Introduction
Glioblastoma is the most common type of malignant
brain tumor. The prognosis of glioblastoma is extremely poor, even
with standard treatments, such as chemoradiotherapy.
O6-methylguanine DNA methyltransferase (MGMT) promoter
methylation is associated with favorable outcomes after
temozolomide (TMZ) chemotherapy in patients with newly diagnosed
glioblastoma (1). Thus, the
evaluation of MGMT methylation status is important for the
treatment of these patients.
Several radiological studies have shown the
potential of conventional magnetic resonance imaging (MRI) to
predict the MGMT methylation status using image texture and
deep learning architectures (2–7).
Dynamic susceptibility contrast (DSC) MRI offers insight into tumor
tissue vascularity by analyzing perfusion. Previous radiological
studies using MRI perfusion have aimed to predict MGMT
methylation status noninvasively in gliomas using radiological
characteristics (5,8–11) and
radiomics (12–15). Some reports revealed that DSC-MRI
could be used as a noninvasive technique to predict genetic
mutations preoperatively without surgical specimen, and to
determine molecular characteristics such as IDH mutation
status and methylation status of the MGMT promoter in
glioblastomas (5,8–10,16).
In contrast, other reports have indicated that cerebral blood
volume (CBV) did not differ significantly between tumors with
methylated or unmethylated MGMT (17,18).
The possibility of predicting MGMT methylation status from
DSC-MRI perfusion using a radiological approach remains
controversial, and there is no expert consensus regarding clinical
use.
The potential impact of DSC-MRI perfusion in the
prediction of MGMT methylation status in glioblastoma
remains disputed. In previous reports, elevated CBV has been
associated with decreased survival of glioblastoma patients
(19–22). In contrast, MGMT methylation
status is highly correlated with survival in glioblastomas with
moderate vascularity, but not in those with high vascularity
(17,23). Furthermore, patients with
glioblastomas showing stable or increasing CBV following
chemoradiotherapy experienced significantly improved PFS,
particularly in those cases presenting MGMT methylation
(24).
This study aimed to evaluate the possibility of
using DSC-MRI perfusion as a non-invasive method to predict
MGMT methylation status and prognosis in newly diagnosed
glioblastoma patients.
Materials and methods
Study design and patient
selection
This retrospective study was approved by the
Clinical Research Review Committee of Osaka University (Approval
No. 22302). The inclusion criteria were as follows: patients who i)
have definite pathological results; ii) have MRI images available,
including conventional MRI and DSC-MRI, before surgery; iii) did
not undergo radiotherapy or chemotherapy before MRI examination;
iv) have an available MGMT promoter methylation status.
Patients with recurrent tumors, tumors with unsatisfactory images,
and young patients aged less than 18 years old were excluded from
the study. Fifty patients with histologically confirmed primary
glioblastoma, IDH-wildtype (according to the 2021 World
Health Organization International Histological Classification of
Tumors) who underwent tumor resection at our institution between
January 2017 and January 2023 were included in the study (34 men
and 16 women; median age, 70.5 years; Table I). All patients were diagnosed
according to the 2021 guidelines, regardless of the resection date.
DSC-MRI and conventional MRI pulse sequences were acquired
preoperatively for all patients. All patients underwent surgical
resection with concomitant TMZ treatment and radiotherapy, followed
by adjuvant TMZ treatment. Tumor samples were collected after
resection.
 | Table I.Characteristics of patients with
glioblastoma. |
Table I.
Characteristics of patients with
glioblastoma.
| Characteristic | Number of
patients | Values | % |
|---|
| Sex |
|
|
|
|
Male | 34 |
| 68.0 |
|
Female | 16 |
| 32.0 |
| Age, years |
|
|
|
|
Median |
| 70.5 |
|
|
Range |
| 24-88 |
|
| MGMT
promoter methylation, % |
|
|
|
|
Median |
| 0.41 |
|
|
Range |
| 0.00–92.07 |
|
| CBV ratio |
|
|
|
|
Mean |
| 1.66 |
|
|
Range |
| 0.51–5.14 |
|
| CBF ratio |
|
|
|
|
Mean |
| 2.39 |
|
|
Range |
| 0.49–11.03 |
|
| MTT ratio |
|
|
|
|
Mean |
| 0.81 |
|
|
Range |
| 0.44–1.46 |
|
| PFS, months |
|
|
|
|
Median |
| 8.6 |
|
|
Range |
| 2.2–60.1 |
|
| Overall survival,
months |
|
|
|
|
Median |
| 19.5 |
|
|
Range |
| 2.5–60.1 |
|
Magnetic resonance imaging
All images, including axial T1-, T2-, and
T2*-weighted images, fluid-attenuated inversion recovery, and
contrast-enhanced T1-weighted sequences (T1Gd) were obtained using
a 3-T MR unit (DISCOVERY MR 750; GE Healthcare, Milwaukee, WI, USA)
with a 32-channel head coil. Perfusion MRI was performed using a
T2*-weighted, single-shot, gradient-recalled, echo-planar imaging
(GRE EPI) sequence. The perfusion MRI sequence parameters were as
follows: repetition time/echo time, 2000/13.3 ms; matrix, 128×128;
flip angle, 60; section thickness, 5 mm; and acquisition time, 90
sec. The contrast, a standard dose of 0.1 mmol/kg body weight of
meglumine gadoterate (Guerbet Japan, Tokyo, Japan), was injected at
a rate of 2–3 ml/s, followed by saline flush using a power
injector.
Imaging analysis
Imaging analysis was performed using Synapse Vincent
(Fuji Medical Systems, Tokyo, Japan) in perfusion mode. A single
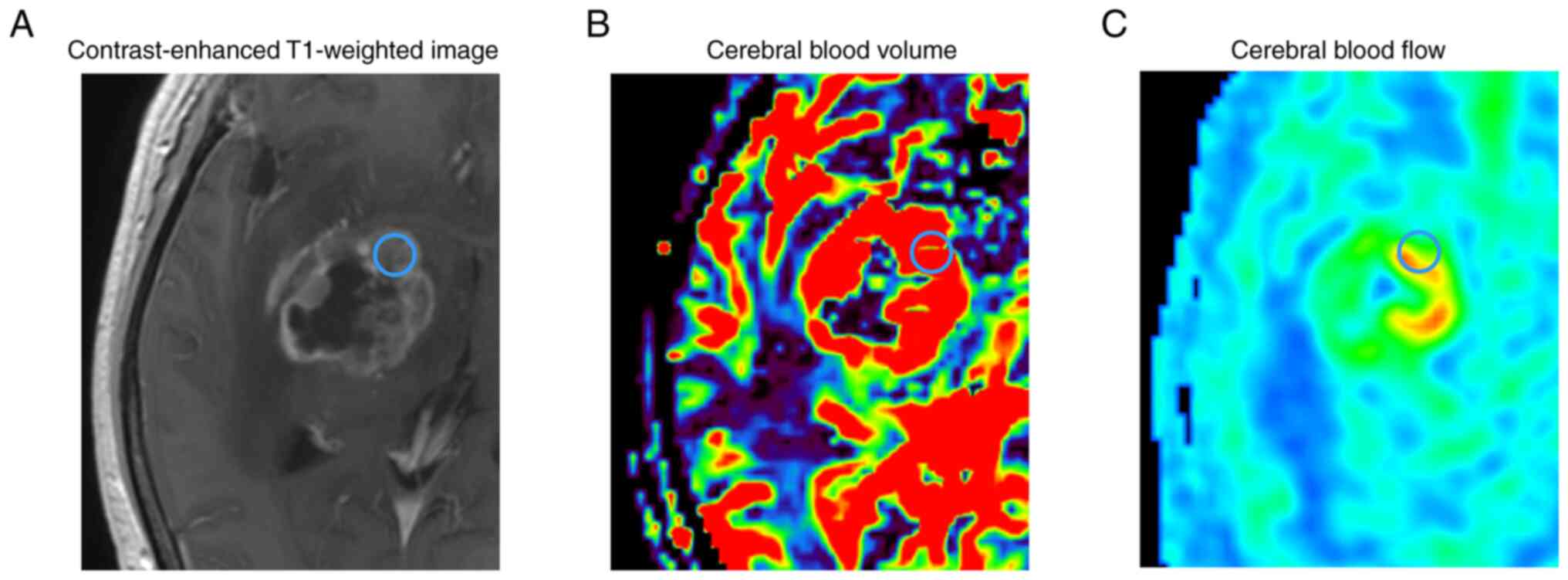
region of interest (ROI) with a diameter of 5 mm (Fig. 1) was set manually on the solid part
in each of the enhancing tumor regions from every patient, avoiding
areas of cyst formation, hemorrhage, and large vessels, as per
previous reports (16,25,26),
and the values of CBV, cerebral blood flow (CBF), and mean transit
time (MTT) were calculated. The ROI was manually set in the
contralateral normal area for each enhanced tumor. The
contralateral area was normal and contained no tumor
infiltration.
