Introduction
Hepatocellular carcinoma (HCC) is a common type of
liver cancer, which is a growing global public health problem
(1). The majority of patients with
HCC are diagnosed at an advanced stage with a <12.5% 5-year
survival (2,3). Currently, surgery is the cornerstone
treatment of HCC; however, due to the metastasis and recurrence,
the prognosis of HCC remains unsatisfactory (4). In particular, treatment options for
advanced HCC are limited. The mechanisms underlying the
tumorigenesis and development of HCC are still unclear, which is a
key reason for the lack of effective treatments for HCC (5). Thus, it is crucial that the mechanisms
and novel therapeutic targets of HCC are uncovered.
M-phase phosphoprotein 8 (MPP8), encoded by
MPHOSPH8, was first identified as an M-phase phosphoprotein in 1996
(6). This protein has been found to
bind methylated histone H3 lysine 9 (H3K9m3) (7), and to mediate the recruitment of the
human silencing hub, a complex containing MPP8, to genomic loci
rich in H3K9me3 (8). In previous
years, the role of MPP8 in cancers has been gradually gaining
attention (9). Previous studies
have shown that MPP8 plays a cancer-promoting role in non-small
cell lung cancer, gastric cancer and melanoma (10–13);
however, its role in HCC remains unclear.
MicroRNAs (miRNAs) are a large class of small
non-coding RNAs with a length of ~22 nucleotides (14). miRNAs can attenuate target gene
expression through specifically binding to the 3′ untranslated
region (3′ UTR) of the mRNA of the target gene (15). MiR-576-3p has been reported to be a
tumor suppressor that plays a suppressive role in the
proliferation, migration and invasion of HCC cells (16); however, the molecular mechanisms
have yet to be elucidated, and at present there have been no
studies on the association between miR-576-3p and MPP8.
The PI3K/Akt signaling pathway plays a crucial role
in multiple malignant phenotypes of HCC cells including
proliferation, migration and invasion (17). Akt, the key node of this signaling
pathway, becomes activated by phosphorylation and performs
functions by regulating a series of downstream effectors.
Activation of the PI3K/Akt pathway can be determined by assessing
the level of Akt phosphorylation (18). At present, the relationship between
MPP8 and the PI3K/Akt signaling pathway in HCC remains unclear.
In the present study, the role of MPP8 was evaluated
by modulating malignant phenotypes of HCC cells. Furthermore, the
upstream and downstream regulatory mechanisms of MPP8 were
investigated.
Materials and methods
Bioinformatics analysis
MPP8 expression analysis and related survival
analysis was performed using The University of Alabama at
Birmingham Cancer data analysis Portal (UALCAN; http://ualcan.path.uab.edu/) (19) based on the data from The Cancer
Genome Atlas (TCGA; ID: TCGA-LIHC; http://portal.gdc.cancer.gov/projects/TCGA-LIHC).
Significance cut-off level was set at P<0.05. TargetScanHuman
(version 7.2; http://www.targetscan.org/vert_72/) (20) and miRDB (http://mirdb.org/mirdb) (21) were used to predict the potential
miRNAs that target the 3′ UTR of MPP8 mRNA and specific binding
sites.
Cell culture
The normal liver cell line THLE-2 was purchased from
American Type Culture Collection. The liver cancer cell lines Hepg2
and Huh7 were purchased from The Cell Bank of Type Culture
Collection of The Chinese Academy of Sciences. The liver cancer
cell line MHCC97-H was purchased from Shanghai Zhong Qiao Xin Zhou
Biotechnology Co., Ltd. The cells were cultured in DMEM containing
10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37°C
with 5% CO2. All cell culture reagents were purchased
from Gibco (Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was performed to detect the levels of mRNA
and miRNA. For mRNA detection, total RNA of each sample (n=3) was
extracted using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). cDNA for detecting MPP8 (MPHOSPH8) and
β-actin (ACTB) were synthesized by the reverse transcription method
with a 1st Strand cDNA Synthesis SuperMix Kit (cat. no. 11141ES;
Shanghai Yeasen Biotechnology Co., Ltd.). RT reaction was conducted
as follows: 25°C for 5 min, 55°C for 15 min, and 85°C for 5 min.
qPCR for mRNA detection was performed with a qPCR SYBR Green Master
Mix (cat. no. 11201ES; Shanghai Yeasen Biotechnology Co., Ltd.).
qPCR was performed as follows: 95°C for 5 min, 40 cycles of 95°C
for 10 sec, 60°C for 20 sec, and 72°C for 20 sec. For miRNA
detection, total miRNA of each sample was extracted using a MiPure
Cell/Tissue miRNA Kit (cat. no. RC201; Vazyme Biotech Co., Ltd.).
cDNA for detecting miR-576-3p and U6 were synthesized using a miRNA
1st Strand cDNA Synthesis Kit (by stem-loop; cat. no. MR101; Vazyme
Biotech Co., Ltd.). The RT reaction was conducted as follows: 25°C
for 5 min, 55°C for 15 min, and 85°C for 5 min. qPCR for miRNA
detection was performed with a miRNA Universal SYBR qPCR Master Mix
(cat. no. MQ101; Vazyme Biotech Co., Ltd.). The qPCR was performed
as follows: 95°C for 5 min, 40 cycles of 95°C for 10 sec and 60°C
for 30 sec. MPHOSPH8 expression was normalized to ACTB, and
miR-576-3p expression was normalized to U6 (22). The 2−∆∆Cq method was used
to calculate the relative RNA expression levels (23). The primers used in qPCR are listed
in Table I and triplicates of each
sample were run.
 | Table I.Sequences of primers used for reverse
transcription-quantitative PCR. |
Table I.
Sequences of primers used for reverse
transcription-quantitative PCR.
| Target | Forward
(5′-3′) | Reverse
(5′-3′) | NCBI
accession/miRBase accession |
|---|
| MPHOSPH8 |
GCGAAGCAGTCTAACAATGTG |
AGTCGATGACATGCGATTGG | NM_017520.4 |
| ACTB |
CTCCATCCTGGCCTCGCTGT |
GCTGTCACCTTCACCGTTCC | NM_001101.5 |
|
microRNA-576-3p |
CGCGAAGATGTGGAAAAATT |
AGTGCAGGGTCCGAGGTATT | MI0003583 |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT | NR_004394 |
Antibodies and drugs
Primary antibodies for western botting were as
follows: Anti-MPP8 (1:10,000; cat. no. 16796–1-AP; Proteintech
Group, Inc.), anti-β-actin (1:10,000; cat. no. 60008-1-Ig;
Proteintech Group, Inc.), anti-phospho-pan-Akt (1:500; cat. no.
AF0016; Affinity Biosciences, Ltd.) and anti-pan-Akt (1:500; cat.
no. AF6261; Affinity Biosciences, Ltd.). Secondary antibodies for
western blotting (1:10,000; goat anti-mouse, cat. no. SA00001-1;
goat anti-rabbit, cat. no. SA00001-2) were purchased from
Proteintech Group, Inc. Recombinant human insulin-like growth
factor-1 (IGF-1) protein (cat. no. 291-G1; R&D Systems, Inc.),
the agonist of the PI3K/Akt signaling pathway used for rescue
experiments, was dissolved in culture medium and used at a
concentration of 100 ng/ml.
