Introduction
Gynecological malignancies, encompassing numerous
types of cancer of the female reproductive system, represent a
notable global health burden. Among these malignancies, ovarian,
endometrial and cervical cancer are the most prevalent. According
to GLOBOCAN 2020 statistics, ovarian, endometrial and cervical
cancer accounted for 313,959, 417,367 and 604,127 new global cases,
respectively, and ranked 19, 17 and 9th, in terms of global cancer
incidence. In addition, regarding the mortality rates, ovarian
cancer, endometrial cancer and cervical cancer were responsible for
207,252, 97,370 and 341,831 deaths respectively, collectively
representing 6.5% of all female cancer-related deaths (1).
Despite advances in medical science, the diagnosis
and treatment of gynecological malignancies continue to pose
notable challenges. Early detection methods remain elusive for a
number of patients, and the current therapeutic approaches often
fall short in managing advanced or recurrent disease. However,
recent research into circular RNAs (circRNAs) has emerged as a
promising option for both diagnostic and therapeutic innovations in
gynecological oncology (2,3). circRNAs are a class of non-coding RNAs
characterized by their covalently closed loop structure, which have
gained considerable attention in cancer research due to their
stability, tissue-specific expression and diverse regulatory
functions (4). In the context of
gynecological malignancies, circRNAs have been implicated in
various aspects of tumor biology, including cell proliferation,
metastasis and drug resistance (5).
For example, in ovarian cancer, circWHSC1 has been shown to promote
tumor progression by sponging microRNA (miRNA/miR)-145 and
miR-1182, ultimately upregulating mucin-1 (MUC1) and human
telomerase reverse transcriptase (hTERT) (6). In endometrial cancer, homo
sapiens (hsa)_circRNA_079422 has been found to enhance tumor
growth and metastasis by modulating the miR-136-5p/high-mobility
group AT-hook 2 axis (7).
Similarly, in cervical cancer, circRNAs serve a key regulatory role
in cervical cancer progression, showing potential as therapeutic
agents and novel biomarkers; however, further clinical research is
needed to fully understand their therapeutic benefits (8). The unique properties of circRNAs,
including their abundance, stability in bodily fluids and
tissue-specific expression patterns, make them promising candidates
for biomarker development in gynecological cancer (9). Moreover, the diverse regulatory
functions of circRNAs offer potential targets for novel therapeutic
interventions (10).
The present review aimed to provide a comprehensive
overview of recent advances in circRNA research within the context
of gynecological malignancies. The biogenesis and functions of
circRNAs, their roles in the pathogenesis of ovarian, endometrial
and cervical cancer were explored, and as well as potential
applications in diagnosis, prognosis and treatment. By summarizing
current knowledge and identifying future research directions, the
present review highlighted the potential of circRNAs as a novel
avenue for improving the management of gynecological malignancies
(Table I).
 | Table I.Key circRNAs and their functions in
ovarian, endometrial and cervical cancer. |
Table I.
Key circRNAs and their functions in
ovarian, endometrial and cervical cancer.
