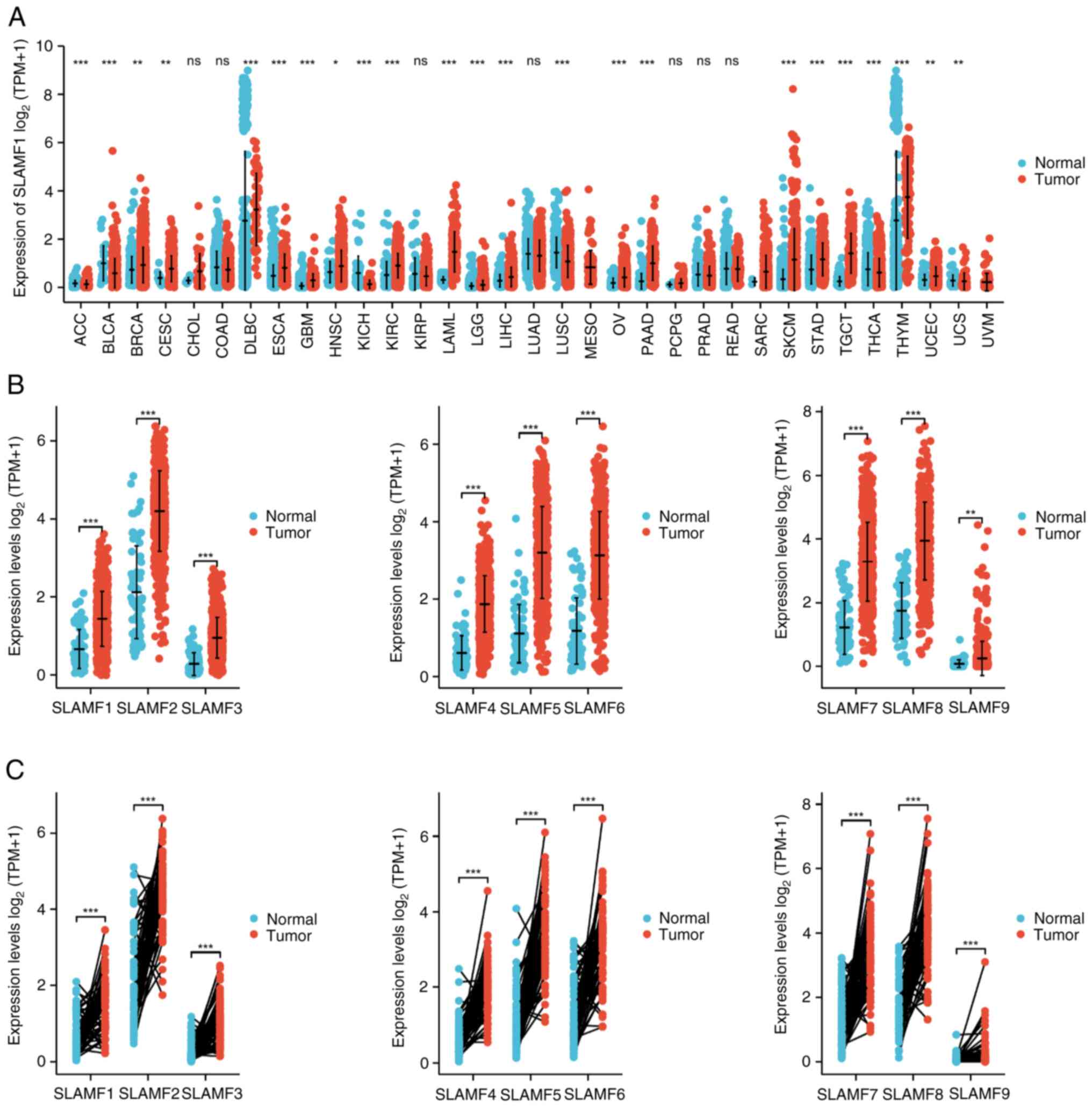
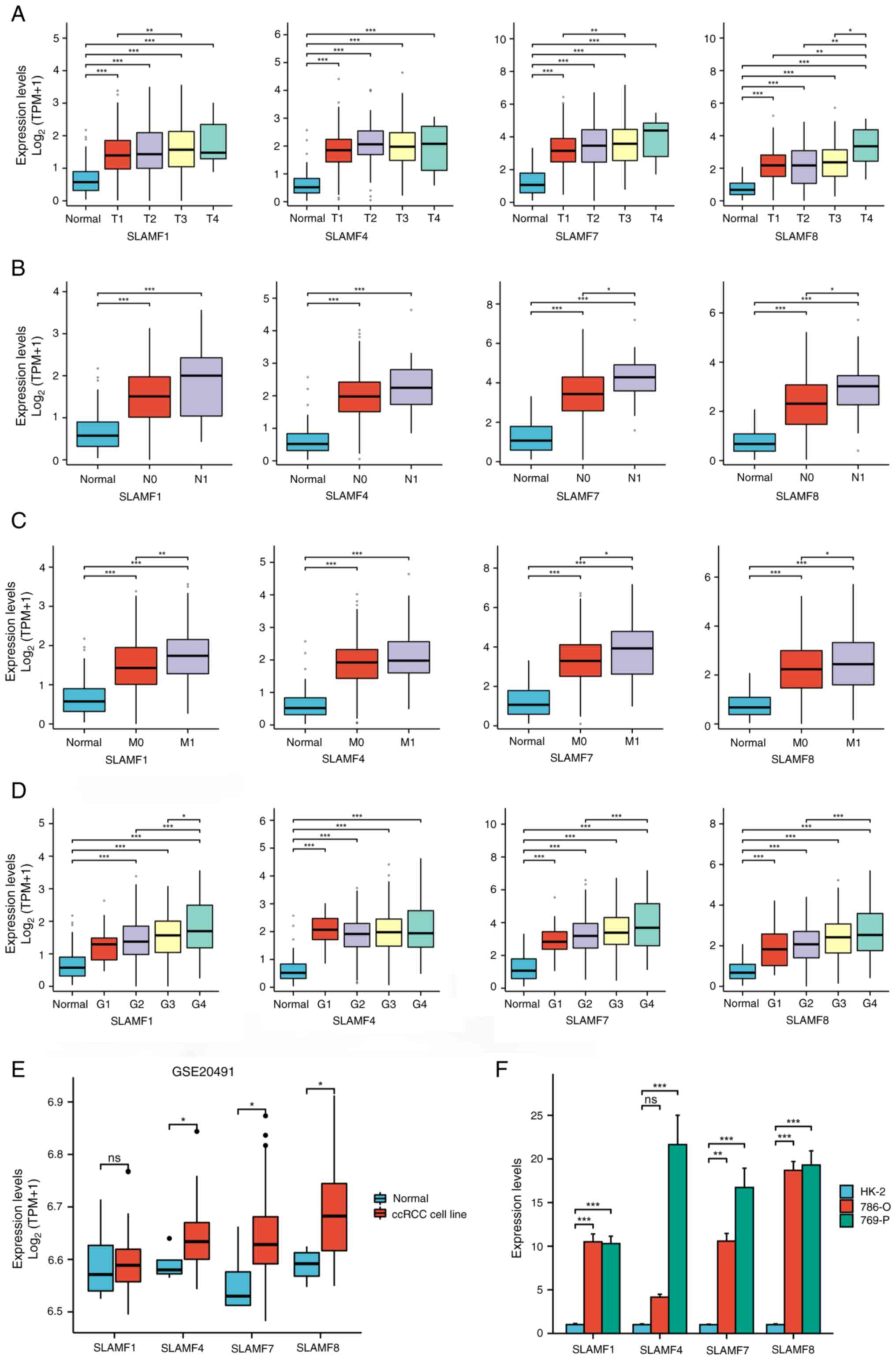
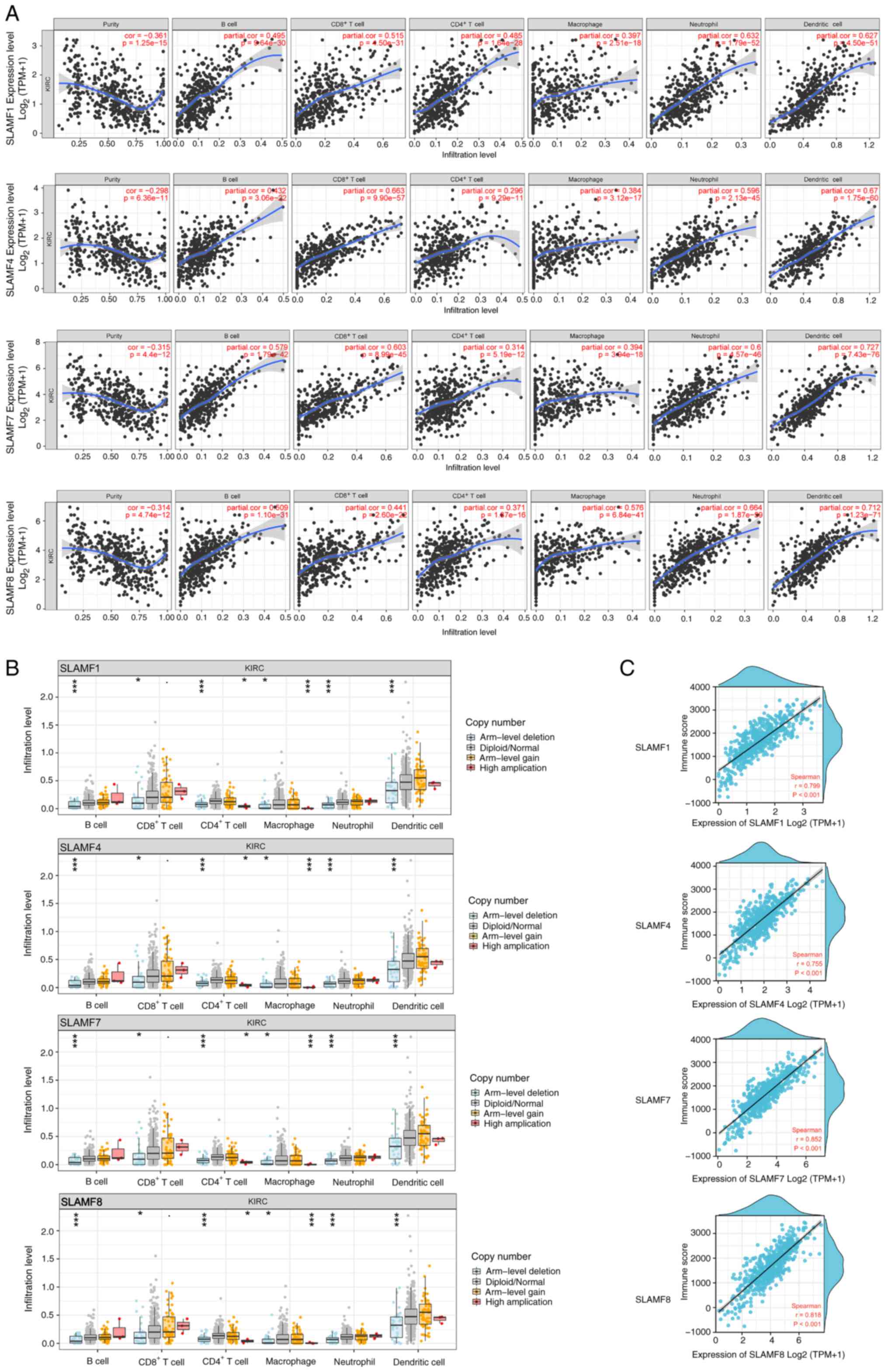
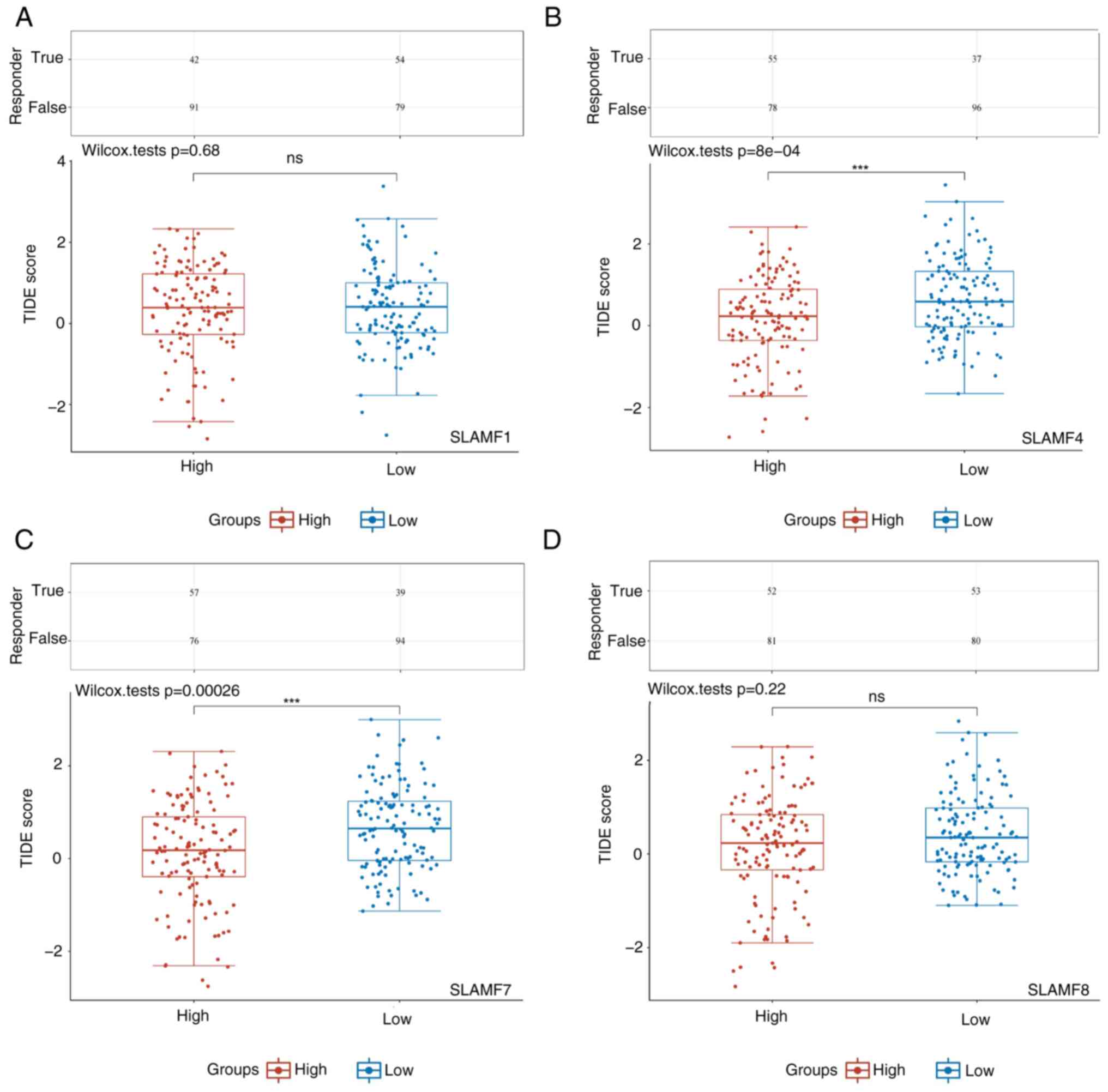
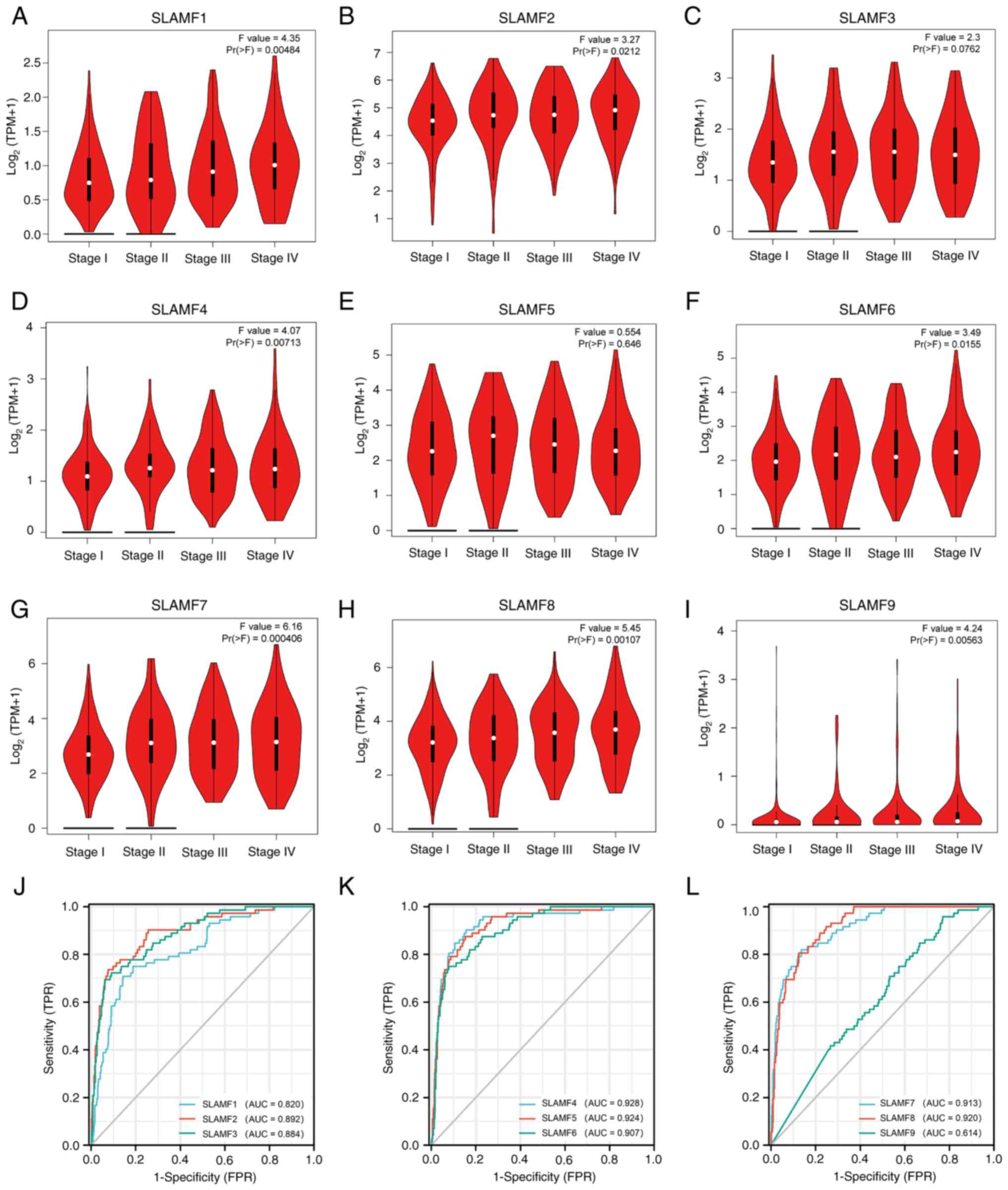
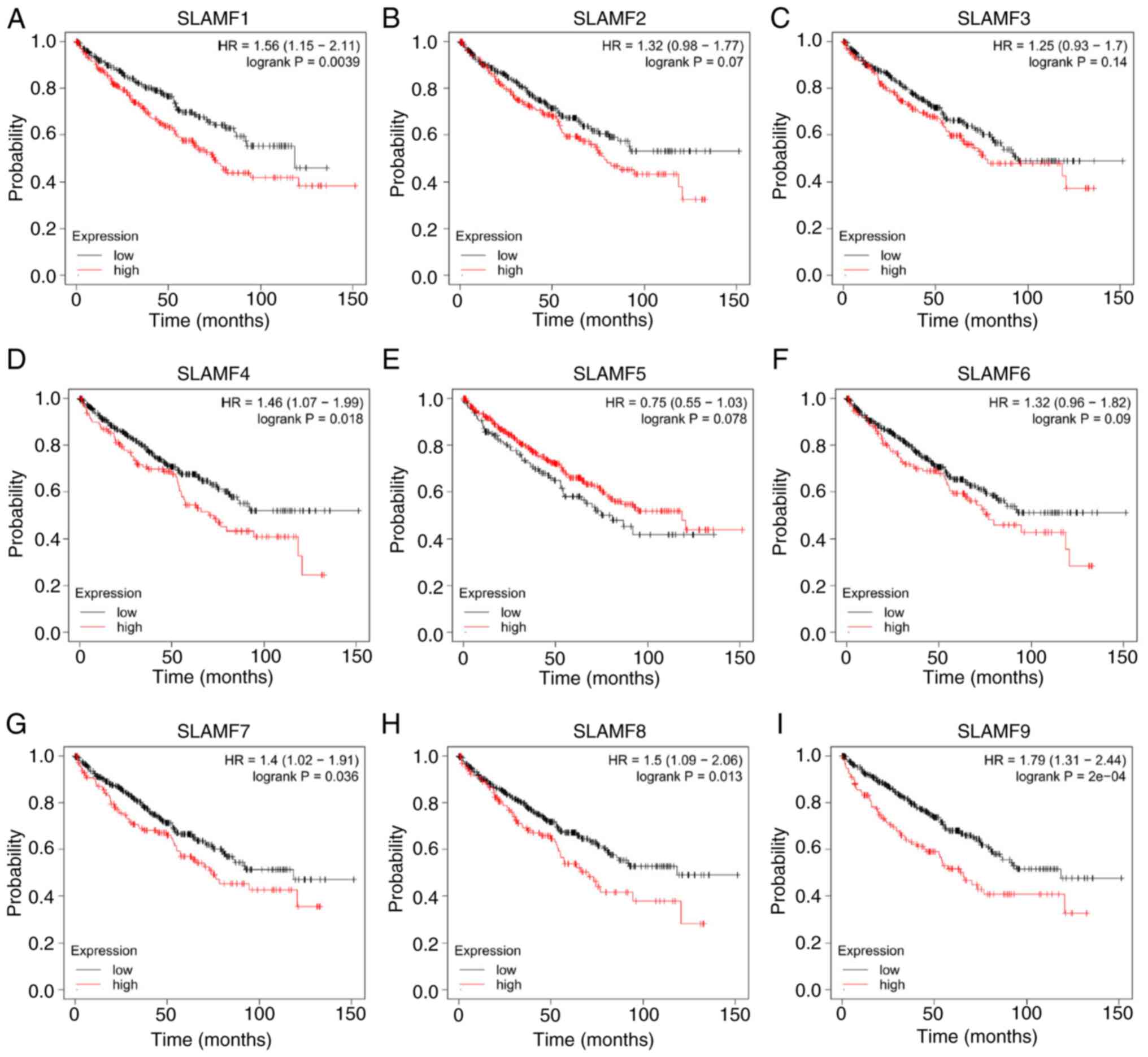
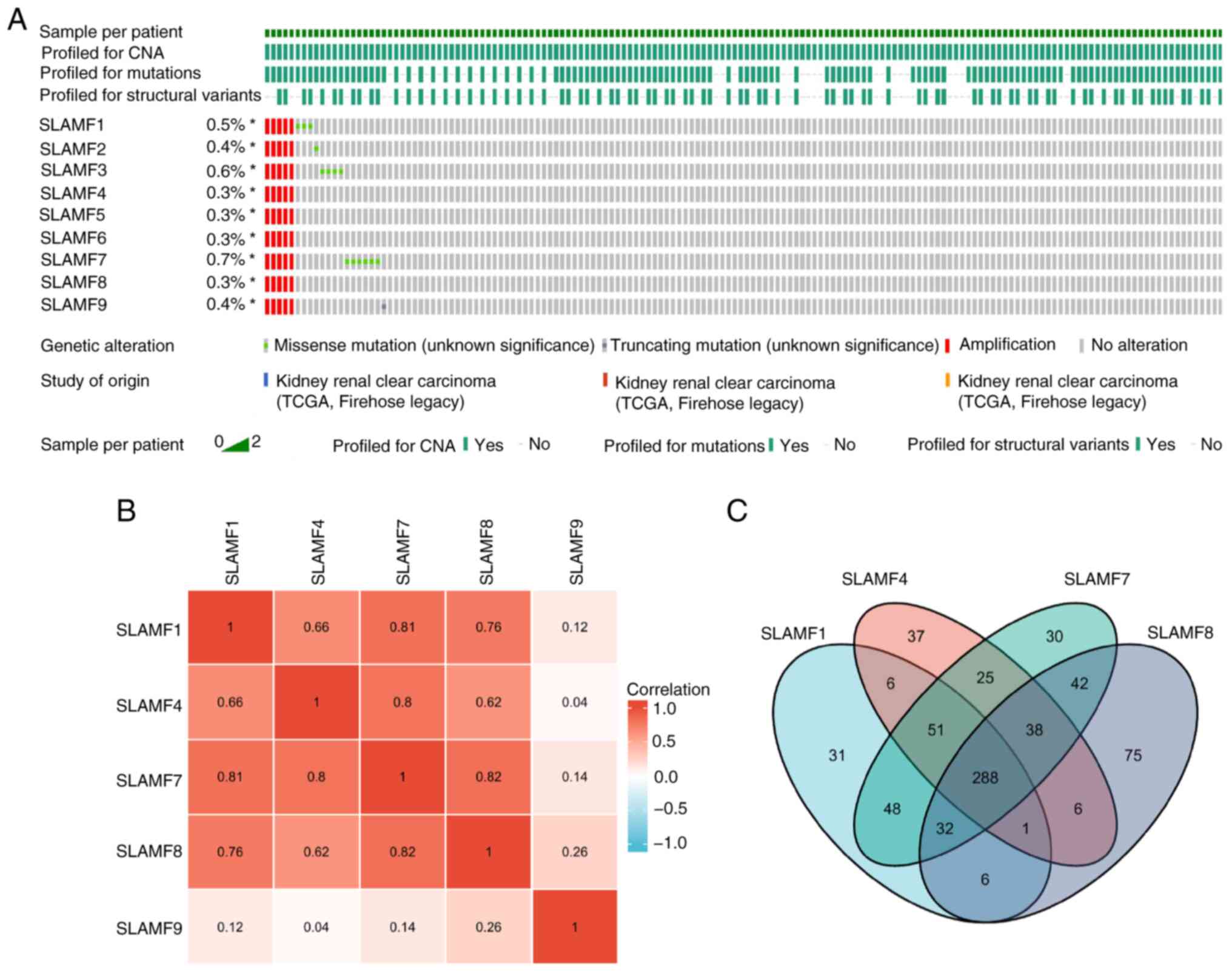
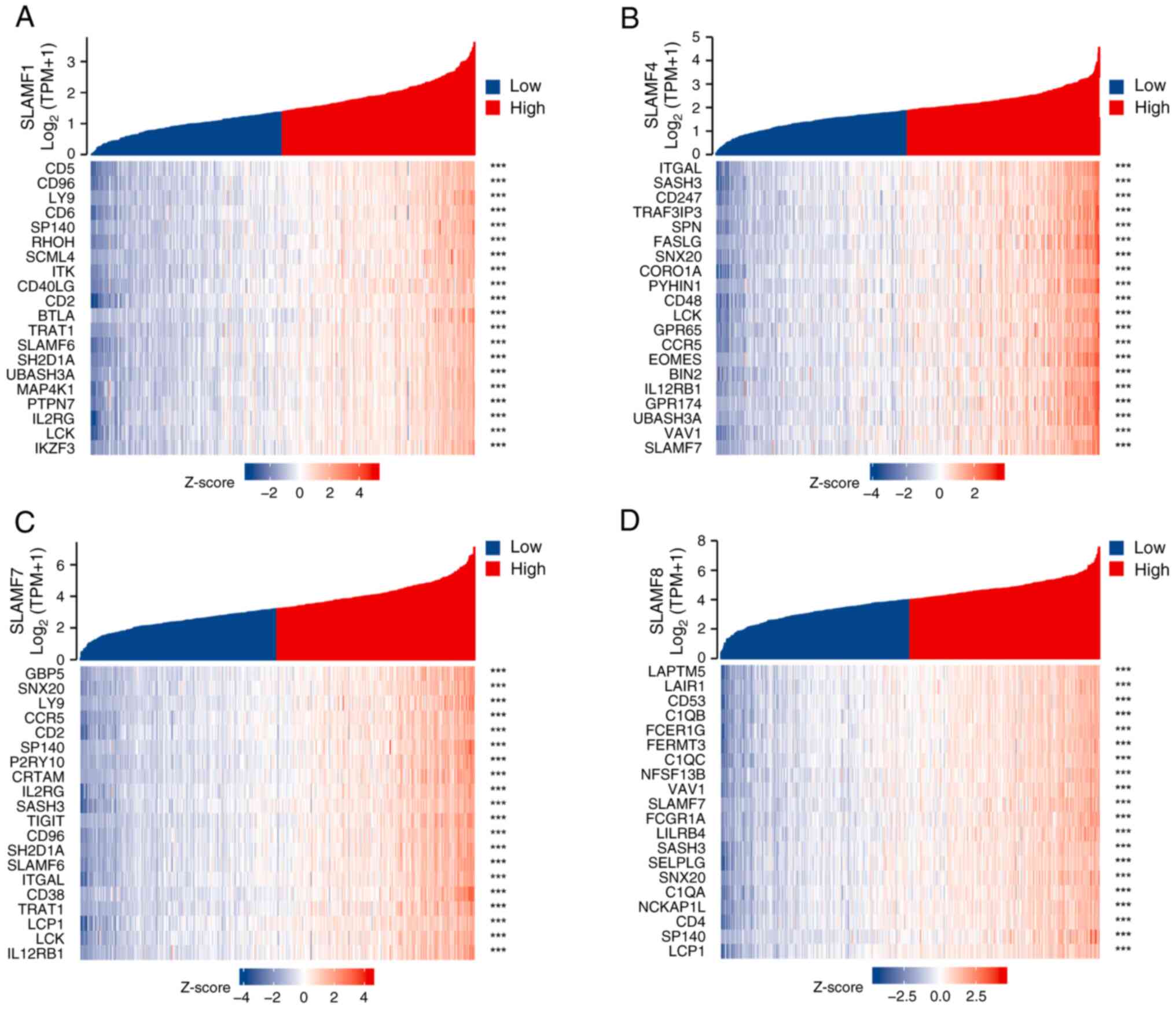
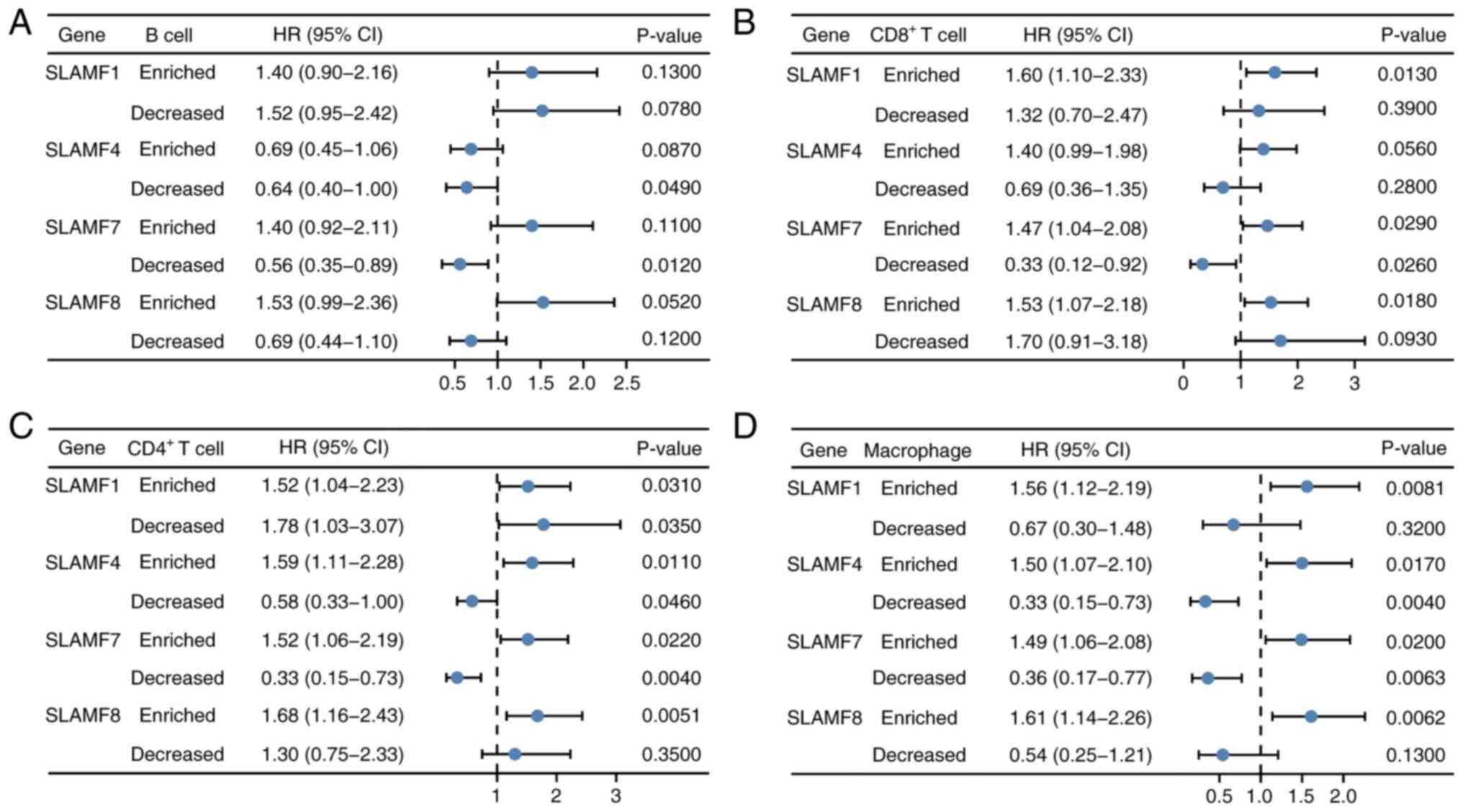
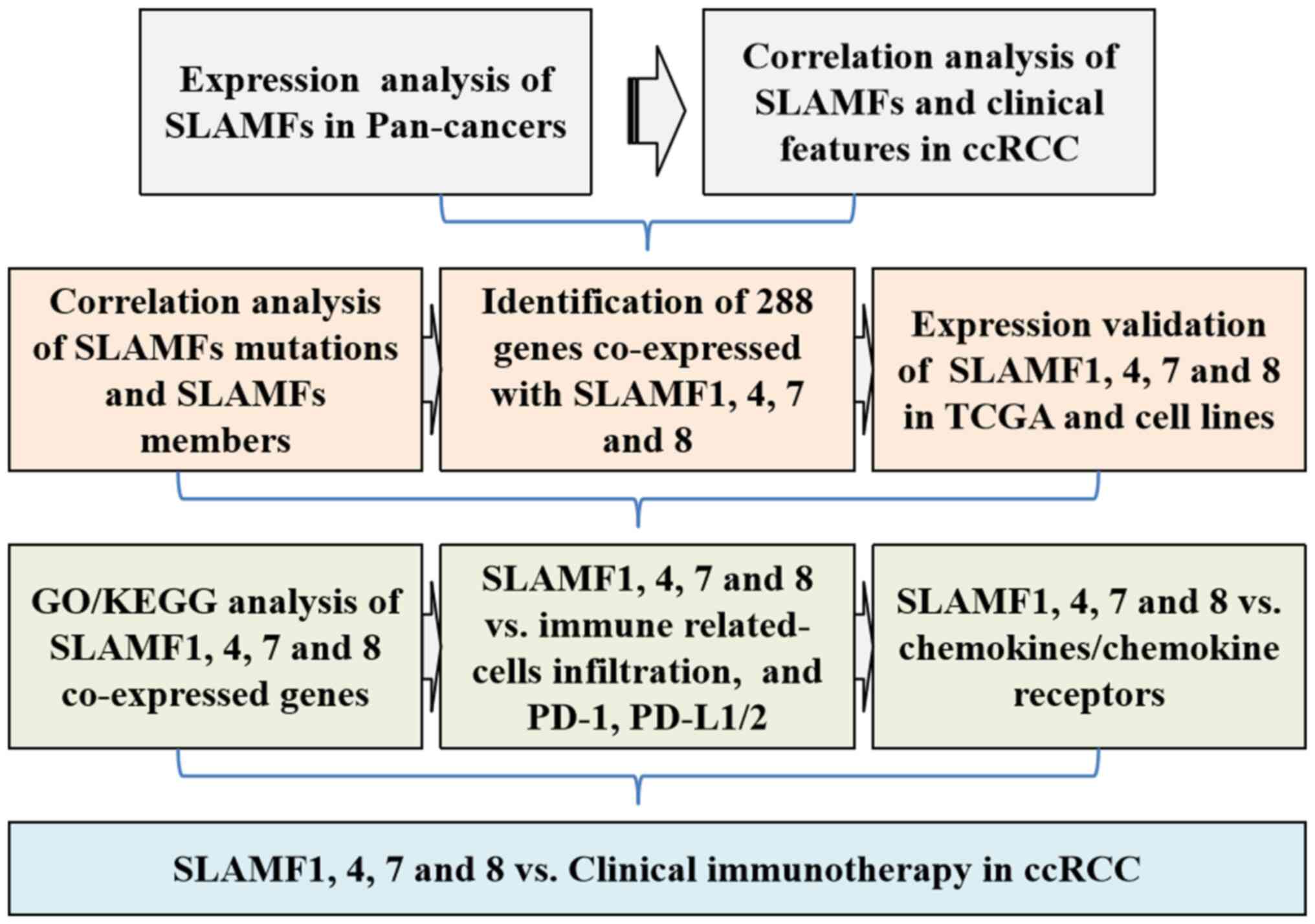
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moch H, Cubilla AL, Humphrey PA, Reuter VE
and Ulbright TM: The 2016 WHO classification of tumours of the
