|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang W, Zhang R, Zeng Y, Li Y, Chen Y,
Zhou J, Zhang Y, Wang A, Zhu J, Liu Z, et al: ALCAP2 inhibits lung
adenocarcinoma cell proliferation, migration and invasion via the
ubiquitination of β-catenin by upregulating the E3 ligase NEDD4L.
Cell Death Dis. 12:7552021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim JW, Marquez CP, Kostyrko K, Koehne AL,
Marini K, Simpson DR, Lee AG, Leung SG, Sayles LC, Shrager J, et
al: Antitumor activity of an engineered decoy receptor targeting
CLCF1-CNTFR signaling in lung adenocarcinoma. Nat Med.
25:1783–1795. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oudkerk M, Liu S, Heuvelmans MA, Walter JE
and Field JK: Lung cancer LDCT screening and mortality
reduction-evidence, pitfalls and future perspectives. Nat Rev Clin
Oncol. 18:135–151. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fang C, Liang Y, Huang Y, Jiang D, Li J,
Ma H, Guo L, Jiang W and Feng Y: P3H4 promotes malignant
progression of lung adenocarcinoma via interaction with EGFR.
Cancers (Basel). 14:32432022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pan D, Klare K, Petrovic A, Take A,
Walstein K, Singh P, Rondelet A, Bird AW and Musacchio A:
CDK-regulated dimerization of M18BP1 on a Mis18 hexamer is
necessary for CENP-A loading. Elife. 6:e233522017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fujita Y, Hayashi T, Kiyomitsu T, Toyoda
Y, Kokubu A, Obuse C and Yanagida M: Priming of centromere for
CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1.
Dev Cell. 12:17–30. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nardi IK, Zasadzińska E, Stellfox ME,
Knippler CM and Foltz DR: Licensing of centromeric chromatin
assembly through the Mis18α-Mis18β heterotetramer. Mol Cell.
61:774–787. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sullivan KF, Hechenberger M and Masri K:
Human CENP-A contains a histone H3 related histone fold domain that
is required for targeting to the centromere. J Cell Biol.
127:581–592. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
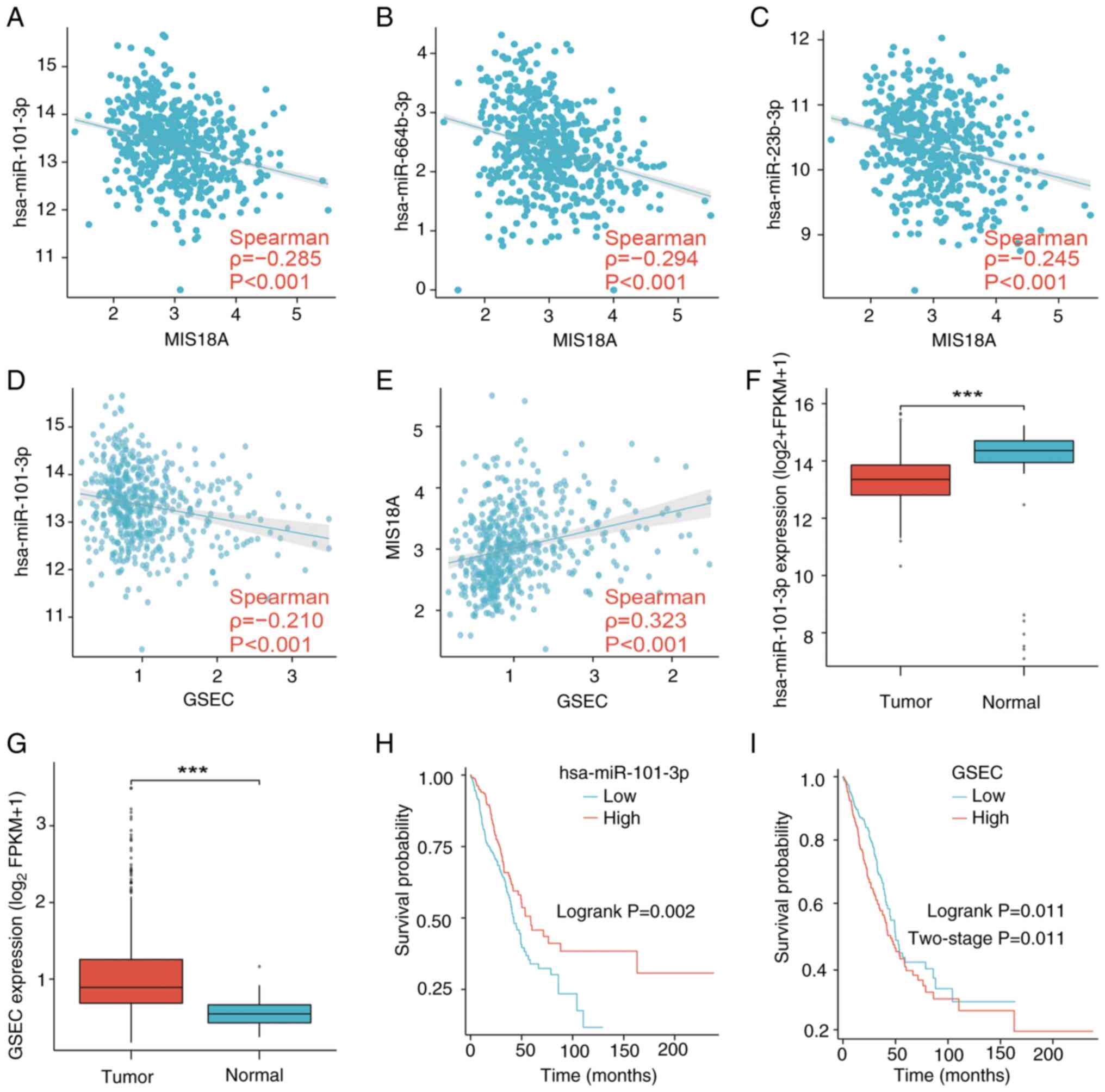
11
|
Liu WT, Wang Y, Zhang J, Ye F, Huang XH,
Li B and He QY: A novel strategy of integrated microarray analysis
identifies CENPA, CDK1 and CDC20 as a cluster of diagnostic
