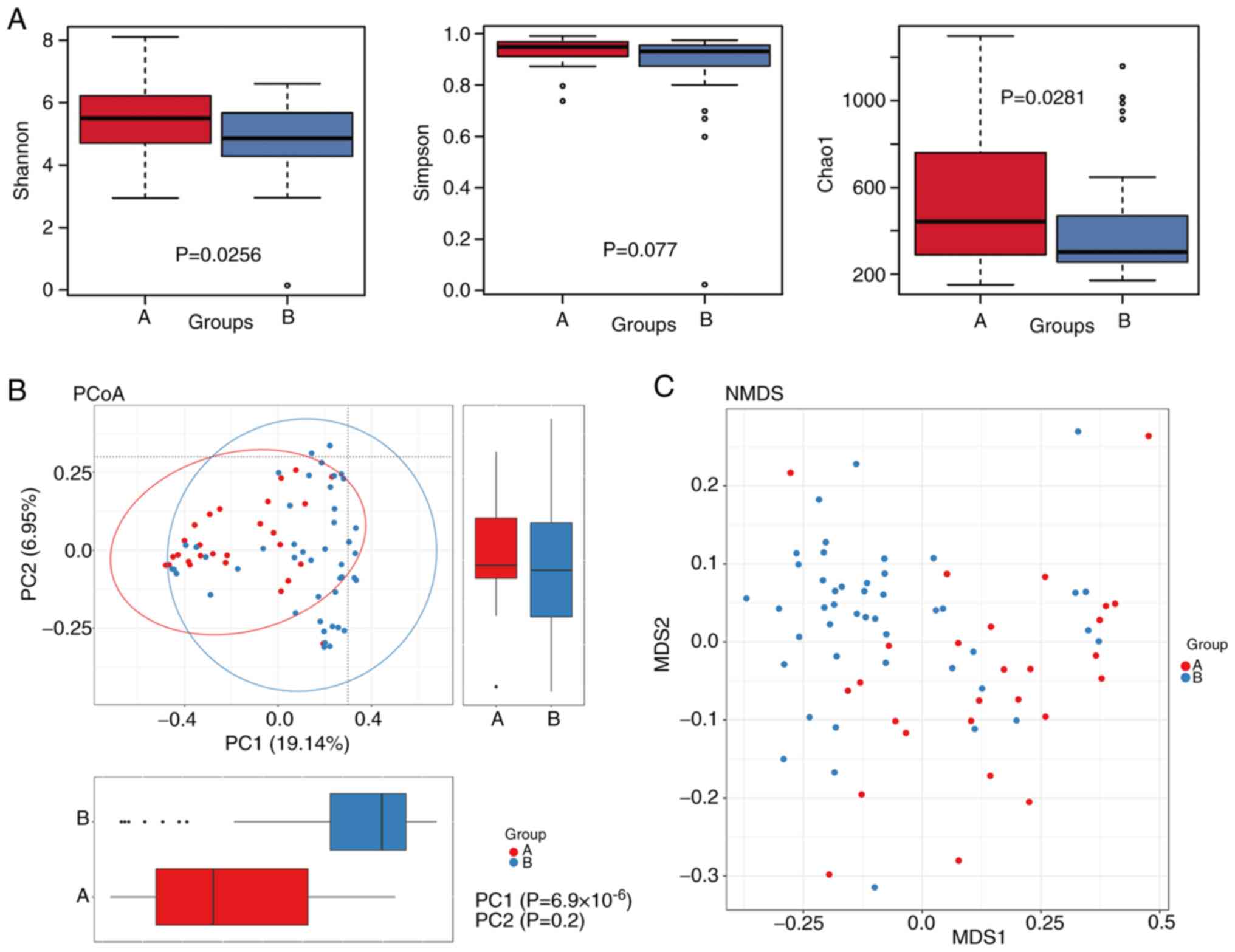
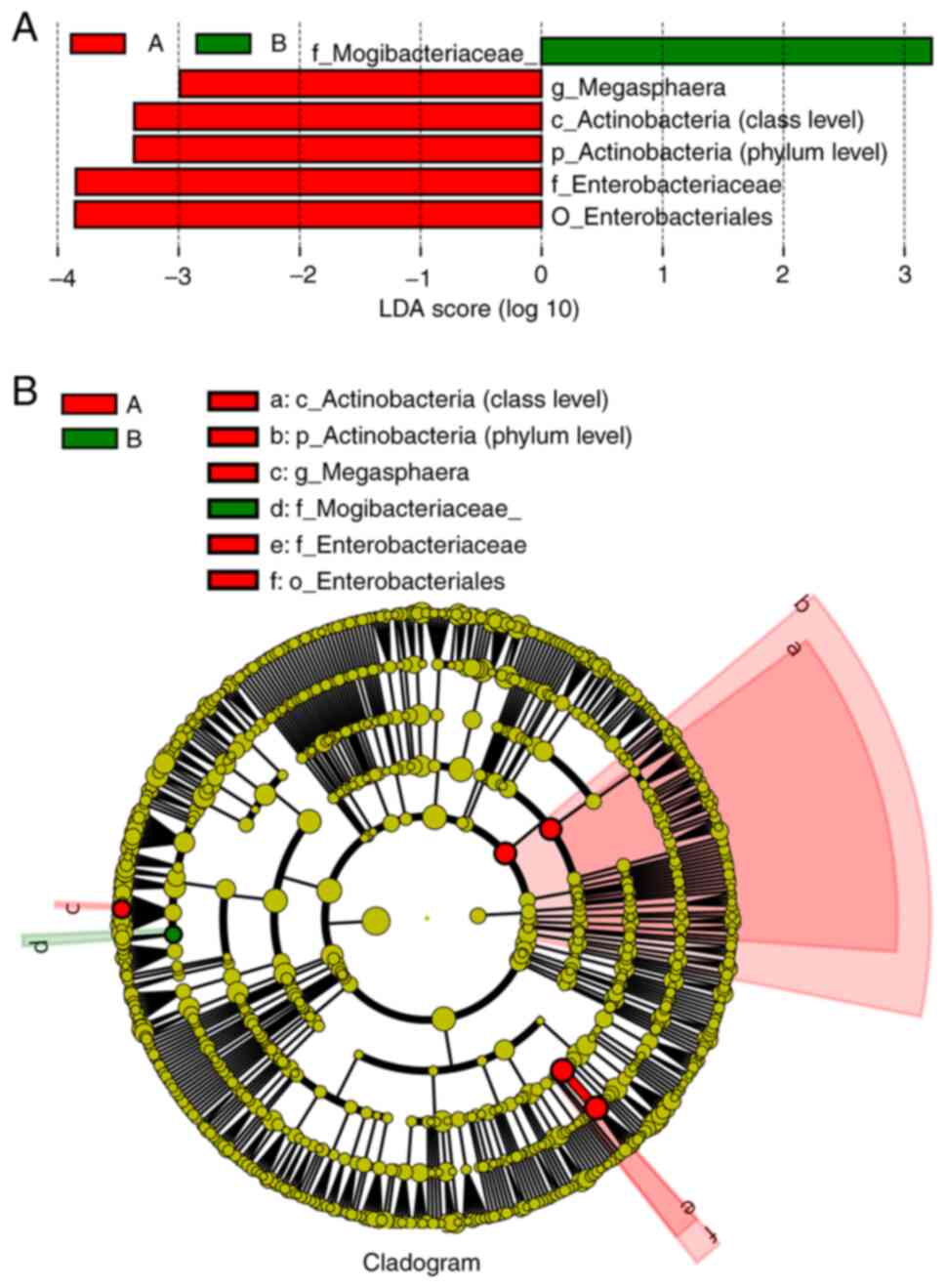
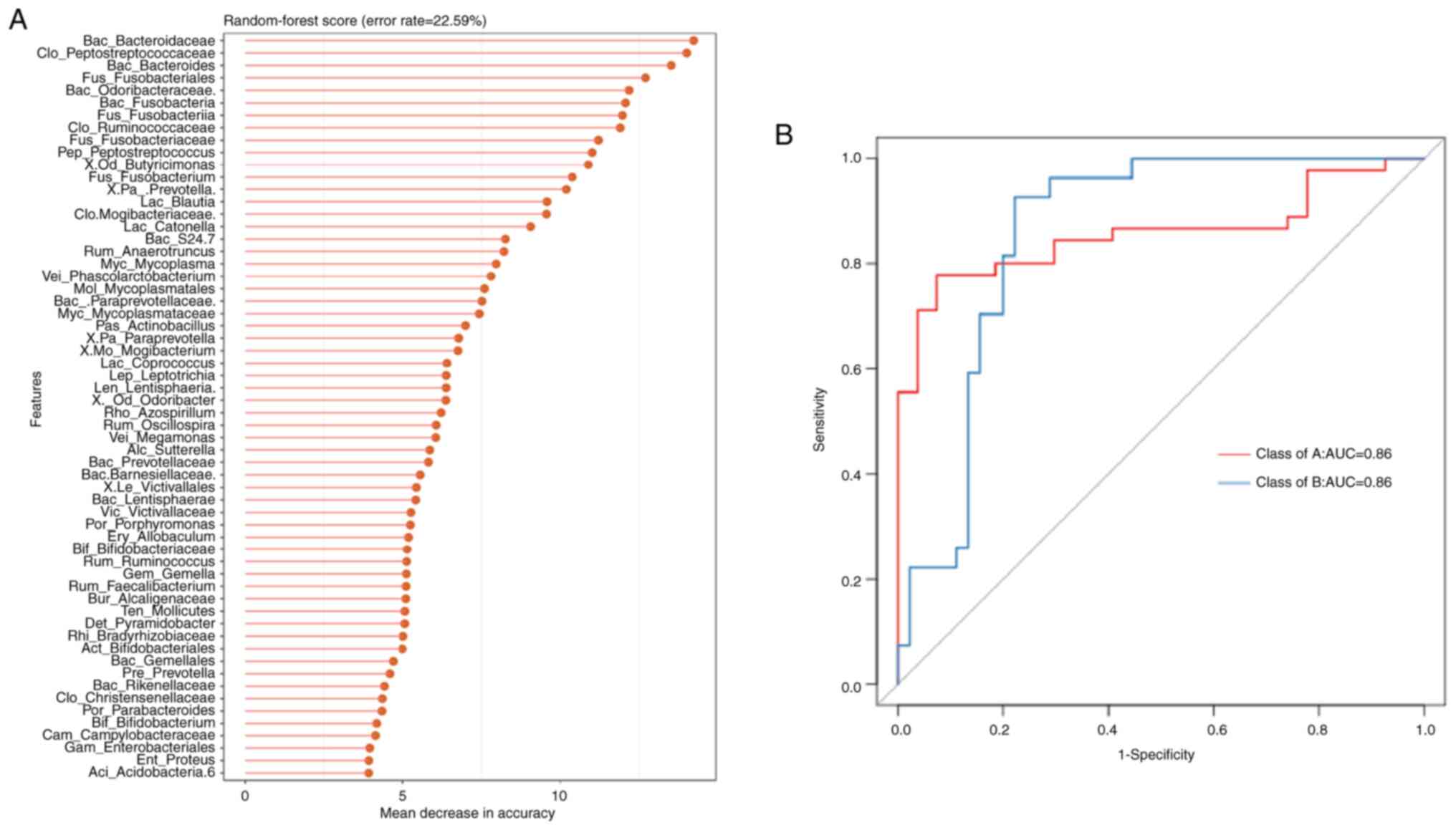
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baba Y, Iwatsuki M, Yoshida N, Watanabe M
and Baba H: Review of the gut microbiome and esophageal cancer:
Pathogenesis and potential clinical implications. Ann Gastroenterol
Surg. 1:99–104. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen R, Zheng RS, Zhang SW, Zeng HM, Wang
SM, Sun KX, Gu XY, Wei WW and He J: Analysis of incidence and
mortality of esophageal cancer in China, 2015. Zhonghua Yu Fang Yi
Xue Za Zhi. 53:1094–1097. 2019.(In Chinese). PubMed/NCBI
|
|
5
|
Wen X, Wen D, Yang Y, Chen Y, Wang G and
Shan B: Urban-rural disparity in helicobacter pylori
infection-related upper gastrointestinal cancer in China and the
decreasing trend in parallel with socioeconomic development and
urbanization in an endemic area. Ann Glob Health. 83:444–462. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Q, Rao Y, Guo X, Liu N, Liu S, Wen P,
Li S and Li Y: Oral microbiome in patients with oesophageal
squamous cell carcinoma. Sci Rep. 9:190552019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kwon YJ, Kwak HJ, Lee HK, Lim HC and Jung
DH: Comparison of bacterial community profiles from large intestine
specimens, rectal swabs, and stool samples. Appl Microbiol
Biotechnol. 105:9273–9284. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zoetendal EG, Raes J, van den Bogert B,
Arumugam M, Booijink CC, Troost FJ, Bork P, Wels M, de Vos WM and
Kleerebezem M: The human small intestinal microbiota is driven by
rapid uptake and conversion of simple carbohydrates. ISME J.
