Introduction
Cervical cancer accounts for a considerable
proportion of gynecological malignancies. Cervical cancer ranked
fourth among female cancer-related morbidity and mortality, with
661,021 new cases and 348,189 deaths in 2022 (1). According to GLOBOCAN 2022, cases in
China accounted for 22.7% and 16.0% of global morbidity and
mortality rates, respectively, with 150,659 new cases and 55,694
deaths (2). In addition, due to the
low population coverage rate (21.4%) for cervical cytology and the
low human papillomavirus (HPV) vaccination rate, eliminating
cervical cancer is a challenging task (3). Currently, cervical cancer is treated
with a comprehensive conventional approach that includes surgery,
radiotherapy and chemotherapy (4).
However, this strategy is not effective in patients with advanced
cancer, and there is a high incidence of recurrence and metastasis.
The 5-year disease-free survival is approaches 100% for patients
with stage IA cervical cancer, 70–85% for those with stage IB1 and
smaller IIA lesions, 50–70% for those with stages IB2 and IIB,
30–50% for those with stage III, and 5–15% for those with stage IV
(5). Therefore, it is important to
elucidate the mechanisms of cervical cancer development and
identify specific tumor markers.
It has been established that the activation of
proto-oncogenes and the inactivation or mutation of tumor
suppressor genes are involved in the tumorigenesis of cancer
(6). Therefore, genes related to
cervical cancer were screened out using the Gene Expression
Profiling Interactive Analysis (GEPIA) database. CHMP4C, as a
subunit of endosomal sorting complex required for transport-III, is
essential for the Aurora B-mediated abscission checkpoint (NoCut)
and can delay the abscission time and reduce the accumulation of
DNA damage (7,8). A previous study also reported that
CHMP4C is undetectable in normal tissues but overexpressed in
ovarian cancer tissues (9).
Moreover, there are higher expression levels of CHMP4C in the
extracellular vesicles of prostate cancer with a high Gleason
score, which may promote the progression of prostate cancer
(10). However, the relationship
between CHMP4C and cervical cancer remains to be studied.
Therefore, the present study aimed to assess the role of CHMP4C in
the development of cervical cancer and evaluate the regulatory
pathways in which CHMP4C may be involved.
Materials and methods
Bioinformatics analysis
GEPIA (gepia.cancer-pku.cn/index.html) was used to
screen genes overexpressed in cervical squamous cell cancer,
developed survival curve, and automatically calculate Log-rank and
HR. Additionally, targetscan (https://www.targetscan.org/vert_72/), mirDIP
(http://ophid.utoronto.ca/mirDIP/),
and miR-Gator (http://mirgator.kobic.re.kr/), to suggest miRNA with
binding sites to target genes.
Specimens
The present study obtained 19 normal cervical
tissues, 88 cervical squamous cell carcinoma tissues and 8 cervical
adenocarcinoma tissues from hospitalized patients in the Third
Affiliated Hospital Guangzhou Medical University (Guangzhou, China)
between 2014–2019. Patients diagnosed with metastatic cervical
cancer or other malignant tumors, or with a history of radiotherapy
and chemotherapy before surgery were excluded. The patients agreed
to use the tissue removed during surgery for general scientific
research and signed an informed consent form. The Ethics Committee
of the Third Affiliated Hospital Guangzhou Medical University
approved the present study (approval no. 2019-037). The tissues
were scored according to the degree of staining and the percentage
of positively stained cells, and the final scores were added.
Cell culture
The HPV16-positive cervical cancer SiHa cell line
(National Infrastructure of Cell Line Resource) was cultured in
RPMI1640 medium (HyClone; Cytiva), and the human cervical
epithelial immortalized H8 (Jennio Biotech Co., Ltd.),
HPV16-positive cervical cancer Caski (National Infrastructure of
Cell Line Resource) and HPV18-positive cervical cancer HeLa
(Pricella®; Wuhan Elabscience Biotechnology Co., Ltd.)
cell lines were cultured in DMEM medium (HyClone; Cytiva),
containing 10% fetal bovine serum (Shanghai Excell Biological
Technology Co., Ltd.). The cells were plated in petri dishes and
cultured in an incubator at 37°C and 5% CO2
atmosphere.
Small interfering (si)RNA
transfection
siRNAs (Sigma-Aldrich; Merck KGaA) against target
genes were used to knockdown the expression of the target genes,
and Lipofectamine™ 3000 (Thermo Fisher Scientific, Inc.) was used
as a cell transfection reagent. A total of 50 nM siRNA was
transfected into SiHa and HeLa cells at 37°C for 48 h immediately
before subsequent experiment. The target genes and corresponding
siRNA sequences were as follows: CHMP4C,
5′-CAGAUUGAUGGCACACUUUdTdT-3′ (forward) and
5′-AAAGUGUGCCAUCAAUCUGdTdT-3′ (reverse); HPV16 E6,
5′-GAGUAUAGACAUUAUUGUUdTdT-3′ (forward) and
5′-AACAAUAAUGUCUAUACUCdTdT-3′ (reverse); and HPV18 E6,
5′-CAGACUCUGUGUAUGGAGAdTdT-3′ (forward) and
5′-UCUCCAUACACAGAGUCUGdTdT-3′ (reverse). The scrambled siRNA
sequences used for the control were as follows:
5′-UUCUCCGAACGUGUCACGUTTdTdT-3′ (forward) and
5′-ACGUGACACGUUCGGAGAATTdTdT-3′ (reverse).
