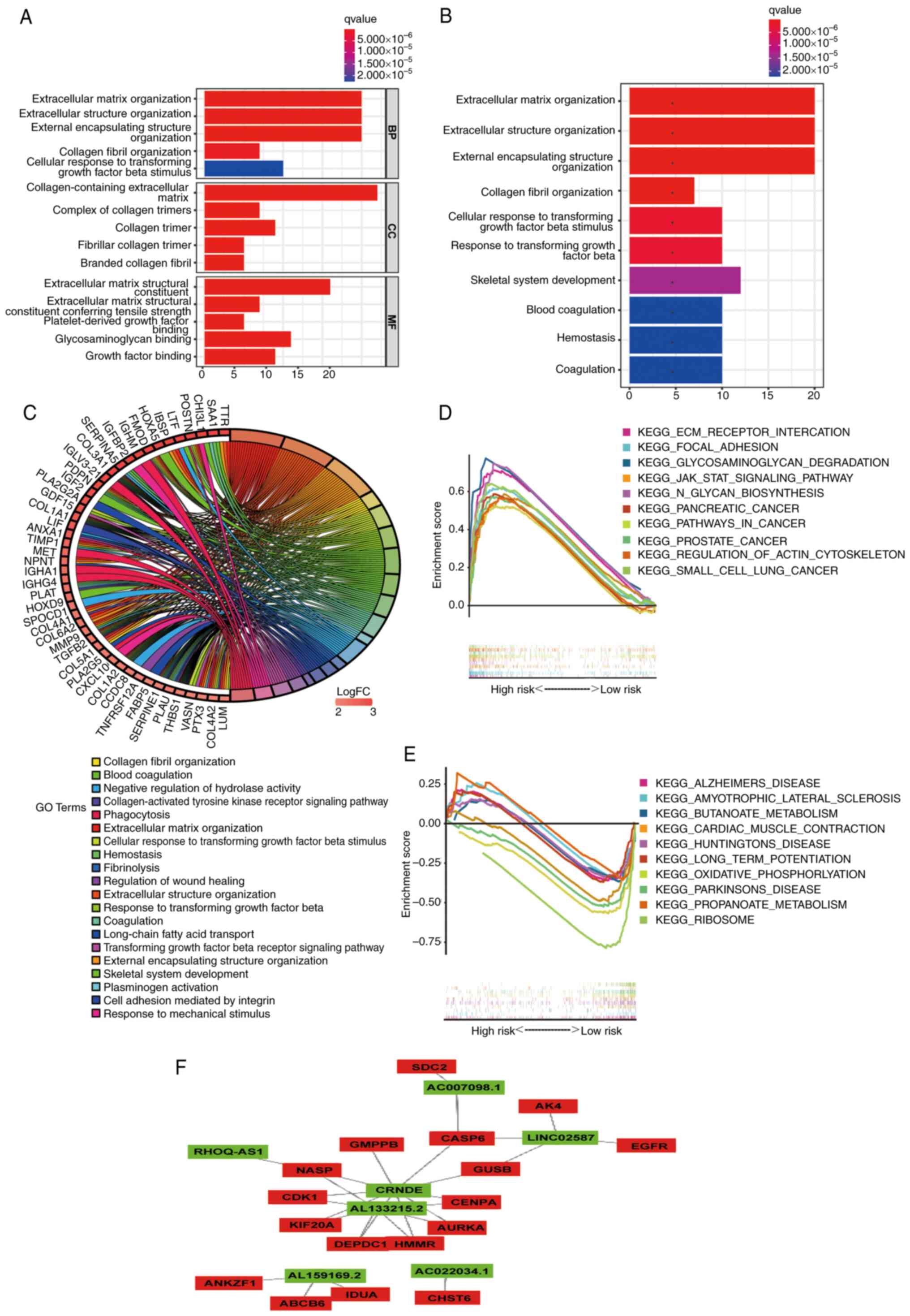
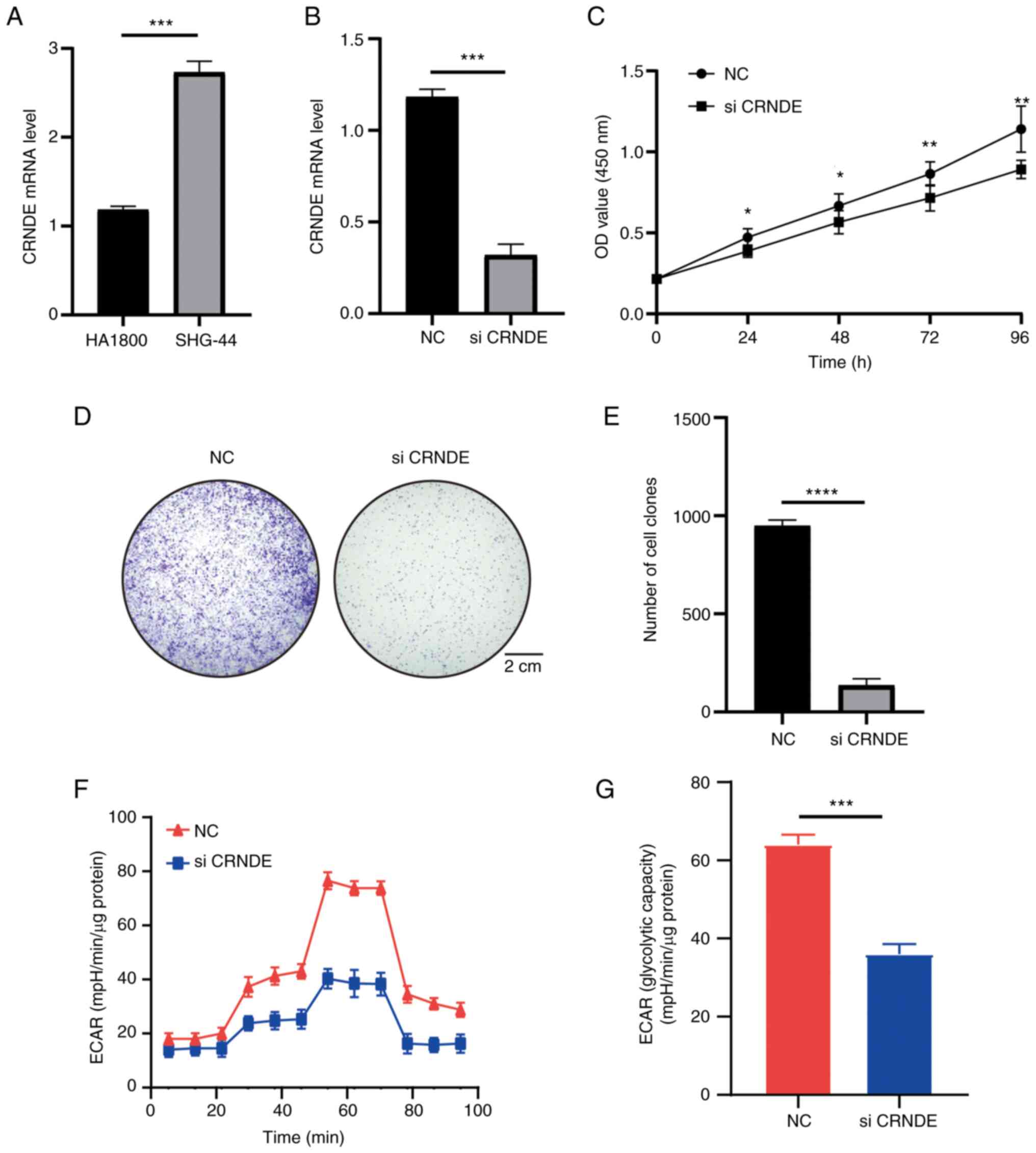
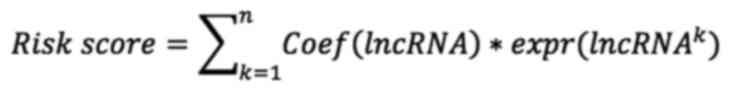|
1
|
Ostrom QT, Patil N, Cioffi G, Waite K,
Kruchko C and Barnholtz-Sloan JS: CBTRUS Statistical Report:
Primary Brain and Other Central Nervous System Tumors Diagnosed in
the United States in 2013–2017. Neuro Oncol. 22 (12 Suppl
2):iv1–iv96. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: A ‘state of the
science’ review. Neuro Oncol. 16:896–913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cancer Genome Atlas Research Network, .
Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA,
Rheinbay E, Miller CR, Vitucci M, et al: Comprehensive, Integrative
Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med.
372:2481–2498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Okita Y, Narita Y, Miyahara R, Miyakita Y,
Ohno M and Shibui S: Health-related quality of life in long-term
survivors with Grade II gliomas: The contribution of disease
recurrence and Karnofsky Performance Status. Jpn J Clin Oncol.