Western blotting
Western blotting was used to detect protein levels.
First, total protein of each sample (n=3) was extracted using RIPA
lysis buffer (CST Biological Reagents Co., Ltd.). Protein
concentration determination was performed using a BCA Protein Assay
kit. The protein samples (20 µg of total protein per lane) were
separated on 10% SDS-PAGE gels (Shanghai Yeasen Biotechnology Co.,
Ltd.), and then electro-transferred onto 0.45 µm PVDF membranes
(MilliporeSigma). After blocking with 5% bovine serum albumin
(MilliporeSigma) in Tris-buffered saline containing 0.05% Tween-20
(Beijing Solarbio Science & Technology Co., Ltd.) for 1 h at
room temperature, incubation with primary antibodies overnight at
4°C, and incubation with corresponding secondary antibodies for 1 h
at room temperature, protein chemiluminescence was detected using
an ECL kit (Tanon Science and Technology Co., Ltd.). The gray value
of the band was quantified using ImageJ (version 1.51; National
Institutes of Health). β-actin was used as the control.
Cell transfection
Small interfering RNAs (siRNAs) for MPP8 knockdown
and scrambled siRNAs as a control were synthesized by Sangon
Biotech Co., Ltd., and the sequences were as follows: MPP8-siRNA
forward, CCAAAGCAGUCAGGAAGGAUAUUCA, and reverse,
UGAAUAUCCUUCCUGACUGCUUUGG; control-siRNA forward,
UUCUCCGAACGUGUCACGUTT, and reverse, ACGUGACACGUUCGGAGAATT.
MiR-576-3p-mimic (cat. no. 4464066) for up-regulating miR-576-3p
expression and corresponding control-mimic (cat. no. 4464058) were
purchased from Thermo Fisher Scientific, Inc. (sequences not
available). The human MPHOSPH8 gene (NM_017520.4) was cloned into
pcDNA 3.1/His B (cat. no. V385-20; Thermo Fisher Scientific, Inc.)
for protein overexpression, and the empty vector was used as a
control. Lipofectamineâ 3000 transfection reagent (cat.
no. L3000075; Thermo Fisher Scientific, Inc.) was used to transfect
the siRNAs (50 nM), miRNA-mimics (50 nM) and plasmids (1 µg/ml)
into HCC cells following the manufacturer's instructions. After
transfection for 6 h at 37°C, culture medium was replaced with
fresh medium, and cells were cultured for another 24 h.
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8) assay was performed
to assess cell proliferation. Liver cancer cells (n=3) were seeded
at 2,500 cells/well in a 96-well culture plate. After cell
attachment, the cells were processed according to experimental
requirements, and timing was started. After this, at 24, 48 and 72
h timepoints, 10 µl of CCK-8 (cat. no. 40210ES; Shanghai Yeasen
Biotechnology Co., Ltd.) solution was added to each well. After 2 h
of incubation at 37°C, the absorbance was measured at 450 nm with a
microplate reader (Thermo Fisher Scientific, Inc.). The CCK-8 assay
results were expressed as optical density values, which reflect the
number of viable cells.
The plate colony formation assay was used to assess
cell proliferation. Liver cancer cells (n=3) were seeded at 200
cells/well in a 12-well culture plate. After incubation for 10
days, the colonies were fixed in 4% paraformaldehyde (Beijing
Solarbio Science & Technology Co., Ltd.) for 20 min at room
temperature, and stained with 1% Crystal Violet Ammonium Oxalate
Solution (Beijing Solarbio Science & Technology Co., Ltd.) for
15 min at room temperature. Colony (>50 cells/colony) numbers
were counted using ImageJ software.
Cell migration assay
The wound healing assay was used to assess cell
migration (24). Liver cancer cells
(n=3) were seeded at 1×106 cells/well in a 6-well
culture plate in complete DMEM containing 10% FBS. After cell
attachment, cells were serum-starved and a 200-µl pipette tip was
used to make a scratch in each well. Images were captured at four
random fields of view using a light microscope (magnification, ×40;
Olympus Corporation) at 0 and 48 h timepoints. Wound area was
measured using Image J software. Wound-healing assay results were
presented as migration rate (%)=(initial wound area-wound area at
48 h)/initial wound area ×100.
Cell invasion assay
Transwell invasion assay was performed to assess
cell invasion. Liver cancer cells (n=3) were seeded at
5×104 cells/well in the upper chambers of Transwell
plates (24-well, 8-µm pore size; Labgic Technology Co., Ltd.),
which were precoated with Matrigel (Corning, Inc.) at 37°C for 1 h.
The upper chamber was supplied with FBS-free DMEM, and the lower
chamber was supplied with DMEM containing 10% FBS. Following 24 h
incubation at 37°C, non-invasive liver cancer cells remaining on
the upper side of the membrane were removed using a cotton swab.
After which, the invaded cells were fixed in 4% paraformaldehyde
for 20 min at room temperature, and stained with 1% Crystal Violet
Ammonium Oxalate Solution for 15 min at room temperature. Images
were captured at four random fields of view using a light
microscope (magnification, ×200). The number of invaded cells were
counted using ImageJ software.
Dual-luciferase reporter gene
assay
The luciferase reporter plasmids containing the
predicted miR-576-3p binding site of wild-type MPP8 3′UTR (Luc-WT)
or mutant MPP8 3′UTR (Luc-MUT) were constructed using pEZX-MT06
(Guangzhou iGene Biotechnology Co., Ltd.). The empty vectors,
Luc-WT and Luc-MUT were co-transfected with control-mimic or
miR-576-3p-mimic into liver cancer cells using the transfection
agent Lipofectamine® 3000 as aforementioned. After 48 h,
the results expressed as relative luciferase activity (Firefly
luciferase/Renilla luciferase) were determined using a
Duo-Luciferase HS Assay kit (cat. no. LF005; Guangzhou iGene
Biotechnology Co., Ltd.) following the manufacturer's
instructions.
