Introduction
Breast cancer is the most prevalent malignancy and
the most commonly diagnosed disease in women (11.7% of total cancer
cases), and its incidence has increased rapidly in recent years
(1,2). Cancer recurrence and metastases are
the primary factors accounting for high mortality in patients with
breast cancer (3). Metastasis
occurs when tumor cells acquire the ability to spread from the
primary tumor site and to invade surrounding stromal tissue
(4). Approximately 90% of
cancer-related mortalities are due to metastasis, and in the
context of tumorigenesis, epithelial-mesenchymal transition (EMT)
is a critical step in the early metastatic cascade (5,6). To
address these challenges, targeted therapy and anticancer drugs are
still the two most promising avenues for breast cancer treatment.
Alongside the well-known BRCA1 and BRCA2 genes, other
genes have been reported to be related to breast cancer, including
FOXQ1, FSIP1 and LTBP1 (7–9).
Therefore, exploring the influence of genes on the ability of
breast cancer to metastasize is a common research topic.
Our previous study (Geng et al, unpublished
data) demonstrated that iroquois homeobox 5 (IRX5) could inhibit
the migration and invasion of breast cancer cells and the
overexpression of IRX5 may lead to high expression of tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein β
(YWHAB). This suggests that IRX5 may regulate the expression
of YWHAB. Promoters regulate gene transcription at the most
basic level, and thus, affect gene expression (10). Therefore, IRX5 may regulate
the expression of YWHAB by affecting the promoter of
YWHAB. YWHAB is a member of the 14-3-3 protein family, which
is crucial for cell proliferation, and has seven isoforms in
mammals, including β, γ, ε, η, σ, τ/θ and ζ, which can assemble
into homodimers or heterodimers to regulate the activities of their
binding partners (11). These
proteins not only serve as signaling integration points for cell
cycle control and apoptosis, but also serve an essential role in
health, disease and drug development. Furthermore, these proteins
regulate the proliferation and migration of cancer and are
upregulated in numerous types of cancer (12–14).
The YWHAB gene encodes the 14-3-3β protein, which is
involved in cell cycle control and apoptosis, and functions
differently in numerous cancer types. The 14-3-3β protein has been
reported to be involved in the induction of bladder cancer cell
phase arrest (15), in the
tumorigenesis and metastasis of lung cancer (16) and in the proliferation and apoptosis
of glioma cells (17,18). Furthermore, YWHAB is a
biomarker in hepatocellular carcinoma (19). Although YWHAB serves
different roles in different cancer types, its specific mechanism
requires further investigation. Notably, to the best of our
knowledge, there are no studies assessing the role of YWHAB
in breast cancer.
The present study explored the regulatory effect of
IRX5 on YWHAB using a dual-luciferase reporter gene
assay. YWHAB expression was altered by lentiviral
transduction in vitro and the biological function of
YWHAB was assessed.
Materials and methods
Bioinformatics
The University of Alabama at Birmingham Cancer data
analysis portal (UALCAN; http://ualcan.path.uab.edu/analysis.html) was used to
analyze the expression levels of IRX5 and YWHAB in
different subtypes of breast cancer cells in The Cancer Genome
Atlas (TCGA) database. The dataset used was TCGA dataset: Breast
invasive carcinoma in UALCAN. Gene Expression Profiling Interactive
Analysis (GEPIA; http://gepia.cancer-pku.cn/index.html) was used to
analyze the correlation between YWHAB and IRX5 expression in breast
cancer. The dataset used was breast invasive carcinoma (BRCA) tumor
(BRCA: breast cancer) in GEPIA.
Cell culture
MDA-MB-231, MCF7 and 293FT cell lines were purchased
from Procell Life Science & Technology Co. Ltd. The MDA-MB-231
cells were cultured in DMEM/F12 (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% FBS (MilliporeSigma), 100 U/ml penicillin and
100 µg/ml streptomycin. The MCF7 and 293FT cells were cultured in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS, 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco; Thermo
Fisher Scientific, Inc.). All cells were cultured in a saturated
humidity incubator containing 5% CO2 at 37°C.
Dual luciferase assay
The promoter sequence upstream of the human
YWHAB transcription start site (NC_000020.11:
44883702-44885654) was obtained by PCR amplification. 293FT cell
genomic DNA as the PCR template was extracted using the Genomic DNA
Mini-Preps Kit (Beijing Baiolaibo Technology Co., Ltd.). Prime STAR
GXL DNA Polymerase, dNTP Mixture and 5X PrimeSTAR GXL Buffer were
purchased from Takara Biotechnology Co., Ltd. The primers used are
shown in Table I. The PCR reaction
procedure was as follows: 98°C for 2 min, followed by 30 cycles of
98°C for 10 sec, 50°C for 15 sec and 68°C for 2 min; and final
extension at 68°C for 10 min. PCR products were detected using a 1%
agarose gel containing ethidium bromide and bands were observed
using a gel imaging system (ProteinSimple). Finally, the
correctness of the sequence was verified by Sanger sequencing
(Sangon Biotech Co., Ltd.). The YWHAB-promoter-PGL3 vector was
obtained by ligating the promoter sequence into the pGL3-basic
vector (Promega Corporation). The open reading frames of the
IRX5 (NM_005853.6) gene were cloned into a pcDNA3.1(+)
vector (Invitrogen; Thermo Fisher Scientific, Inc.). For the
luciferase assay, 5×104 293FT cells were plated and
cultured in 24-well plates overnight at 37°C until they reached 80%
confluence, after which, they were co-transfected with
YWHAB-promoter-PGL3 vector, pcDNA3.1-IRX5 vector and PGMR-TK
Renilla luciferase reporter plasmid (Genomeditech) using
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). Firefly and
Renilla luciferase activity were measured after transfection
for 24 h using a Dual Luciferase Reporter Gene Assay Kit (Beyotime
Institute of Biotechnology).
 | Table I.Primers and shRNA sequences used in
the present study. |
Table I.