| Type of cancer | circRNA | Function | (Refs.) |
|---|
| Ovarian | circMUC16 | Promotes
proliferation and invasion by sponging miR-199a-5p to regulate
beclin-1 and RUNX1 expression | (74) |
| Ovarian | circCSPP1 | Promotes cancer
progression by counteracting the tumor-suppressive effects of
miR-1236-3p on ZEB1 | (75) |
| Ovarian | circFBXW7 | Encodes FBXW7-185aa
protein, which acts as a tumor suppressor by inhibiting cancer cell
proliferation and migration | (61) |
| Ovarian | circRNA-encoded
peptide SHPRH-146aa | Inhibits tumor
growth by protecting full-length SHPRH from degradation | (63) |
| Endometrial | circTNFRSF21 | Serves a crucial
role in cancer cell growth and cell cycle progression by
competitively binding with miR-1227, activating the MAPK13/ATF2
signaling pathway | (104) |
| Endometrial | circWHSC1 | Promotes cell
proliferation, migration and invasion while inhibiting
apoptosis | (91) |
| Endometrial |
hsa_circ_0061140 | Promotes
progression and migration through a regulatory axis involving
miR-149-5p and STAT3 | (107) |
| Cervical | circE7 | Encodes E7
oncoprotein, contributing to transforming properties of HPV | (62) |
| Cervical |
hsa_circ_0023404 | Promotes cancer
development by sponging miR-136, leading to increased expression of
TFCP2 and subsequent activation of the YAP pathway | (126) |
| Cervical | circ_0005576 | Promotes cell
proliferation and metastasis by sponging miR-153-3p and promoting
kinesin family member 20A expression | (128) |
Introduction to circRNA
circRNAs represent a unique class of non-coding RNA
molecules characterized by their covalently closed loop structure,
which distinguishes them from linear RNAs by the absence of 5′-3′
polarity and polyadenylated tails (11). This distinctive structure, first
identified in viruses by Sanger et al (12) in 1976, confers properties of
enhanced stability, cooperativity and self-complementarity. The
presence of circRNAs in eukaryotic cells was subsequently
demonstrated by Hsu and Coca-Prados in 1979 (13), marking the beginning of a new era in
RNA biology.
Initially, the importance of circRNAs was
underappreciated due to limitations in RNA sequencing technologies
and bioinformatics tools. For a number of years, these molecules
were dismissed as non-functional by-products of aberrant splicing
events (14); however, advances in
high-throughput sequencing and computational analysis have led to a
paradigm shift in the understanding of circRNAs and their potential
roles in cellular processes (15).
A key breakthrough in circRNA research came with the discovery that
these molecules often harbor numerous miRNA binding sites, enabling
them to function as competitive endogenous RNAs (ceRNAs) or ‘miRNA
sponges’ (16). This mechanism
allows circRNAs to regulate gene expression by modulating the
availability of miRNAs, thereby influencing various cellular
processes and pathways (17). In
the context of gynecological malignancies, this regulatory function
has been implicated in key aspects of tumor biology, including cell
proliferation, metastasis and drug resistance (3). Beyond their role as miRNA sponges,
circRNAs have been shown to interact with RNA-binding proteins
(RBPs), potentially modulating protein function and localization
(18). In addition, some circRNAs
have been reported to encode functional peptides, such as
circZNF609 (19) and circFBXW7
(20), adding another layer of
complexity to their regulatory potential. These diverse functions
underscore the importance of circRNAs in both physiological and
pathological processes, including the development and progression
of gynecological cancer. As research in this field continues to
evolve, circRNAs are emerging as promising biomarkers and potential
therapeutic targets in gynecological oncology (21). The unique properties of circRNAs,
including tissue-specific expression patterns and stability in
bodily fluids, make them attractive candidates for non-invasive
diagnostic and prognostic applications (22). Moreover, the ability to manipulate
circRNA expression levels or function offers novel options for
therapeutic interventions in gynecological malignancies (23).
Classification
The diversity of circRNAs is reflected in their
classification, which is primarily based on their genomic origin
and structure. Understanding these classifications is essential for
elucidating their roles in gynecological malignancies. circRNAs can
be categorized into three main subclasses based on their genomic
origin and structure. Exonic circRNA (EcircRNA) is the most
prevalent form, accounting for >80% of known circRNAs (24); these are generated through the
process of ‘backsplicing’, where a downstream 5′ splice site
(donor) is joined to an upstream 3′ splice site (acceptor).