Statistical analysis
All quantified data were expressed as the mean ±
standard deviation. Statistical analysis was performed using
GraphPad Prism statistical package (version 8.0.1; Dotmatics). Data
normality was analyzed using Shapiro-Wilk test. CCK-8 assay data
was analyzed by two-way ANOVA followed by post hoc Sidak's or
Tukey's test. In addition, statistical differences between multiple
groups were analyzed by one-way ANOVA followed by post hoc Sidak's
test. Statistical differences were determined using Student's
unpaired t-test for two-group comparisons. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of MPP8 in HCC
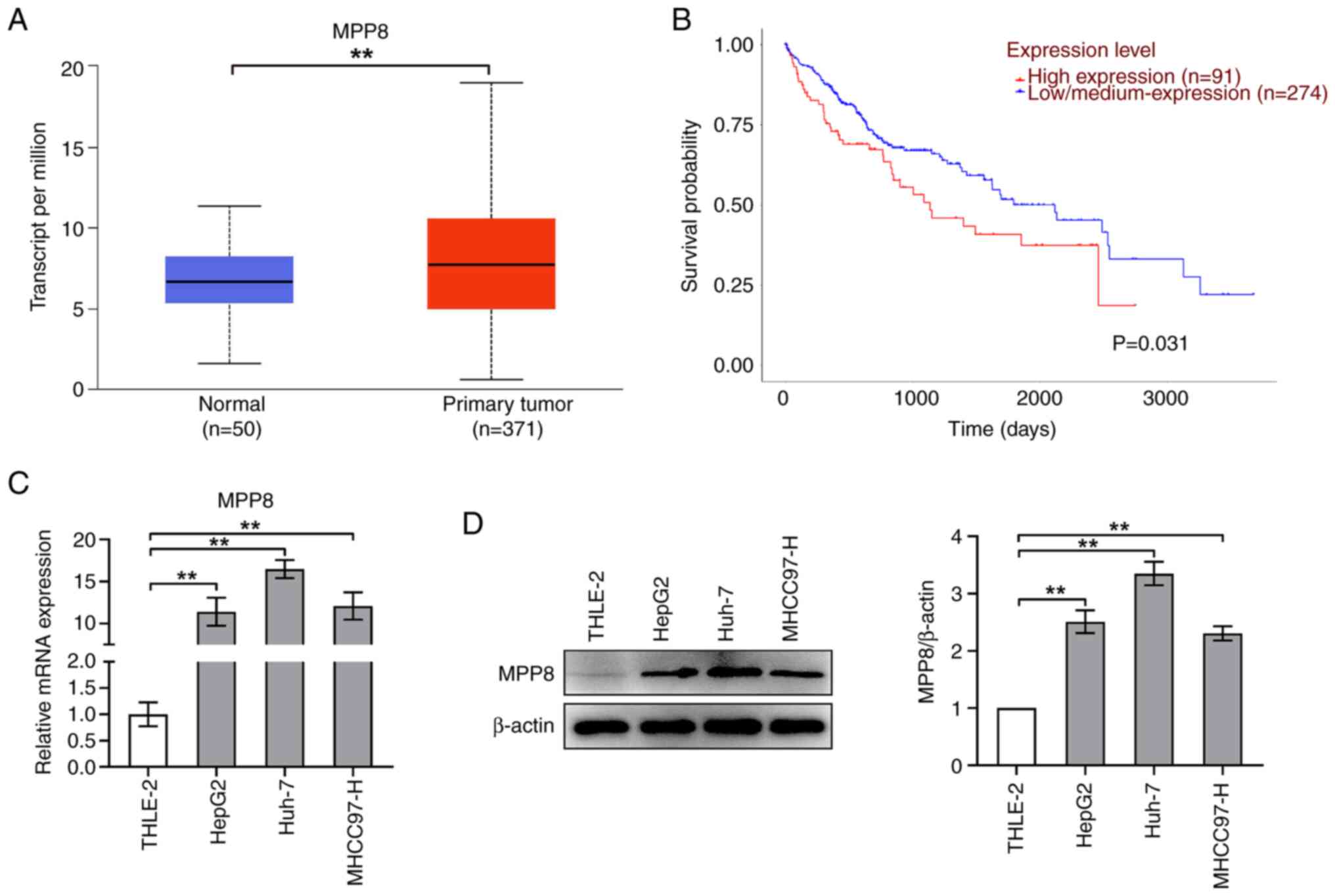
First, MPP8 expression and survival analysis were
performed using the UALCAN database. As shown in Fig. 1A, the expression level of MPP8 in
normal samples was significantly higher than that in primary tumor
samples. The results of the survival analysis showed that patients
with low/medium MPP8 expression had improved survival time than
those with high MPP8 expression (Fig.
1B). Next, to verify the expression of MPP8 in cells, RT-qPCR
and western blotting were performed to assess the levels of mRNA
and protein, respectively. As shown in Fig. 1C, the mRNA level of MPP8 in liver
cancer cell lines (HepG2, Huh-7 or MHCC97-H) was significantly
higher than that in a normal liver cell line (THLE-2), which was
consistent with the expression results obtained by the database
analysis. Furthermore, western blotting revealed similar trends to
those of the RT-qPCR (Fig. 1D).
Overall, these data demonstrated that MPP8 was upregulated in liver
cancer cell lines and was a risk factor for HCC. Additionally,
based on the expression results, the Huh-7 cell line was chosen for
subsequent experiments.
Role of MPP8 in the proliferation,
migration, and invasion of HCC cells
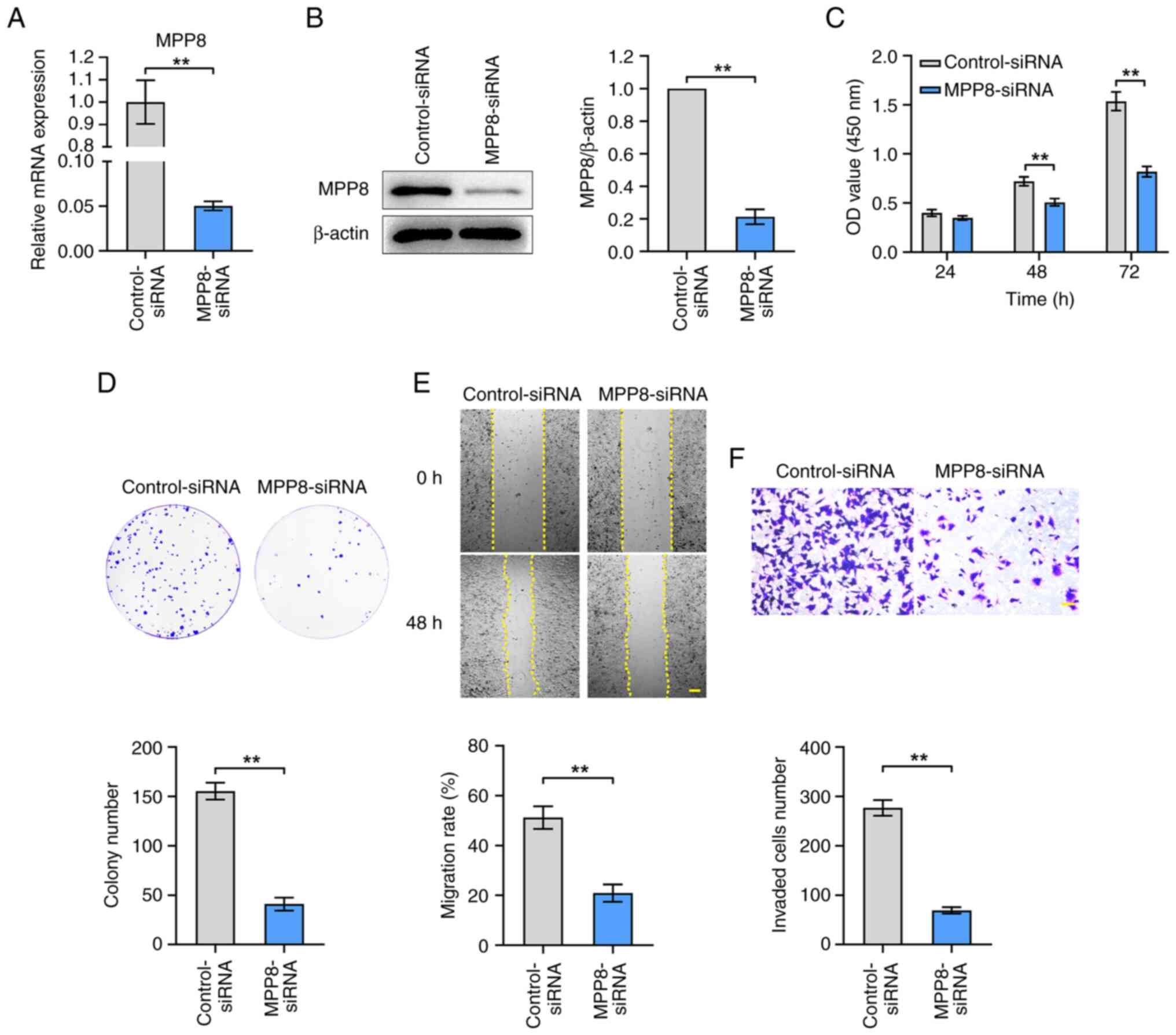
To evaluate the role of MPP8 in regulating the
malignant phenotypes of HCC cells, the effects of MPP8 knockdown on
cell proliferation, migration and invasion were measured.