Primers and shRNA sequences used in
the present study.
| Assay | Gene | Sequence
(5′-3′) |
|---|
| PCR | YWHAB-promoter |
GGGGTACCGTGGCACAGTACAGGGGTCA |
|
|
|
CCCAAGGCTTCCTCGCCTTCACCGCTAGCC |
| RT-PCR | YWHAB | F:
GCTCTAGACACCATGACAATGGATAAAAGTGAGC |
|
|
| R:
CGGGATCCGTTCTCTCCCTCCCCAGCGT |
| RT-qPCR | GAPDH | F:
ACACCCACTCCTCCACCTTT |
|
|
| R:
TTACTCCTTGGAGGCCATGT |
|
| YWHAB | F:
CACAGAACAGGGGCATGAAC |
|
|
| R:
GTCCTGCAGTTCTGCCTCT |
|
| FAM20C | F:
TGTTCAAACCCATGAAACAAACG |
|
|
| R:
GTAGTAGGAACACTCGCCGT |
|
| PLEC | F:
AGCACCTCATCAAGGCCCAG |
|
|
| R:
TTGTCCAGATGAGGCCAAGG |
|
| MMP1 | F:
TACCTGGAAAAATACTACAACCTGA |
|
|
| R:
GGCGTGTAATTTTCAATCCTGTAG |
|
| DDIT3 | F:
AACCTGAGGAGAGAGTGTTCA |
|
|
| R:
TAATGGGGAGTGGCTGGAAC |
|
| HERPUD1 | F:
CGAGATTGGTTGGATTGGACCT |
|
|
| R:
TTTCAGGATCAGTGCCTTCCTGT |
|
| MMP3 | F:
TGGACAAAGGATACAACAGGGAC |
|
|
| R:
ATCTTGAGACAGGCGGAACC |
|
| IL24 | F:
ACAGGACCAGAGGGACAAGA |
|
|
| R:
AGGGTAAAACCCAGGCAAGG |
|
| TNFSF15 | F:
GGATCTGGGACTGAGCTTTGG |
|
|
| R:
CCTGTCCTTTTAGAGCCTGGAAC |
| shRNA | shYWHAB | CCGGCCAATGCTACACAACCAGAAACTCGAGTTT |
|
|
| CTGGTTGTGTAGCATTGGTTTTTGAATTC |
|
| sh-scramble | CCGGTCCTAAGGTTAAGTCGCCCTCGCTCGAGCGA |
|
|
| GGGCGACTTAACCTTAGGTTTTTGAATTC |
Lentivirus transduction and stable
cell line construction
The coding sequences (CDSs) of the YWHAB gene
(GenBank NM_139323.4) were obtained by reverse transcription-PCR
(RT-PCR) and were checked by Sanger sequencing (Sangon Biotech Co.,
Ltd.). RNAiso Plus reagent (Takara Biotechnology Co., Ltd.) was
used to extracted RNA from 293FT cells, and the PrimeScriptTMRT
reagent Kit with gDNA Eraser (Takara Biotechnology Co., Ltd.) was
used to reverse transcribe RNA into cDNA. The primers used are
shown in Table I. The PCR reaction
procedure used was as follows: 98°C for 2 min; 98°C for 30 sec,
50°C for 30 sec and 68°C for 1 min for 30 cycles; 68°C for 1 min.
The verified CDSs were ligated into the lentiviral vector
PEB-3XFlag-GP (Guangzhou Huijun Biotechnology Co., Ltd.) and were
packaged using the GM easy™ lentiviral Packaging Kit (Genomeditech;
third-generation lentiviral packaging system). The lentiviral
transduction plasmid:packaging plasmid:envelope plasmid ratio was
2:1:1. Briefly, 10 µg of the aforementioned recombinant plasmid and
10 µg of the GM easy™ Lentiviral Mix plasmid (Genomeditech) were
co-transfected into 293FT cells (10-cm Petri dish; 80% confluence)
using 60 µl HG Transgene™ Reagent (Genomeditech). The cells were
cultured at 37°C with 5% CO2 for 18 h and then the
culture medium was replaced. After culturing for 48 h, the
supernatant was collected and contained the lentiviral particles.
The lentiviral particles (MOI, 20) were added to MDA-MB-231 cells
and the culture medium was replaced 12 h later. Empty PEB-3XFlag-GP
was packaged and transduced simultaneously as a control. After 48
h, puromycin (2 µg/ml) was used for selection for 2 weeks and 0.5
µg/ml puromycin was used for maintenance. The time interval between
transfection and subsequent experimentation was 14 days. Western
blotting and RT-quantitative PCR (RT-qPCR) were used to detect the
YWHAB transduction efficiency. MDA-MB-231 cells were
transduced with the empty lentiviral vector or YWHAB-linked
lentiviral vector and were named MDA-MB-231-VC and
MDA-MB-231-YWHAB, respectively. The lentiviral vector used to knock
down YWHAB was purchased from Guangzhou IGE Biotechnology
Co., Ltd. The short hairpin RNA (sh) sequence targeting
YWHAB and the scrambled negative control are shown in
Table I. Lentiviral packaging and
transduction were performed in the aforementioned manner. The MCF7
cells with knockdown of YWHAB were referred to as
MCF7-shYWHAB cells, and the control cells were referred to as
MCF7-CT cells.
RNA extraction and RT-qPCR
Total RNA was extracted from MDA-MB-231, MCF7 and
293FT cells using RNAiso Plus reagent (Takara Biotech Co., Ltd.)
and was reverse transcribed into cDNA using the PrimeScript™ RT
reagent Kit with gDNA Eraser (Takara Biotechnology Co., Ltd.)
according to the manufacturer's instructions. qPCR was performed
using the 2X SG Fast qPCR Master Mix Kit (Sangon Biotech Co., Ltd.)
on a LightCycler96 (Roche Diagnostics) with the following
thermocycling conditions: Initial denaturation at 95°C for 5 min,
followed by 40 cycles of 95°C for 3 sec, 60°C for 30 sec and 97°C
for 1 sec. mRNA levels were quantified using the 2−ΔΔCq
method and normalized to the internal reference gene GAPDH
(20). The primers used in the
present study are shown in Table
I.
Western blotting
Proteins were extracted from cells (MDA-MB-231-VC,
MDA-MB-231-YWHAB, MCF7-CT and MCF7-shYWHAB cells) using RIPA lysis
buffer (Beyotime Institute of Biotechnology). Total protein was
quantified using the BCA Protein Assay Kit (Beyotime Institute of
Biotechnology) and the concentration was then adjusted to 4 µg/µl.