biomarkers in lung adenocarcinoma. Cancer Lett. 425:43–53. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim IS, Lee M, Park KC, Jeon Y, Park JH,
Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH and Kim KI: Roles of Mis18α
in epigenetic regulation of centromeric chromatin and CENP-A
loading. Mol Cell. 46:260–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun SY, Hu XT, Yu XF, Zhang YY, Liu XH,
Liu YH, Wu SH, Li YY, Cui SX and Qu XJ: Nuclear translocation of
ATG5 induces DNA mismatch repair deficiency (MMR-D)/microsatellite
instability (MSI) via interacting with Mis18α in colorectal cancer.
Br J Pharmacol. 178:2351–2369. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra1662013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Landi MT, Dracheva T, Rotunno M, Figueroa
JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et
al: Gene expression signature of cigarette smoking and its role in
lung adenocarcinoma development and survival. PLoS One.
3:e16512008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei TYW, Hsia JY, Chiu SC, Su LJ, Juan CC,
Lee YC, Chen JM, Chou HY, Huang JY, Huang HM and Yu CT: Methylosome
protein 50 promotes androgen- and estrogen-independent
tumorigenesis. Cell Signal. 26:2940–2950. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor- infiltrating immune cells. Cancer Res. 77:e108–e110.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Therneau T: A package for survival
analysis in R. R package version 3.6–4. 2024.
|
|
22
|
Kassambara A, Kosinski M and Biecek P:
Survminer: Drawing survival curves using ‘ggplot2’. R package
version 0.4.9. 2021.
|
|
23
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mostafavi S, Ray D, Warde-Farley D,
Grouios C and Morris Q: GeneMANIA: A real-time multiple association
network integration algorithm for predicting gene function. Genome
Biol. 9 (Suppl 1):S42008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mayakonda A, Lin DC, Assenov Y, Plass C
and Koeffler HP: Maftools: Efficient and comprehensive analysis of
somatic variants in cancer. Genome Res. 28:1747–1756. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang W, Soares J, Greninger P, Edelman EJ,
Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et
al: Genomics of drug sensitivity in cancer (GDSC): A resource for
therapeutic biomarker discovery in cancer cells. Nucleic Acids Res.