MPP8-siRNA was used to down-regulate the expression of MPP8. As
shown in Fig. 2A and B, the results
of RT-qPCR and western blotting confirmed the efficacy of
MPP8-siRNA in inhibiting MPP8 expression. Then related assays were
performed to examine the malignant phenotypes of HCC cells. The
results of the CCK-8 assay showed that the viability of HCC cells
in the knockdown group was significantly lower than that in the
control group at 48 and 72 h timepoints (Fig. 2C). In the plate colony formation
assay, the knockdown group showed significantly lower colony
numbers than the control group (Fig.
2D). Moreover, in the wound healing assay, the knockdown group
showed a significantly lower migration rate than the control group
(Fig. 2E). Knockdown group showed a
significantly lower number of invaded cells than the control group
(Fig. 2F). Overall, these results
demonstrated that MPP8 played a promoting role in the
proliferation, migration, and invasion of HCC cells.
Role of the PI3K/Akt signaling pathway
in MPP8-mediated regulatory effects on the malignant phenotypes of
HCC cells
To further investigate whether the PI3K/Akt pathway
was involved in MPP8-mediated regulation of HCC cells, the level of
Akt phosphorylation was detected to evaluate the activation level
of this pathway, and IGF-1, an agonist of the PI3K/Akt pathway, was
used for the subsequent rescue experiments. As shown in Fig. 3A, down-regulating MPP8 expression
significantly inhibited the ratio of phosphorylated (p-) Akt/total
(t-) Akt. Meanwhile, the results also showed that IGF-1 stimulation
raised the ratio of p-Akt/t-Akt and reversed the inhibited ratio by
MPP8 knockdown. These results indicated that MPP8 promoted the
PI3K/Akt pathway in HCC cells, and the IGF-1 as a pathway agonist
was effective. Next, related rescue experiments were carried out.
The CCK-8 assay results revealed that activating the PI3K/Akt
pathway significantly reversed the inhibited viability of HCC cells
by MPP8 knockdown at 48 and 72 h timepoints (Fig. 3B). The results of plate colony
formation assay showed that activating the pathway significantly
reversed MPP8 knockdown-induced reduction in the colony number of
Huh-7 cells (Fig. 3C). As for cell
migration, the results of wound-healing assay revealed that
activating the pathway significantly reversed the inhibited
migration rate of HCC cells by MPP8 knockdown (Fig. 3D). The results of the Transwell
assay showed that activating the pathway significantly reversed the
decrease in invaded cells number induced by MPP8 knockdown
(Fig. 3E). Overall, these data
demonstrated that MPP8 promoted the proliferation, migration and
invasion of HCC cells by activating the PI3K/Akt signaling
pathway.
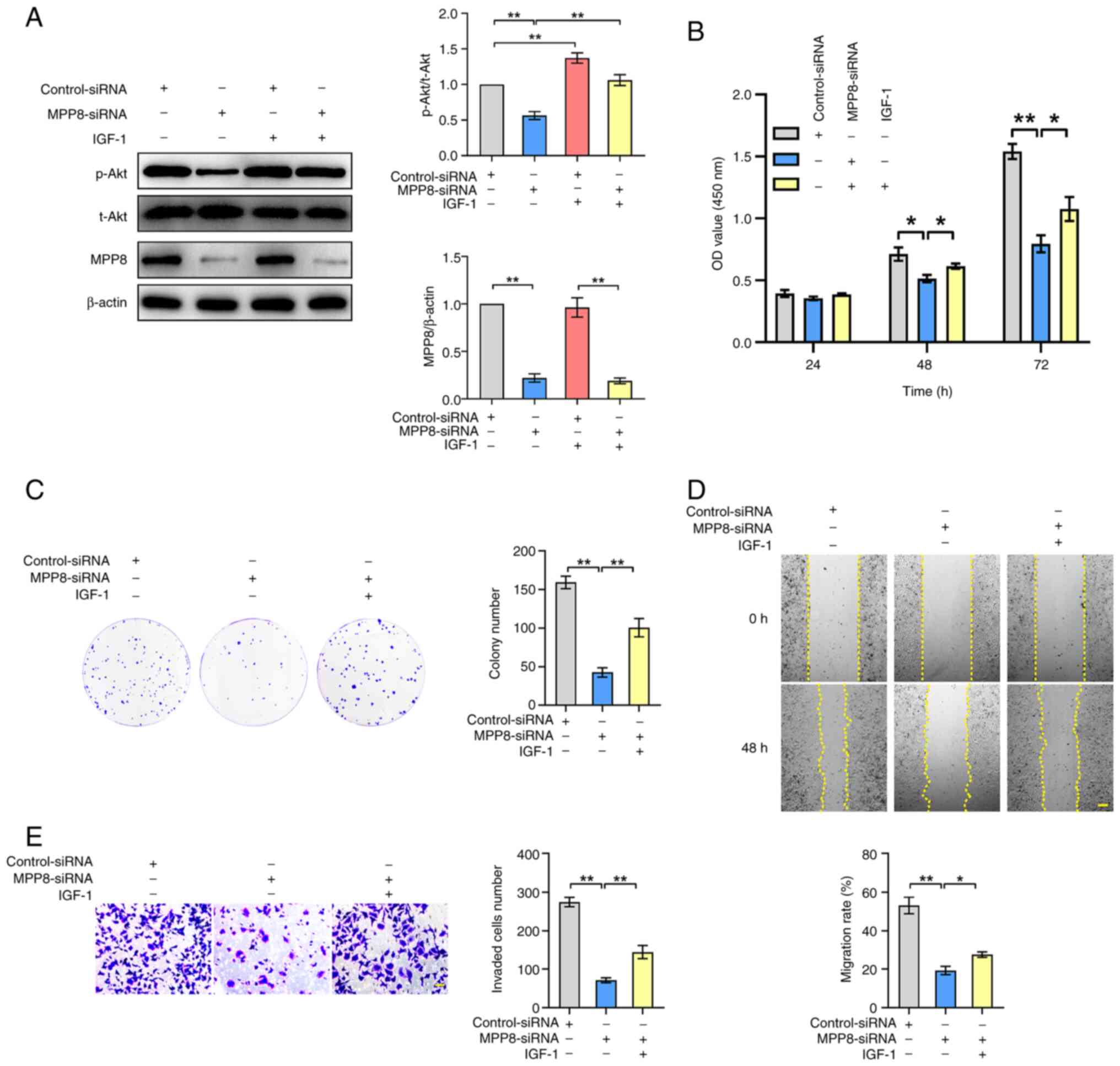 | Figure 3.Role of the PI3K/Akt signaling
pathway in MPP8-mediated regulatory effects on the malignant
phenotypes of HCC cells. (A) Western blotting was used to detect
the effect of MPP8 knockdown on the PI3K/Akt pathway, and to
confirm the validity of IGF-1 as the agonist of this pathway.