urinary system and male genital organs-part A: Renal, penile, and
testicular tumours. Eur Urol. 70:93–105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marchetti A, Rosellini M, Mollica V, Rizzo
A, Tassinari E, Nuvola G, Cimadamore A, Santoni M, Fiorentino M,
Montironi R and Massari F: The molecular characteristics of
non-clear cell renal cell carcinoma: What's the story morning
glory? Int J Mol Sci. 22:62372021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang J, Han P, Qian J, Zhang S, Wang S,
Cao Q and Shao P: Knockdown of ALPK2 blocks development and
progression of renal cell carcinoma. Exp Cell Res. 392:1120292020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He YH, Chen C and Shi Z: The biological
roles and clinical implications of microRNAs in clear cell renal
cell carcinoma. J Cell Physiol. 233:4458–4465. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao S, Yan L, Zhang H, Fan X, Jiao X and
Shao F: Identification of a metastasis-associated gene signature of
clear cell renal cell carcinoma. Front Genet. 11:6034552021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cochetti G, Cari L, Nocentini G, Maulà V,
Suvieri C, Cagnani R, Rossi De Vermandois JA and Mearini E:
Detection of urinary miRNAs for diagnosis of clear cell renal cell
carcinoma. Sci Rep. 10:212902020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Obeng RC, Arnold RS, Ogan K, Master VA,
Pattaras JG, Petros JA and Osunkoya AO: Molecular characteristics
and markers of advanced clear cell renal cell carcinoma: Pitfalls
due to intratumoral heterogeneity and identification of genetic
alterations associated with metastasis. Int J Urol. 27:790–797.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cros J, Sbidian E, Posseme K, Letierce A,
Guettier C, Benoît G and Ferlicot S: Nestin expression on tumour
vessels and tumour-infiltrating macrophages define a poor prognosis
subgroup of pt1 clear cell renal cell carcinoma. Virchows Arch.
469:331–337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Janowitz T, Welsh SJ, Zaki K, Mulders P
and Eisen T: Adjuvant therapy in renal cell carcinoma-past,
present, and future. Semin Oncol. 40:482–491. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Motzer RJ, Jonasch E, Agarwal N, Alva A,
Baine M, Beckermann K, Carlo MI, Choueiri TK, Costello BA, Derweesh
IH, et al: Kidney cancer, version 3.2022, NCCN clinical practice
guidelines in oncology. J Natl Compr Canc Netw. 20:71–90. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rosellini M, Marchetti A, Mollica V, Rizzo
A, Santoni M and Massari F: Prognostic and predictive biomarkers
for immunotherapy in advanced renal cell carcinoma. Nat Rev Urol.