Disease-to-normal ratios were calculated by dividing
the values of CBV, CBF, and MTT for the tumors by the values of the
contralateral normal area, and described as rCBV, rCBE, and rMTT,
respectively.
Genomic DNA extraction
Tumor samples were immediately frozen and stored at
−80°C or immersed in RNAlater Stabilization Solution (Thermo Fisher
Scientific, Waltham, MA). Genomic DNA was extracted using a DNeasy
Blood & Tissue Kit (Qiagen, Valencia, CA, USA) or NucleoSpin
Tissue (Macherey-Nagel, Düren, Germany), as described previously
(27).
MGMT promoter methylation
analysis
The methylation status of the MGMT promoter
(accession number: NM_002412.5) was assessed using quantitative
methylation-specific PCR (qMSP). Purified DNA was subjected to
bisulfite modification by an EZ DNA Methylation-Gold Kit (Zymo
Research, Irvine, CA), according to the manufacturer's
instructions. The qMSP was performed on a QuantStudio12K Flex
Real-Time PCR System (Thermo Fisher Scientific) with POWER
SYBR® Green PCR Master Mix (Thermo Fisher Scientific).
The bisulfite-modified DNA was amplified using specific primers for
each methylated or unmethylated molecule as listed in Table II. Real-time PCR conditions were as
follows: 95°C for 10 min followed by 45 cycles of 95°C for 10 sec,
and 60°C for 60 sec. The quantification of methylated and
unmethylated sequences was performed by employing the standard
curve method as previously described. In the dissociation curve
analysis, heterogeneity of the amplified methylated and
unmethylated molecules was assessed from melting temperature. The
mean ± standard deviation of methylation value was calculated from
triplicate PCRs. We used a 1% cut-off value for the determination
of MGMT methylation based on an outcome-based study of newly
diagnosed GBMs as mentioned in our previous publications (27,28).
Sequences of primers used for quantitative methylation-specific PCR
are provided in Table II.
 | Table II.Sequences of primers used for
quantitative methylation specific PCR. |
Table II.
Sequences of primers used for
quantitative methylation specific PCR.
| Gene | Primer | Sequence
(5′-3′) |
|---|
| MGMT
promoter | M-forward |
TTTCGACGTTCGTAGGTTTTCGC |
|
| M-reverse |
GCACTCTTCCGAAAACGAAACG |
|
| U-forward |
TTTGTGTTTTGATGTTTGTAGGTTTTTGT |
|
| U-reverse |
AACTCCACACTCTTCCAAAAACAAAACA |
Statistical analysis
Statistical analyses were performed using Prism
version 9 (GraphPad Software, San Diego, CA, USA). Results were
considered statistically significant at a P-value of <0.05. The
unpaired t-test was used for comparisons between two groups.
Receiver operating characteristic (ROC) curve analysis was
performed to compare the performance of each imaging parameter
based on each ROI in distinguishing tumors with MGMT
methylation from those without MGMT methylation. The
Kaplan-Meier method was used to derive OS and PFS curves.
We also attempted to construct a model based on
three perfusion parameters to determine MGMT methylation
status in glioblastomas by performing multiple logistic
regression.
Results
Perfusion MRI parameters and MGMT
methylation status
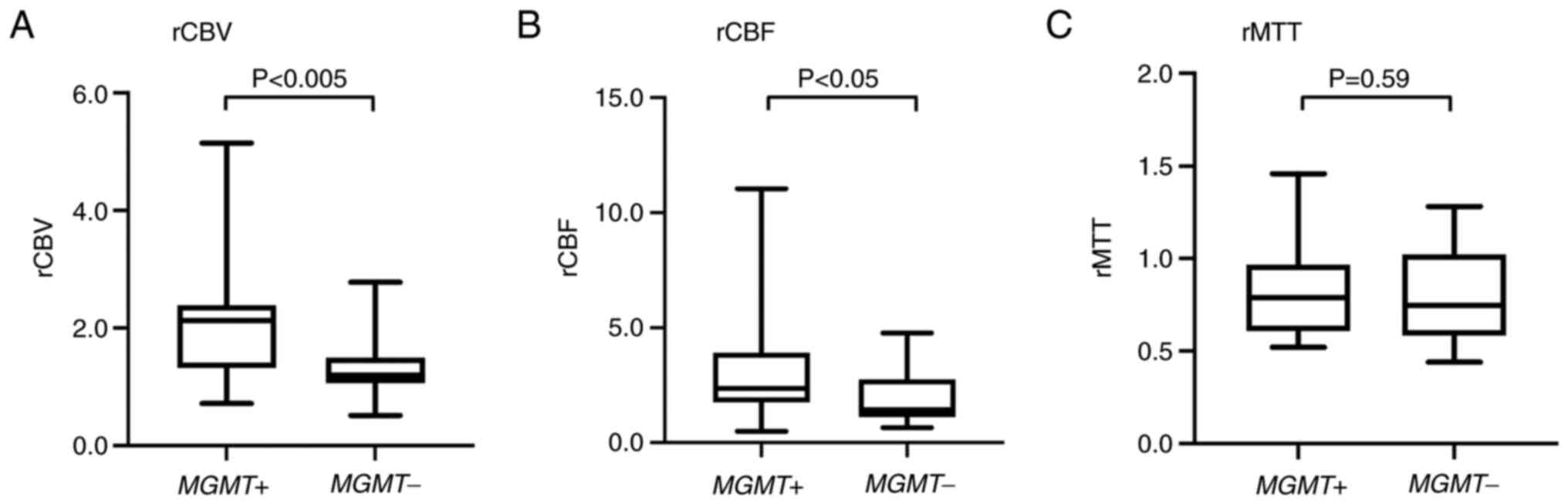
The mean rCBV for tumors with MGMT
methylation (2.09; range, 0.72–5.14) was significantly higher than
that for tumors without MGMT methylation (1.33; range,
0.51–2.78; P<0.005). The mean rCBF for tumors with MGMT
methylation (3.08; range, 0.49–11.03) was significantly higher than
that for tumors without MGMT methylation (1.85; range,
0.65–4.77; P<0.05). In contrast, the rMTT for tumors with and
without MGMT methylation did not differ (Fig. 2, Table
III).
 | Table III.Correlation between MRI perfusion
parameters and MGMT promoter methylation status in patients
with glioblastoma. |
Table III.