45:906–913. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Batsios G, Viswanath P, Subramani E, Najac
C, Gillespie AM, Santos RD, Molloy AR, Pieper RO and Ronen SM:
PI3K/mTOR inhibition of IDH1 mutant glioma leads to reduced 2HG
production that is associated with increased survival. Sci Rep.
9:105212019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang C, Yu R, Li Z, Song H, Zang D, Deng
M, Fan Y, Liu Y, Zhang Y and Qu X: Comprehensive analysis of genes
based on chr1p/19q co-deletion reveals a robust 4-gene prognostic
signature for lower grade glioma. Cancer Manag Res. 11:4971–4984.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ganapathy-Kanniappan S and Geschwind JF:
Tumor glycolysis as a target for cancer therapy: Progress and
prospects. Mol Cancer. 12:1522013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liberti MV and Locasale JW: The warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Locasale JW and Cantley LC: Metabolic flux
and the regulation of mammalian cell growth. Cell Metab.
14:443–451. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ganapathy-Kanniappan S: Molecular
intricacies of aerobic glycolysis in cancer: Current insights into
the classic metabolic phenotype. Crit Rev Biochem Mol Biol.
53:667–682. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu YC, Lin P, Zhao YJ, Wu LY, Wu YQ, Peng
JB, He Y and Yang H: Pan-cancer analysis of clinical significance
and associated molecular features of glycolysis. Bioengineered.
12:4233–4246. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang J, Ren B, Yang G, Wang H, Chen G, You
L, Zhang T and Zhao Y: The enhancement of glycolysis regulates
pancreatic cancer metastasis. Cell Mol Life Sci. 77:305–321. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu Z, Liu Z, Zhou X, Lu Y, Yao Y, Wang W,
Lu S, Wang B, Li F and Fu W: A glycolysis-related two-gene risk
model that can effectively predict the prognosis of patients with
rectal cancer. Hum Genomics. 16:52022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Reuss AM, Groos D, Buchfelder M and
Savaskan N: The Acidic Brain-Glycolytic switch in the
microenvironment of malignant glioma. Int J Mol Sci. 22:55182021.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alexander RP, Fang G, Rozowsky J, Snyder M
and Gerstein MB: Annotating non-coding regions of the genome. Nat
Rev Genet. 11:559–571. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan C, Tang Y, Wang J, Xiong F, Guo C,
Wang Y, Zhang S, Gong Z, Wei F, Yang L, et al: Role of long
non-coding RNAs in glucose metabolism in cancer. Mol Cancer.
16:1302017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao N, Zhang J, Zhao Q, Chen C and Wang
H: Mechanisms of long Non-Coding RNAs in biological characteristics
and aerobic glycolysis of glioma. Int J Mol Sci. 22:111972021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao L, Ji G, Le X, Wang C, Xu L, Feng M,
Zhang Y, Yang H, Xuan Y, Yang Y, et al: Long noncoding RNA
LINC00092 acts in Cancer-Associated fibroblasts to drive glycolysis
and progression of ovarian cancer. Cancer Res. 77:1369–1382. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun X, Huang X, Sun X, Chen S, Zhang Z, Yu
Y and Zhang P: Oxidative Stress-Related lncRNAs are potential
biomarkers for predicting prognosis and immune responses in
patients With LUAD. Front Genet. 13:9097972022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen C, Liu YQ, Qiu SX, Li Y, Yu NJ, Liu K
and Zhong LM: Five metastasis-related mRNAs signature predicting
the survival of patients with liver hepatocellular carcinoma. BMC
Cancer. 21:6932021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao J, Wang L and Wei B: Identification
and validation of an energy Metabolism-Related lncRNA-mRNA
signature for Lower-Grade glioma. Biomed Res Int.
2020:37082312020.PubMed/NCBI
|
|
22
|
Sun X, Song J, Lu C, Sun X, Yue H, Bao H,
Wang S and Zhong X: Characterization of cuproptosis-related lncRNA
landscape for predicting the prognosis and aiding immunotherapy in
lung adenocarcinoma patients. Am J Cancer Res. 13:778–801.