Plasmid and mimics transfection
Full-length cDNAs of human CHMP4C were synthesized
by Guangzhou Dahong Biotechnology Co., Ltd. and cloned into the
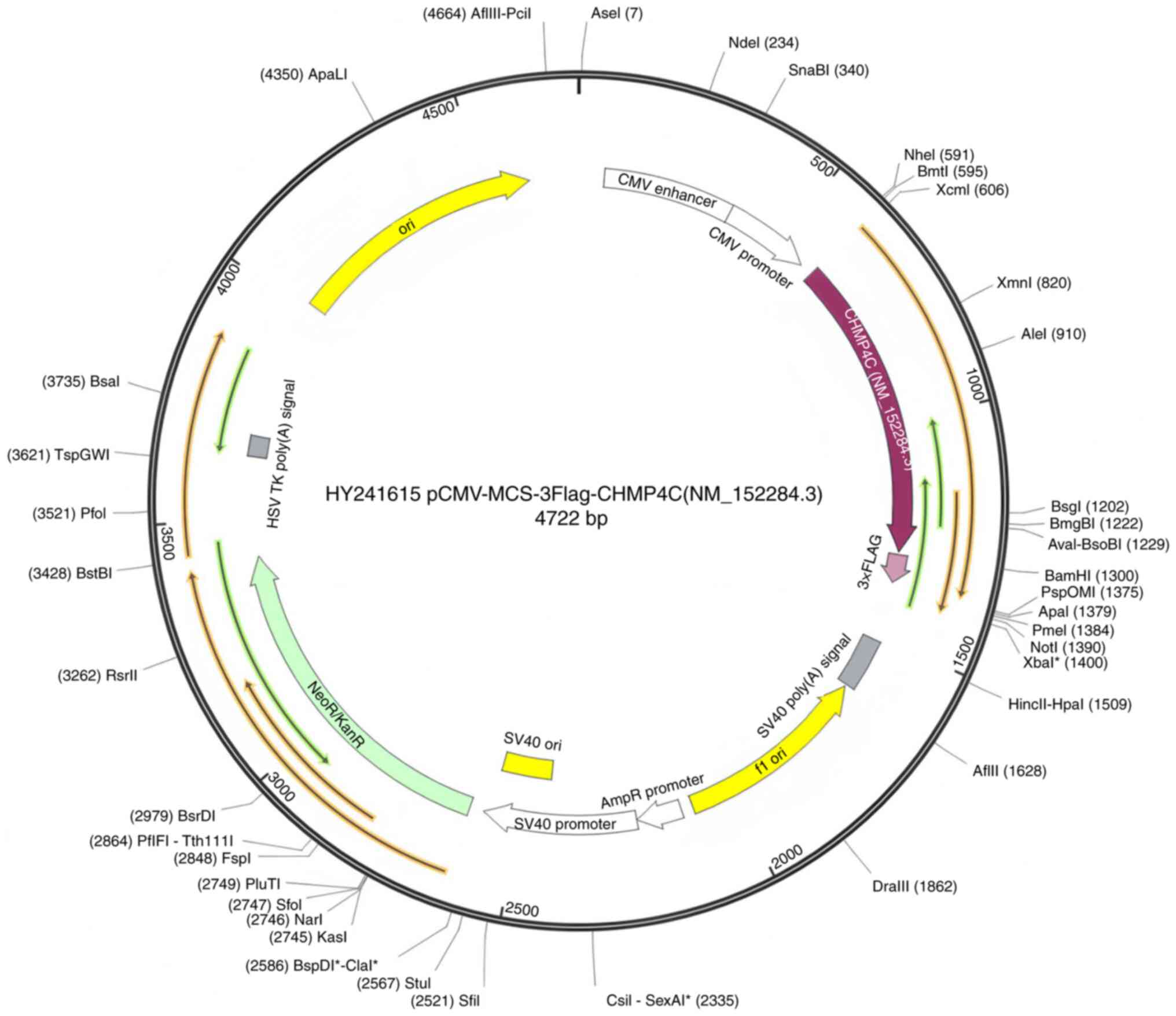
expression vector pCMV-MCS-3Flag (Guangzhou Dahong Biotechnology
Co., Ltd) (Fig. 1). When cell
confluence reached 80–90%, the plasmids were transfected using
Lipofectamine 3000 and p3000 reagents (Thermo Fisher Scientific,
Inc.), following the manufacturer's protocol. A total of 50 nM
microRNA (miRNA/miR) and negative control mimics (Guangzhou RiboBio
Co., Ltd.) were transfected into SiHa and HeLa cells using
Lipofectamine 3000 reagent. A total of 48 h after transfection at
37°C, the transfection efficiency was immediately assessed using
reverse transcription-quantitative PCR (RT-qPCR). The sequences of
the miR-543 mimic were 5′-AAACAUUCGCGGUGCACUUCUU-3′ (sense),
5′-UUUGUAAGCGCCACGUGAAGAA-3′ (antisense). The sequences of the
negative control mimic were 5′-UUUGUACUACACAAAAGUACUG-3′ (sense),
5′-AAACAUGAUGUGUUUUCAUGAC-3′ (antisense).
MTT assay
After the SiHa and HeLa cells were digested and
collected, 3×103 cells and 100 µl medium were added to
each well in a 96-well plate. After the cells had adhered, the
experimental group was transfected with CHMP4C siRNA, whereas the
negative control group was transfected with scramble siRNA. A total
of 20 µl MTT solution was then added to each well, followed by
incubation at 37°C for 4 h. The liquid was then aspirated from the
well, and 150 µl dimethyl sulfoxide was added. Finally, a
microplate reader (BioTek; Agilent Technologies, Inc.) was used to
measure the optical density value of each well at 490 nm.
Wound-healing assay
SiHa and HeLa cells were inoculated in a six-well
plate. When the cell density reached ~80%, a 200 µl pipette tip was
used to scratch the cells, and then serum-free medium was added.
Images of the scratches were captured under a microscope (Olympus
IX73; fluorescence microscope; Olympus Corporation and Leica DMi1;
light microscope; Leica Microsystems GmbH) and then processed using
cellSens Version 2.1 (Olympus Corporation) and Leica Application
Suite X Version 4.12 software (Leica Microsystems GmbH). The wound
area was measured using Image J software Version 1.52a (National
Institutes of Health). The following formula was used to assess the
level of wound-healing: Mobility=(starting area-final
area)/starting area ×100%.
Apoptosis assay
The transfected SiHa and HeLa cells were collected
using EDTA-free trypsin and washed once with PBS. The cells were
resuspended in 100 µl binding buffer (1X) and 5 µl Annexin
V-FITC/PI staining solution (BD Biosciences) and placed in the
dark. After the addition of 400 µl binding buffer (1X) to each
tube, apoptosis was detected using flow cytometry (BD FACScanto™ II
Clinical Flow Cytometry System; BD Biosciences, and Attune NxT Flow
Cytometer; Thermo Fisher Scientific, Inc.). FlowJo software
(version 10.8; BD Biosciences) and Attune NxT Flow Cytometry
Software (version 2.6; Thermo Fisher Scientific, Inc.) were used
for analysis.
Cell invasion assay
Matrigel (BD Bioscience) was diluted at a 1:15 ratio
with cold serum-free medium (RPMI-1640 or DMEM; HyClone; Cytiva)
refrigerated at 4°C and 30 µl was plated into the filters of the
Transwell chambers (Corning, Inc.). Then the chambers were placed
in a 24-well plate in a 37°C incubator for 4 h. Cells were
suspended at a density of 4×104 in 200 µl serum-free
medium (RPMI1640 or DMEM, HyClone; Cytiva) and plated in the upper
chamber, whilst 600 µl medium supplemented with 10% fetal bovine
serum (Shanghai Excell Biological Technology Co., Ltd. Bio) was
added to the lower chamber. After incubation at 37°C for 48 h, the
chamber was fixed with 4% formaldehyde for 30 min and stained with
1% crystal violet for 15 min both at room temperature. Photographs
were captured under a microscope (Olympus IX73, fluorescence
microscope, Olympus Corporation), and the invasive cells were
manually counted. Each experiment was performed in triplicate, and
images of 3 fields of view were captured for each replicate.
Immunohistochemistry
The normal and cervical cancer tissues were fixed in
4% paraformaldehyde (at room temperature for 24 h, embedded in
paraffin wax, and cut into 4 mm slices. The paraffin-embedded
tissue sections were heated in a 65°C incubator for 4 h and dewaxed
with xylene (Foshan Bodi Chemical Reagent Co., Ltd) for 20 min
twice at room temperature. Then they were immersed into 100%
alcohol (Foshan Bodi Chemical Reagent Co., Ltd) for 5 min, 95%
alcohol for 2 min, 85% alcohol for 2 min, 75% alcohol for 1 min,
and finally turned into distilled water for 3 min 3 times, so as to
fully hydrate. The sections were immersed in an EDTA antigen
retrieval solution and heated in a pressure cooker for 5 min with
the cover unlocked. Then the cooker was locked, removed from the
heat source 3 min after the pressure reaches the highest. After
then, the sections allowed to cool at room temperature. After
inactivation of endogenous catalase activity using 3% hydrogen
peroxide, the sections were blocked with 5% BSA (BEIJING SOLARBIO
Technology Co., Ltd.) in a 37°C incubator for 40 min. The sections
were then incubated with anti-CHMP4C antibodies (1:400; #GTX122876;
GeneTex, Inc.) overnight at 4°C. The following day, the sections
were re-warmed at room temperature for 30 min and washed three
times with PBS. Horseradish peroxidase-conjugated anti-rabbit
antibodies (1:800; cat. no. ab97051; Abcam) were then added and the
sections were incubated for 45 min in a 37°C incubator. After
diaminobenzidine coloration and hematoxylin staining at room
temperature for 10 min, the sections were dehydrated, sealed with a
neutral resin and imaged under a microscope (Leica Microsystems
GmbH).