6:1415–1426. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cuevas-Sierra A, Riezu-Boj JI, Guruceaga
E, Milagro FI and Martinez JA: Sex-Specific associations between
gut prevotellaceae and host genetics on adiposity. Microorganisms.
8:9382020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
La-Ongkham O, Nakphaichit M, Nakayama J,
Keawsompong S and Nitisinprasert S: Age-related changes in the gut
microbiota and the core gut microbiome of healthy Thai humans. 3
Biotech. 10:2762020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eck A, Rutten N, Singendonk M, Rijkers GT,
Savelkoul P, Meijssen CB, Crijns CE, Oudshoorn JH, Budding AE and
Vlieger AM: Neonatal microbiota development and the effect of early
life antibiotics are determined by two distinct settler types. PLoS
One. 15:e2281332020. View Article : Google Scholar
|
|
12
|
Neckovic A, van Oorschot R, Szkuta B and
Durdle A: Investigation of direct and indirect transfer of
microbiomes between individuals. Forensic Sci Int Genet.
45:1022122020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pan HW, Du LT, Li W, Yang YM, Zhang Y and
Wang CX: Biodiversity and richness shifts of mucosa-associated gut
microbiota with progression of colorectal cancer. Res Microbiol.
171:107–114. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matsumoto H, Kuroki Y, Higashi S, Goda K,
Fukushima S, Katsumoto R, Oosawa M, Murao T, Ishii M, Oka K, et al:
Analysis of the colonic mucosa associated microbiota (MAM) using
brushing samples during colonic endoscopic procedures. J Clin
Biochem Nutr. 65:132–137. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu AQ, Vogtmann E, Shao DT, Abnet CC, Dou
HY, Qin Y, Su Z, Wei WQ and Chen W: A comparison of biopsy and
mucosal swab specimens for examining the microbiota of upper
gastrointestinal carcinoma. Cancer Epidemiol Biomarkers Prev.
28:2030–2037. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peters BA, Wu J, Pei Z, Yang L, Purdue MP,
Freedman ND, Jacobs EJ, Gapstur SM, Hayes RB and Ahn J: Oral
microbiome composition reflects prospective risk for esophageal
cancers. Cancer Res. 77:6777–6787. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Deshpande NP, Riordan SM,
Castaño-Rodríguez N, Wilkins MR and Kaakoush NO: Signatures within
the esophageal microbiome are associated with host genetics, age,
and disease. Microbiome. 6:2272018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Elliott D, Walker AW, O'Donovan M,
Parkhill J and Fitzgerald RC: A non-endoscopic device to sample the
oesophageal microbiota: A case-control study. Lancet Gastroenterol
Hepatol. 2:32–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu G, Gail MH, Shi J, Klepac-Ceraj V,
Paster BJ, Dye BA, Wang GQ, Wei WQ, Fan JH, Qiao YL, et al:
Association between upper digestive tract microbiota and
cancer-predisposing states in the esophagus and stomach. Cancer
Epidemiol Biomarkers Prev. 23:735–741. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gall A, Fero J, Mccoy C, Claywell BC,
Sanchez CA, Blount PL, Li X, Vaughan TL, Matsen FA, Reid BJ and
Salama NR: Bacterial composition of the human upper
gastrointestinal tract microbiome is dynamic and associated with
genomic instability in a Barrett's esophagus cohort. PLoS One.