Circular intronic RNA (ciRNA) is derived from intron lariats that
evade the typical linear splicing debranching step, retaining a
2′-5′ linkage at the branch point (25). Exon-intron circRNA (EIciRNA)
contains both exonic and intronic sequences and has been shown to
enhance the expression of their parent genes in cis,
suggesting a potential regulatory role (26). Other types of circRNAs include
antisense circRNAs, which are derived from antisense transcripts,
and intergenic circRNAs, which originate from intergenic regions or
other unannotated genomic sequences (27). As research progresses, and new
biotechnologies and bioinformatics strategies emerge, the
understanding of circRNA classification and function could continue
to evolve (28). Future studies may
identify additional subtypes or refine the current classification
system, providing deeper insights into the diverse roles of these
unique RNA molecules in cellular processes and disease
mechanisms.
The classification of circRNAs is not merely an
academic exercise but has significant implications for
understanding their functions in gynecological cancer. For example,
EcircRNAs often act as miRNA sponges, modulating gene expression
and influencing tumor progression (29). By contrast, ciRNAs and EIciRNAs tend
to regulate gene expression through interactions with the
transcriptional machinery (30).
The ongoing exploration of circRNA classification in gynecological
cancer promises to uncover novel biomarkers for early detection and
prognosis, as well as potential therapeutic targets. For example,
the tissue-specific expression patterns of certain circRNA classes
may be exploited in the development of targeted therapies or
diagnostic tools specific to ovarian, endometrial or cervical
cancer (3).
Formation mechanisms of circRNA
The biogenesis of circRNAs in gynecological
malignancies remains an area of active investigation, with several
proposed models offering insights into their formation (4,27,31). A
prominent hypothesis posits that during the splicing of mRNA
precursors, a process critical in gene expression regulation,
non-contiguous exons may be brought into proximity through partial
folding of the transcript. This spatial rearrangement can lead to
exon skipping, resulting in the formation of a lariat intermediate
structure encompassing both exonic and intronic sequences.
Subsequently, this lariat undergoes a reverse splicing event,
culminating in the generation of a stable circRNA molecule. This
mechanism may be particularly relevant in the context of
gynecological tumors, where aberrant splicing events are frequently
observed and could contribute to the dysregulation of circRNA
formation, potentially influencing tumor progression and treatment
response (32).
In the context of gynecological malignancies, an
alternative model of circRNA biogenesis has gained traction,
emphasizing the role of intronic sequences in facilitating
circularization. This mechanism, known as intron pair-driven
circularization, involves the interaction of specific complementary
sequences within flanking introns. These sequences, often found at
the termini of exons, engage in reverse complementary base pairing,
effectively bridging distant genomic regions. This intronic
‘kissing’ interaction brings downstream splice donor sites into
close proximity with upstream splice acceptor sites, thereby
promoting the circularization of the intervening exonic sequences.
In gynecological tumors, where genomic instability and chromosomal
rearrangements are common, this mechanism may be particularly
relevant, potentially contributing to the aberrant expression of
circRNAs observed in these malignancies. Understanding this process
could provide insights into tumor-specific circRNA profiles, and
their potential diagnostic or therapeutic implications in
gynecological oncology (33).
Another emerging model of circRNA formation
highlights the pivotal role of RBPs in the landscape of
gynecological malignancies. These versatile proteins exhibit a
marked capacity to influence circRNA biogenesis through specific
interactions with both exonic and intronic sequences. RBPs can
selectively bind to motifs located at the termini of exons and
introns, subsequently forming dimeric complexes. These RBP dimers
serve as molecular bridges, effectively reducing the spatial
distance between distal intronic regions. This RBP-mediated
proximity facilitates the circular connection of exons, promoting
circRNA formation. In the context of gynecological tumors, where
the aberrant expression and function of RBPs is frequently
observed, this mechanism may contribute to the dysregulation of
circRNA profiles. Understanding the interplay between RBPs and
circRNA biogenesis in these malignancies may indicate novel
diagnostic biomarkers and potential therapeutic targets, thereby
advancing the understanding of tumor biology and treatment
strategies in gynecological oncology (34).