Dual-luciferase reporter assay
As the presence of a binding site between CHMP4C and
miR-543 was predicted (data not shown), a CHMP4C 3′-untranslated
region (3′-UTR) containing a miR-543-binding site sequence was
designed. 293T cells (Pricella®; Wuhan Elabscience
Biotechnology Co., Ltd.) were seeded into 24-well plates and
transfected with 600 ng SV40-dual-luciferase vector (Shanghai
Genechem Co., Ltd.) expressing a wild type CHMP4C 3′-UTR using
Lipofectamine™ 3000 (Thermo Fisher Scientific, Inc.), whilst the
mutant type acted as a control. At the same time, 50 nM miR-543 or
negative mimics (Shanghai Genechem Co., Ltd.) were co-transfected.
The sequence of the miR-543 mimics were
5′-AAACAUUCGCGGUGCACUUCUU-3′ (sense) and
5′-AAGAAGUGCACCGCGAAUGUUU-3′ (antisense). The sequences of the
negative control mimic were 5′-UUUGUACUACACAAAAGUACUG-3′ (sense),
5′-AAACAUGAUGUGUUUUCAUGAC-3′ (antisense). After 48 h, the
Dual-Luciferase® Reporter Assay System (Promega
Corporation) was used to detect the level of luciferase activity.
The relative luciferase intensity was normalized to renilla
luciferase activity.
RT-qPCR
Total RNA was extracted from SiHa and HeLa cervical
cancer cells using the TRIzol reagent (Takara Bio, Inc.). After
separation, precipitation, washing and dissolution, the
concentration of RNA was detected using a spectrophotometer [Unico
(Shanghai) Instrument Co., Ltd.]. For mRNA analysis, 2 µg total RNA
was reverse-transcribed to synthesize complementary (c)DNA using
random primers and avian myeloblastosis virus transcriptase
(PrimeScript™ RT reagent Kit (Perfect Real Time); Takara Bio,
Inc.). The reverse transcription program was set at 37°C for 15
min, 85°C for 5 min, and finally cooled to 4°C. qPCR amplification
of the cDNA was then performed using the SYBR® Premix Ex
Taq™ II kit (Takara Bio, Inc.) with 18S as an internal control. The
amplification conditions were as follows: Initial denaturation at
95°C for 30 s, followed by 40 cycles of 95°C for 5 s and an 60°C
for 30 sec. The Hairpin-it™ miRNAs qPCR Quantitation Kit (Shanghai
GenePharma Co., Ltd.) was used to detect miRNA expression according
to the manufacturer's protocols, and U6 was used as the internal
reference. The relative expression of CHMP4C or miR-543 was
calculated using the 2−ΔΔCq method (11). The sequences of the primers used
were as follows: CHMP4C, 5′-GAGAAGCCCTGGAGAAC-3′ (forward) and
5′-CACCAAAGCCAACCC-3′ (reverse); 18S, 5′-CTTAGTTGGTGGAGCGATTTGTC-3′
(forward) and 5′-CGGACATCTAAGGGCATCACA-3′ (reverse); miR-543,
5′-GAGAAGTTGCCCGTGTT-3′ (forward) and 5′-CGCGAATGTTTCGTCA-3′
(reverse); and U6, 5′-CGCTTCGGCAGCACATATAC-3′ (forward) and
5′-AAATATGGAACGCTTCACGA-3′ (reverse).
Western blotting
SiHa and HeLa cell proteins were extracted using
RIPA lysis buffer (Shanghai Bestbio Biotechnology Co., Ltd,) after
transfection, and protein determination was performed by the BCA
method. Proteins of different molecular weights were separated
using 10% SDS-PAGE in which 40 µg protein was loaded to each lane,
and subsequently electrotransferred to Hybond membranes
(MilliporeSigma; Merck KGaA). Subsequently, 3% BSA (BEIJING
SOLARBIO TECHNOLOGY CO., LTD) was used to block non-specific sites
at room temperature for 2 h, and anti-CHMP4C (1:1,000; #16256-1-AP;
Proteintech Group, Inc.), anti-Bcl-XL (1:2,000; #10783-1-AP;
Proteintech Group, Inc.), anti-Bcl2 (1:500; #66799-1-Ig;
Proteintech Group, Inc.), anti-Survivin (1:1,000; #10508-1-AP;
Proteintech Group, Inc.), anti-Caspase-7 (1:1,000; #27155-1-AP;
Proteintech Group, Inc.) and anti-HPV16/18 E6 (1:50; #sc-460; Santa
Cruz Biotechnology, Inc.) antibodies were added and incubated
overnight at 4°C. Anti-β-actin (1:10,000; #66009-1-Ig; Proteintech
Group, Inc.), GAPDH (1:1,000; #60004-1-Ig; Proteintech Group, Inc.)
or anti-α-tubulin (1:1,000; #80762-1-RR; Proteintech Group, Inc.)
antibodies were used as controls. On the second day, the membrane
was incubated with anti-rabbit IgG (1:10,000; cat. no. #SA00001-2;
Proteintech Group, Inc.) or anti-mouse (1:10,000; #SA00001-1;
Proteintech Group, Inc.) antibodies at room temperature for 2 h.
The protein bands were visualized using the Enhanced
Chemiluminescence kit (SuperSignal™ West Pico PLUS
Chemiluminescence substrate; Thermo Fisher Scientific, Inc.).
Data analysis
SPSS 24.0 software (IBM Corp.) was used to perform
the statistical analysis. All data are expressed as the mean ±
standard deviation. The immunohistochemistry results were analyzed
using Fisher's exact test. Comparisons between the experimental and
control groups were performed using an unpaired double-tailed
t-test, and multiple groups were compared using one-way ANOVA and
Dunnett's posttest. P<0.05 was considered to indicate a
statistically significant difference.