41((Database Issue)): D955–D961. 2013.PubMed/NCBI
|
|
31
|
Stridfeldt F, Cavallaro S, Hååg P,
Lewensohn R, Linnros J, Viktorsson K and Dev A: Analyses of single
extracellular vesicles from non-small lung cancer cells to reveal
effects of epidermal growth factor receptor inhibitor treatments.
Talanta. 259:1245532023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang K, Zhang L, Mi Y, Tang Y, Ren F, Liu
B, Zhang Y and Zheng P: A ceRNA network and a potential regulatory
axis in gastric cancer with different degrees of immune cell
infiltration. Cancer Sci. 111:4041–4050. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX,
Wu ZH, Chen XW, Gao P, Miao ZF and Wang ZN: Non-coding RNAs
participate in the regulatory network of CLDN4 via ceRNA mediated
miRNA evasion. Nat Commun. 8:2892017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Malhotra J, Malvezzi M, Negri E, La
Vecchia C and Boffetta P: Risk factors for lung cancer worldwide.
Eur Respir J. 48:889–902. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cucchiara F, Petrini I, Romei C, Crucitta
S, Lucchesi M, Valleggi S, Scavone C, Capuano A, De Liperi A,
Chella A, et al: Combining liquid biopsy and radiomics for
personalized treatment of lung cancer patients. State of the art
and new perspectives. Pharmacol Res. 169:1056432021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang L, Zhang M, Pan X, Zhao M, Huang L,
Hu X, Wang X, Qiao L, Guo Q, Xu W, et al: Integrative serum
metabolic fingerprints based multi-modal platforms for lung
adenocarcinoma early detection and pulmonary nodule classification.
Adv Sci (Weinh). 9:e22037862022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen X, Shu W, Zhao L and Wan J: Advanced
mass spectrometric and spectroscopic methods coupled with machine
learning for in vitro diagnosis. VIEW. 4:202200382023. View Article : Google Scholar
|
|
39
|
Liang D, Wang Y and Qian K: Nanozymes:
Applications in clinical biomarker detection. Interdiscip Med.
1:e202300202023. View Article : Google Scholar
|
|
40
|
Baumann K: Keeping centromeric identity.
Nat Rev Mol Cell Biol. 13:3402012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pussila M, Törönen P, Einarsdottir E,
Katayama S, Krjutškov K, Holm L, Kere J, Peltomäki P, Mäkinen MJ,
Linden J and Nyström M: Mlh1 deficiency in normal mouse colon
mucosa associates with chromosomally unstable colon cancer.
Carcinogenesis. 39:788–797. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ji X, Bossé Y, Landi MT, Gui J, Xiao X,
Qian D, Joubert P, Lamontagne M, Li Y, Gorlov I, et al:
Identification of susceptibility pathways for the role of
chromosome 15q25.1 in modifying lung cancer risk. Nat Commun.
9:32212018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Carroll B and Korolchuk VI: Nutrient
sensing, growth and senescence. FEBS J. 285:1948–1958. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kamal MA, Al-Zahrani MH, Khan SH, Al-Subhi
HA, Kuerban A, Aslam M, Al-Abbasi FA and Anwar F: Tubulin proteins
in cancer resistance: A review. Curr Drug Metab. 21:178–185. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen L, Zeng C, Yan L, Liao W, Zhen C and
Yao J: Prognostic value of holliday junction-recognizing protein
and its correlation with immune infiltrates in lung adenocarcinoma.
Oncol Lett. 24:2322022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yin Q, Chen W, Zhang C and Wei Z: A
convolutional neural network model for survival prediction based on
prognosis-related cascaded Wx feature selection. Lab Invest.