The proteins (60 µg/lane) were separated by SDS-PAGE on a 10% gel
and then transferred onto PVDF membranes (MilliporeSigma) for 90
min at 4°C at 150 mAh, and the membranes were blocked with 5%
non-fat milk for 1 h at room temperature and incubated with the
following primary antibodies overnight at 4°C: Mouse anti-Flag
(1:1,000; cat. no. D191041; Sangon Biotech Co., Ltd.), rabbit
anti-vimentin (1:1,000; cat. no. 5741T; Cell Signaling Technology,
Inc.), rabbit anti-YWHAB (1:1,000; cat. no. BM4752; Boster
Biological Technology) and mouse anti-GAPDH (1:1,000; cat. no.
D190090; Sangon Biotech Co., Ltd.). The membranes were then washed
three times with TBS with 0.1% Tween-20 and incubated with the
following HRP-conjugated secondary antibodies at 4°C for 2 h:
HRP-conjugated Goat Anti-Mouse (1:5,000; cat. no. D110087; Sangon
Biotech Co., Ltd.) and HRP-conjugated Goat Anti-Rabbit (1:5,000;
cat. no. D110058; Sangon Biotech Co., Ltd.). The antibody against
the Flag tag was used to examine the exogenous YWHAB protein with
the Flag tag. Finally, an HRP-DAB color development kit (Tiangen
Biotech Co., Ltd.) or ultrasensitive luminescence reagent (Sangon
Biotech Co., Ltd.) was used for visualization. Images were captured
using an image analysis system (5200; Tanon Science and Technology
Co., Ltd.) and protein bands were semi-quantified using ImageJ 1.8
(National Institutes of Health).
MTT cell proliferation assay
The MTT assay was performed to examine cell
proliferation. MDA-MB-231-VC and MDA-MB-231-YWHAB cells were seeded
at a density of 5×103 cells/well in 96-well plates and
cultured for 1, 2, 3, 4 and 5 days. The cells were then incubated
with MTT reagent (Beijing Solarbio Science & Technology Co.,
Ltd.) for 4 h. DMSO (Beijing Solarbio Science & Technology Co.,
Ltd.) was added to each well to dissolve formazan crystals, and the
absorbance was determined at 490 nm using a microplate reader
(Super Max 3100; Shanghai Shanpu Biotechnology Co., Ltd.).
Cell colony formation assay
Cell colony formation was measured using a plate
colony formation assay. MDA-MB-231-VC and MDA-MB-231-YWHAB cells
were seeded into 6-well plates at a density of 200 cells/well and
were cultured at 37°C in 5% CO2 for 2 weeks until
visible cell colonies were formed. Cells were washed with PBS and
fixed in 4% paraformaldehyde for 15 min at room temperature. Before
counting, the cell colonies were gently washed with PBS and then
stained with 0.1% crystal violet for 15 min at room temperature.
Viable colonies containing ≥50 cells were counted manually using an
inverted light microscope (DP72; Olympus Corporation).
Wound healing assay
To assess breast cancer cell migration, a wound
healing assay was performed. MDA-MB-231-VC and MDA-MB-231-YWHAB
cells were seeded in 6-well plates at a density of 2×105
cells/well and were cultured at 37°C for 24 h. When the cells
achieved stable attachment and reached 90% confluence, a parallel
linear scratch was generated using a sterile 200-µl pipette tip.
After rinsing with PBS, the wounded cells were cultured in
serum-free DMEM (Gibco; Thermo Fisher Scientific, Inc.). Images
were captured using an inverted light microscope (DP72; Olympus
Corporation) after 0, 24 and 48 h of incubation at 37°C. The areas
were measured using ImageJ 1.8 (National Institutes of Health). The
cell wound healing rate can reflect migration, and was calculated
as follows: Relative migration rate (%)=closure area/initial area
×100%.
Cell migration and invasion
assays
Cell invasion and migration were assessed using
24-well Transwell plates (8-µm pore size; Corning, Inc.) and a Cell
Invasion Chamber (8-µm pore size; Beijing Labselect). The
pre-coating with Matrigel of the Cell Invasion Chamber was
completed by the manufacturer. Cells (5×104;
MDA-MB-231-VC, MDA-MB-231-YWHAB, MCF7-CT and MCF7-shYWHAB cells)
were collected and seeded into the upper chambers in serum-free
culture medium. Medium containing 10% FBS was added to the lower
chambers to attract cells migrating and invading through the
membranes. After incubation at 37°C for 24 h, the cells in the
upper chambers were removed by washing with PBS. The cells that had
migrated and invaded through the membranes of the chambers were
fixed with paraformaldehyde at room temperature for 15 min, stained
with crystal violet at room temperature for 15 min and counted
under an inverted light microscope (DP72; Olympus Corporation).
Kyoto Encyclopedia of Genes and
Genomes (KEGG) and Gene Ontology (GO) enrichment analyses
The transcriptome analysis of MDA-MB-231-YWHAB and
MDA-MB-231-VC cells was performed by Novogene Co., Ltd. Total RNA
was isolated from cells using TRIzol reagent (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols. Total
RNA was identified and quantified as follows: 1% agarose gel
electrophoresis was used to analyze the degree of RNA degradation
and DNA contamination in the sample; a spectrophotometer
(NanoPhotometer; Implen GmbH) was used to measure RNA purity and
concentration; RNA integrity and quantity were measured using the
RNA Nano 6000 Assay Kit (Agilent Technologies, Inc.) on the
Bioanalyzer 2100 system (Agilent Technologies, Inc.). The kits used
to Library preparation included: ABclonal First Strand synthesis
module (cat. no. RK20353L; ABclonal Biotech Co., Ltd.), ABclonal
Second Strand synthesis module (cat. no. RK20346L; ABclonal Biotech
Co., Ltd.) and KC-Digital Stranded mRNA Library Prep Kit for
Illumina-UMI (cat. no. DR085-01/02; Seqhealth). Library
quantification was performed using a Qubit2.0 fluorometer
(Invitrogen; Thermo Fisher Scientific, Inc.). Libraries were
sequenced with a NovaSeq 6000 S4 Reagent Kit v1.5 (300 cycles;
(cat. no. 20028312; Illumina, Inc.). The final library loading
concentration was 5 nM. An Illumina Novaseq 6000 (Illumina, Inc.)
was used to conduct paired-end sequencing with the PE150 mode
according to the manufacturer's protocol. UMI-tools (v1.1.2) was
used to identify the unique molecular identifier sequence on each
sequence in the clean reads (21).
Cleaning of reads was performed by deleting reads that contain
adapter reads, plot-N reads and low-quality reads. HISAT2 v2.1.0
was used to construct a reference genome index and perform
reference genome alignment on paired-end clean reads (22). StringTie (v2.2.1) was used for new
gene prediction (23).