Results
Characteristics of CHMP4C in the
tumorigenesis of cervical cancer
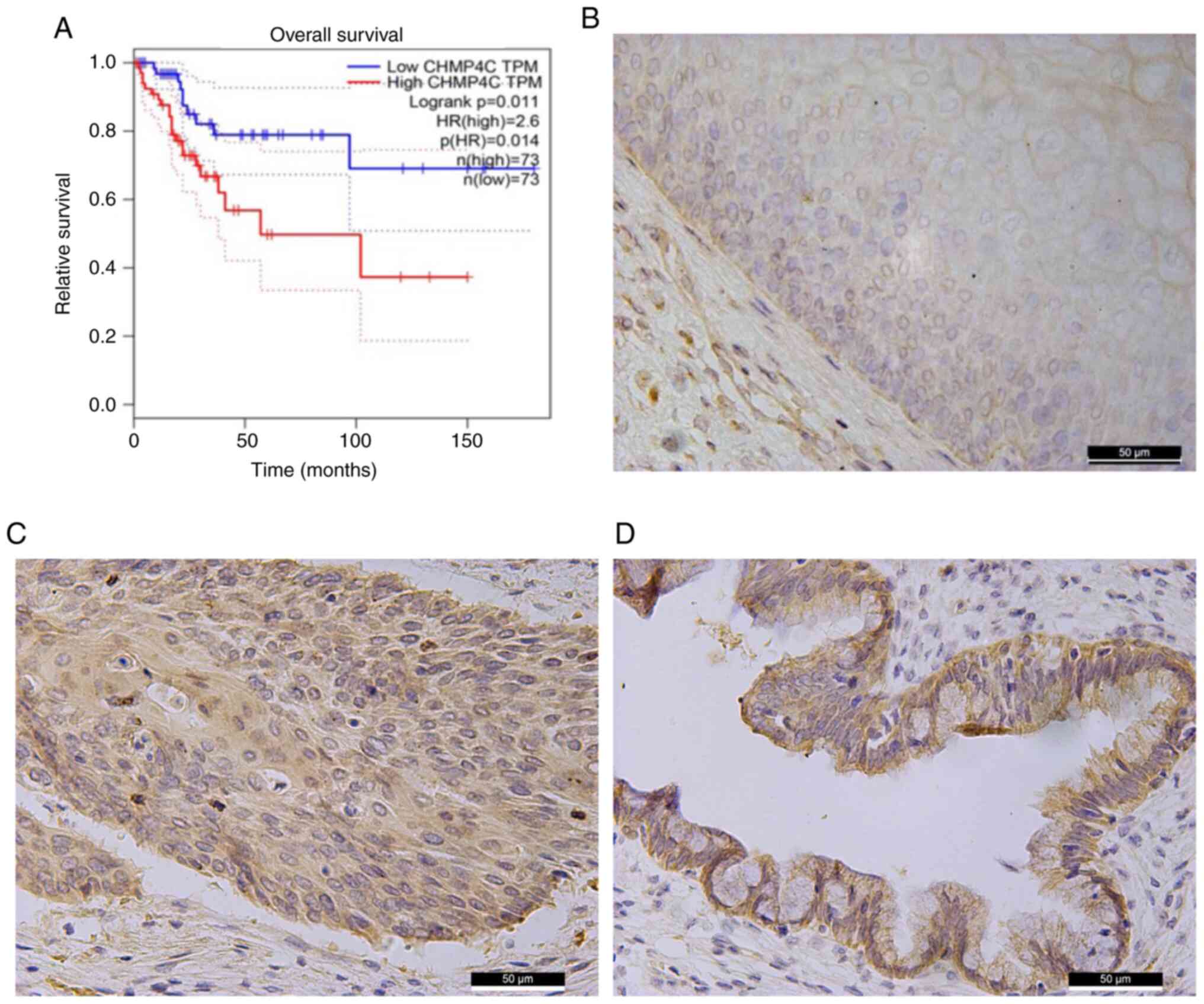
Bioinformatics analysis was used to assess the role
of CHMP4C in cervical cancer. The GEPIA database and log-rank tests
demonstrated that a higher CHMP4C expression may have a worse
clinical prognosis than those with a lower CHMP4C expression
(Fig. 2A). Following this,
immunohistochemistry was performed using normal cervical and
cervical cancer tissues to further assess the expression of CHMP4C
in cervical cancer. The results revealed that the expression level
of CHMP4C in malignant cervical tumor tissues was significantly
higher than that in normal tissues (Fig. 2B-D; Table SI; P<0.05). This finding
indicates that CHMP4C may participate in cervical cancer
tumorigenesis. Moreover, the level of CHMP4C was positively
associated with the age of the patients (Table SII; P<0.05).
Effect of CHMP4C expression on the
phenotype of cervical cancer cells
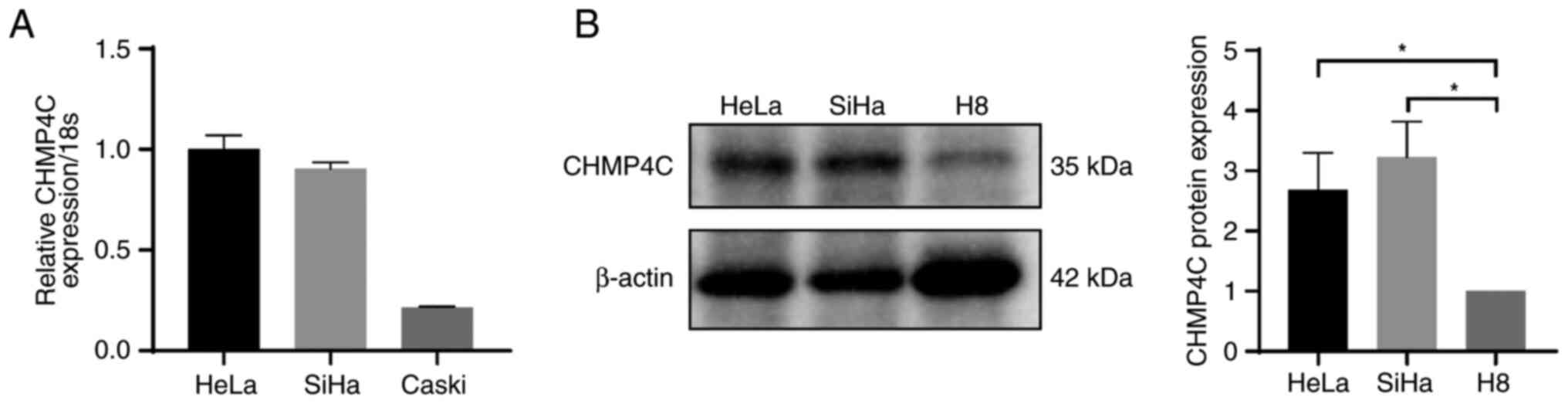
Firstly, expression of CHMP4C in each HPV-positive
cervical cancer cell line was detected using RT-qPCR, and SiHa and
HeLa cells were selected as the experimental objects as they had
the highest expression of CHMP4C out of the cervical cancer cell
lines assessed (Fig. 3A).
Furthermore, western blotting demonstrated that the expression
level of CHMP4C was significantly higher in cervical cancer SiHa
and HeLa cells compared with that in the normal cervical H8 cells
(Fig. 3B).