102:1064–1074. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wei Y, Ouyang GL, Yao WX, Zhu YJ, Li X,
Huang LX, Yang XW and Jiang WJ: Knockdown of HJURP inhibits
non-small cell lung cancer cell proliferation, migration, and
invasion by repressing Wnt/β-catenin signaling. Eur Rev Med
Pharmacol Sci. 23:3847–3856. 2019.PubMed/NCBI
|
|
48
|
Afsharpad M, Nowroozi MR, Mobasheri MB,
Ayati M, Nekoohesh L, Saffari M, Zendehdel K and Modarressi MH:
Cancer-testis antigens as new candidate diagnostic biomarkers for
transitional cell carcinoma of bladder. Pathol Oncol Res.
25:191–199. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Naetar N, Hutter S, Dorner D, Dechat T,
Korbei B, Gotzmann J, Beug H and Foisner R: LAP2alpha-binding
protein LINT-25 is a novel chromatin-associated protein involved in
cell cycle exit. J Cell Sci. 120:737–747. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Abdel-Maksoud MA, Hassan F, Mubarik U,
Mubarak A, Farrag MA, Alghamdi S, Atuahene SA, Almekhlafi S and
Aufy M: An in-silico approach leads to explore six genes as a
molecular signatures of lung adenocarcinoma. Am J Cancer Res.
13:727–757. 2023.PubMed/NCBI
|
|
51
|
Zhou H, Bian T, Qian L, Zhao C, Zhang W,
Zheng M, Zhou H, Liu L, Sun H, Li X, et al: Prognostic model of
lung adenocarcinoma constructed by the CENPA complex genes is
closely related to immune infiltration. Pathol Res Pract.
228:1536802021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wu Q, Chen YF, Fu J, You QH, Wang SM,
Huang X, Feng XJ and Zhang SH: Short hairpin RNA-mediated
down-regulation of CENP-A attenuates the aggressive phenotype of
lung adenocarcinoma cells. Cell Oncol (Dordr). 37:399–407. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang Y, Wang Y, Ren C, Wang H, Zhang Y and
Xiu Y: Upregulation of centromere protein K is crucial for lung
adenocarcinoma cell viability and invasion. Adv Clin Exp Med.
30:691–699. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lai H, Wen X, Peng Y and Zhang L:
Identification of stem cell-related gene markers by comprehensive
transcriptome analysis to predict the prognosis and immunotherapy
of lung adenocarcinoma. Curr Stem Cell Res Ther. 19:743–754. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Monteran L, Zait Y and Erez N: It's all
about the base: Stromal cells are central orchestrators of
metastasis. Trends Cancer. 10:208–229. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gao D, Fang L, Liu C, Yang M, Yu X, Wang
L, Zhang W, Sun C and Zhuang J: Microenvironmental regulation in
tumor progression: Interactions between cancer-associated
fibroblasts and immune cells. Biomed Pharmacother. 167:1156222023.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hebeisen M, Oberle SG, Presotto D, Speiser
DE, Zehn D and Rufer N: Molecular insights for optimizing T cell
receptor specificity against cancer. Front Immunol. 4:1542013.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Speiser DE, Chijioke O, Schaeuble K and
Münz C: CD4+ T cells in cancer. Nat Cancer. 4:317–329.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cardenas MA, Prokhnevska N and Kissick HT:
Organized immune cell interactions within tumors sustain a
productive T-cell response. Int Immunol. 33:27–37. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Laurent V, Guérard A, Mazerolles C, Le
Gonidec S, Toulet A, Nieto L, Zaidi F, Majed B, Garandeau D,
Socrier Y, et al: Periprostatic adipocytes act as a driving force
for prostate cancer progression in obesity. Nat Commun.