10:e1290552015. View Article : Google Scholar
|
|
21
|
Okereke IC, Miller AL, Hamilton CF, Booth
AL, Reep GL, Andersen CL, Reynolds ST and Pyles RB: Microbiota of
the oropharynx and endoscope compared to the esophagus. Sci Rep.
9:102012019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fillon SA, Harris JK, Wagner BD, Kelly CJ,
Stevens MJ, Moore W, Fang R, Schroeder S, Masterson JC, Robertson
CE, et al: Novel device to sample the esophageal microbiome-the
esophageal string test. PLoS One. 7:e429382012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kageyama S, Takeshita T, Takeuchi K,
Asakawa M, Matsumi R, Furuta M, Shibata Y, Nagai K, Ikebe M, Morita
M, et al: Characteristics of the salivary microbiota in patients
with various digestive tract cancers. Front Microbiol. 10:17802019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lederer AK, Rasel H, Kohnert E, Kreutz C,
Huber R, Badr MT, Dellweg P, Bartsch F and Lang H: Gut microbiota
in diagnosis, therapy and prognosis of cholangiocarcinoma and
gallbladder carcinoma-a scoping review. Microorganisms.
11:23632023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zeng R, Gou H, Lau HC and Yu J: Stomach
microbiota in gastric cancer development and clinical implications.
Gut. 17:gutjnl-2024-332815. 2024.
|
|
26
|
Li Z, Shi C, Zheng J, Guo Y, Fan T, Zhao
H, Jian D, Cheng X, Tang H and Ma J: Fusobacterium nucleatum
predicts a high risk of metastasis for esophageal squamous cell
carcinoma. BMC Microbiol. 21:3012021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li M, Shao D, Zhou J, Gu J, Qin J, Chen W
and Wei W: Signatures within esophageal microbiota with progression
of esophageal squamous cell carcinoma. Chin J Cancer Res.
32:755–767. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rognes T, Flouri T, Nichols B, Quince C
and Mahé F: VSEARCH: A versatile open source tool for metagenomics.
PeerJ. 4:e25842016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Quast C, Pruesse E, Yilmaz P, Gerken J,
Schweer T, Yarza P, Peplies J and Glöckner FO: The SILVA ribosomal
RNA gene database project: Improved data processing and web-based
tools. Nucleic Acids Res. 41:D590–D596. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Caporaso JG, Kuczynski J, Stombaugh J,
Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich
JK, Gordon JI, et al: QIIME allows analysis of high-throughput
community sequencing data. Nat Methods. 7:335–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huson DH and Weber N: Microbial community
analysis using MEGAN. Methods Enzymol. 531:465–485. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mitra S, Stärk M and Huson DH: Analysis of
16S rRNA environmental sequences using MEGAN. BMC Genomics. 12
(Suppl 3):S172011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Asnicar F, Weingart G, Tickle TL,
Huttenhower C and Segata N: Compact graphical representation of
phylogenetic data and metadata with GraPhlAn. PeerJ. 3:e10292015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Segata N, Izard J, Waldron L, Gevers D,
Miropolsky L, Garrett WS and Huttenhower C: Metagenomic biomarker
discovery and explanation. Genome Biol. 12:R602011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Breiman L: Random forests. Machine
Learning. 45:5–32. 2001. View Article : Google Scholar
|
|
36
|
Liaw A and Wiener M: Classification and
regression by randomforest. R News. 2:18–22. 2002.
|
|
37
|
Ward T, Larson J, Meulemans J, Hillmann B,
Lynch J, Sidiropoulos D, Spear J, Caporaso G, Blekhman R, Knight R,
et al: BugBase predicts organism level microbiome phenotypes:
bioRxiv. 2:doi.org/10.1101/133462. 2017.
|
|
38
|
Thomas AM, Jesus EC, Lopes A, Aguiar S Jr,
Begnami MD, Rocha RM, Carpinetti PA, Camargo AA, Hoffmann C,
Freitas HC, et al: Tissue-associated bacterial alterations in
rectal carcinoma patients revealed by 16S rRNA community profiling.