Correlation between MRI perfusion
parameters and MGMT promoter methylation status in patients
with glioblastoma.
|
| MGMT
promotor methylation status | Univariate |
|---|
|
|
|
|
|---|
| Perfusion
parameters | Methylated
(n=22) | Unmethylated
(n=28) | P-value |
|---|
| Mean CBV ratio | 2.09 | 1.33 | 0.002 |
| Mean CBF ratio | 3.08 | 1.85 | 0.020 |
| Mean MTT ratio | 0.83 | 0.79 | 0.590 |
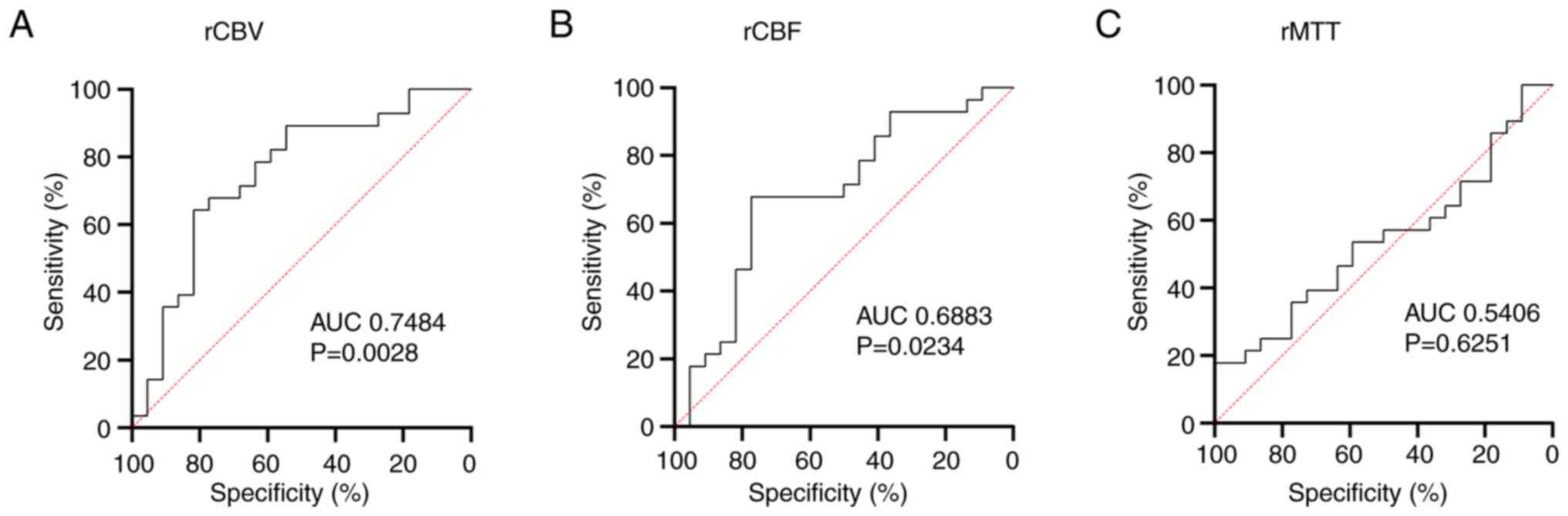
Receiver operating characteristic (ROC) analysis
showed that the rCBV [area under the curve (AUC)=0.7484] and rCBF
(AUC=0.6883) were more effective in distinguishing between tumors
with and without MGMT methylation than the rMTT (AUC=0.5406;
Fig. 3).
We attempted to construct a model based on three
perfusion parameters to determine MGMT methylation status in
glioblastomas by performing multiple logistic regression. The
following predictive formula, created using parameters derived from
the multiple logistic regression, was obtained to estimate the
probability of MGMT methylation (probability range: 0 to 1)
for each ROI:
log_odds=0.01832 + 4.743 * rCBV + 1.034 * rCBF +
4.214 * rMTT odds=exp(log_odds)
Probability=odds/(ones(size(odds)) + odds)
Prognosis according to MGMT
methylation status
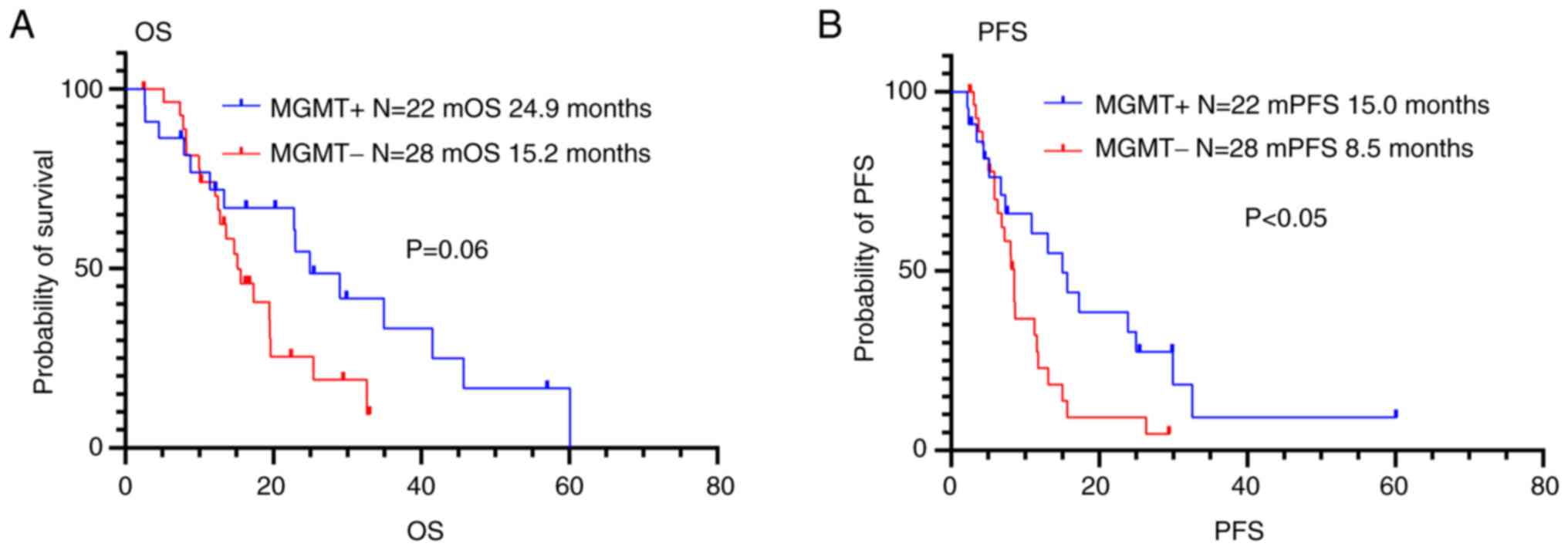
The PFS and OS were 15.0 months and 24.9 months,
respectively, in the patients with MGMT methylation, and 8.5
months and 15.2 months, respectively, in the patients without
MGMT methylation (Fig. 4 and
Table IV). Patients with
MGMT methylation had longer PFS than those without
MGMT methylation (P<0.05), but there was no significant
difference in OS between patients with and without MGMT
methylation (P=0.06).
 | Table IV.Univariate analyses of median
survival time and PFS of patients with glioblastoma. |
Table IV.