FeatureCounts (v2.0.1) was used to count reads mapped to each gene
(24). Differential expression
analysis of the two cell groups was performed using the edgeR
software package (v3.22.5) (25).
The genes with a log2 fold change value >1 and
P<0.05 were considered significantly differentially expressed
genes (DEGs). GO (http://geneontology.org/) and KEGG (https://www.genome.jp/kegg/) enrichment analyses were
performed to evaluate the biological function of YWHAB and
to investigate the pathways in which the DEGs were involved. GO and
KEGG enrichment analysis was performed using the clusterProfiler R
package (3.8.1) (26).
Statistical analysis
All data were processed using the statistical
software SPSS 20.0 (IBM Corp.) and GraphPad Prism 8.0 (Dotmatics).
The data are presented as the mean ± SD. Differences between means
were evaluated using unpaired Student's t-tests or one-way analysis
of variance with Tukey's post hoc test. All experiments were
repeated three times. P<0.05 was considered to indicate a
statistically significant difference.
Results
IRX5 binds to the promoter of YWHAB
and regulates its expression
Using TCGA, it was demonstrated that the expression
levels of IRX5 and YWHAB were both decreased with the
increase in the metastatic ability of breast cancer cell subtypes
(metastatic ability of breast cancer cell subtypes: Luminal <
HER2-positive < triple-negative) (Fig. 1A). This showed that the higher the
metastatic ability of breast cancer subtypes, the lower the
expression levels of YWHAB and IRX5. High expression
of YWHAB may inhibit the metastatic ability of breast
cancer. The very weak positive association between YWHAB and
IRX5 gene expression was determined by Pearson correlation
analysis using GEPIA (Fig. 1B). The
correctness of the YWHAB promoter sequences was verified
using Sanger sequencing (Fig. 1C).
To determine whether IRX5 interacts with the YWHAB promoter, a
dual-luciferase assay was used to detect the interaction between
IRX5 and the YWHAB promoter. Compared with PCDNA3.1-IRX5 or
PGL3-YWHAB-promoter alone, the simultaneous addition of both
significantly increased the luciferase activity (Fig. 1D). This indicated that IRX5
significantly increased luciferase activity by affecting the YWHAB
promoter. 293FT cells were transfected with pcDNA3.1-IRX5 plasmid
at numerous concentrations to confirm the regulatory effect on the
promoter. As the concentration of pcDNA3.1-IRX5 plasmid increased,
the luciferase activity also increased (Fig. 1E). These results suggested that IRX5
may regulate YWHAB gene expression by affecting the
YWHAB promoter.
YWHAB does not affect breast cancer
cell proliferation
To elucidate the function of YWHAB in breast
cancer cells, YWHAB was overexpressed in MDA-MB-231 cells.
Sanger sequencing was performed to verify the correctness of YWHAB
coding sequences (Fig. 2A). The
overexpression of YWHAB was verified by RT-qPCR and western
blotting. YWHAB mRNA expression was significantly increased in the
cells transfected with the YWHAB overexpression plasmid
compared with those transfected with the empty vector (Fig. 2B). Western blotting also
demonstrated successful transduction using the Flag antibody to
detect Flag-tagged YWHAB protein (Fig.
2C). The MTT assay was performed to identify the effect of
YWHAB overexpression on cell proliferation. Cell
proliferation curves demonstrated that there was no significant