20:133–157. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang B, Chen D and Hua H: TBC1D3 family is
a prognostic biomarker and correlates with immune infiltration in
kidney renal clear cell carcinoma. Mol Ther Oncolytics. 22:528–538.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Santoni M, Rizzo A, Mollica V, Matrana MR,
Rosellini M, Faloppi L, Marchetti A, Battelli N and Massari F: The
impact of gender on the efficacy of immune checkpoint inhibitors in
cancer patients: The MOUSEION-01 study. Crit Rev Oncol Hematol.
170:1035962022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rizzo A, Mollica V, Dall'Olio FG, Ricci
AD, Maggio I, Marchetti A, Rosellini M, Santoni M, Ardizzoni A and
Massari F: Quality of life assessment in renal cell carcinoma phase
II and III clinical trials published between 2010 and 2020: A
systematic review. Future Oncol. 17:2671–2681. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mollica V, Rizzo A, Marchetti A, Tateo V,
Tassinari E, Rosellini M, Massafra R, Santoni M and Massari F: The
impact of ECOG performance status on efficacy of immunotherapy and
immune-based combinations in cancer patients: The MOUSEION-06
study. Clin Exp Med. 23:5039–5049. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dragovich MA and Mor A: The SLAM family
receptors: Potential therapeutic targets for inflammatory and
autoimmune diseases. Autoimmun Rev. 17:674–682. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gunes M, Rosen ST, Shachar I and Gunes EG:
Signaling lymphocytic activation molecule family receptors as
potential immune therapeutic targets in solid tumors. Front
Immunol. 15:12974732024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Farhangnia P, Ghomi SM, Mollazadehghomi S,
Nickho H, Akbarpour M and Delbandi AA: SLAM-family receptors come
of age as a potential molecular target in cancer immunotherapy.
Front Immunol. 14:11741382023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tojjari A, Giles FJ, Vilbert M, Saeed A
and Cavalcante L: SLAM modification as an immune-modulatory
therapeutic approach in cancer. Cancers (Basel). 15:48082023.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su R, Jin C, Zhou L, Cao Y, Kuang M, Li L
and Xiang J: Construction of a ceRNA network of hub genes affecting
immune infiltration in ovarian cancer identified by WGCNA. BMC
Cancer. 21:9702021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lewinsky H, Gunes EG, David K, Radomir L,
Kramer MP, Pellegrino B, Perpinial M, Chen J, He TF, Mansour AG, et
al: CD84 is a regulator of the immunosuppressive microenvironment
in multiple myeloma. JCI Insight. 6:e1416832021.PubMed/NCBI
|
|
23
|
O'Connell P, Hyslop S, Blake MK, Godbehere
S, Amalfitano A and Aldhamen YA: SLAMF7 Signaling reprograms t
cells toward exhaustion in the tumor microenvironment. J Immunol.
206:193–205. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Agresta L, Hoebe KHN and Janssen EM: The
emerging role of CD244 signaling in immune cells of the tumor
microenvironment. Front Immunol. 9:28092018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yigit B, Wang N, Ten Hacken E, Chen SS,
Bhan AK, Suarez-Fueyo A, Katsuyama E, Tsokos GC, Chiorazzi N, Wu
CJ, et al: SLAMF6 as a regulator of exhausted CD8+ T cells in
cancer. Cancer Immunol Res. 7:1485–1496. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
GTEx Consortium: The genotype-tissue
expression (GTEx) project. Nat Genet. 45:580–585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vivian J, Rao AA, Nothaft FA, Ketchum C,
Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD,
Musselman-Brown A, et al: Toil enables reproducible, open source,
big biomedical data analyses. Nat Biotechnol. 35:314–316. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47((W1)): W556–W560.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Obuchowski NA and Bullen JA: Receiver
operating characteristic (ROC) curves: Review of methods with
applications in diagnostic medicine. Phys Med Biol. 63:07TR012018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lánczky A and Győrffy B: Web-based
survival analysis tool tailored for medical research (KMplot):
Development and implementation. J Med Internet Res. 23:e276332021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: an open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48((W1)): W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hughes CE and Nibbs RJB: A guide to
chemokines and their receptors. FEBS J. 285:2944–2971. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kakinuma T and Hwang ST: Chemokines,
chemokine receptors, and cancer metastasis. J Leukoc Biol.
79:639–651. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kotsias F, Cebrian I and Alloatti A:
Antigen processing and presentation. Int Rev Cell Mol Biol.
348:69–121. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang X, Spandidos A, Wang H and Seed B:
PrimerBank: A PCR primer database for quantitative gene expression
analysis, 2012 update. Nucleic Acids Res. 40((Database Issue)):
D1144–D1149. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang H, Song C, Ding Y, Pan X, Ge Z, Tan
BH, Gowda C, Sachdev M, Muthusami S, Ouyang H, et al:
Transcriptional regulation of JARID1B/KDM5B histone demethylase by
ikaros, histone deacetylase 1 (HDAC1), and casein kinase 2 (CK2) in
B-cell acute lymphoblastic leukemia. J Biol Chem. 291:4004–4018.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chan BKC: Data analysis using R
programming. Adv Exp Med Biol. 1082:47–122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Han Y, Liu D and Li L: PD-1/PD-L1 pathway:
current researches in cancer. Am J Cancer Res. 10:727–742.
2020.PubMed/NCBI
|
|
48
|
Wen M, Li Y, Qin X, Qin B and Wang Q:
Insight into cancer immunity: MHCs, immune cells and commensal
microbiota. Cells. 12:18822023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ma R, Qu X, Che X, Yang B, Li C, Hou K,
Guo T, Xiao J and Liu Y: Comparative analysis and in vitro
experiments of signatures and prognostic value of immune checkpoint
genes in colorectal cancer. Onco Targets Ther. 14:3517–3534. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Agresta L, Lehn M, Lampe K, Cantrell R,
Hennies C, Szabo S, Wise-Draper T, Conforti L, Hoebe K and Janssen
EM: CD244 represents a new therapeutic target in head and neck
squamous cell carcinoma. J Immunother Cancer. 8:e0002452020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lewinsky H, Barak AF, Huber V, Kramer MP,
Radomir L, Sever L, Orr I, Mirkin V, Dezorella N, Shapiro M, et al:
CD84 regulates PD-1/PD-L1 expression and function in chronic
lymphocytic leukemia. J Clin Invest. 128:5465–5478. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wu D, Zhang P, Ma J, Xu J, Yang L, Xu W,
Que H, Chen M and Xu H: Serum biomarker panels for the diagnosis of
gastric cancer. Cancer Med. 8:1576–1583. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Quan Q, Xiong X, Wu S and Yu M:
Identification of immune-related key genes in ovarian cancer based
on WGCNA. Front Genet. 12:7602252021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Veillette A and Latour S: The SLAM family
of immune-cell receptors. Curr Opin Immunol. 15:277–285. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wu N and Veillette A: SLAM family
receptors in normal immunity and immune pathologies. Curr Opin
Immunol. 38:45–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Smart JA, Oleksak JE and Hartsough EJ:
Cell adhesion molecules in plasticity and metastasis. Mol Cancer
Res. 19:25–37. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ishihara M, Hu J, Zhang X, Choi Y, Wong A,
Cano-Ruiz C, Zhao R, Tan P, Tso JL and Wu L: Comparing metastatic
clear cell renal cell carcinoma model established in mouse kidney
and on chicken chorioallantoic membrane. J Vis Exp. 10.3791/60314.