Univariate analyses of median
survival time and PFS of patients with glioblastoma.
| Variable | No. of cases | MST | P-value
(log-rank) | PFS | P-value
(log-rank) |
|---|
| MGMT
promoter methylation status |
|
|
|
|
|
|
Methylated | 22 | 24.9 | 0.06 | 15.0 | 0.03 |
|
Unmethylated | 28 | 15.2 |
| 8.5 |
|
| CBV ratio |
|
|
|
|
|
|
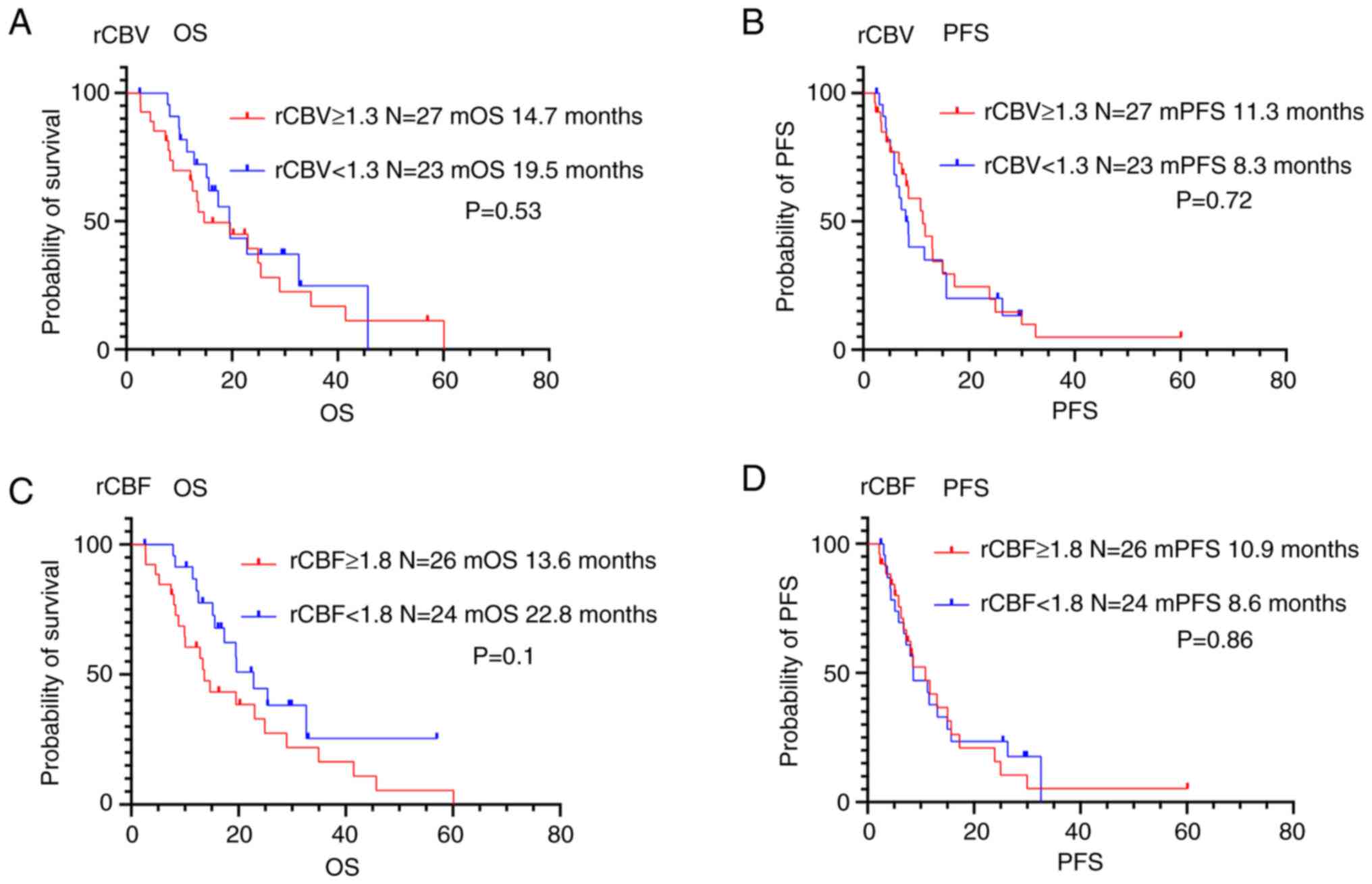
<1.3 | 23 | 19.5 | 0.53 | 8.3 | 0.72 |
|
≥1.3 | 27 | 14.7 |
| 11.3 |
|
| CBF ratio |
|
|
|
|
|
|
<1.8 | 24 | 22.8 | 0.10 | 8.6 | 0.86 |
|
≥1.8 | 26 | 13.6 |
| 10.9 |
|
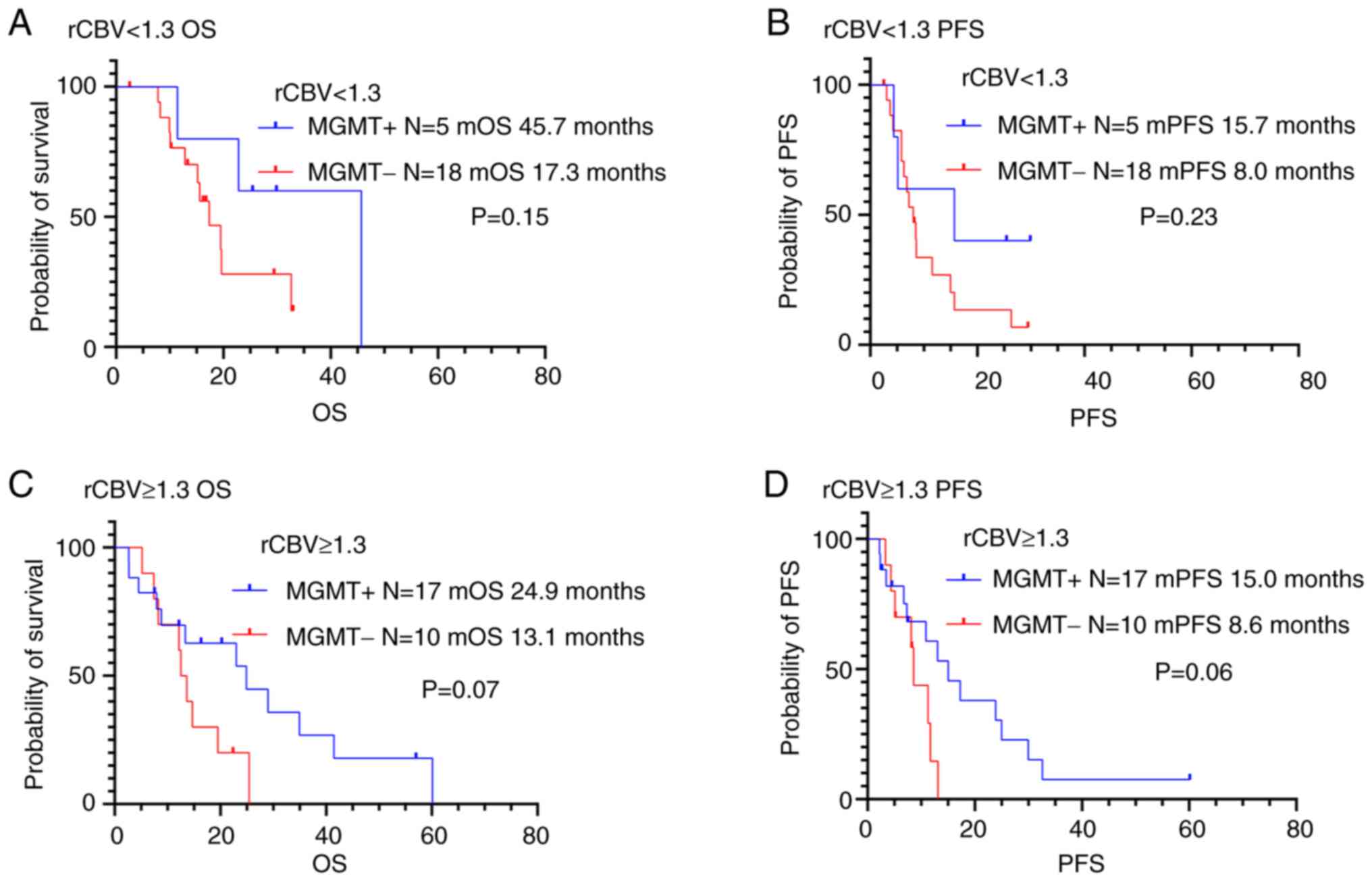
| CBV ratio
<1.3 |
|
|
|
|
|
| MGMT
methylated | 5 | 45.7 | 0.15 | 15.7 | 0.23 |
| MGMT
unmethylated | 18 | 17.3 |
| 8.0 |
|
| CBV ratio ≥1.3 |
|
|
|
|
|
| MGMT
methylated | 17 | 24.9 | 0.07 | 15.0 | 0.06 |
| MGMT
unmethylated | 10 | 13.1 |
| 8.6 |
|
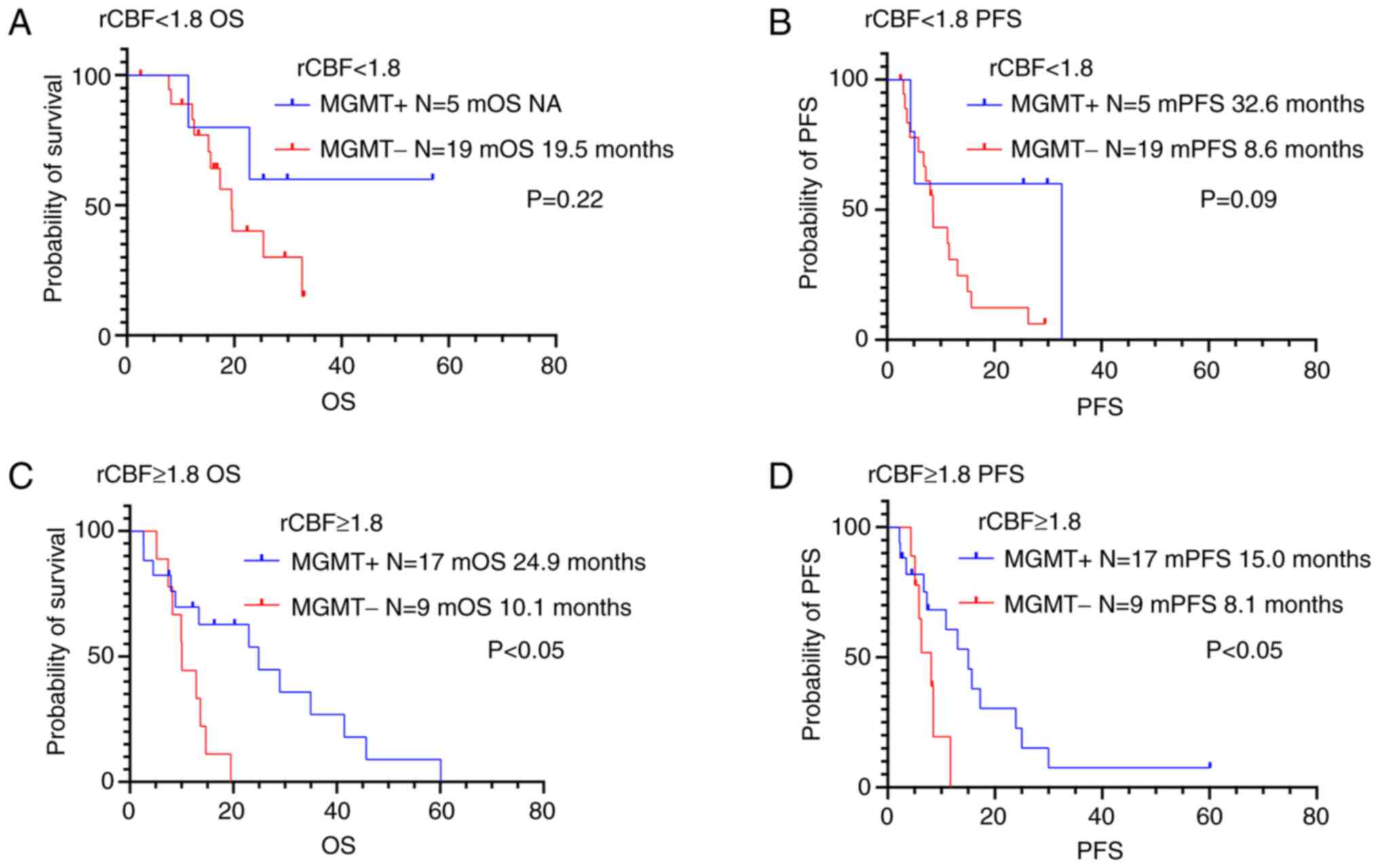
| CBF ratio
<1.8 |
|
|
|
|
|
| MGMT
methylated | 5 | NA | 0.22 | 32.6 | 0.09 |
| MGMT
unmethylated | 19 | 19.5 |
| 8.6 |
|
| CBF ratio ≥1.8 |
|
|
|
|
|
| MGMT
methylated | 17 | 24.9 | 0.01 | 15.0 | 0.04 |
| MGMT
unmethylated | 9 | 10.1 |
| 8.1 |
|
Prognosis according to perfusion MRI
parameters
In contrast, there was no association between
perfusion MRI parameters and OS or PFS in patients with
glioblastoma (Fig. 5 and Table IV).
Prognosis according to MGMT
methylation status and perfusion MRI parameters
The study investigated the significance in PFS and
OS differences between the following two groups: low vascularity
tumors with MGMT methylation and low vascularity tumors
without MGMT methylation (Figs.
6, 7 and Table IV). Juan-Albarracín et al
reported that significant differences were observed in the
Kaplan-Meier estimated survival functions for populations divided