difference in the proliferation of cells between the YWHAB-OE and
VC groups (Fig. 2D). No significant
difference in colony numbers was observed between the YWHAB-OE and
VC groups (Fig. 2E). These findings
indicated that YWHAB overexpression may have no effect on
breast cancer cell proliferation.
YWHAB inhibits breast cancer cell
migration and invasion
The characteristics of cancer cells also include
migration and invasion. The aforementioned results demonstrated
that YWHAB had no significant effect on cell proliferation.
The effect of overexpression of YWHAB on cell migration and
invasion was assessed using wound healing and Transwell assays. In
the wound healing assay, the relative migration rate of the
YWHAB-OE group was significantly reduced at 48 h compared with that
of the VC group (Fig. 3A). The
Transwell assay demonstrated that YWHAB-OE significantly reduced
cell migration and invasion compared with that of the VC group
(Fig. 3B and C). Furthermore,
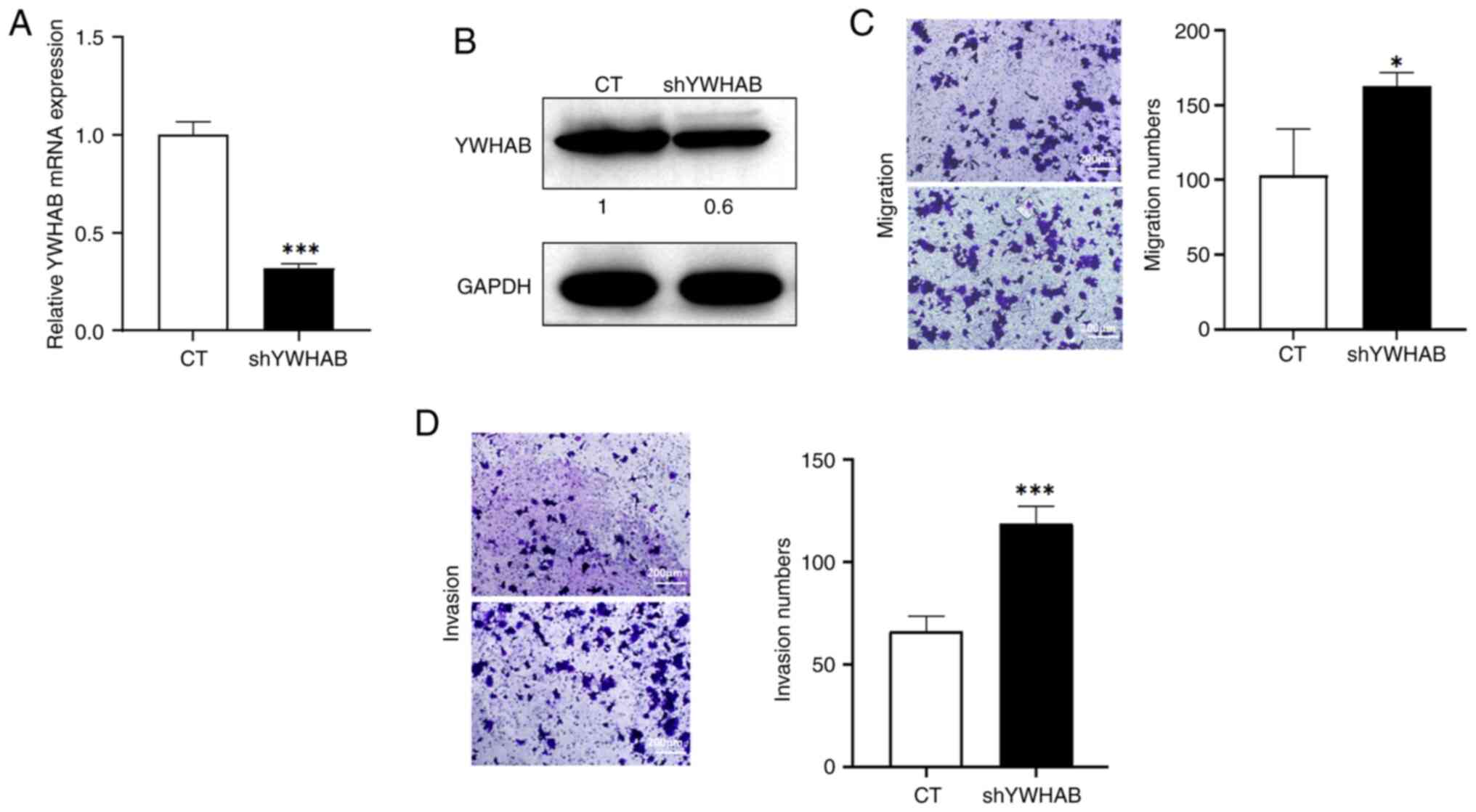
YWHAB expression was knocked down in MCF7 breast cancer
cells, which was verified by RT-qPCR and western blotting. The
results demonstrated that in MCF7-shYWHAB cells, the mRNA levels of
YWHAB were significantly reduced compared with those in the
control group (Fig. 4A). Western
blotting demonstrated a notable decrease in the protein levels of
YWHAB in MCF7-shYWHAB cells (Fig.
4B). Furthermore, the Transwell assays demonstrated that
YWHAB knockdown significantly increased cell migration and
invasion compared with those in the control group (Fig. 4C and D). These results indicated
that YWHAB could inhibit cell migration in vitro,
which suggests that YWHAB may act as a tumor suppressor
gene.
Functional enrichment analyses
To clarify the molecular pathways that are involved
in YWHAB-induced suppression of cell migration and invasion,
transcriptome analysis was performed in YWHAB-overexpressing
cells and control cells. The data from this assay have been
uploaded to the Gene Expression Omnibus database (GSE218458). A
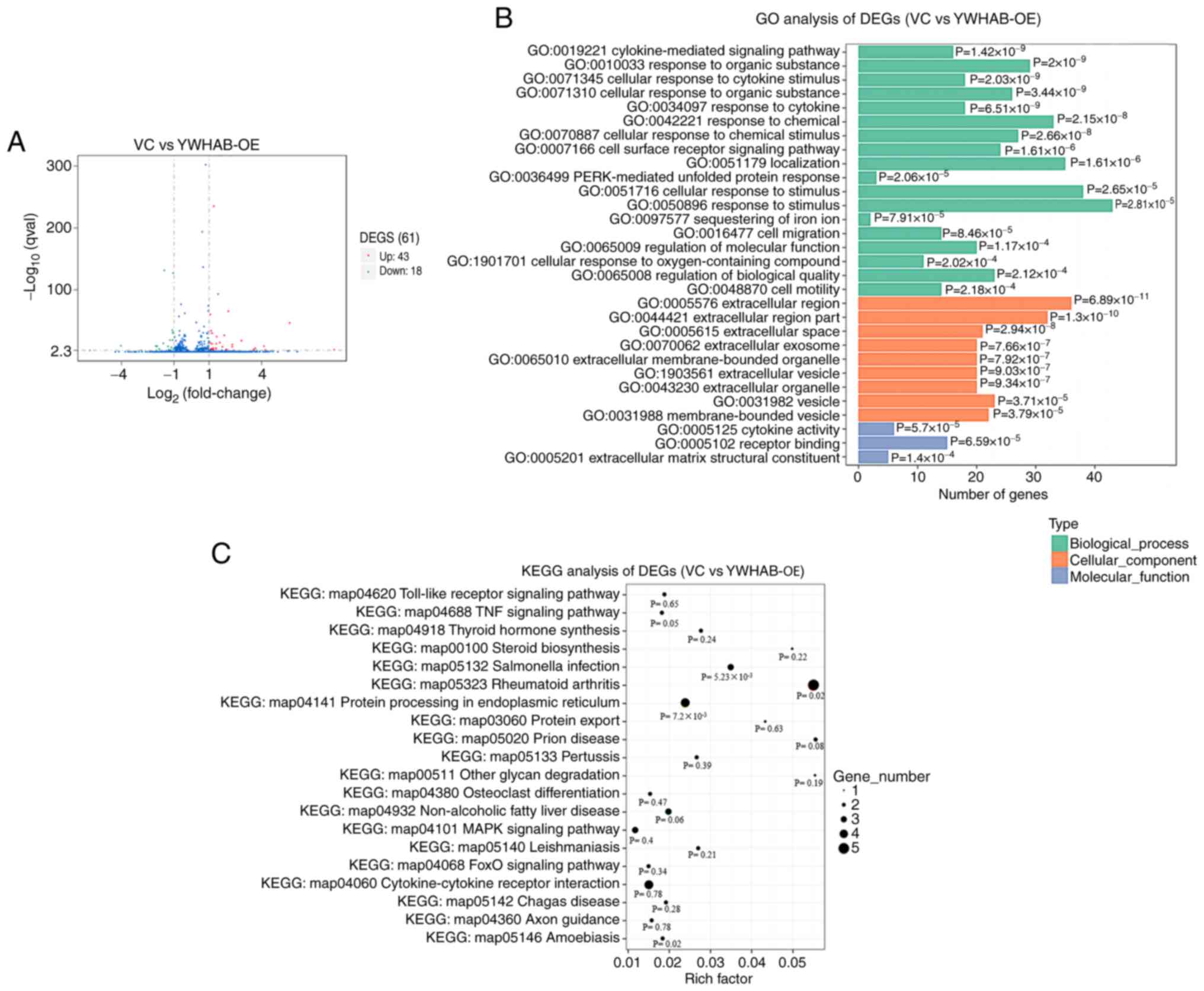
total of 61 genes were demonstrated to be differentially expressed,
among which 43 genes were upregulated and 18 genes were
downregulated when YWHAB was overexpressed in MDA-MB-231
cells (Fig. 5A). GO and KEGG
pathway enrichment analyses were used to assess the pathways and
molecular functions in which the DEGs were involved, in order to
better understand the functions of YWHAB overexpression. GO
was used to classify gene functions where each DEG could be
assigned to one or more GO terms within three domains. ‘Response to
stimulus’ (GO:0050896), ‘cellular response to stimulus’
(GO:0051716) and ‘localization’ (GO:0051179) were enriched in the
biological process domain. Within the cellular component domain,
the three most enriched categories were ‘extracellular region’
(GO:0005576), ‘extracellular region part’ (GO:0044421) and
‘vesicle’ (GO:0031982). In terms of molecular functions, the
enriched categories were ‘cytokine activity’ (GO:0005125),
‘receptor binding’ (GO:0005102) and ‘extracellular matrix
structural constituent’ (GO:0005201) (Fig. 5B). The DEGs were mainly associated
with the terms relating to the extracellular region, environmental
stimuli and binding to receptors, suggesting that YWHAB may be
involved in these signaling pathways affecting metastasis of cells.