Rescue experiments with activation of the PI3K/Akt pathway were
performed to determine whether this pathway was involved in
MPP8-mediated regulation of the proliferation assessed by (B) CCK-8
and (C) colony formation assay, (D) migration was assessed by
wound-healing assay (scale bar, 200 µm), and (E) invasion was
assessed by Transwell invasion assay (scale bar, 50 µm) in HCC
cells. *P<0.05 and **P<0.01 vs. corresponding control. MPP8,
M-phase phosphoprotein 8; HCC, hepatocellular carcinoma; siRNA,
small interfering RNA; p-, phosphorylated; t-, total; CCK-8, Cell
Counting Kit-8; OD, optical density; IGF-1, insulin-like growth
factor-1. |
Targeted regulatory relationship
between miR-576-3p and MPP8 in HCC cells
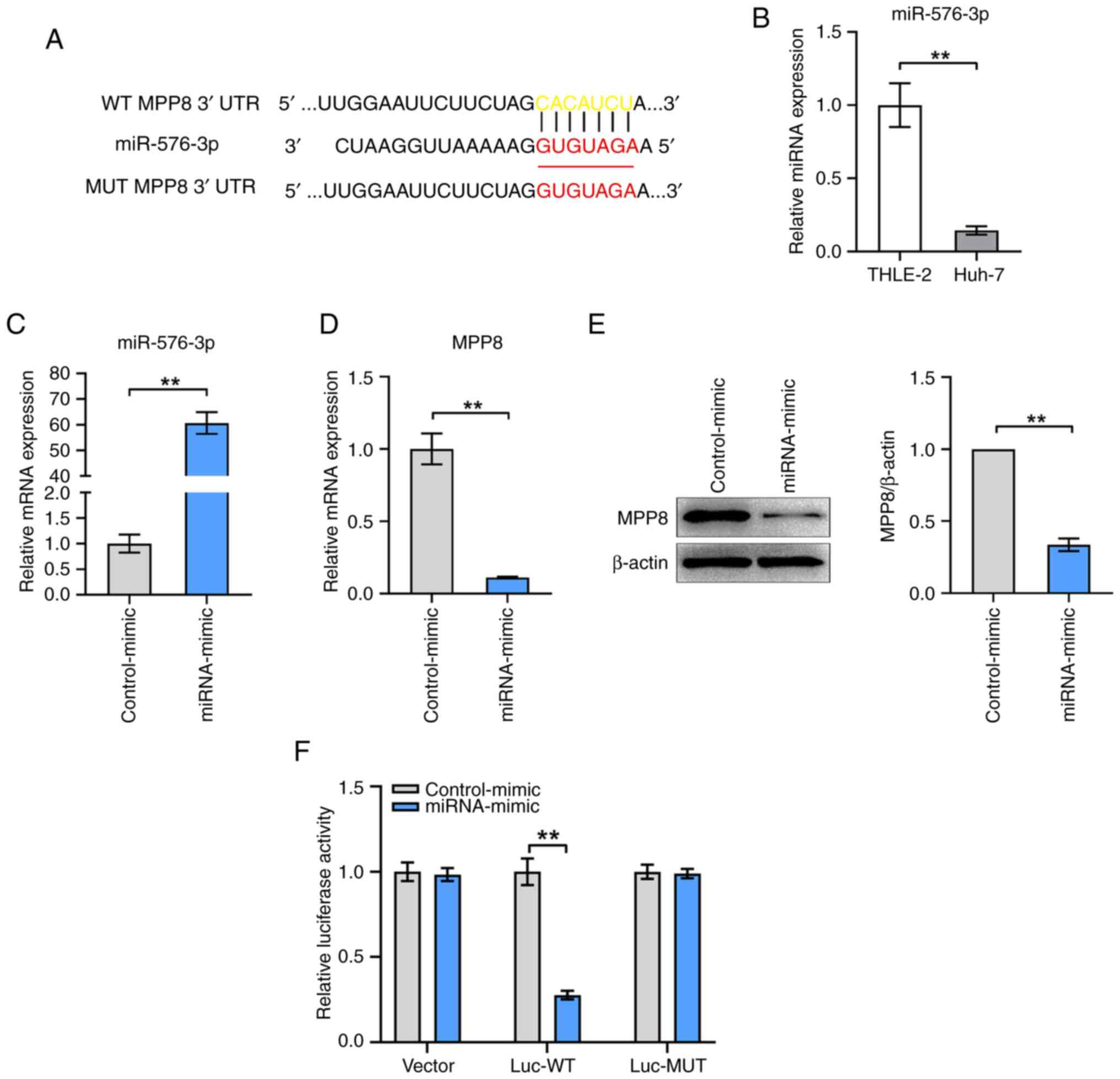
To explore the upstream regulator of MPP8,
TargetScanHuman and miRDB were used to predict the potential miRNAs
targeting MPP8 mRNA. As shown in Fig.
4A, miR-576-3p and the binding site between it and the mRNA
3′UTR of MPP8 were predicted. First, the expression levels of
miR-576-3p in THLE-2 and Huh-7 cells were measured, the results
confirmed that miR-576-3p expression was lower in HCC cells
compared with that in normal liver cells (Fig. 4B). Next, miRNA mimics for miR-576-3p
were used to up-regulate miR-576-3p expression in HCC cells. The
results of the RT-qPCR showed that the level of miR-576-3p in the
mimic group was significantly increased compared with that in the
control group (Fig. 4C), which
confirmed that the mimic was effective. The results of further
experiments for detecting MPP8 expression showed that miR-576-3p
overexpression significantly inhibited the mRNA (Fig. 4D) and protein (Fig. 4E) levels of MPP8 in HCC cells.