based on the median rCBV and rCBF (29). They indicated that the median rCBV
and rCBF were found to be the relevant prognostic markers in
patients with glioblastoma. Previous studies assessed the combined
role of tumor vascularity, estimated from perfusion MRI, and
MGMT methylation status on OS in patients with glioblastoma
(17,23). The classification of tumor
vascularity was based on the median rCBV and rCBF values reported
by Juan-Albarracín' et al (17,23).
We validated thresholds calculated from the current study cohort
based on previous reports (17,23,29)
and defined the vascular groups using the median rCBV and rCBF.
There was no significant association between MGMT
methylation status and prognosis in patients with low vascularity
tumors (rCBV <1.3 or rCBF <1.8). We also evaluated
differences in PFS and OS in high vascularity tumors (rCBV ≥1.3 or
rCBF ≥1.8) with methylated and unmethylated MGMT promoters.
There was no association between MGMT methylation status and
OS or PFS in patients with high rCBV (rCBV ≥1.3). On the other
hand, high vascularity tumors (rCBF ≥1.8) with MGMT
methylation were associated to longer OS and PFS compared to those
without MGMT methylation (P<0.05).
Discussion
Our study indicates that CBV and CBF can be used to
predict the MGMT methylation status in glioblastomas.
According to our results, the rCBV and rCBF in tumors with
MGMT methylation were higher than those in tumors without
MGMT methylation. The possibility of predicting the
MGMT methylation status from DSC-MRI using a radiological
approach remains controversial. In previous reports, the CBV
derived from DSC-MRI of glioblastomas with a methylated MGMT
promoter were reported to be lower than those corresponding to
glioblastomas with unmethylated MGMT (8,10,16).