Front Cell Infect Microbiol. 6:1792016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Human Microbiome Project Consortium, .
Structure, function and diversity of the healthy human microbiome.
Nature. 486:207–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lloyd-Price J, Abu-Ali G and Huttenhower
C: The healthy human microbiome. Genome Med. 8:512016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vuik F, Dicksved J, Lam SY, Fuhler GM, van
der Laan L, van de Winkel A, Konstantinov SR, Spaander M,
Peppelenbosch MP, Engstrand L and Kuipers EJ: Composition of the
mucosa-associated microbiota along the entire gastrointestinal
tract of human individuals. United European Gastroenterol J.
7:897–907. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rius-Rocabert S, Llinares PF, Pozuelo MJ,
Garcia A and Nistal-Villan E: Oncolytic bacteria: Past, present and
future. Fems Microbiol Lett. 366:fnz1362019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nejman D, Livyatan I, Fuks G, Gavert N,
Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E,
et al: The human tumor microbiome is composed of tumor
type-specific intracellular bacteria. Science. 368:973–980. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang H, Fu L, Leiliang X, Qu C, Wu W, Wen
R, Huang N, He Q, Cheng Q, Liu G and Cheng Y: Beyond the gut: The
intratumoral microbiome's influence on tumorigenesis and treatment
response. Cancer Commun (Lond). 44:1130–1167. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Langille MG, Zaneveld J, Caporaso JG,
Mcdonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega
Thurber RL, Knight R, et al: Predictive functional profiling of
microbial communities using 16S rRNA marker gene sequences. Nat
Biotechnol. 31:814–821. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Garcia MD, Sanabria J, Wist J and Holmes
E: Effect of operational parameters on the cultivation of the gut
microbiome in continuous bioreactors inoculated with feces: A
systematic review. J Agric Food Chem. 71:6213–6225. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang S, Cao X and Huang H: Sampling
strategies for three-dimensional spatial community structures in
IBD microbiota research. Front Cell Infect Microbiol. 7:512017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jandhyala SM, Talukdar R, Subramanyam C,
Vuyyuru H, Sasikala M and Reddy DN: Role of the normal gut
microbiota. World J Gastroenterol. 21:8787–8803. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kashiwagi S, Naito Y, Inoue R, Takagi T,
Nakano T, Inada Y, Fukui A, Katada K, Mizushima K, Kamada K, et al:
Mucosa-associated microbiota in the gastrointestinal tract of
healthy Japanese subjects. Digestion. 101:107–120. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Saffarian A, Mulet C, Regnault B, Amiot A,
Tran-Van-Nhieu J, Ravel J, Sobhani I, Sansonetti PJ and Pedron T:
Crypt- and mucosa-associated core microbiotas in humans and their
alteration in colon cancer patients. mBio. 10:e01315–e01319. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Vasapolli R, Schütte K, Schulz C, Vital M,
Schomburg D, Pieper DH, Vilchez-Vargas R and Malfertheiner P:
Analysis of transcriptionally active bacteria throughout the
gastrointestinal tract of healthy individuals. Gastroenterology.
157:1081–1092. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Richard ML and Sokol H: The gut mycobiota:
Insights into analysis, environmental interactions and role in
gastrointestinal diseases. Nat Rev Gastroenterol Hepatol.
16:331–345. 2019.PubMed/NCBI
|
|
53
|
Yuan X, Chang C, Chen X and Li K: Emerging
trends and focus of human gastrointestinal microbiome research from
2010–2021: A visualized study. J Transl Med. 19:3272021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Budding AE, Grasman ME, Eck A, Bogaards
JA, Vandenbroucke-Grauls CM, van Bodegraven AA and Savelkoul PH:
Rectal swabs for analysis of the intestinal microbiota. PLoS One.