7:102302016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhu M, Xu W, Wei C, Huang J, Xu J, Zhang
Y, Zhao Y, Chen J, Dong S, Liu B and Liang C: CCL14 serves as a
novel prognostic factor and tumor suppressor of HCC by modulating
cell cycle and promoting apoptosis. Cell Death Dis. 10:7962019.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen J, Rajasekaran M, Xia H, Zhang X,
Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BK, Ooi LL,
et al: The microtubule-associated protein PRC1 promotes early
recurrence of hepatocellular carcinoma in association with the
Wnt/β-catenin signalling pathway. Gut. 65:1522–1534. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jiang N, Zou C, Zhu Y, Luo Y, Chen L, Lei
Y, Tang K, Sun Y, Zhang W, Li S, et al: HIF-1α-regulated miR-1275
maintains stem cell-like phenotypes and promotes the progression of
LUAD by simultaneously activating Wnt/β-catenin and Notch
signaling. Theranostics. 10:2553–2570. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cao Y, Geng J, Wang X, Meng Q, Xu S, Lang
Y, Zhou Y, Qi L, Wang Z, Wei Z, et al: RNA-binding motif protein 10
represses tumor progression through the Wnt/β-catenin pathway in
lung adenocarcinoma. Int J Biol Sci. 18:124–139. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Itakura M, Terashima Y, Shingyoji M, Yokoi
S, Ohira M, Kageyama H, Matui Y, Yoshida Y, Ashinuma H, Moriya Y,
et al: High CC chemokine receptor 7 expression improves
postoperative prognosis of lung adenocarcinoma patients. Br J
Cancer. 109:1100–1108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Choucair K, Morand S, Stanbery L, Edelman
G, Dworkin L and Nemunaitis J: TMB: A promising immune-response
biomarker, and potential spearhead in advancing targeted therapy
trials. Cancer Gene Ther. 27:841–853. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hellmann MD, Nathanson T, Rizvi H, Creelan
BC, Sanchez-Vega F, Ahuja A, Ni A, Novik JB, Mangarin LMB,
Abu-Akeel M, et al: Genomic features of response to combination
immunotherapy in patients with advanced non-small-cell lung cancer.
Cancer Cell. 33:843–852.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Samstein RM, Lee CH, Shoushtari AN,
Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ,
Omuro A, et al: Tumor mutational load predicts survival after
immunotherapy across multiple cancer types. Nat Genet. 51:202–206.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Goodman AM, Kato S, Bazhenova L, Patel SP,
Frampton GM, Miller V, Stephens PJ, Daniels GA and Kurzrock R:
Tumor mutational burden as an independent predictor of response to
immunotherapy in diverse cancers. Mol Cancer Ther. 16:2598–2608.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Rizvi H, Sanchez-Vega F, La K, Chatila W,
Jonsson P, Halpenny D, Plodkowski A, Long N, Sauter JL, Rekhtman N,
et al: Molecular determinants of response to anti-programmed cell
death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in
patients with non-small-cell lung cancer profiled with targeted
next-generation sequencing. J Clin Oncol. 36:633–641. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Hellmann MD, Ciuleanu TE, Pluzanski A, Lee
JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers
S, Salman P, et al: Nivolumab plus ipilimumab in lung cancer with a
high tumor mutational burden. N Engl J Med. 378:2093–2104. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Havel JJ, Chowell D and Chan TA: The
evolving landscape of biomarkers for checkpoint inhibitor
immunotherapy. Nat Rev Cancer. 19:133–150. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Jardim DL, Goodman A, de Melo Gagliato D
and Kurzrock R: The challenges of tumor mutational burden as an
immunotherapy biomarker. Cancer Cell. 39:154–173. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chan TA, Yarchoan M, Jaffee E, Swanton C,
Quezada SA, Stenzinger A and Peters S: Development of tumor
mutation burden as an immunotherapy biomarker: Utility for the
oncology clinic. Ann Oncol. 30:44–56. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yang Y, Adebali O, Wu G, Selby CP, Chiou
YY, Rashid N, Hu J, Hogenesch JB and Sancar A: Cisplatin-DNA adduct
repair of transcribed genes is controlled by two circadian programs
in mouse tissues. Proc Natl Acad Sci USA. 115:E4777–E4785.