The DEGs were also assigned to the biological pathways described in
KEGG to better understand their specific metabolic pathways in
cells (Fig. 5C). The ‘TNF signaling
pathway’ (KEGG: map04688) is related to cancer processes. The
pathway with the largest number of DEGs was ‘Rheumatoid arthritis’
(KEGG: map05323). These results indicated that YWHAB and
DEGs may affect the metastasis of breast cancer cells by
participating in multiple signaling pathways.
By searching existing studies, three genes (MMP1,
MMP3 and IL24) related to breast cancer or EMT were
selected (27–32). In the present study, both
transcriptome analysis results and RT-qPCR detection indicated that
MMP1, MMP3 and IL24 were downregulated (Fig. 6A). In addition, five DEGs
(FAM20C, PLEC, TNFSF15, DDIT3 and HERPUD1) were
randomly selected for RT-qPCR detection to verify whether their
expression changes were consistent with the transcriptome analysis
results. The results showed that FAM20C and PLEC were
upregulated in MDA-MB-231-YWHAB cells. TNFSF15, DDIT3 and
HERPUD1 were downregulated (Fig.
6A). The upregulation and downregulation trends of these genes
detected by RT-qPCR were consistent with the trends in
transcriptome analysis.
YWHAB can reduce vimentin
expression
Vimentin is a mesenchymal marker that is associated
with EMT (33). The changes in
vimentin expression after overexpression and knockdown of
YWHAB were assessed. After the overexpression of
YWHAB, the protein levels of vimentin were decreased,
whereas after the knockdown of YWHAB, the protein levels of
vimentin were increased (Fig. 6B).
The aforementioned results suggest that there may be an association
between YWHAB and vimentin.
Discussion
The incidence of breast cancer is increasing each
year (1). Therefore, there is a
need to evaluate the mechanism underlying breast cancer, since few
molecular targets of breast cancer are known. In the present study
a weak positive association between YWHAB and IRX5
expression was demonstrated using GEPIA. Furthermore, the
expression levels of IRX5 and YWHAB were decreased
with the increase in metastatic ability of breast cancer cell
subtypes (luminal < HER2-positive < triple-negative).
Finally, it was demonstrated that IRX5 regulates YWHAB gene
expression via the promoter of YWHAB.
The IRX family of transcription factors are upstream
factors identified to regulate neural gene expression in
Drosophila (34). The IRX
family contains six genes, IRX1, IRX2, IRX3, IRX4, IRX5 and
IRX6, in mice and humans, which encode highly conserved
homologous framework structural domains (34,35),
and serve pivotal roles in cell development and human cancer
progression (36,37). IRX5 functions in a variety of
tumors through different signaling pathways, such as prostate
cancer, hepatocellular carcinoma, tongue squamous cell carcinoma
and colorectal cancer (38–41). In our previous study (Geng et
al, unpublished data), it was demonstrated that IRX5 could
affect breast cancer cell invasion. Previous studies have reported
that YWHAB is aberrantly expressed during cancer
development. YWHAB is upregulated in gastric cancer, lung
cancer and hepatocellular carcinoma, and is downregulated in B cell
lymphoma (42–44). These findings indicated that
YWHAB likely serves distinct roles in different malignancies
and may be considered a therapeutic target for some types of
cancer. YWHAB is also associated with proliferation,
migration and invasion in hepatocellular carcinoma, gastric cancer
and cervical cancer (45–47). Based on this, we hypothesized that
YWHAB may serve a role in the development of breast cancer
and may be an effective molecular therapeutic target. Therefore,
MDA-MB-231 cells were selected to assess the impact of
overexpression of YWHAB.
The results of the present study demonstrated that
YWHAB had no effect on cell proliferation, but it inhibited
the migration and invasion of cells, as determined by wound healing
and Transwell assays. In general, the occurrence of apoptosis and
changes in the cell cycle are accompanied by changes in cell
proliferation (48). In the present
study, no significant changes in cell proliferation were observed,
suggesting that YWHAB most likely does not impact pathways
involved in the cell cycle and apoptosis. Transcriptome analysis
was performed to evaluate DEGs in MDA-MB-231 cells overexpressing
YWHAB. The most significantly enriched pathway with the largest
number of DEGs was ‘Rheumatoid arthritis’ (KEGG: map05323). The
‘TNF signaling pathway’ (KEGG: map04688) is closely related to
cancer. According to the functional enrichment results,
YWHAB may be involved in multiple signaling pathways.
However, the pathways affected by YWHAB still require
further research. Furthermore, it has been reported that a number
of DEGs are associated with EMT. For example, MMP1 improves
the metastasis of MDA-MB-231 cells by promoting the EMT process
(27). MMP1 knockdown
suppresses colorectal cancer progression by inhibiting EMT
(28). IL24 promotes
bronchial EMT transformation in chronic asthma (29). MMP3 is similar to MMP1
in terms of affecting the EMT process, and MMP3 promotes EMT
in mouse mammary epithelial cells (30). The hemopexin domain of MMP3
is responsible for mammary epithelial invasion, and the
downregulation of MMP3 inhibits breast tumor development
(31,32). This indicates that downregulation of
MMP1 and MMP3 inhibits EMT progression in breast
cancer. In the present study, the expression levels of MMP1,
MMP3 and IL24 were all downregulated after
overexpression of YWHAB. Therefore, the effect of YWHAB on
EMT was evaluated. The protein expression levels of the mesenchymal
marker vimentin, epithelial marker E-cadherin and EMT transcription
factor Slug were examined following overexpression of YWHAB.