In contrast, other reports have indicated that CBV does not differ
significantly between tumors with methylated and unmethylated
MGMT (17,18). Using stereotactic image-based
histological validation, Song et al reported that CBF showed
no statistically significant differences between gliomas with and
without MGMT promoter methylation (30). Perfusion parameters are influenced
by the location of the tumor in relation to major blood vessels,
heterogeneous vascularization of the tumor, tumor necrosis, and
intratumoral cystic changes. The DSC-MR perfusion technique is
known to be affected by the partial volume effect caused by
adjacent tissues. Contouring ROI, excluding necrosis and proximate
vascular structures, reduces the partial volume effect caused by
adjacent tissues. As mentioned in previous reports (16,25,26),
ROIs were drawn to avoid calcification, blood products, dense bone,
or large vessels to ensure the accuracy of the measurements. The
size of the ROI for the solid part in our study was smaller than
that in previous studies (16,25,26).
As glioblastomas are heterogeneous tumors, the ROIs in our study
were accurately set on the solid part, which contained only
enhancing tumor core lesions in each tumor region (Fig. 1) to exclude the effect of tumor
heterogeneity. Therefore, rCBV and rCBF may be affected by the
definition of the size of ROI. There is a possibility that the
prediction of MGMT methylation status could be heavily
affected by the method used for ROI design. However, it is still
unclear whether rCBV and rCBF were affected by the small ROI or the
MGMT methylation. Although it is desirable to perform a
regression analysis to clarify whether ROI or MGMT
methylation factors were corrected, this makes it very difficult to
perform the mentioned analysis, since multiple ROIs or ROIs of
different sizes were not set throughout the course of our
study.
Meanwhile, Hegi et al have suggested that the
methylation status of the MGMT promoter may have prognostic
value and, additionally, may be a clinically relevant predictor of
the benefit of TMZ chemotherapy (1). HIF-1 was discovered as a molecular
target associated with intratumoral hypoxia (31). As previously demonstrated, HIF-1α
silencing dramatically increases sensitivity to TMZ in vivo
(32). Tang et al showed
that the inhibition of HIF-1α through knock-down sensitizes glioma
cells to TMZ, with a decrease in MGMT expression (33). Persano er al. showed that HIF-1α
suppression promotes the downregulation of MGMT, and this is
sufficient to override glioblastoma resistance to TMZ (34). In the present study, glioblastomas
with MGMT promoter methylation showed higher rCBV and rCBF
than those without. Glioblastomas with maintained perfusion and
oxygenation levels may have suppressed HIF-1α expression and
downregulated MGMT expression, and may be susceptible to TMZ
treatment.
In contrast, whether MRI perfusion parameters
correlate with the prognosis of glioblastoma remains controversial.
Previous studies have shown that CBV (19–22,35)
and CBF (36) have prognostic
value. However, no significant association between overall survival
time and CBV has been reported in previous studies (37,38).
The prognostic correlation between CBV and MGMT methylation
status may be influenced by conditions such as tumor vascularity
and treatment-induced changes over time. Previous studies have
shown a highly significant impact of MGMT status on the
prognosis of patients with moderately vascularized tumors, but not
in patients with highly vascularized tumors (17,23).
Goldman et al reported that treatment-induced changes in CBV
affect the prognosis of glioblastoma (24). They reported that glioblastomas that
showed stable or increasing CBV following chemoradiotherapy were
associated to a significantly improved PFS compared to those with
decreased CBV following chemoradiotherapy, particularly in those
exhibiting MGMT methylation (24). Batchelor et al found that
patients with glioblastoma treated with chemoradiotherapy plus
cediranib demonstrated an increase in perfusion and significantly
improved survival compared to patients treated with
chemoradiotherapy alone. This effect may be due to anti-angiogenic
therapy, normalization of blood flow, and enhancement of drug
delivery (39). It has been assumed
that the methylation of the MGMT promoter induced by the
maintained CBV and improved oxygenation enhanced the therapeutic
benefits of alkylating agents. In our study, highly vascularized
tumors based on rCBF with MGMT methylation were associated
to longer OS and PFS than those without MGMT methylation.
High CBF tumors may be less hypoxic, leading to MGMT
promoter methylation, and improved prognosis with TMZ treatment.
The failure to observe a significant difference in OS with and
without MGMT methylation can be attributed to the small
sample size, which reduced the power (40,41).
This can be seen from the P-value of 0.06, which is very close to
the significance level. Conversely, even with such a low detection
power, a significant difference in OS can be confirmed between
patients with and without MGMT methylation who have more
highly vascular tumors (rCBF ≥1.8), which may suggest the idea that
MGMT methylation status has a stronger effect on OS in cases
with higher rCBF. Radiological diagnosis using rCBV and rCBF has
the potential to predict MGMT methylation status
preoperatively, without reliance on surgical specimens. In our
study, there was no association between perfusion MRI parameters
and OS or PFS in patients with glioblastoma. Furthermore, there was
no significant association between MGMT methylation status
and prognosis in patients with lower vascularity tumors based on
both the rCBV and the rCBF and those with more highly vascularized
tumors based on rCBV. The measurement of cerebral blood perfusion
in DSC-MRI is based on the assumption that gadolinium-based
contrast agents do not cross the blood-brain barrier. CBV is
calculated by the tissue signal change caused by the
gadolinium-based contrast agent and the arterial input function.
Based on the assumption that the gadolinium-based contrast agents
do not cross the blood-brain barrier, the CBV changes caused by the
gadolinium-based contrast agents are thought to be due to the
gadolinium-based contrast agent stored in the capillaries. However,
this assumption does not hold in glioblastoma tumor tissues where
the blood-brain barrier has been disrupted. The value of the CBV
calculated by the model described above is ambiguous (42). Conversely, CBF is calculated by
dividing CBV by MTT (CBF=CBV/MTT), where MTT is the time taken for
the tracer to pass through the region of interest. Thus, CBF
compensates somewhat for the blood-brain barrier breakdown. It is
possible that calculated values with such technical ‘corrections’
more sensitively reflect tumor characteristics. It is not certain
that the combination of MRI perfusion parameters with MGMT
methylation status can be used to predict the prognosis of
glioblastomas. We are skeptical that the combination of perfusion
MRI parameters with MGMT methylation status can be used to
predict the prognosis of glioblastomas.
A few limitations and caveats in the current study
should be noted and addressed. As previously mentioned, this study
was limited by its small sample size, leading to potential bias in
our results. First, as the ROIs in our study were accurately set on
the solid part in each enhancing tumor region to exclude the effect
of tumor heterogeneity while avoiding areas of cyst formation,
hemorrhage and large vessels, the size of the ROIs was smaller than
that in previous studies (16,25,26).
Therefore, there is a possibility that the prediction for
MGMT methylation status may have been heavily affected by
the method used for ROI design. Second, while the methylation
status of the MGMT promoter may have prognostic value, there
was no significant difference in the OS of patients with and
without MGMT methylation in our study. Future large-scale
studies are required to validate the proposed prognostic value of
CBF and MGMT methylation status.
In conclusion, we aimed to evaluate whether DSC-MRI
could be employed as a non-invasive method to predict MGMT
methylation status and prognosis in newly diagnosed glioblastoma
patients. Our study indicates that rCBV and rCBF can be used to
predict the MGMT methylation status preoperatively, offering
the possibility to change clinical management in patients affected
by glioblastoma. However, we are not certain that the combination
of MRI perfusion parameters with MGMT methylation status can
be used to predict prognosis in these patients.
Acknowledgements
Not applicable.
Funding
This work was supported by Japan Science and Technology Agency
(grant no. JPMJPF2009).
Availability of data and materials
The data generated in the present study are not
publicly available due to them containing information that could
compromise research participant privacy/consent but may be
requested from the corresponding author.