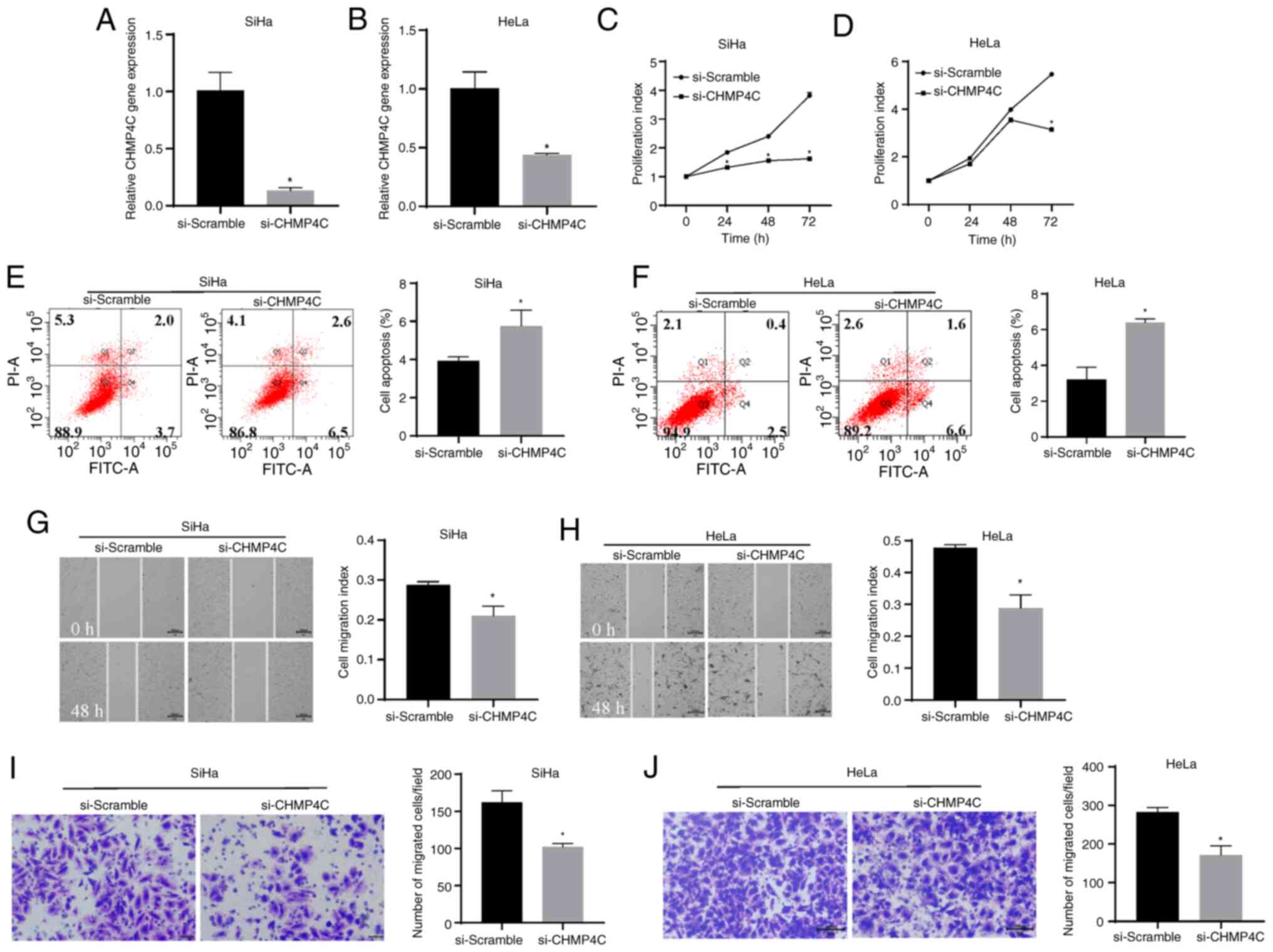
To evaluate the biological function of CHMP4C, SiHa
and HeLa cells were transfected with si-CHMP4C or si-Scramble, and
the transfection efficiency was determined using RT-qPCR. The
results demonstrated that the expression level of CHMP4C
significantly decreased following siRNA transfection, compared with
the control (Fig. 4A and B;
P<0.05). Silencing of CHMP4C expression was also associated with
a significant decrease in cell proliferation, compared with the
control (Fig. 4C and D; P<0.05).
This indicates that CHMP4C promotes cell proliferation. Moreover,
the cell apoptosis assay revealed that after 48 h of transfection,
the si-CHMP4C cells had a significantly higher apoptosis rate than
the control cells (Fig. 4E and F;
P<0.05). A significant reduction in cell migration (Fig. 4G and H; P<0.05) and invasion
(Fig. 4I and J; P<0.05) was also
demonstrated in cells with downregulated CHMP4C expression, in
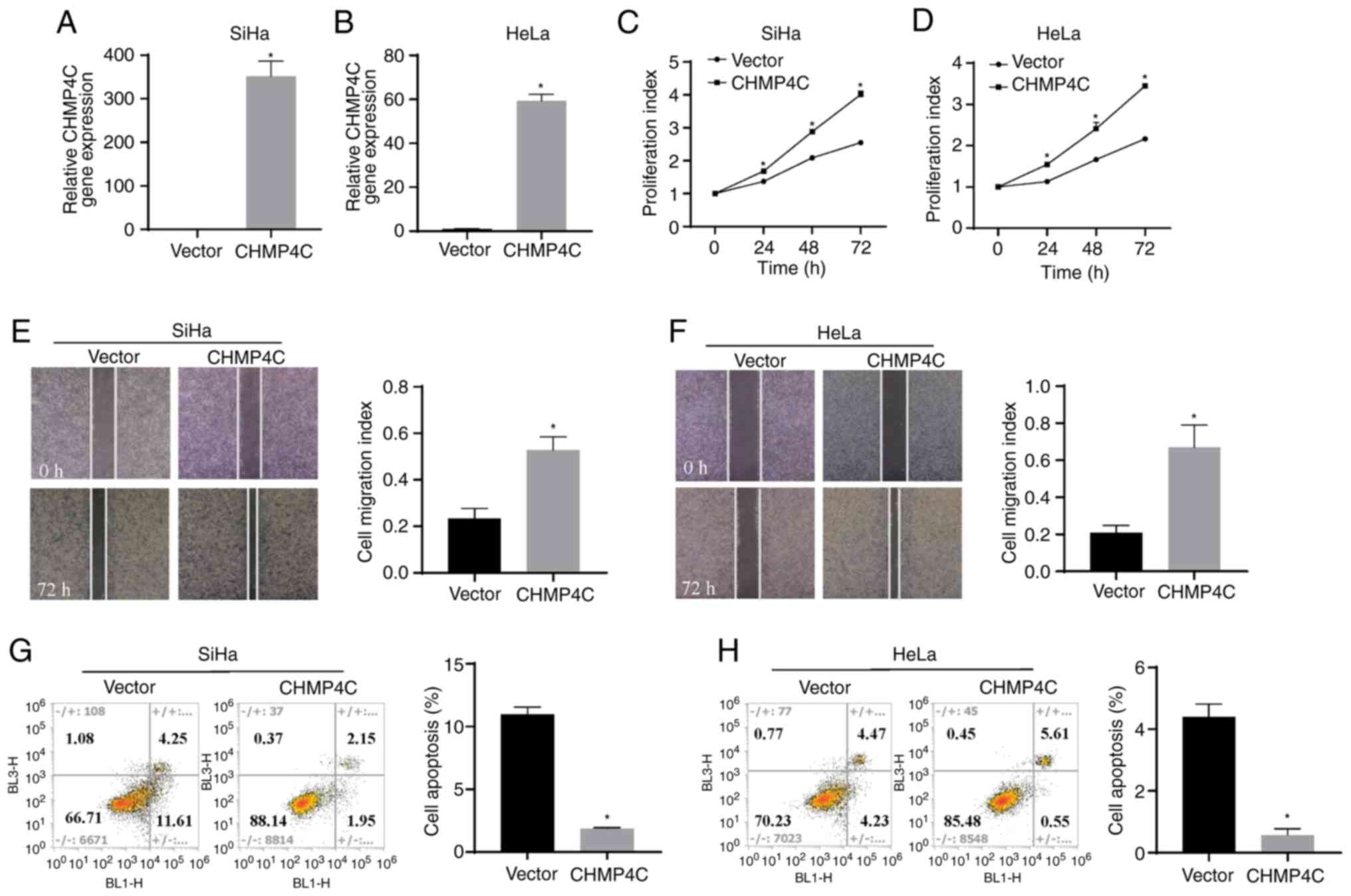
comparison with the control cells. Conversely, the plasmid was
transfected in the SiHa and HeLa cells to overexpress CHMP4C and
assess the transfection efficiency of the plasmid using RT-qPCR
(Fig. 5A and B; P<0.05). The
results revealed that, in comparison with the vector control,
overexpression of CHMP4C resulted in a significant increase in cell
proliferation (Fig. 5C and D;
P<0.05) and migration (Fig. 5E and
F; P<0.05), and a significant reduction in the rate of cell
apoptosis (Fig. 5G and H;
P<0.05). The observed changes in phenotype indicate that CHMP4C
acts as a proto-oncogene in cervical cancer cells.