It was demonstrated that the expression of vimentin was decreased
with the overexpression of YWHAB and increased after
YWHAB knockdown. However, no significant changes were
observed in the expression levels of E-cadherin and Slug (data not
shown).
Vimentin is the major intermediate filament protein
of mesenchymal cells and is ubiquitously expressed in normal
mesenchymal cells (49). Reduction
in mesenchymal characteristics can increase cell adhesion and
ultimately reduce cell motility (50). Furthermore, another study reported
that reduced vimentin levels can inhibit breast cancer cell
metastasis (51). Consequently, the
upregulation of YWHAB in breast cells could potentially
result in the loss of mesenchymal cell characteristics and inhibit
cell motility. There may be an association between YWHAB and
vimentin, which requires further study.
In conclusion, the present study highlighted the
inhibitory effect of YWHAB on breast cancer cell migration
and invasion. We hypothesized that YWHAB, a regulatory gene
associated with breast cancer, may reduce the mesenchymal
characteristics of breast cancer cells and could serve as a
promising therapeutic target. Furthermore, the results of the
present study suggest that IRX5 may regulate YWHAB gene
expression by affecting the YWHAB promoter. However, as the
experiments performed in the present study were all performed in
vitro, the effect of YWHAB on breast cancer metastasis
in vivo also requires verification. Finally, the specific
regulatory roles of IRX5 and YWHAB in breast cancer also
require further study.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Anhui Provincial Natural
Science Foundation (grant no. 2308085MC79) and the Natural Science
Foundation of Anhui Higher Education Institutions (grant no.
2023AH040055).
Availability of data and materials
The transcriptome data generated in the present
study may be found in the Gene Expression Omnibus database under
accession number GSE218458 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE218458.
The other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
JL designed the study and supervised the data
collection. JL and XG confirm the authenticity of all the raw data.
XG, WX and YS performed the experiments. JY and DZ performed the
data analysis. XG and DZ wrote the manuscript, and JY, YS and XG
revised the manuscript. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang L, Zhang S and Wang X: The metabolic
mechanisms of breast cancer metastasis. Front Oncol. 10:6024162020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gui P: Evolution of metastasis: New tools
and insights. Trends Cancer. 8:98–109. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mehlen P and Puisieux A: Metastasis: A
question of life or death. Nat Rev Cancer. 6:449–458. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zeisberg M and Neilson EG: Biomarkers for
epithelial-mesenchymal transitions. J Clin Invest. 119:1429–1437.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu
F, Ethier SP, Miller F and Wu G: Forkhead transcription factor
foxq1 promotes epithelial-mesenchymal transition and breast cancer
metastasis. Cancer Res. 71:1292–1301. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yan M, Wang J, Ren Y, Li L, He W, Zhang Y,
Liu T and Li Z: Over-expression of FSIP1 promotes breast cancer
progression and confers resistance to docetaxel via MRP1
stabilization. Cell Death Dis. 10:2042019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang J, Deng H and Wang J: LTBP1 promotes
the progression of triple negative breast cancer via activating the
RhoA/ROCK signaling pathway. Cancer Insight. 3:1–3. 2023.
View Article : Google Scholar
|
|
10
|
Blazeck J and Alper HS: Promoter
engineering: Recent advances in controlling transcription at the
most fundamental level. Biotechnol. 8:46–58. 2013.
|
|
11
|
Petosa C, Masters SC, Bankston LA, Pohl J,
Wang B, Fu H and Liddington RC: 14-3-3zeta binds a phosphorylated
Raf peptide and an unphosphorylated peptide via its conserved
amphipathic groove. J Biol Chem. 273:16305–16310. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aghazadeh Y and Papadopoulos V: The role
of the 14-3-3 protein family in health, disease, and drug
development. Drug Discov Today. 21:278–287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gardino AK and Yaffe MB: 14-3-3 proteins
as signaling integration points for cell cycle control and
apoptosis. Semin Cell Dev Biol. 22:688–695. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sime W, Niu Q, Abassi Y, Masoumi KC,
Zarrizi R, Køhler JB and Massoumi R: BAP1 induces cell death via
interaction with 14-3-3 in neuroblastoma. Cell Death Dis.
9:4582018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ou TT, Wang CJ, Lee YS, Wu CH and Lee HJ:
Gallic acid induces G2/M phase cell cycle arrest via regulating
14-3-3β release from Cdc25C and Chk2 activation in human bladder
transitional carcinoma cells. Mol Nutr Food Res. 54:1781–1790.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Komiya Y, Akiyama H, Sakumoto R and
Tashiro F: FBI1/Akirin2 promotes tumorigenicity and metastasis of
Lewis lung carcinoma cells. Biochem Biophys Res Commun.
444:382–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao L, Lei H, Chang MZ, Liu ZQ and Bie XH:
Down-regulation of 14-3-3β exerts anti-cancer effects through
inducing ER stress in human glioma U87 cells: Involvement of
CHOP-Wnt pathway. Biochem Biophys Res Commun. 462:389–395. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gong F, Wang G, Ye J, Li T, Bai H and Wang
WW: 14-3-3beta regulates the proliferation of glioma cells through
the GSK3beta/beta-catenin signaling pathway. Oncol Rep.
30:2976–2982. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu X, Bao M, Huang J, Zhou L and Zheng S:
Identification and validation of novel biomarkers for diagnosis and
prognosis of hepatocellular carcinoma. Front Oncol. 10:5414792020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smith T, Heger A and Sudbery L: UMI-tools:
Modeling sequencing errors in Unique Molecular Identifiers to
improve quantification accuracy. Genome Res. 27:491–499. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liao Y, Smyth GK and Shi W: featureCounts:
An efficient general purpose program for assigning sequence reads
to genomic features. Bioinformatics. 30:923–930. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu YH, Tao ZH, Chen Y, Lin SC, Zhu MG, Ji
W, Liu XJ, Li T and Hu X: Exosomal MMP-1 transfers metastasis
potential in triple-negative breast cancer through PAR1-mediated
EMT. Breast Cancer Res Treat. 193:65–81. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang K, Zheng J, Yu J, Wu Y, Guo J, Xu Z
and Sun X: Knockdown of MMP1 inhibits the progression of colorectal
cancer by suppressing the PI3K/Akt/cmyc signaling pathway and EMT.