Authors' contributions
YO conceived and designed the study. DC, YO, RU,
HiK, RH, NoK, NaK, YK and HaK acquired the data. DC, YO and SY
analyzed and interpreted the data and drafted the manuscript. AA
and NT contributed to the methodology for radiological analysis. DC
and YO confirmed the authenticity of all the raw data. All authors
provided critical revision of the manuscript for important
intellectual content. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was conducted according to the guidelines
of the Declaration of Helsinki and approved by the Ethics Committee
of Osaka University Hospital (approval no. 22302). Written informed
consent was obtained from all patients.
Patient consent for publication
Patients provided written informed consent for
publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hegi ME, Diserens AC, Gorlia T, Hamou MF,
de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani
L, et al: MGMT gene silencing and benefit from temozolomide in
glioblastoma. N Engl J Med. 352:997–1003. 2005. View Article : Google Scholar
|
|
2
|
Ahn SS, Shin NY, Chang JH, Kim SH, Kim EH,
Kim DW and Lee SK: Prediction of methylguanine methyltransferase
promoter methylation in glioblastoma using dynamic
contrast-enhanced magnetic resonance and diffusion tensor imaging.
J Neurosurg. 121:367–373. 2014. View Article : Google Scholar
|
|
3
|
Doniselli FM, Pascuzzo R, Agro M, Aquino
D, Anghileri E, Farinotti M, Pollo B, Paterra R, Cuccarini V,
Moscatelli M, et al: Development of A Radiomic Model for MGMT
Promoter Methylation Detection in Glioblastoma Using Conventional
MRI. Int J Mol Sci. 25:1382023. View Article : Google Scholar
|
|
4
|
Drabycz S, Roldan G, de Robles P, Adler D,
McIntyre JB, Magliocco AM, Cairncross JG and Mitchell JR: An
analysis of image texture, tumor location, and MGMT promoter
methylation in glioblastoma using magnetic resonance imaging.
Neuroimage. 49:1398–1405. 2010. View Article : Google Scholar
|
|
5
|
Han Y, Yan LF, Wang XB, Sun YZ, Zhang X,
Liu ZC, Nan HY, Hu YC, Yang Y, Zhang J, et al: Structural and
advanced imaging in predicting MGMT promoter methylation of primary
glioblastoma: A region of interest based analysis. BMC Cancer.
18:2152018. View Article : Google Scholar
|
|
6
|
Kanas VG, Zacharaki EI, Thomas GA, Zinn
PO, Megalooikonomou V and Colen RR: Learning MRI-based
classification models for MGMT methylation status prediction in
glioblastoma. Comput Methods Programs Biomed. 140:249–257. 2017.
View Article : Google Scholar
|
|
7
|
Sanada T, Kinoshita M, Sasaki T, Yamamoto
S, Fujikawa S, Fukuyama S, Hayashi N, Fukai J, Okita Y, Nonaka M,
et al: Prediction of MGMT promotor methylation status in
glioblastoma by Contrast-enhanced T1-weighted intensity image.
Neurooncol Adv. 6:vdae0162024.
|
|
8
|
Ozturk K, Soylu E and Cayci Z: Correlation
between dynamic susceptibility contrast perfusion MRI and genomic
alterations in glioblastoma. Neuroradiology. 63:1801–1810. 2021.
View Article : Google Scholar
|
|
9
|
Paech D, Windschuh J, Oberhollenzer J,
Dreher C, Sahm F, Meissner JE, Goerke S, Schuenke P, Zaiss M,
Regnery S, et al: Assessing the predictability of IDH mutation and
MGMT methylation status in glioma patients using
relaxation-compensated multipool CEST MRI at 7.0 T. Neuro Oncol.
20:1661–1671. 2018. View Article : Google Scholar
|
|
10
|
Ryoo I, Choi SH, Kim JH, Sohn CH, Kim SC,
Shin HS, Yeom JA, Jung SC, Lee AL, Yun TJ, et al: Cerebral blood
volume calculated by dynamic susceptibility contrast-enhanced
perfusion MR imaging: Preliminary correlation study with
glioblastoma genetic profiles. PLoS One. 8:e717042013. View Article : Google Scholar
|
|
11
|
Shen N, Zhang S, Cho J, Li S, Zhang J, Xie
Y, Wang Y and Zhu W: Application of cluster analysis of time
evolution for magnetic resonance Imaging-derived oxygen extraction
fraction mapping: A Promising strategy for the genetic profile
prediction and grading of glioma. Front Neurosci. 15:7368912021.
View Article : Google Scholar
|
|
12
|
Do DT, Yang MR, Lam LHT, Le NQK and Wu YW:
Improving MGMT methylation status prediction of glioblastoma
through optimizing radiomics features using genetic algorithm-based
machine learning approach. Sci Rep. 12:134122022. View Article : Google Scholar
|
|
13
|
Li ZC, Bai H, Sun Q, Li Q, Liu L, Zou Y,
Chen Y, Liang C and Zheng H: Multiregional radiomics features from
multiparametric MRI for prediction of MGMT methylation status in
glioblastoma multiforme: A multicentre study. Eur Radiol.
28:3640–3650. 2018. View Article : Google Scholar
|
|
14
|
Sasaki T, Kinoshita M, Fujita K, Fukai J,
Hayashi N, Uematsu Y, Okita Y, Nonaka M, Moriuchi S, Uda T, et al:
Radiomics and MGMT promoter methylation for prognostication of
newly diagnosed glioblastoma. Sci Rep. 9:144352019. View Article : Google Scholar
|
|
15
|
Xi YB, Guo F, Xu ZL, Li C, Wei W, Tian P,
Liu TT, Liu L, Chen G, Ye J, et al: Radiomics signature: A
potential biomarker for the prediction of MGMT promoter methylation
in glioblastoma. J Magn Reson Imaging. 47:1380–1387. 2018.
View Article : Google Scholar
|
|
16
|
Lu J, Li X and Li H: Perfusion parameters
derived from MRI for preoperative prediction of IDH mutation and
MGMT promoter methylation status in glioblastomas. Magn Reson
Imaging. 83:189–195. 2021. View Article : Google Scholar
|
|
17
|
Fuster-Garcia E, Lorente Estelles D,
Alvarez-Torres MDM, Juan-Albarracin J, Chelebian E, Rovira A,
Acosta CA, Pineda J, Oleaga L, Molla-Olmos E, et al: MGMT
methylation may benefit overall survival in patients with
moderately vascularized glioblastomas. Eur Radiol. 31:1738–1747.
2021. View Article : Google Scholar
|
|
18
|
Moon WJ, Choi JW, Roh HG, Lim SD and Koh
YC: Imaging parameters of high grade gliomas in relation to the
MGMT promoter methylation status: The CT, diffusion tensor imaging,
and perfusion MR imaging. Neuroradiology. 54:555–563. 2012.
View Article : Google Scholar
|
|
19
|
Bonekamp D, Deike K, Wiestler B, Wick W,
Bendszus M, Radbruch A and Heiland S: Association of overall
survival in patients with newly diagnosed glioblastoma with
contrast-enhanced perfusion MRI: Comparison of intraindividually
matched T1-and T2 (*)-based bolus techniques. J Magn Reson Imaging.
42:87–96. 2015. View Article : Google Scholar
|
|
20
|
Hirai T, Murakami R, Nakamura H, Kitajima
M, Fukuoka H, Sasao A, Akter M, Hayashida Y, Toya R, Oya N, et al:
Prognostic value of perfusion MR imaging of high-grade
astrocytomas: Long-term Follow-up study. AJNR Am J Neuroradiol.