CHMP4C expression is regulated by
E6
The occurrence of cervical cancer is related to HPV
(namely, high-risk HPV16 and HPV18) infection. Viral DNA is
integrated into the host genome to encode oncogenic proteins, such
as E6, which interfere with the cell cycle and apoptosis via
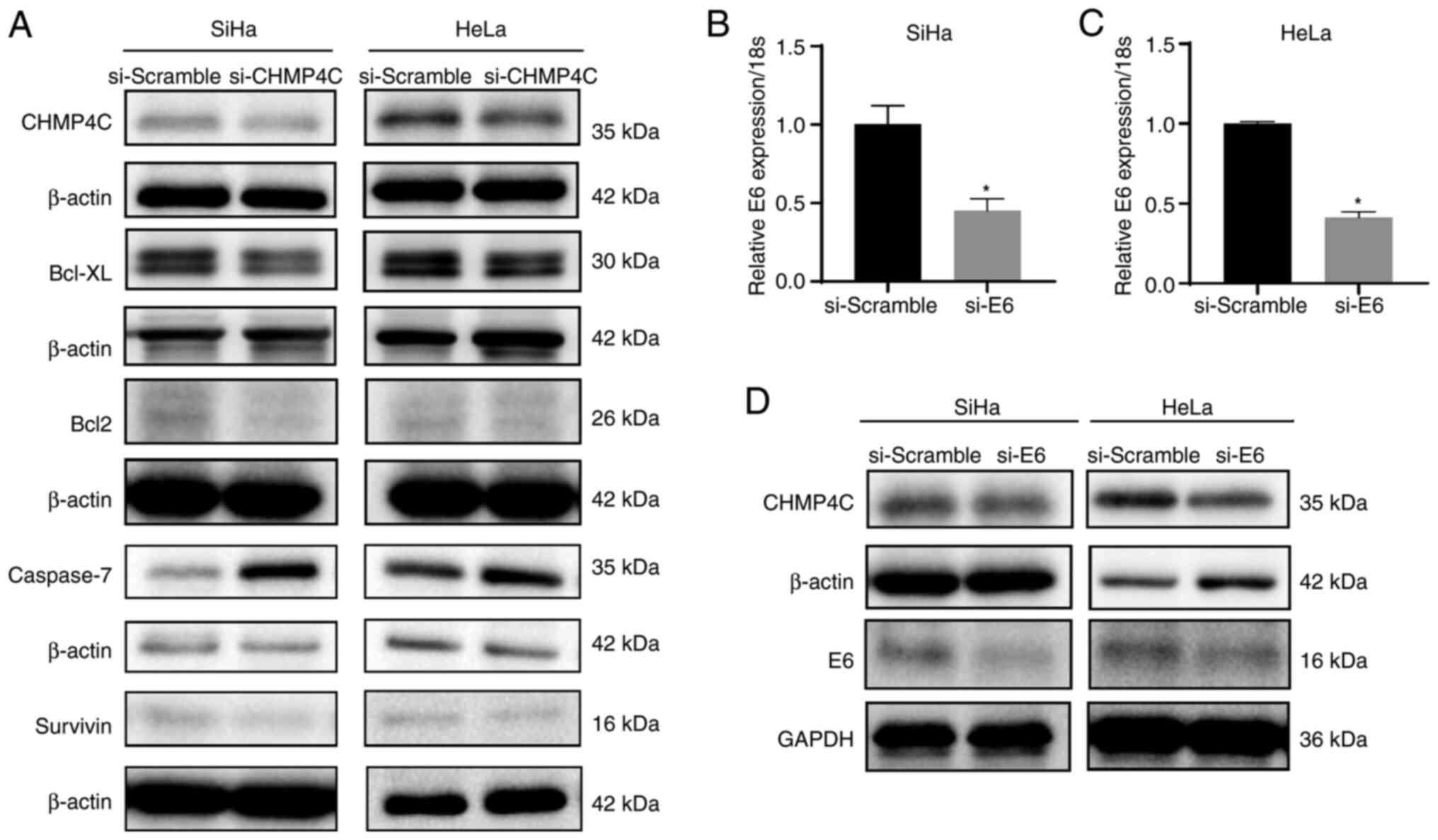
regulation of Bcl2 and Bcl-XL (12–14).
Western blotting showed that the expression of Bcl2, Bcl-xL, and
Survivin was decreased when CHMP4C expression was silenced, but
Caspase-7 expression was increased (Fig. 6A). Therefore, CHMP4C increased the
susceptibility of cervical cancer through the regulation of Bcl2,
Bcl-XL, Survivin and Caspase-7. Furthermore, to determine whether
CHMP4C expression was associated with cervical HPV infection, HPV16
E6 expression was silenced in SiHa cells, and HPV18 E6 expression
was silenced in HeLa cells through siRNA transfection. This
resulted in a significant decrease in the expression of CHMP4C,
compared with the controls (Fig.
6B-D; P<0.05), and demonstrated that HPV infection may lead
to increased expression of CHMP4C.
miR-543 directly targets CHMP4C
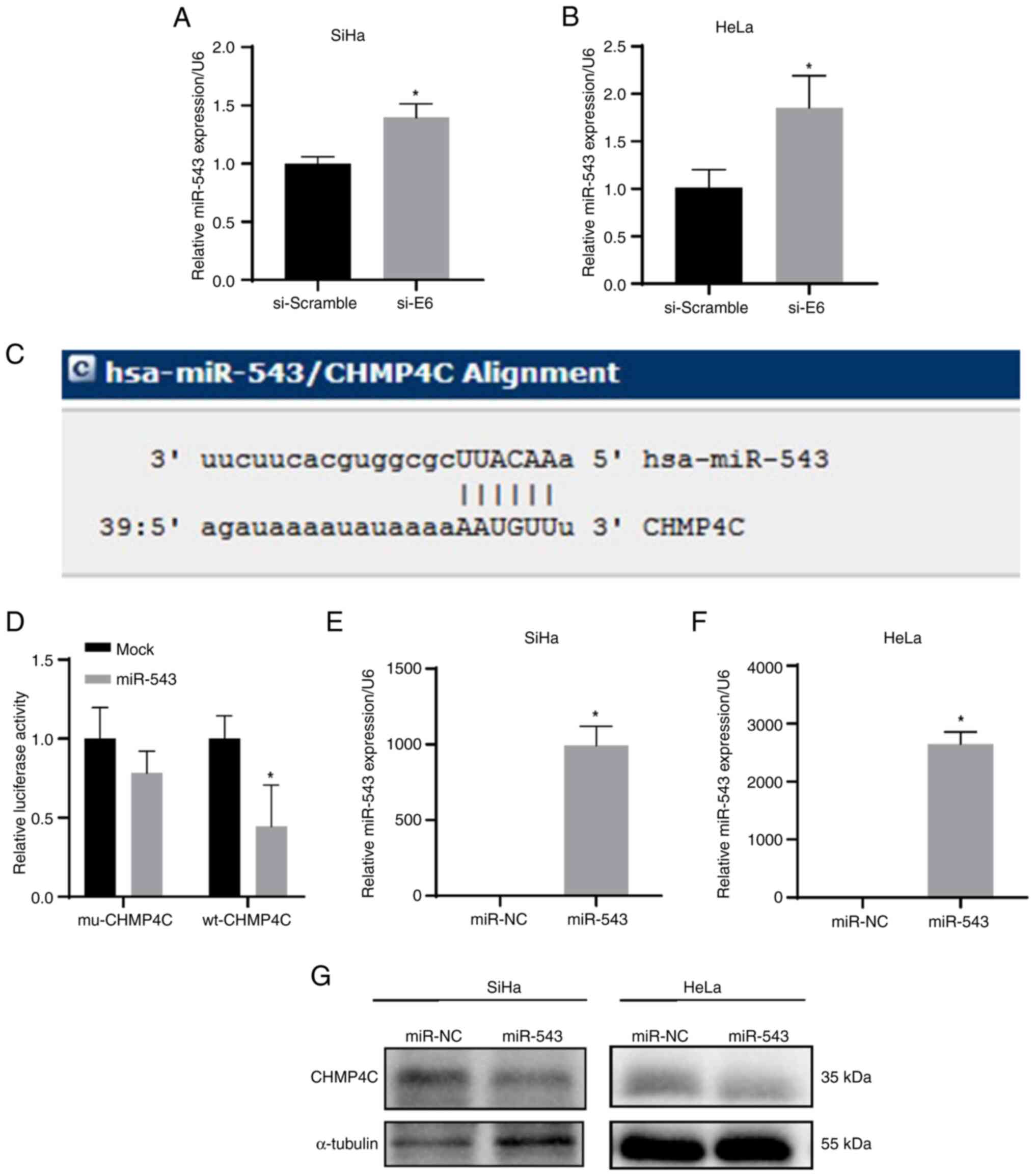
E6 induction of the expression of CHMP4C was
assessed. A previous study reported that E6 could inhibit miR-543
expression in human foreskin keratinocytes (HFKs) (15). Accordingly, the findings of the
present study demonstrated significant increases in miR-543
expression after E6 siRNA transfection in cervical cancer cells,
compared with that of the controls (Fig. 7A and B; P<0.05). Moreover, the
results of bioinformatics analysis revealed that miR-543 could bind
with the 3′-UTR of CHMP4C (Fig.