2023.PubMed/NCBI
|
|
23
|
Jerome Friedman TH and RT: Regularization
paths for generalized linear models via coordinate descent. J Stat
Softw. 33:1–22. 2010.PubMed/NCBI
|
|
24
|
Wu X, Sui Z, Zhang H, Wang Y and Yu Z:
Integrated analysis of lncRNA-Mediated ceRNA network in lung
adenocarcinoma. Front Oncol. 10:5547592020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He Y, Zhang J, Chen Z, Sun K, Wu X, Wu J
and Sheng L: A seven-gene prognosis model to predict biochemical
recurrence for prostate cancer based on the TCGA database. Front
Surg. 9:9234732022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Iasonos A, Schrag D, Raj GV and Panageas
KS: How to build and interpret a nomogram for cancer prognosis. J
Clin Oncol. 26:1364–1370. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhuang W, Sun H, Zhang S, Zhou Y, Weng W,
Wu B, Ye T, Huang W, Lin Z, Shi L and Shi K: An immunogenomic
signature for molecular classification in hepatocellular carcinoma.
Mol Ther Nucleic Acids. 25:105–115. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chakraborty H and Hossain A: R package to
estimate intracluster correlation coefficient with confidence
interval for binary data. Comput Methods Programs Biomed.
155:85–92. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mayakonda A, Lin DC, Assenov Y, Plass C
and Koeffler HP: Maftools: Efficient and comprehensive analysis of
somatic variants in cancer. Genome Res. 28:1747–1756. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Z, Li J, Zhang P, Zhao L, Huang B, Xu
Y, Wu G and Xia Q: The Role of ERBB signaling pathway-related genes
in kidney renal clear cell carcinoma and establishing a prognostic
risk assessment model for patients. Front Genet. 13:8622102022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Geeleher P, Cox N and Huang RS:
pRRophetic: An R package for prediction of clinical
chemotherapeutic response from tumor gene expression levels. PLoS
One. 9:e1074682014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Brunson JC: Ggalluvial: Layered grammar
for alluvial plots. J Open Source Softw. 5:20172020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li XN, Du ZW and Huang Q: Modulation
effects of hexamethylene bisacetamide on growth and differentiation
of cultured human malignant glioma cells. J Neurosurg. 84:831–838.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gulhan PY, Eroz R, Ataoglu O, İnce N,
Davran F, Öztürk CE, Gamsızkan Z and Balbay OA: The evaluation of
both the expression and serum protein levels of Caspase-3 gene in
patients with different degrees of SARS-CoV2 infection. J Med
Virol. 94:897–905. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Feng Q, Qian C and Fan S: A
hypoxia-related long non-coding RNAs signature associated with
prognosis in lower-grade glioma. Front Oncol. 11:7715122021.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu Y, Sha H, Sun X, Zhang Y, Wu Y, Zhang
J, Zhang H, Wu J and Feng J: CRNDE: An oncogenic long non-coding
RNA in cancers. Cancer Cell Int. 20:1622020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang J, Yin M, Peng G and Zhao Y: CRNDE:
An important oncogenic long non-coding RNA in human cancers. Cell
Prolif. 51:e124402018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Choi S, Yu Y, Grimmer MR, Wahl M, Chang SM
and Costello JF: Temozolomide-associated hypermutation in gliomas.
Neuro Oncol. 20:1300–1309. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bale TA and Rosenblum MK: The 2021 WHO
classification of tumors of the central nervous system: An update
on pediatric low-grade gliomas and glioneuronal tumors. Brain
Pathol. 32:e130602022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li J, Qian Y, Zhang C, Wang W, Qiao Y,
Song H, Li L, Guo J, Lu D and Deng X: LncRNA LINC00473 is involved
in the progression of invasive pituitary adenoma by upregulating
KMT5A via ceRNA-mediated miR-502-3p evasion. Cell Death Dis.
12:5802021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang X, Gejman R, Mahta A, Zhong Y, Rice
KA, Zhou Y, Cheunsuchon P, Louis DN and Klibanski A: Maternally
expressed gene 3, an imprinted noncoding RNA gene, is associated
with meningioma pathogenesis and progression. Cancer Res.
70:2350–2358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ma X, Jin W, Zhao C, Wang X and Wang K:
CRNDE: A valuable long noncoding RNA for diagnosis and therapy of
solid and hematological malignancies. Mol Ther Nucleic Acids.
28:190–201. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu
XY, Yu ZW, Jia YH, Bai XF, Li L, et al: The lncRNA CRNDE promotes
colorectal cancer cell proliferation and chemoresistance via
miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol
Cancer. 16:92017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang F, Wang H, Yu J, Yao X, Yang S, Li
W, Xu L and Zhao L: LncRNA CRNDE attenuates chemoresistance in
gastric cancer via SRSF6-regulated alternative splicing of PICALM.