Functions of circRNA
circRNAs have emerged as key regulators of gene
expression and are implicated in the pathogenesis of various human
diseases, including gynecological malignancies. These versatile
molecules exhibit multiple functional modalities that contribute to
cellular processes and disease progression. Current research has
elucidated several key functions of circRNAs: i) They can modulate
transcriptional activity; ii) act as ceRNAs by sequestering miRNAs;
iii) serve as scaffolds for protein complexes; and iv) possess
protein-coding potential under specific cellular conditions
(35). These diverse functions
highlight the significance of circRNAs in the complex landscape of
gynecological tumor biology and present potential avenues for
diagnostic and therapeutic interventions.
Transcriptional regulation by circRNA
The intricate interplay between circRNA biogenesis
and transcriptional regulation has emerged as a notable area of
interest in gynecological oncology (36,37).
This complex relationship indicates novel mechanisms of gene
expression modulation that may be key in the development and
progression of gynecological cancer. In a pioneering study,
Ashwal-Fluss et al (38)
reported a competitive relationship between circRNA splicing and
pre-mRNA processing. This competition was shown to occur at the 5′
and 3′ splice sites, suggesting that circRNA formation may serve as
a regulatory checkpoint in the gene expression cascade. In the
context of gynecological tumors, where aberrant gene expression is
a hallmark feature, this circRNA-mediated regulation of linear
transcript levels could represent a novel layer of transcriptional
control. Further elucidating this concept, research has
demonstrated that optimized circRNA expression systems can markedly
enhance backsplicing efficiency (39). This process effectively sequesters
exons into circular structures, consequently attenuating the
production of linear mRNA transcripts. For example, in ovarian
cancer, the formation of circ-mucin 16 (MUC16) has been shown to
reduce the expression of its linear counterpart, CA125, a
well-known biomarker for ovarian cancer (40). This finding highlights the potential
impact of circRNA biogenesis on the expression of clinically
relevant genes in gynecological malignancies. In endometrial cancer
the circRNA, circ-rho GTPase activating protein 12 (ARHGAP12), has
been shown to regulate the expression of its parental gene
ARHGAP12, a tumor suppressor, by competing for splicing machinery
(41). This regulatory mechanism
can influence cell proliferation and invasion, underscoring the
functional significance of circRNA-mediated transcriptional control
in gynecological cancer.
Moreover, some circRNAs have been shown to interact
directly with RNA polymerase II or other components of the
transcriptional machinery. For example, the ciRNA ci-ankyrin repeat
domain 52 has been reported to accumulate at its sites of
transcription and modulate the elongation of its parent gene by
associating with RNA polymerase II (42). While this specific circRNA has not
been studied in gynecological cancer, similar mechanisms could
occur, potentially influencing the expression of key oncogenes or
tumor suppressors. The ability of circRNAs to influence linear mRNA
production through competitive splicing mechanisms may have notable
implications for tumor biology, potentially affecting key oncogenic
or tumor-suppressive pathways. These findings underscore the
potential of circRNAs as key modulators of gene expression in
gynecological malignancies. The complex interplay between circRNA
biogenesis and transcriptional regulation offers new perspectives
on the molecular mechanisms underlying these types of cancer.
Further investigation into this regulatory axis could identify new
targets for therapeutic intervention and contribute to the
understanding of the complex transcriptional landscape in
gynecological cancer. As research in this field progresses, it will
be necessary to elucidate the tissue-specific and cancer-specific
aspects of circRNA-mediated transcriptional regulation. This
knowledge may lead to the development of novel diagnostic tools and
therapeutic strategies tailored to specific gynecological
malignancies, potentially improving patient outcomes.
miRNA sponges
circRNAs have emerged as key regulators of gene
expression in gynecological malignancies, primarily through their
function as miRNA sponges. This mechanism is characterized by the
presence of multiple miRNA binding sites within circRNA molecules,
enabling them to competitively sequester miRNAs (43). By preventing miRNAs from binding to
their target mRNAs, circRNAs effectively reduce the inhibitory
effect of miRNAs on translation, leading to the increased
expression of target genes (44).