Oncol Rep. 43:1103–1112. 2020.PubMed/NCBI
|
|
29
|
Feng KN, Meng P, Zou XL, Zhang M, Li H,
Yang HL, Li HT and Zhang TT: IL-37 protects against airway
remodeling by reversing bronchial epithelial-mesenchymal transition
via IL-24 signaling pathway in chronic asthma. Resp Res.
23:2442022. View Article : Google Scholar
|
|
30
|
Nelson CM, Khauv D, Bissell MJ and Radisky
DC: Change in cell shape is required for matrix
metalloproteinase-induced epithelial-mesenchymal transition of
mammary epithelial cells. J Cell Biochem. 105:25–33. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chu C, Liu X, Bai X, Zhao T, Wang M, Xu
RC, Li MQ, Hu YY, Li WH, Liu H, et al: MiR-519d suppresses breast
cancer tumorigenesis and metastasis via targeting MMP3. Int J Biol
Sci. 14:228–236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Correia AL, Mori H, Chen EI, Schmitt FC
and Bissell MJ: The hemopexin domain of MMP3 is responsible for
mammary epithelial invasion and morphogenesis through extracellular
interaction with HSP90beta. Genes Dev. 27:805–817. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Iwatsuki M, Mimori K, Yokobori T, Ishi H,
Beppu T, Nakamori S, Baba H and Mori M: Epithelial-mesenchymal
transition in cancer development and its clinical significance.
Cancer Sci. 101:293–299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gómez-Skarmeta JL and Modolell J: Iroquois
genes: Genomic organization and function in vertebrate neural
development. Curr Opin Genet Dev. 12:403–408. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim KH, Rosen A, Bruneau BG, Hui CC and
Backx PH: Iroquois homeodomain transcription factors in heart
development and function. Circ Res. 110:1513–1524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Leyns L, Gómez-Skarmeta JL and
Dambly-Chaudière C: iroquois: A prepattern gene that controls the
formation of bristles on the thorax of Drosophila. Mech Develop.
59:63–72. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Marcinkiewicz KM and Gudas LJ: Altered
epigenetic regulation of homeobox genes in human oral squamous cell
carcinoma cells. Exp Cell Res. 320:128–143. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang L, Song F, Sun H, Zhang L and Huang
C: IRX5 promotes NF-κB signalling to increase proliferation,
migration and invasion via OPN in tongue squamous cell carcinoma. J
Cell Mol Med. 22:3899–3910. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Myrthue A, Rademacher BL, Pittsenbarger J,
Kutyba-Brooks B, Gantner M, QianD Z and Beer TM: The iroquois
homeobox gene 5 is regulated by 1,25-dihydroxyvitamin D3 in human
prostate cancer and regulates apoptosis and the cell cycle in LNCaP
prostate cancer cells. Clin Cancer Res. 14:3562–3570. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhu L, Dai L, Yang N, Liu M, Ma S, Li CC,
Shen J, Lin T, Wang D, Pan W and Li X: Transcription factor IRX5
promotes hepatocellular carcinoma proliferation and inhibits
apoptosis by regulating the p53 signalling pathway. Cell Biochem
Funct. 38:621–629. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhu Q, Wu Y, Yang M, Wang Z, Zhang H,
Jiang XL, Chen M, Jin TY and Wang T: IRX5 promotes colorectal
cancer metastasis by negatively regulating the core components of
the RHOA pathway. Mol Carcinog. 58:2065–2076. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hua Y, Wang H, Wang H, Wu X, Yang L, Wang
CL, Li X, Jin YH, Li M, Wang L, et al: Circular RNA Circ_0006282
promotes cell proliferation and metastasis in gastric cancer by
regulating MicroRNA-144-5p/Tyrosine 3-Monooxygenase/Tryptophan
5-Monooxygenase activation protein β axis. Cancer Manag Res.
13:815–827. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li C, Li Z and Zhang M: Low Expression of
14-3-3beta is associated with adverse survival of diffuse large
B-Cell lymphoma patients. Front Med (Lausanne). 6:2372019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu YJ, Jan YJ, Ko BS, Liang SM and Liou
JY: Involvement of 14-3-3 proteins in regulating tumor progression
of hepatocellular carcinoma. Cancers. 7:1022–1036. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu TA, Jan YJ, Ko BS, Chen SC, Liang SM,
Hung YL, Chiun H, Shen TL, Lee YM, Chen PF, et al: Increased
expression of 14-3-3β promotes tumor progression and predicts
extrahepatic metastasis and worse survival in hepatocellular
carcinoma. Am J Pathol. 179:2698–2708. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tang Y, Lv P, Sun Z, Han L and Zhou W:
14-3-3β promotes migration and invasion of human hepatocellular
carcinoma cells by modulating expression of MMP2 and MMP9 through
PI3K/Akt/NF-κB pathway. PLoS One. 11:e01460702016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang X, Zhang Q, Zhang K, Wang F, Qiao X
and Cui JQ: Circ SMARCA5 inhibited tumor metastasis by interacting
with SND1 and downregulating the YWHAB gene in cervical
cancer. Cell Transplant. 30:9636897209837862021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Driskell RR, Lichtenberger BM, Hoste E,
Kretzschmar K, Simons BD, Charalambous M, Ferron SR, Herault Y,
Pavlovic G, Ferguson-Smith AC and Watt FM: Distinct fibroblast
lineages determine dermal architecture in skin development and
repair. Nature. 504:277–281. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lohneis P, Sinn M, Klein F, Bischoff S,
Striefler JK, Wislocka L, Sinn BV, Pelzer U, Oettle H, Riess H, et
al: Tumour buds determine prognosis in resected pancreatic ductal
adenocarcinoma. Br J Cancer. 118:1485–1491. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Calaf GM, Balajee AS, Montalvo-Villagra
MT, Leon M, Navarrete M D, Alvarez RG, Roy D, Narayan G and Jorge
AQ: Vimentin and Notch as biomarkers for breast cancer progression.
Oncol Lett. 7:721–727. 2014. View Article : Google Scholar : PubMed/NCBI
|