Furthermore, dual-luciferase reporter gene assay was performed to
verify the binding relationship between miR-576-3p and MPP8 mRNA.
The results showed that the relative luciferase activity of cells
transfected with the mimic for miR-576-3p was significantly lower
than that of cells transfected with the control-mimic in the Luc-WT
group, and no significant change was observed in the Luc-MUT group
(Fig. 4F). Overall, these results
demonstrated that MPP8 was negatively regulated by miR-576-3p
through direct targeting.
Role of the miR-576-3p/MPP8 axis in
HCC cells
To further investigate the role of the
miR-576-3p/MPP8 axis in HCC cells, rescue experiments were carried
out to verify whether miR-576-3p regulated the PI3K/Akt signaling
pathway and tumor cell phenotypes via MPP8. First, the
overexpression of MPP8 was validated by RT-qPCR (Fig. 5A) and western blotting (Fig. 5B). Next, the detection results of
the PI3K/Akt pathway showed that up-regulating miR-576-3p
significantly inhibited the level of the pathway, and up-regulating
the decreased MPP8 expression mediated by miR-576-3p significantly
reversed the inhibited level of the PI3K/Akt pathway (Fig. 5C). As for detection of cell
proliferation, the CCK-8 assay results showed that MPP8
overexpression significantly reversed the decreased viability of
HCC cells by up-regulating miR-576-3p expression at 48 or 72 h
timepoints (Fig. 5D). The plate
colony formation assay results revealed that MPP8 overexpression
significantly reversed the reduced colony number of HCC cells by
up-regulating miR-576-3p expression (Fig. 5E), which was consistent with the
trend of the CCK-8 results. In terms of cell migration, as shown in
Fig. 5F, MPP8 overexpression
significantly reversed the inhibited migration rate of HCC cells by
up-regulating miR-576-3p expression. In parallel, Transwell assay
results revealed that up-regulating MPP8 expression significantly
reversed the reduced invaded cells number of HCC cells by
upregulating miR-576-3p expression (Fig. 5G). Overall, these results indicated
that miR-576-3p inhibited the PI3K/Akt pathway and the malignant
phenotypes of HCC cells by downregulating MPP8 expression.
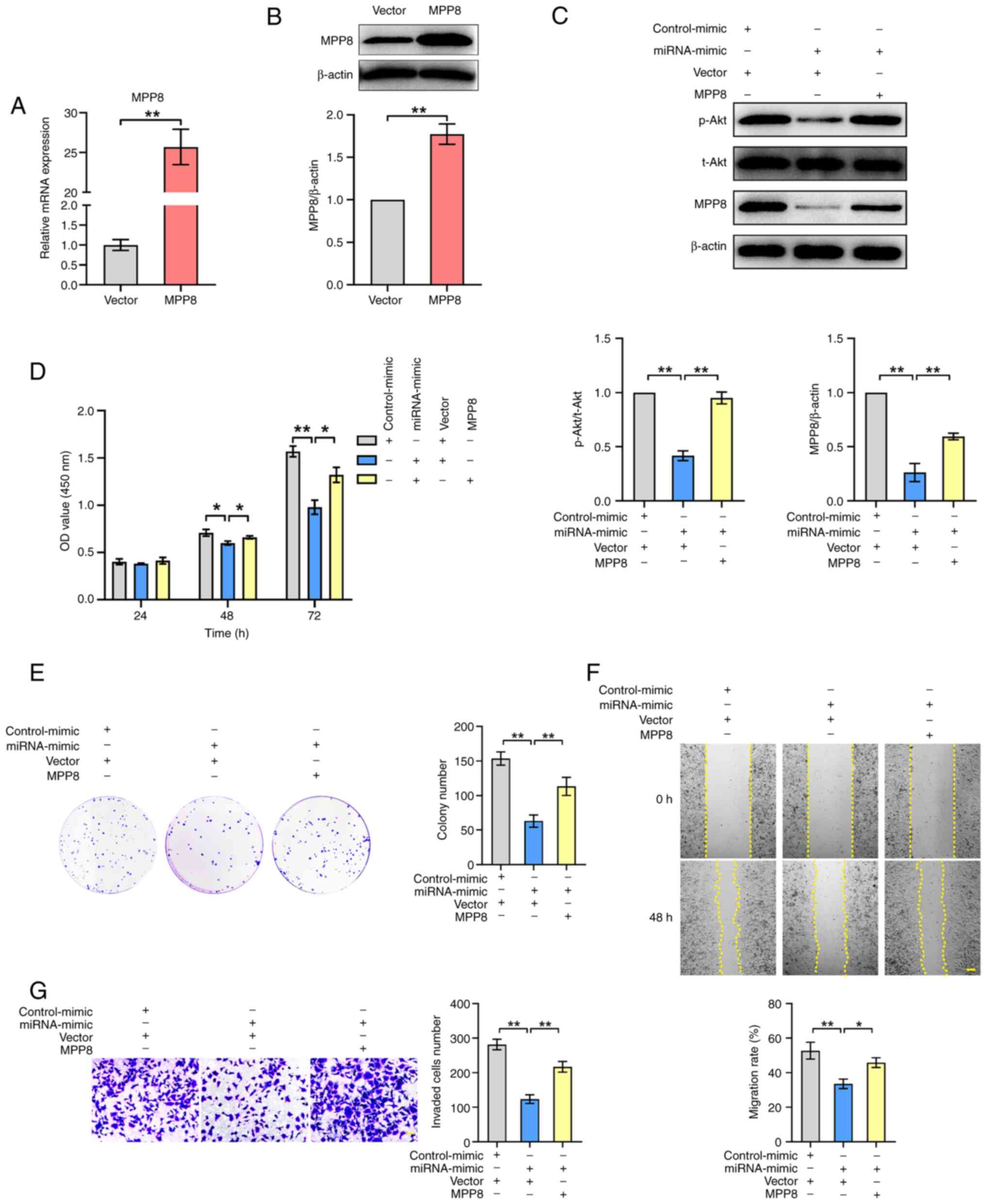 | Figure 5.Role of miR-576-3p/MPP8 axis in HCC
cells. The validity of MPP8 overexpression plasmid was confirmed by
(A) RT-qPCR for mRNA expression and (B) western blotting for
protein expression. (C) Western blotting used to detect the effect
of up-regulating miR-576-3p on the PI3K/Akt pathway, and to detect
the effect of MPP8 overexpression on miR-576-3p-mediated regulation
of the PI3K/Akt pathway. Rescue experiments with MPP8
overexpression were performed to determine whether MPP8 was
involved in miR-576-3p-mediated regulation of the proliferation
assessed by (D) CCK-8 and (E) colony formation assay, (F) migration
assessed by wound-healing assay (scale bar, 200 µm) and (G)
invasion assessed by Transwell invasion assay (scale bar, 50 µm) in
HCC cells. *P<0.05 and **P<0.01 vs. corresponding control.
MPP8, M-phase phosphoprotein 8; HCC, hepatocellular carcinoma; p-,
phosphorylated; t-, total; CCK-8, Cell Counting Kit-8; OD, optical
density; RT-qPCR, reverse transcription-quantitative PCR. |
Discussion
After surgical treatment, the 5-year survival rate
of patients with early stage HCC can reach 70% (25); however, patients with advanced HCC
are unfit for surgery (26).