7C). In addition, the dual-luciferase reporter assay
demonstrated that the relative luciferase activity was
significantly reduced in the group co-transfected with wild-type
CHMP4C 3′-UTR and miR-543, compared with that of the control groups
(Fig. 7D; P<0.05). These results
imply that miR-543 directly targets the 3′-UTR of CHMP4C.
Furthermore, after overexpression of miR-543 by transfection of the
miR-543 mimics in SiHa and HeLa cells, western blotting indicated
that CHMP4C expression was significantly reduced compared with that
in control cells (Fig. 7E-G;
P<0.05).
Discussion
The abscission checkpoint can inhibit the abscission
of cells with spindle-midzone defects and the lagging chromosomes
promote CHMP4C phosphorylation by Aurora B. This affects its
activity, leading to the failure of cytokinesis (16,17).
It has been reported that radiation induces the expression of
Aurora B and CHMP4C in non-small cell lung cancer cells, thereby
maintaining cell cycle checkpoints and cell viability, and
resisting apoptosis. Inhibition of CHMP4C was reported to promote
the proliferation of cancer cells by inhibiting DNA repair by
reducing phosphorylated H2A histone family member X and p53-binding
protein 1 (53BP1) foci reacting to radiation stress (18). It has also been reported that the
absence of CHMP4C-dependent abscission checkpoints increases the
accumulation of 53BP1-associated DNA damage foci (6). The role of CHMP4C has received
increasing attention in the field of cancer research. Pharoah et
al (19) implicated CHMP4C as
the most likely susceptibility gene candidate in the 8q21 locus of
a new locus associated with the risk of epithelial ovarian cancer.
Moreover, human CHMP4C polymorphisms also increase the risk of
prostate and skin cancer (8).
With regards to the relationship between CHMP4C and
cervical cancer, the immunohistochemistry results of the present
study demonstrated that the expression level of CHMP4C was higher
in cervical cancer tissues compared with normal tissues. Analysis
of the GEPIA database revealed that cervical cancer is associated
with a poor clinical prognosis due to the increased expression of
CHMP4C. Accordingly, in the present study, CHMP4C silencing in SiHa
and HeLa cells resulted in a reduction in the level of cell
proliferation, migration and invasion, and an increase in the rate
of apoptosis. Conversely, CHMP4C overexpression had the opposite
effect on cell proliferation, cell migration and apoptosis. In
addition, silencing of CHMP4C reduced the level of Bcl2, Bcl-XL and
Survivin expression, and increased the expression level of
Caspase-7. The process of apoptosis involves several genes,
including those of the Bcl-2 family, the caspase family and the
inhibitor of apoptosis proteins (IAP) family (20). The Bcl2 protein family is the key to
the regulation and execution of mitochondrial-mediated intrinsic
apoptosis, among which Bcl2 and Bcl-XL belong to a group of
pro-survival proteins (21,22). Moreover, studies have revealed that
the expression of Bcl2 may be associated with the development of
cervical cancer and provide prognostic information (22,23).
Survivin is a member of the IAP family of proteins and inhibits
apoptosis by binding directly to Caspase-3 and Caspase-7 of the
caspase family to inhibit their activity (24). It is overexpressed in many cancers
and serves an important role in promoting tumor cell proliferation,
progression, angiogenesis, therapeutic resistance and poor
prognosis (25). A previous study
also reported that overexpression of CHMP4C in cervical cancer
cells accelerates cell migration and invasion by activating
epithelial-mesenchymal transition (26). The aforementioned results
demonstrate that CHMP4C increases the susceptibility of cervical
cancer and promotes tumorigenesis and progression of cervical
cancer.
Infection with HPV is the underlying cause of
invasive cervical cancer (27–29).
Viral DNA is integrated into the host genome to encode the
oncogenic E6 and E7 proteins. E6 and E7 can interfere with cell
cycle and apoptosis, which are critical factors in cellular
transformation and carcinogenesis (12,13).
Therefore, occurrence of cervical cancer is related to HPV
infection, and CHMP4C increases the susceptibility to cervical
cancer. To determine whether CHMP4C was related to cervical HPV
infection, the present study knocked down E6 expression in cervical
cancer cells and the results revealed that the expression of CHMP4C
was decreased in the E6-knockdown cells. Thus, the E6 protein
encoded by the HPV during cervical infection may regulate CHMP4C
expression.
According to a previous report, miR-543 expression
decreases after HPV16 E6 expression increased in HFKs (15). miRNAs are non-protein-encoding RNAs
with a length of ~21 nucleotides. They inhibit the expression of
their target genes primarily through interaction with the
corresponding 3′-UTR of the mRNA, thereby regulating biological
events, including cell proliferation, differentiation and apoptosis
(30,31). Moreover, miRNA dysregulation is
associated with the occurrence, development, metastasis and drug
resistance to cancer (32).
Bioinformatics analysis revealed a potential connection site
between the 3′-UTR of CHMP4C and miR-543, and this implies that E6
may regulate CHMP4C expression through miR-543. After E6 was
knocked down in SiHa and HeLa cells, the expression level of
miR-543 was elevated. Meanwhile, elevated miR-543 expression was
associated with reduced CHMP4C expression. Moreover, the
dual-luciferase reporter assay demonstrated that CHMP4C was the
target of miR-543. In addition, studies have reported that miR-543
expression is decreased in cervical cancer, and its expression
level is associated with tumor size, FIGO stage and lymph node
metastasis. In line with the results of the present study, miR-543
is known to inhibit tumor cell proliferation, invasion and
migration, and also promote apoptosis, thereby inhibiting the
development of cervical cancer (33). Based on these findings, we
hypothesize that CHMP4C promotes the progression of cervical cancer
through the HPV E6/miR-543 axis.