Mol Cancer. 20:62021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Momtazmanesh S and Rezaei N: Long
non-coding RNAs in diagnosis, treatment, prognosis, and progression
of glioma: A State-of-the-Art review. Front Oncol. 11:7127862021.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao Z, Liu M, Long W, Yuan J, Li H, Zhang
C, Tang G, Jiang W, Yuan X, Wu M and Liu Q: Knockdown lncRNA CRNDE
enhances temozolomide chemosensitivity by regulating autophagy in
glioblastoma. Cancer Cell Int. 21:4562021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
DeBerardinis RJ, Lum JJ, Hatzivassiliou G
and Thompson CB: The biology of cancer: Metabolic reprogramming
fuels cell growth and proliferation. Cell Metab. 7:11–20. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Deberardinis RJ, Sayed N, Ditsworth D and
Thompson CB: Brick by brick: Metabolism and tumor cell growth. Curr
Opin Genet Dev. 18:54–61. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang Y, Liu J, Ren F, Chu Y and Cui B:
Identification and validation of a four-long non-coding RNA
signature associated with immune infiltration and prognosis in
colon cancer. Front Genet. 12:6711282021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang G, Liu P, Li J, Jin K, Zheng X and
Xie L: Novel prognosis and therapeutic response model of
immune-related lncRNA pairs in clear cell renal cell carcinoma.
Vaccines (Basel). 10:11612022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Vitale I, Manic G, Coussens LM, Kroemer G
and Galluzzi L: Macrophages and Metabolism in the Tumor
Microenvironment. Cell Metab. 30:36–50. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang ZH, Peng WB, Zhang P, Yang XP and
Zhou Q: Lactate in the tumour microenvironment: From immune
modulation to therapy. EBioMedicine. 73:1036272021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Peng M, Yin N, Chhangawala S, Xu K, Leslie
CS and Li MO: Aerobic glycolysis promotes T helper 1 cell
differentiation through an epigenetic mechanism. Science.
354:481–484. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
D'Angelo F, Ceccarelli M, Tala, Garofano
L, Zhang J, Frattini V, Caruso FP, Lewis G, Alfaro KD, Bauchet L,
et al: The molecular landscape of glioma in patients with
Neurofibromatosis 1. Nat Med. 25:176–187. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Guo X, Pan Y, Xiong M, Sanapala S,
Anastasaki C, Cobb O, Dahiya S and Gutmann DH: Midkine activation
of CD8+ T cells establishes a neuron-immune-cancer axis
responsible for low-grade glioma growth. Nat Commun. 11:21772020.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Tao B, Song Y, Wu Y, Peng T, Peng L, Xia
K, Xia X, Chen L and Zhong C: Matrix stiffness promotes glioma cell
stemness by activating BCL9L/Wnt/β-catenin signaling. Aging (Albany
NY). 13:5284–5296. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Jiang Y, Han Q, Zhao H and Zhang J:
Promotion of epithelial-mesenchymal transformation by
hepatocellular carcinoma-educated macrophages through
Wnt2b/β-catenin/c-Myc signaling and reprogramming glycolysis. J Exp
Clin Cancer Res. 40:132021. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fan Q, Yang L, Zhang X, Ma Y, Li Y, Dong
L, Zong Z, Hua X, Su D, Li H and Liu J: Autophagy promotes
metastasis and glycolysis by upregulating MCT1 expression and
Wnt/β-catenin signaling pathway activation in hepatocellular
carcinoma cells. J Exp Clin Cancer Res. 37:92018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhou M, He J, Li Y, Jiang L, Ran J, Wang
C, Ju C, Du D, Xu X, Wang X, et al: N6-methyladenosine
modification of REG1α facilitates colorectal cancer progression via
β-catenin/MYC/LDHA axis mediated glycolytic reprogramming. Cell
Death Dis. 14:5572023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Dong S, Liang S, Cheng Z, Zhang X, Luo L,
Li L, Zhang W, Li S, Xu Q, Zhong M, et al: ROS/PI3K/Akt and
Wnt/β-catenin signalings activate HIF-1α-induced metabolic
reprogramming to impart 5-fluorouracil resistance in colorectal
cancer. J Exp Clin Cancer Res. 41:152022. View Article : Google Scholar : PubMed/NCBI
|