Exploring new therapeutic targets for HCC treatment is of great
significance and in continuous progression. The present study
indicated that high expression of MPP8 may be associated with poor
prognosis in patients with HCC, as demonstrated through the exerted
promotive effects on the proliferation, migration and invasion of
HCC cells. This was similar to several previous studies on the role
of MPP8 in certain cancers. One study on non-small cell lung cancer
showed that MPP8 is expressed highly in cancer tissues and cells,
and promoted the proliferation of cancer cells (10). A total of two studies on gastric
cancer showed that MPP8 played promotive roles in the growth and
metastasis of cancer cells (11,12).
Furthermore, one study on melanoma showed that MPP8 is expressed
highly in cancer tissues, and enhanced the proliferation, migration
and invasion of cancer cells (13).
The findings of the present study further support that MPP8 is a
cancer-promoting gene; however, more studies into the roles of MPP8
in cancers are required.
As for the downstream mechanism driving
MPP8-mediated modulating malignant phenotypes of HCC cells, the
present study demonstrated that MPP8 exerted its effects by
activating the PI3K/Akt pathway. This pathway is commonly activated
in human cancers (27), including
in HCC, as demonstrated by the present study; however, the specific
regulatory mechanism between MPP8 and the PI3K/Akt pathway needs to
be further elucidated. Based on the reported function of MPP8 as a
transcriptional repressor (28), it
is hypothesized that MPP8 may activate the PI3K/Akt pathway by
down-regulating an inhibitor of this pathway, such as E-cadherin
(9,29). This will be a direction of our
future work. As for the upstream regulator of MPP8 in HCC, the
present work, through bioinformatics analysis, predicted the
potential of miR-576-3p to bind to MPP8 mRNA, and further
demonstrated that miR-576-3p suppressed the PI3K/Akt pathway and
malignant cell phenotypes through directly targeting and negatively
regulating MPP8. MiR-576-3p as a tumor suppressor has been reported
to be down-regulated in several cancers. Previous studies found
that miR-576-3p inhibited the proliferation, migration and invasion
of breast cancer cells through targeting SRY-box transcription
factor 4 (30), the proliferation,
migration, invasion and glycolysis of gastric cancer cells by
targeting hypoxia inducible factor-1α (31), the proliferation of bladder cancer
cells by targeting cyclin D1 (32),
and the migration and invasion of lung adenocarcinoma cells by
targeting serum/glucocorticoid-regulated kinase 1 (33). Song et al (16) found that miR-576-3p inhibited the
ability of HCC cell proliferation, migration and invasion through
targeting hypoxia inducible factor-1α. The findings of the present
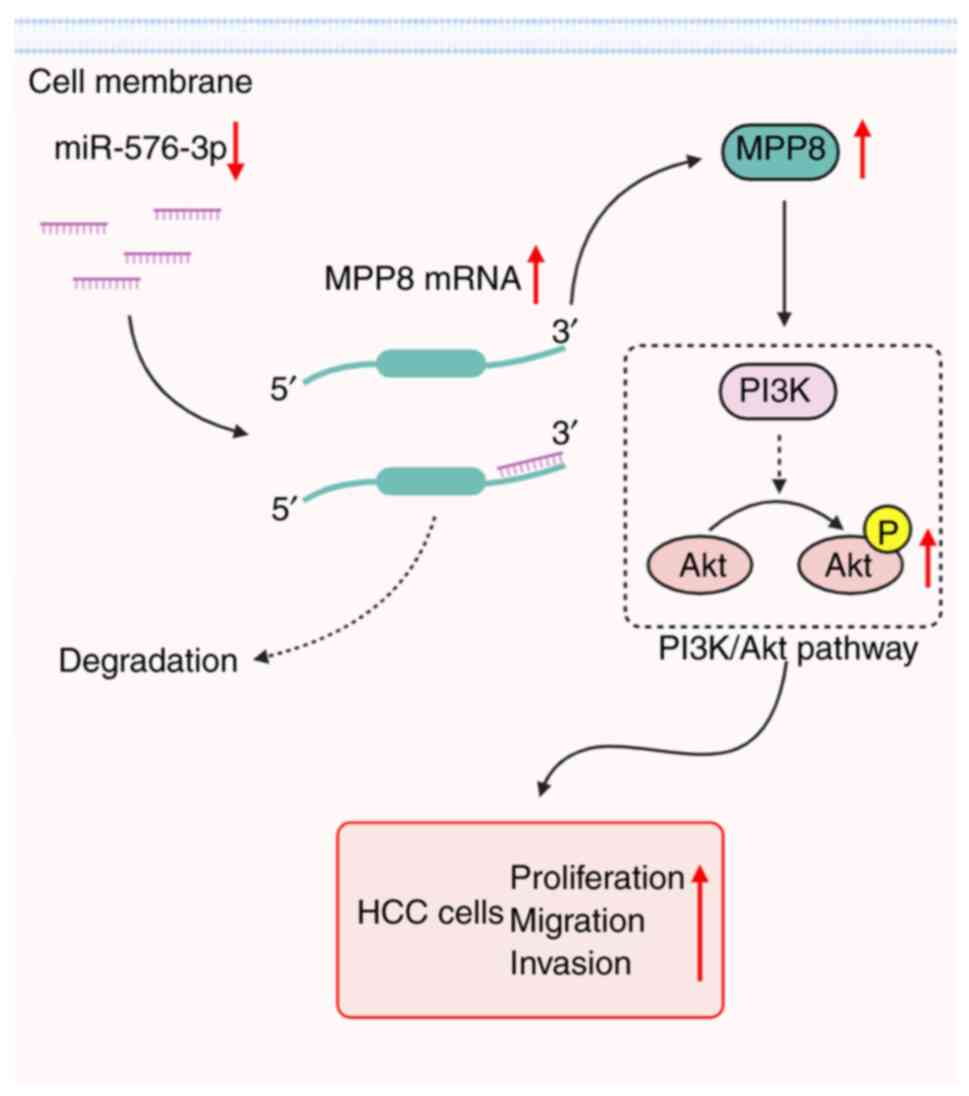
study not only provided a novel miR-576-3p target but also showed
the regulatory mechanism of the miR-576-3p/MPP8 axis via the
PI3K/Akt signaling pathway in HCC cells (Fig. 6); however, further studies are still
required to explore the effects and mechanisms of miR-576-3p in
cancers.
Taken together, the present study demonstrated that
up-regulated MPP8 promoted HCC cell proliferation, migration and
invasion, which was directly modulated by the down-regulated
miR-576-3p, and the activation of the PI3K/Akt pathway was involved
in this axis-mediated regulation. These findings not only extend
our understanding of the pathogenesis of HCC, but also provide a
potential new therapeutic target for HCC treatment.
Acknowledgements
Not applicable.