In conclusion, the present research indicates that
the HPV-encoded E6 oncoprotein may reduce the expression level of
miR-543 and relieve the inhibitory effect of miR-543 on CHMP4C. By
contrast, high expression level of CHMP4C may inhibit apoptosis by
regulating Bcl2, Bcl-XL, Survivin and Caspase-7 expression, thereby
promoting the tumorigenesis and progression of cervical cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present research was supported by Guangdong Medical Science
and Technology Research (grant no. A2023337), the Guangzhou Science
and Technology Project (grant nos. 2024A03J0182 and 2023A03J0375)
and the Clinical High-tech and Major Technology Projects in
Guangzhou (grant no. 2024PL-ZD06).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
RCL, YMJ, JH, YPD, YMZ and XJS conceived the study
and wrote the manuscript. RCL, YMJ, JH, YPD and YMZ performed the
experiments. RCL, YMJ and JH analyzed the data. RCL and XJS
confirmed the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Third Affiliated Hospital Guangzhou Medical
University (approval no. 2019-037). Written informed consent was
obtained from all individual participants included in the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CHMP4C
|
charged multivesicular body protein
4C
|
|
GEPIA
|
Gene Expression Profiling Interactive
Analysis
|
|
HPV
|
human papillomavirus
|
|
miRNA/miR
|
microRNA
|
|
siRNA
|
small interfering RNA
|
|
3′-UTR
|
3′-untranslated region
|
|
IAP
|
inhibitor of apoptosis proteins
|
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
World Health Organization (WHO), .
International agency for research on cancer. Global Cancer
Observatory: Cancer Today. WHO; Geneva: 2022, https://gco.iarc.who.int/media/globocan/factsheets/populations/160-china-fact-sheet.pdfMarch
10–2025
|
|
3
|
Bao H, Zhang L and Wang L, Zhang M, Zhao
Z, Fang L, Cong S, Zhou M and Wang L: Significant variations in the
cervical cancer screening rate in China by individual-level and
geographical measures of socioeconomic status: A multilevel model
analysis of a nationally representative survey dataset. Cancer Med.
7:2089–2100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Small W Jr, Bacon MA, Bajaj A, Chuang LT,
Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR,
Viswanathan AN and Gaffney DK: Cervical cancer: A global health
crisis. Cancer. 123:2404–2412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cohen PA, Jhingran A, Oaknin A and Denny
L: Cervical cancer. Lancet. 393:169–182. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carlton JG, Caballe A, Agromayor M, Kloc M
and Martin-Serrano J: ESCRT-III governs the Aurora B-mediated
abscission checkpoint through CHMP4C. Science. 336:220–225. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sadler JBA, Wenzel DM, Williams LK,
Guindo-Martínez M, Alam SL, Mercader JM, Torrents D, Ullman KS,
Sundquist WI and Martin-Serrano J: A cancer-associated polymorphism
in ESCRT-III disrupts the abscission checkpoint and promotes genome
instability. Proc Natl Acad Sci USA. 115:E8900–E8908. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nikolova DN, Doganov N, Dimitrov R,
Angelov K, Low SK, Dimova I, Toncheva D, Nakamura Y and Zembutsu H:
Genome-wide gene expression profiles of ovarian carcinoma:
Identification of molecular targets for the treatment of ovarian
carcinoma. Mol Med Rep. 2:365–384. 2009.PubMed/NCBI
|
|
10
|
Fujita K, Kume H, Matsuzaki K, Kawashima
A, Ujike T, Nagahara A, Uemura M, Miyagawa Y, Tomonaga T and
Nonomura N: Proteomic analysis of urinary extracellular vesicles
from high Gleason score prostate cancer. Sci Rep. 7:429612017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tommasino M: The human papillomavirus
family and its role in carcinogenesis. Semin Cancer Biol. 26:13–21.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Narisawa-Saito M and Kiyono T: Basic
mechanisms of high-risk human papillomavirus-induced
carcinogenesis: Roles of E6 and E7 proteins. Cancer Sci.
98:1505–1511. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miyashita T, Krajewski S, Krajewska M,
Wang HG, Lin HK, Liebermann DA, Hoffman B and Reed JC: Tumor
suppressor p53 is a regulator of bcl-2 and bax gene expression in
vitro and in vivo. Oncogene. 9:1799–1805. 1994.PubMed/NCBI
|
|
15
|
Harden ME, Prasad N, Griffiths A and
Munger K: Modulation of microRNA-mRNA target pairs by human
papillomavirus 16 oncoproteins. mBio. 8:e02170–16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Norden C, Mendoza M, Dobbelaere J,
Kotwaliwale CV, Biggins S and Barral Y: The NoCut pathway links
completion of cytokinesis to spindle midzone function to prevent
chromosome breakage. Cell. 125:85–98. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Capalbo L, Montembault E, Takeda T, Bassi
ZI, Glover DM and D'Avino PP: The chromosomal passenger complex
controls the function of endosomal sorting complex required for
transport-III Snf7 proteins during cytokinesis. Open Biol.
2:1200702012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li K, Liu J, Tian M, Gao G, Qi X, Pan Y,
Ruan J, Liu C and Su X: CHMP4C disruption sensitizes the human lung
cancer cells to irradiation. Int J Mol Sci. 17:182015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pharoah PD, Tsai YY, Ramus SJ, Phelan CM,
Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H, et
al: GWAS meta-analysis and replication identifies three new
susceptibility loci for ovarian cancer. Nat Genet. 45:362–370.
370e1-22013. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang Y and Yu X: Regulation of apoptosis:
The ubiquitous way. FASEB J. 17:790–799. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chipuk JE, Moldoveanu T, Llambi F, Parsons
MJ and Green DR: The BCL-2 family reunion. Mol Cell. 37:299–310.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ter Harmsel B, Smedts F, Kuijpers J,
Jeunink M, Trimbos B and Ramaekers F: BCL-2 immunoreactivity
increases with severity of CIN: A study of normal cervical
epithelia, CIN, and cervical carcinoma. J Pathol. 179:26–30. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dimitrakakis C, Kymionis G, Diakomanolis
E, Papaspyrou I, Rodolakis A, Arzimanoglou I, Leandros E and
Michalas S: The possible role of p53 and bcl-2 expression in
cervical carcinomas and their premalignant lesions. Gynecol Oncol.
77:129–136. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shin S, Sung BJ, Cho YS, Kim HJ, Ha NC,
Hwang JI, Chung CW, Jung YK and Oh BH: An anti-apoptotic protein
human survivin is a direct inhibitor of caspase-3 and −7.
Biochemistry. 40:1117–1123. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen X, Duan N, Zhang C and Zhang W:
Survivin and tumorigenesis: Molecular mechanisms and therapeutic
strategies. J Cancer. 7:314–323. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin SL, Wang M, Cao QQ and Li Q: Chromatin
modified protein 4C (CHMP4C) facilitates the malignant development
of cervical cancer cells. FEBS Open Bio. 10:1295–1303. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Crosbie EJ, Einstein MH, Franceschi S and
Kitchener HC: Human papillomavirus and cervical cancer. Lancet.
382:889–899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Muñoz N, Bosch FX, de Sanjosé S, Herrero
R, Castellsagué X, Shah KV, Snijders PJ and Meijer CJ;
International Agency for Research on Cancer Multicenter Cervical
Cancer Study Group, : Epidemiologic classification of human
papillomavirus types associated with cervical cancer. N Engl J Med.
348:518–527. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Smith JS, Lindsay L, Hoots B, Keys J,
Franceschi S, Winer R and Clifford GM: Human papillomavirus type
distribution in invasive cervical cancer and high-grade cervical
lesions: A meta-analysis update. Int J Cancer. 121:621–632. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Davis BN and Hata A: Regulation of
MicroRNA biogenesis: A miRiad of mechanisms. Cell Commun Signal.
7:182009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Acunzo M, Romano G, Wernicke D and Croce
CM: MicroRNA and cancer-a brief overview. Adv Biol Regul. 57:1–9.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu X, Gan L and Zhang J: miR-543
inhibites cervical cancer growth and metastasis by targeting TRPM7.
Chem Biol Interact. 302:83–92. 2019. View Article : Google Scholar : PubMed/NCBI
|