9:e1013442014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Araujo-Perez F, Mccoy AN, Okechukwu C,
Carroll IM, Smith KM, Jeremiah K, Sandler RS, Asher GN and Keku TO:
Differences in microbial signatures between rectal mucosal biopsies
and rectal swabs. Gut Microbes. 3:530–535. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kotar T, Pirš M, Steyer A, Cerar T, Soba
B, Skvarc M, Poljsak PM and Lejko ZT: Evaluation of rectal swab use
for the determination of enteric pathogens: A prospective study of
diarrhoea in adults. Clin Microbiol Infect. 25:733–738. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Fair K, Dunlap DG, Fitch A, Bogdanovich T,
Methé B, Morris A, Mcverry BJ and Kitsios GD: Rectal swabs from
critically Ill patients provide discordant representations of the
gut microbiome compared to stool samples. mSphere. 4:e00358–e00319.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hernandez-Arriaga A, Baumann A, Witte OW,
Frahm C, Bergheim I and Camarinha-Silva A: Changes in oral
microbial ecology of C57BL/6 mice at different ages associated with
sampling methodology. Microorganisms. 7:2832019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dong L, Yin J, Zhao J, Ma SR, Wang HR,
Wang M, Chen W and Wei WQ: Microbial similarity and preference for
specific sites in healthy oral cavity and esophagus. Front
Microbiol. 9:16032018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
El-Zimaity H, Di Pilato V, Novella RM,
Brcic I, Rajendra S, Langer R, Dislich B, Tripathi M, Guindi M and
Riddell R: Risk factors for esophageal cancer: Emphasis on
infectious agents. Ann N Y Acad Sci. 1434:319–332. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shao D, Vogtmann E, Liu A, Qin J, Chen W,
Abnet CC and Wei W: Microbial characterization of esophageal
squamous cell carcinoma and gastric cardia adenocarcinoma from a
high-risk region of China. Cancer. 125:3993–4002. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Okereke I, Hamilton C, Reep G, Krill T,
Booth A, Ghouri Y, Jala V, Andersen C and Pyles R: Microflora
composition in the gastrointestinal tract in patients with
Barrett's esophagus. J Thorac Dis. 11:S1581–S1587. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li W, Huang Y, Tong S, Wan C and Wang Z:
The characteristics of the gut microbiota in patients with
pulmonary tuberculosis: A systematic review. Diagn Microbiol Infect
Dis. 109:1162912024. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Maki KA, Kazmi N, Barb JJ and Ames N: The
oral and gut bacterial microbiomes: Similarities, differences, and
connections. Biol Res Nurs. 23:7–20. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Gao S, Li S, Ma Z, Liang S, Shan T, Zhang
M, Zhu X, Zhang P, Liu G, Zhou F, et al: Presence of porphyromonas
gingivalis in esophagus and its association with the
clinicopathological characteristics and survival in patients with
esophageal cancer. Infect Agent Cancer. 11:32016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu Y, Lin Z, Lin Y, Chen Y, Peng XE, He
F, Liu S, Yan S, Huang L, Lu W, et al: Streptococcus and prevotella
are associated with the prognosis of oesophageal squamous cell
carcinoma. J Med Microbiol. 67:1058–1068. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Soroush A, Etemadi A and Abrams JA:
Non-acid fluid exposure and esophageal squamous cell carcinoma. Dig
Dis Sci. 67:2754–2762. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Song X, Greiner-Tollersrud OK and Zhou H:
Oral microbiota variation: A risk factor for development and poor
prognosis of esophageal cancer. Dig Dis Sci. 67:3543–3556. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Gao S, Zhang Z, Sun K, Li MX and Qi YJ:
Upper gastrointestinal tract microbiota with oral origin in
relation to oesophageal squamous cell carcinoma. Ann Med.
55:22954012023. View Article : Google Scholar : PubMed/NCBI
|