Funding
This study was supported by The Project of the Department of
Science and Technology of Inner Mongolia Autonomous Region (grant
no. 2020BS03021) and The Department of Science and Technology of
Jilin Province (grant no. 20220202076NC).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XL and CB designed and conceived the study. XL and
CB guided and supervised all the experiments. NZ, MC, WP, CZ, YW
and XG performed the experiments. NZ and XL performed the data
analysis. XL, CB and NZ confirmed the authenticity of all the raw
data. NZ wrote the first draft of the manuscript. XL, CB and NZ
completed the final version of the manuscript. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chapiro J, Duran R, Lin MD, Schernthaner
RE, Wang Z, Gorodetski B and Geschwind JF: Identifying staging
markers for hepatocellular carcinoma before transarterial
chemoembolization: Comparison of three-dimensional quantitative
versus non-three-dimensional imaging markers. Radiology.
275:438–447. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang XJ, Zhang AH and Sun H: Power of
metabolomics in diagnosis and biomarker discovery of hepatocellular
carcinoma. Hepatology. 57:2072–2077. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang WY and Wei C: Advances in the early
diagnosis of hepatocellular carcinoma. Genes Dis. 7:308–319. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tabrizian P, Jibara G, Shrager B, Schwartz
M and Roayaie S: Recurrence of hepatocellular cancer after
resection: Patterns, treatments, and prognosis. Ann Surg.
261:947–955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Z, Wu G, Li J, Wang Y, Ju X and Jiang
W: lncRNA CRNDE promotes the proliferation and metastasis by acting
as sponge miR-539-5p to regulate POU2F1 expression in HCC. BMC
Cancer. 20:2822020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Matsumoto-Taniura N, Pirollet F, Monroe R,
Gerace L and Westendorf JM: Identification of novel M phase
phosphoproteins by expression cloning. Mol Biol Cell. 7:1455–1469.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li J, Li Z, Ruan J, Xu C, Tong Y, Pan PW,
Tempel W, Crombet L, Min J and Zang J: Structural basis for
specific binding of human MPP8 chromodomain to histone H3
methylated at lysine 9. PLoS One. 6:e251042011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tchasovnikarova IA, Timms RT, Matheson NJ,
Wals K, Antrobus R, Göttgens B, Dougan G, Dawson MA and Lehner PJ:
GENE SILENCING. Epigenetic silencing by the HUSH complex mediates
position-effect variegation in human cells. Science. 348:1481–1485.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kokura K, Sun LD, Bedford MT and Fang J:
Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing
and promotes tumour cell motility and invasion. EMBO J.
29:3673–3687. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao XY, Qiao YL, Zhang Y, Wang J, Shen X
and Xu CW: Knockdown of MPP8 suppresses cell proliferation via
regulation of HOXA5 in non-small cell lung cancer cells. Cell Mol
Biol (Noisy-le-grand). 64:27–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu JG, Yan PY, Zhao XM, Xing C, Wang L,
Wang X and Wang2 X: Mechanism of MPP8 in the regulating of growth
and metastasis of gastric cancer by P53/BCL-2 signaling pathway and
emt. Acta Med Mediterr. 35:1331–1335. 2019.
|
|
12
|
Wang Y, Xiao H, Wang C, Wu H, He H, Yao C,
Cui J and Li W: M-phase phosphoprotein 8 promotes gastric cancer
growth and metastasis via p53/Bcl-2 and EMT-related signaling
pathways. J Cell Biochem. 121:2330–2342. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan B, Lin L, Ying ZY, Ying MX, Zhou QY
and Shi L: Repression of M-phase phosphoprotein 8 inhibits melanoma
growth and metastasis in vitro and in vivo. Int J Clin Exp Pathol.
10:12003–12009. 2017.PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song Y, Jin X, Liu Y, Wang S, Bian F, Zhao
Q, Shi H and Gao Z: Long noncoding RNA ZFPM2-AS1 promotes the
proliferation, migration, and invasion of hepatocellular carcinoma
cells by regulating the miR-576-3p/HIF-1α axis. Anticancer Drugs.
32:812–821. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun EJ, Wankell M, Palamuthusingam P,
McFarlane C and Hebbard L: Targeting the PI3K/Akt/mTOR pathway in
hepatocellular carcinoma. Biomedicines. 9:16392021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Downward J: PI 3-kinase, Akt and cell
survival. Semin Cell Dev Biol. 15:177–182. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen YH and Wang XW: miRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48(D1): D127–D131. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lou Z, Gong YQ, Zhou X and Hu GH: Low
expression of miR-199 in hepatocellular carcinoma contributes to
tumor cell hyper-proliferation by negatively suppressing XBP1.
Oncol Lett. 16:6531–6539. 2018.PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu J, Wen X, Liu B, Zhang Q, Zhang J,
Miao H and Zhu R: Diosmetin inhibits the metastasis of
hepatocellular carcinoma cells by downregulating the expression
levels of MMP-2 and MMP-9. Mol Med Rep. 13:2401–2408. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mazzaferro V, Regalia E, Doci R, Andreola
S, Pulvirenti A, Bozzetti F, Montalto F, Ammatuna M, Morabito A and
Gennari L: Liver transplantation for the treatment of small
hepatocellular carcinomas in patients with cirrhosis. N Engl J Med.
334:693–699. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kudo M, Trevisani F, Abou-Alfa GK and
Rimassa L: Hepatocellular carcinoma: Therapeutic guidelines and
medical treatment. Liver Cancer. 6:16–26. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-kinase-AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Murata K, Sato S, Haruta M, Goshima T,
Chiba Y, Takahashi S, Sharif J, Koseki H, Nakanishi M and Shimada
M: Physical interaction between MPP8 and PRC1 complex and its
implication for regulation of spermatogenesis. Biochem Biophys Res
Commun. 458:470–475. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lau MT, Klausen C and Leung PCK:
E-cadherin inhibits tumor cell growth by suppressing PI3K/Akt
signaling via β-catenin-Egr1-mediated PTEN expression. Oncogene.
30:2753–2766. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qiu XM, Zhang Q, Deng QF and Li Q:
Circular RNA hsa_circ_0012673 promotes breast cancer progression
via miR-576-3p/SOX4 axis. Mol Biotechnol. 65:61–71. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li H, Cao B, Zhao R, Li T, Xu X, Cui H,
Deng H, Gao J and Wei B: circDNMT1 promotes malignant progression
of gastric cancer through targeting miR-576-3p/hypoxia inducible
factor-1 alpha axis. Front Oncol. 12:8171922022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liang Z, Li SQ, Xu X, Xu X, Wang X, Wu J,
Zhu Y, Hu Z, Lin Y, Mao Y, et al: MicroRNA-576-3p inhibits
proliferation in bladder cancer cells by targeting cyclin D1. Mol
Cells. 38:130–137. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Greenawalt EJ, Edmonds MD, Jain N, Adams
CM, Mitra R and Eischen CM: Targeting of SGK1 by miR-576-3p
inhibits lung adenocarcinoma migration and invasion. Mol Cancer
Res. 17:289–298. 2019. View Article : Google Scholar : PubMed/NCBI
|