This competitive binding phenomenon, known as the ‘miRNA sponge’
effect, represents an important paradigm in post-transcriptional
gene regulation (45). An example
of this regulatory mechanism is circ-cerebellar
degeneration-related protein 1 antisense (CDR1as, also known as
ciRS-7), which has been extensively studied due to its marked
capacity to bind miR-7 (46). While
initially identified in neurological contexts, recent research has
begun to elucidate its potential roles in various types of cancer,
including gynecological tumors (47). For example, in ovarian cancer,
circ_CDR1 has been shown to sponge miR-1270, leading to the
upregulation of suppressor of cancer cell invasion and subsequent
inhibition of tumor progression (48). In the context of gynecological
oncology, the miRNA sponge function of circRNAs has been implicated
in various aspects of tumor biology. For example, in cervical
cancer, circRNA_0000745 may act as a sponge for miR-3178,
upregulating the expression levels of FOXO4 and inhibiting tumor
growth (49). Similarly, in
endometrial cancer, circ_0002577 has been demonstrated to sponge
miR-625-5p, thereby regulating the insulin-like growth factor 1
signaling pathway and influencing tumor progression (50). Understanding these
circRNA-miRNA-mRNA regulatory axes in gynecological malignancies
offers new insights into the molecular mechanisms underlying tumor
development and progression. Moreover, it presents potential
opportunities for the development of novel diagnostic biomarkers
and therapeutic targets in gynecological oncology (51).
Protein scaffolds or sponges
In addition to their role as miRNA sponges, circRNAs
have emerged as important regulators of protein function in various
types of cancer, including gynecological malignancies. This
function is primarily achieved through their ability to act as
protein scaffolds or sponges, facilitating or inhibiting
protein-protein interactions and modulating protein activity
(36). A well-characterized example
of this mechanism is circFOXO3, which has been shown to form a
ternary complex with CDK2 and the CDK inhibitor 1, p21. This
interaction effectively inhibits CDK2 function, leading to cell
cycle arrest (52). While initially
studied in cardiac senescence, a research has begun to elucidate
the potential roles of circFOXO3 in gynecological cancer (53). For example, in ovarian cancer,
circFOXO3 has been shown to drive ovarian cancer progression by
orchestrating exosome-mediated intercellular communication
targeting the miR-422a/proteolipid protein 2 axis, potentially
serving as a valuable biomarker for early detection and treatment
monitoring in this aggressive malignancy (54).
Another notable example is circACC1, which forms a
ternary complex with the β and γ regulatory subunits of
AMP-activated protein kinase (AMPK); this interaction promotes AMPK
holoenzyme activity, enabling cancer cells to adapt to metabolic
stress and grow under nutrient-limited conditions (55). While the specific role of
circ-acetyl-CoA carboxylase 1 (ACC1) in gynecological tumors
remains to be fully elucidated, its function in modulating cellular
metabolism suggests potential implications for tumor progression
and therapeutic resistance in these types of cancer (56). In addition, circPUM1 has been shown
to interact with the nuclear receptor binding protein and the
androgen receptor, forming a protein complex that promotes ovarian
cancer cell proliferation and invasion (57). This example highlights the diverse
mechanisms by which circRNAs can influence protein function and
cellular processes in gynecological malignancies. The ability of
circRNAs to act as protein scaffolds or sponges adds another layer
of complexity to their regulatory functions in cancer biology. By
modulating protein-protein interactions and enzyme activities,
circRNAs can influence various cellular processes, including cell
cycle progression, metabolism and signal transduction (58). Understanding these interactions in
the context of gynecological tumors may provide novel insights into
disease mechanisms and therapeutic targets (59).
Translation of peptides
Traditionally, circRNAs were considered to be
non-coding RNAs; however, a recent study demonstrated that, under
specific conditions, some circRNAs can be translated into
functional peptides, adding a new dimension to their role in
cellular processes and disease pathogenesis, including
gynecological malignancies (39).