29:1505–1510. 2008. View Article : Google Scholar
|
|
21
|
Jain R, Poisson L, Narang J, Gutman D,
Scarpace L, Hwang SN, Holder C, Wintermark M, Colen RR, Kirby J, et
al: Genomic mapping and survival prediction in glioblastoma:
Molecular subclassification strengthened by hemodynamic imaging
biomarkers. Radiology. 267:212–220. 2013. View Article : Google Scholar
|
|
22
|
Law M, Young RJ, Babb JS, Peccerelli N,
Chheang S, Gruber ML, Miller DC, Golfinos JG, Zagzag D and Johnson
G: Gliomas: Predicting time to progression or survival with
cerebral blood volume measurements at dynamic
Susceptibility-weighted Contrast-enhanced perfusion MR imaging.
Radiology. 247:490–498. 2008. View Article : Google Scholar
|
|
23
|
Alvarez-Torres MDM, Fuster-Garcia E,
Balana C, Puig J and Garcia-Gomez JM: Lack of benefit of extending
temozolomide treatment in patients with high vascular glioblastoma
with methylated MGMT. Cancers (Basel). 13:2021. View Article : Google Scholar
|
|
24
|
Goldman J, Hagiwara A, Yao J, Raymond C,
Ong C, Bakhti R, Kwon E, Farhat M, Torres C, Erickson LG, et al:
Paradoxical association between relative cerebral blood volume
dynamics following chemoradiation and increased Progression-free
survival in newly diagnosed IDH Wild-Type MGMT promoter methylated
glioblastoma with measurable disease. Front Oncol. 12:8499932022.
View Article : Google Scholar
|
|
25
|
Shen N, Zhao L, Jiang J, Jiang R, Su C,
Zhang S, Tang X and Zhu W: Intravoxel incoherent motion
Diffusion-weighted imaging analysis of diffusion and microperfusion
in grading gliomas and comparison with arterial spin labeling for
evaluation of tumor perfusion. J Magn Reson Imaging. 44:620–632.
2016. View Article : Google Scholar
|
|
26
|
Wang X, Chen XZ, Shi L and Dai JP: Glioma
grading and IDH1 mutational status: Assessment by intravoxel
incoherent motion MRI. Clin Radiol. 74:651.e7–651.e14. 2019.
View Article : Google Scholar
|
|
27
|
Okita Y, Nonaka M, Shofuda T, Kanematsu D,
Yoshioka E, Kodama Y, Mano M, Nakajima S and Kanemura Y:
(11)C-methinine uptake correlates with MGMT promoter methylation in
nonenhancing gliomas. Clin Neurol Neurosurg. 125:212–216. 2014.
View Article : Google Scholar
|
|
28
|
Sasaki T, Fukai J, Kodama Y, Hirose T,
Okita Y, Moriuchi S, Nonaka M, Tsuyuguchi N, Terakawa Y, Uda T, et
al: Characteristics and outcomes of elderly patients with diffuse
gliomas: A Multi-institutional cohort study by Kansai Molecular
Diagnosis Network for CNS tumors. J Neurooncol. 140:329–339. 2018.
View Article : Google Scholar
|
|
29
|
Juan-Albarracin J, Fuster-Garcia E,
Perez-Girbes A, Aparici-Robles F, Alberich-Bayarri A,
Revert-Ventura A, Marti-Bonmati L and Garcia-Gomez JM:
Glioblastoma: Vascular habitats detected at preoperative dynamic
susceptibility-weighted Contrast-enhanced perfusion MR imaging
predict survival. Radiology. 287:944–954. 2018. View Article : Google Scholar
|
|
30
|
Song S, Shan Y, Wang L, Cheng Y, Yang H,
Zhao G, Wang Z and Lu J: MGMT promoter methylation status shows no
effect on [(18)F]FET uptake and CBF in gliomas: A stereotactic
Image-based histological validation study. Eur Radiol.
32:5577–5587. 2022. View Article : Google Scholar
|
|
31
|
Onnis B, Rapisarda A and Melillo G:
Development of HIF-1 inhibitors for cancer therapy. J Cell Mol Med.
13:2780–2786. 2009. View Article : Google Scholar
|
|
32
|
Li L, Lin X, Shoemaker AR, Albert DH,
Fesik SW and Shen Y: Hypoxia-inducible Factor-1 inhibition in
combination with temozolomide treatment exhibits robust antitumor
efficacy in vivo. Clin Cancer Res. 12:4747–4754. 2006. View Article : Google Scholar
|
|
33
|
Tang JH, Ma ZX, Huang GH, Xu QF, Xiang Y,
Li N, Sidlauskas K, Zhang EE and Lv SQ: Downregulation of HIF-1a
sensitizes U251 glioma cells to the temozolomide (TMZ) treatment.
Exp Cell Res. 343:148–158. 2016. View Article : Google Scholar
|
|
34
|
Persano L, Pistollato F, Rampazzo E, Della
Puppa A, Abbadi S, Frasson C, Volpin F, Indraccolo S, Scienza R and
Basso G: BMP2 sensitizes glioblastoma Stem-like cells to
Temozolomide by affecting HIF-1α stability and MGMT expression.
Cell Death Dis. 3:e4122012. View Article : Google Scholar
|
|
35
|
Burth S, Kickingereder P, Eidel O, Tichy
D, Bonekamp D, Weberling L, Wick A, Low S, Hertenstein A,
Nowosielski M, et al: Clinical parameters outweigh Diffusion- and
Perfusion-derived MRI parameters in predicting survival in newly
diagnosed glioblastoma. Neuro Oncol. 18:1673–1679. 2016. View Article : Google Scholar
|
|
36
|
Gerstner ER, Zhang Z, Fink JR, Muzi M,
Hanna L, Greco E, Prah M, Schmainda KM, Mintz A, Kostakoglu L, et
al: ACRIN 6684: Assessment of tumor hypoxia in newly diagnosed
glioblastoma using 18F-FMISO PET and MRI. Clin Cancer Res.
22:5079–5086. 2016. View Article : Google Scholar
|
|
37
|
Paik W, Kim HS, Choi CG and Kim SJ:
Pre-operative perfusion skewness and kurtosis are potential
predictors of Progression-free survival after partial resection of
newly diagnosed glioblastoma. Korean J Radiol. 17:117–126. 2016.
View Article : Google Scholar
|
|
38
|
White ML, Zhang Y, Kazmi SAJ, Aizenberg M,
Shonka N, Yu F and Appiah AK: Evaluating survival in subjects with
astrocytic brain tumors by dynamic Susceptibility-weighted
perfusion MR imaging. PLoS One. 16:e02442752021. View Article : Google Scholar
|
|
39
|
Batchelor TT, Gerstner ER, Emblem KE, Duda
DG, Kalpathy-Cramer J, Snuderl M, Ancukiewicz M, Polaskova P, Pinho
MC, Jennings D, et al: Improved tumor oxygenation and survival in
glioblastoma patients who show increased blood perfusion after
cediranib and chemoradiation. Proc Natl Acad Sci USA.
110:19059–19064. 2013. View Article : Google Scholar
|
|
40
|
Willan AR and Pinto EM: The value of
information and optimal clinical trial design. Stat Med.
24:1791–1806. 2005. View Article : Google Scholar
|
|
41
|
Zhang L, Cui L and Yang B: Optimal
flexible sample size design with robust power. Stat Med.
35:3385–3396. 2016. View Article : Google Scholar
|
|
42
|
Nagahama H, Shonai T, Takashima H, Hirano
T, Suzuki J and Sakurai Y: MRI of Perfusion: Principles and
Clinical Applications. Igaku Butsuri. 36:103–109. 2016.(In
Japanese).
|