2020. View Article : Google Scholar
|
|
58
|
Fouquet G, Marcq I, Debuysscher V, Bayry
J, Rabbind Singh A, Bengrine A, Nguyen-Khac E, Naassila M and
Bouhlal H: Signaling lymphocytic activation molecules Slam and
cancers: Friends or foes? Oncotarget. 9:16248–16262. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
De Salort J, Sintes J, Llinàs L,
Matesanz-Isabel J and Engel P: Expression of SLAM (CD150)
cell-surface receptors on human B-cell subsets: From pro-B to
plasma cells. Immunol Lett. 134:129–136. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Karampetsou MP, Comte D, Suárez-Fueyo A,
Katsuyama E, Yoshida N, Kono M, Kyttaris VC and Tsokos GC:
Signaling lymphocytic activation molecule family member 1
engagement inhibits T cell-B cell interaction and diminishes
interleukin-6 production and plasmablast differentiation in
systemic lupus erythematosus. Arthritis Rheumatol. 71:99–108. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang N, Morra M, Wu C, Gullo C, Howie D,
Coyle T, Engel P and Terhorst C: CD150 is a member of a family of
genes that encode glycoproteins on the surface of hematopoietic
cells. Immunogenetics. 53:382–394. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gordiienko I, Shlapatska L, Kholodniuk V,
Sklyarenko L, Gluzman DF, Clark EA and Sidorenko SP: The interplay
of CD150 and CD180 receptor pathways contribute to the pathobiology
of chronic lymphocytic leukemia B cells by selective inhibition of
Akt and MAPK signaling. PLoS One. 12:e01859402017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yurchenko M, Shlapatska LM, Romanets OL,
Ganshevskiy D, Kashuba E, Zamoshnikova A, Ushenin YV, Snopok BA and
Sidorenko SP: CD150-mediated Akt signalling pathway in normal and
malignant B cells. Exp Oncol. 33:9–18. 2011.PubMed/NCBI
|
|
64
|
Li D, Xiong W, Wang Y, Feng J, He Y, Du J,
Wang J, Yang M, Zeng H, Yang YG, et al: SLAMF3 and SLAMF4 are
immune checkpoints that constrain macrophage phagocytosis of
hematopoietic tumors. Sci Immunol. 7:eabj55012022.PubMed/NCBI
|
|
65
|
Mittal R, Wagener M, Breed ER, Liang Z,
Yoseph BP, Burd EM, Farris AB III, Coopersmith CM and Ford ML:
Phenotypic T cell exhaustion in a murine model of bacterial
infection in the setting of pre-existing malignancy. PLoS One.
9:e935232014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Bae J, Song W, Smith R, Daley J, Tai YT,
Anderson KC and Munshi NC: A novel immunogenic CS1-specific peptide
inducing antigen-specific cytotoxic T lymphocytes targeting
multiple myeloma. Br J Haematol. 157:687–701. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Sugimoto A, Kataoka TR, Ito H, Kitamura K,
Saito N, Hirata M, Ueshima C, Takei Y, Moriyoshi K, Otsuka Y, et
al: SLAM family member 8 is expressed in and enhances the growth of
anaplastic large cell lymphoma. Sci Rep. 10:25052020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zou CY, Guan GF, Zhu C, Liu TQ, Guo Q,
Cheng W and Wu AH: Costimulatory checkpoint SLAMF8 is an
independent prognosis factor in glioma. CNS Neurosci Ther.
25:333–342. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang Q, Cheng L, Qin Y, Kong L, Shi X, Hu
J, Li L, Ding Z, Wang T, Shen J, et al: SLAMF8 expression predicts
the efficacy of anti-PD1 immunotherapy in gastrointestinal cancers.
Clin Transl Immunology. 10:e13472021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
De Jaeghere EA, Denys HG and De Wever O:
Fibroblasts fuel immune escape in the tumor microenvironment.
Trends Cancer. 5:704–723. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Langhans B, Nischalke HD, Krämer B, Dold
L, Lutz P, Mohr R, Vogt A, Toma M, Eis-Hübinger AM, Nattermann J,
et al: Role of regulatory T cells and checkpoint inhibition in
hepatocellular carcinoma. Cancer Immunol Immunother. 68:2055–2066.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sumitomo R, Hirai T, Fujita M, Murakami H,
Otake Y and Huang CL: M2 tumor-associated macrophages promote tumor
progression in non-small-cell lung cancer. Exp Ther Med.
18:4490–4498. 2019.PubMed/NCBI
|
|
73
|
Tu D, Dou J, Wang M, Zhuang H and Zhang X:
M2 macrophages contribute to cell proliferation and migration of
breast cancer. Cell Biol Int. 45:831–838. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wu X, Gu Z, Chen Y, Chen B, Chen W, Weng L
and Liu X: Application of PD-1 blockade in cancer immunotherapy.
Comput Struct Biotechnol J. 17:661–674. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ai L, Xu A and Xu J: Roles of PD-1/PD-L1
pathway: Signaling, cancer, and beyond. Adv Exp Med Biol.
1248:33–59. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kohli K, Pillarisetty VG and Kim TS: Key
chemokines direct migration of immune cells in solid tumors. Cancer
Gene Ther. 29:10–21. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Marcuzzi E, Angioni R, Molon B and Calì B:
Chemokines and chemokine receptors: Orchestrating tumor
metastasization. Int J Mol Sci. 20:962018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Dai S, Zeng H, Liu Z, Jin K, Jiang W, Wang
Z, Lin Z, Xiong Y, Wang J, Chang Y, et al: Intratumoral
CXCL13+CD8+T cell infiltration determines poor clinical outcomes
and immunoevasive contexture in patients with clear cell renal cell
carcinoma. J Immunother Cancer. 9:e0018232021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang L, Zhang M, Wang L, Li J, Yang T,
Shao Q, Liang X, Ma M, Zhang N, Jing M, et al: Identification of
CCL4 as an immune-related prognostic biomarker associated with
tumor proliferation and the tumor microenvironment in clear cell
renal cell carcinoma. Front Oncol. 11:6946642021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lin J, Yu M, Xu X, Wang Y, Xing H, An J,
Yang J, Tang C, Sun D and Zhu Y: Identification of biomarkers
related to CD8+ T cell infiltration with gene co-expression network
in clear cell renal cell carcinoma. Aging (Albany NY).
12:3694–3712. 2020. View Article : Google Scholar : PubMed/NCBI
|