In a seminal study from 2017, Yang et al (60) provided the first comprehensive
evidence that circRNAs could undergo protein translation following
N6-methyladenosine (m6A) modification. These findings demonstrated
that m6A-driven circRNA translation is a widespread phenomenon,
with hundreds of endogenous circRNAs possessing translation
potential. This discovery not only expanded the understanding of
the coding capacity of the human transcriptome but also opened
novel avenues for exploring circRNA functions in various diseases,
including gynecological cancer. In the context of gynecological
oncology, the potential for circRNAs to be translated into
functional peptides has significant implications. For example, in
breast cancer, circ-F-box and WD repeat domain containing 7 (FBXW7)
has been shown to encode a novel 21-kDa protein, FBXW7-185aa, which
acts as a tumor suppressor by inhibiting the proliferation and
migration of cancer cells (61).
Similarly, in cervical cancer, circE7 derived from high-risk human
papillomavirus (HPV) has been shown to encode the E7 oncoprotein,
contributing to the transforming properties of HPV (62). The ability of circRNAs to produce
functional peptides adds another layer of complexity to their
regulatory roles in gynecological malignancies. These peptides may
interact with other cellular components, modulate signaling
pathways or directly influence tumor progression. For example, in
ovarian cancer, the circRNA-encoded peptide SNF2 histone linker PHD
RING helicase (SHPRH)-146aa has been reported to suppress tumor
growth by protecting full-length SHPRH from degradation (63). As research in this field progresses,
elucidating the specific functions of circRNA-encoded peptides in
various types of gynecological cancer will be crucial.
Understanding these novel mechanisms may provide insights into
tumor-specific molecular signatures and identify new options to
develop innovative diagnostic and therapeutic strategies in
gynecological oncology (64).
circRNA and ovarian cancer
Ovarian cancer is currently the most lethal type of
gynecological malignancy, with a markedly high mortality rate.
Globally, ovarian cancer is responsible for >200,000 deaths
annually (65). The standard
treatment protocol typically involves surgical resection followed
by platinum-based chemotherapy in combination with paclitaxel (PTX)
(66). Despite these interventions,
the prognosis for patients with advanced-stage ovarian cancer
remains poor, with a 5-year survival rate of ~30% (67). This poor outcome is largely
attributed to late-stage diagnosis and the development of
chemoresistance (68). The
mechanisms underlying chemoresistance in ovarian cancer are
multifaceted, involving both tumor microenvironment factors and
intrinsic cellular resistance pathways (69). Given these challenges, there is an
urgent need to develop novel therapeutic strategies and enhance
treatment efficacy to improve patient outcomes. In recent years,
circRNAs have emerged as potential biomarkers and therapeutic
targets in various types of cancer, including ovarian cancer
(70). These unique non-coding RNAs
have been implicated in diverse biological processes, and have
shown promise in modulating chemoresistance and tumor progression
(71). Understanding the role of
circRNAs in ovarian cancer could potentially lead to innovative
diagnostic tools and targeted therapies, addressing the critical
need for improved management of this disease (72).