2018.PubMed/NCBI
|
|
76
|
Zhu L and Chen L: Progress in research on
paclitaxel and tumor immunotherapy. Cell Mol Biol Lett. 24:402019.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Ahmadi A, Mohammadnejadi E and
Razzaghi-Asl N: Gefitinib derivatives and drug-resistance: A
perspective from molecular dynamics simulations. Comput Biol Med.
163:1072042023. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zou J, Lan H, Li W, Xie S, Tong Z, Song X
and Wang C: Comprehensive analysis of circular RNA expression
profiles in gefitinib-resistant lung adenocarcinoma patients.
Technol Cancer Res Treat. 21:153303382211391672022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang Q and Xu K: Advances in the research
of autophagy in EGFR-TKI treatment and resistance in lung cancer.
Zhongguo Fei Ai Za Zhi. 19:607–614. 2016.(In Chinese). PubMed/NCBI
|
|
80
|
Fukuoka M, Wu YL, Thongprasert S,
Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu
DT, Saijo N, et al: Biomarker analyses and final overall survival
results from a phase III, randomized, open-label, first-line study
of gefitinib versus carboplatin/paclitaxel in clinically selected
patients with advanced non-small-cell lung cancer in Asia (IPASS).
J Clin Oncol. 29:2866–2874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Chen C, Wan M, Peng X, Zhang Q and Liu Y:
GPR37-centered ceRNA network contributes to metastatic potential in
lung adenocarcinoma: Evidence from high-throughput sequencing.
Transl Oncol. 39:1018192024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wu X, Sui Z, Zhang H, Wang Y and Yu Z:
Integrated analysis of lncRNA-Mediated ceRNA network in lung
adenocarcinoma. Front Oncol. 10:5547592020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Feng W, Gong H, Wang Y, Zhu G, Xue T, Wang
Y and Cui G: circIFT80 functions as a ceRNA of miR-1236-3p to
promote colorectal cancer progression. Mol Ther Nucleic Acids.
18:375–387. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Rao X, Cao H, Yu Q, Ou X, Deng R and Huang
J: NEAT1/MALAT1/XIST/PKD-Hsa-Mir-101-3p-DLGAP5 axis as a novel
diagnostic and prognostic biomarker associated with immune cell
infiltration in bladder cancer. Front Genet. 13:8925352022.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Duca RB, Massillo C, Dalton GN, Farré PL,
Graña KD, Gardner K and De Siervi A: MiR-19b-3p and miR-101-3p as
potential biomarkers for prostate cancer diagnosis and prognosis.
Am J Cancer Res. 11:2802–2820. 2021.PubMed/NCBI
|
|
87
|
Chen Z, Lin X, Wan Z, Xiao M, Ding C, Wan
P, Li Q and Zheng S: High expression of EZH2 mediated by ncRNAs
correlates with poor prognosis and tumor immune infiltration of
hepatocellular carcinoma. Genes (Basel). 13:8762022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Tao L, Xu C, Shen W, Tan J, Li L, Fan M,
Sun D, Lai Y and Cheng H: HIPK3 inhibition by exosomal
hsa-miR-101-3p is related to metabolic reprogramming in colorectal
cancer. Front Oncol. 11:7583362022. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Song J, Sun Y, Cao H, Liu Z, Xi L, Dong C,
Yang R and Shi Y: A novel pyroptosis-related lncRNA signature for
prognostic prediction in patients with lung adenocarcinoma.
Bioengineered. 12:5932–5949. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhang J, Du C, Zhang L, Wang Y, Zhang Y
and Li J: lncRNA GSEC promotes the progression of triple negative
breast cancer (TNBC) by targeting the miR-202-5p/AXL axis. Onco
Targets Ther. 14:2747–2759. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Liu R, Ju C, Zhang F, Tang X, Yan J, Sun
J, Lv B, Guo Y, Liang Y, Lv XB and Zhang Z: LncRNA GSEC promotes
the proliferation, migration and invasion by sponging
miR-588/EIF5A2 axis in osteosarcoma. Biochem Biophys Res Commun.
532:300–307. 2020. View Article : Google Scholar : PubMed/NCBI
|