Mechanistic studies of circRNA in
ovarian cancer
Previous investigations have provided information on
the roles of circRNAs in ovarian cancer progression, unveiling
complex regulatory networks that contribute to tumor development
and metastasis. These studies have identified specific circRNAs and
their associated molecular pathways, providing valuable insights
into potential therapeutic targets (40,73). A
notable discovery by Gan et al (74) demonstrated that circMUC16 can
promote ovarian cancer cell proliferation and invasion through a
complex regulatory axis. circMUC16 may modulate the expression of
beclin1 and RUNX1 by sponging miR-199a-5p, thereby influencing key
oncogenic processes. This finding not only elucidates a novel
mechanism of ovarian cancer progression but also highlights the
potential of circMUC16 as a therapeutic target. In a separate
study, Li et al (75)
demonstrated the oncogenic role of circ-centrosome and spindle pole
associated protein 1 (CSPP1) in ovarian cancer. This previous study
demonstrated that circCSPP1 can enhance cancer progression by
counteracting the tumor-suppressive effects of miR-1236-3p on ZEB1,
a key transcription factor involved in epithelial-mesenchymal
transition (EMT). In addition, circCSPP1 depletion was shown to
inhibit tumor growth, whereas its overexpression could upregulate
oncogenic proteins, further emphasizing its significance in ovarian
cancer pathogenesis. These mechanistic studies underscore the
complex interplay between circRNAs and their downstream targets in
ovarian cancer. However, the pathogenesis of ovarian cancer
involves multifaceted mechanisms, and circRNA research, while
promising, represents only one aspect of this intricate disease
landscape. Further comprehensive investigations are required to
fully elucidate the role of circRNAs in ovarian cancer and to
translate these findings into clinically relevant applications
(76). As research in this field
progresses, integrating circRNA studies with other aspects of
cancer biology, such as genomics, proteomics and metabolomics, may
provide a more holistic understanding of ovarian cancer
pathogenesis, and identify novel diagnostic and therapeutic
strategies (77).
Diagnostic value of circRNA in ovarian
cancer
circRNAs have emerged as promising biomarkers for
ovarian cancer diagnosis, offering unique advantages due to their
stability and tissue-specific expression patterns. A previous study
highlighted the potential of circRNAs in early detection, disease
staging and prognostic prediction for patients with ovarian cancer;
Ahmed et al (78) pioneered
the comprehensive analysis of circRNA expression profiles in
ovarian cancer. This study demonstrated a significantly higher
number of differentially expressed circRNAs in metastatic tissues
compared with that in primary lesions, in contrast to linear mRNA
patterns. Notably, circRNA expression showed negative correlation
with linear mRNA in key signaling pathways, such as NF-κB and
PI3K/AKT. These findings underscore the consistency and stability
of circRNA expression, suggesting the potential for circRNAs as
robust biomarkers for ovarian cancer detection and progression
monitoring. Further research by Pei et al (79) identified hsa_circ_0013958 as a
highly expressed circRNA in ovarian cancer tissues and cell lines.
hsa_circ_0013958 demonstrated a strong association with disease
stage and lymph node metastasis, highlighting its potential as a
diagnostic and prognostic marker. Several survival analyses have
corroborated the clinical relevance of circRNA expression in
ovarian cancer (76,80). Abnormal expression levels of
specific circRNAs, including hsa_circ_0078607, circ-la
ribonucleoprotein 4 (LARP4) and circFAM53B, have been closely
linked to pathological stage and poor prognosis (81). These findings suggest that circRNA
expression profiles could serve as valuable prognostic indicators
for patients with ovarian cancer. The current lack of accurate
screening and early diagnostic methods for ovarian cancer
underscores the urgent need for novel biomarkers. In-depth research
on circRNAs offers a promising avenue for developing early
detection strategies for ovarian lesions. By enabling timely
diagnosis and treatment initiation, circRNA-based diagnostics could
potentially improve patient survival rates (82). As research in this field progresses,
integrating circRNA biomarkers with existing diagnostic tools may
enhance the accuracy and reliability of ovarian cancer detection.
Moreover, the tissue-specific nature of circRNA expression could
facilitate the development of personalized diagnostic and
prognostic approaches, tailored to the molecular profiles of
individual patients (83).
Therapeutic value of circRNA in
ovarian cancer
Recent advances in circRNA research have revealed
the promising therapeutic potential of circRNA in ovarian cancer
treatment. These unique non-coding RNAs have been demonstrated to
possess the ability to modulate various oncogenic pathways,
offering new avenues for targeted therapies (84,85).
Notably, Lu et al (86)
investigated the tumor-suppressive role of circLARP4 in ovarian
cancer and elucidated that circLARP4 can inhibit ovarian cancer
progression through the miR-513b-5p/LARP4 axis. This finding not
only sheds light on the complex regulatory networks in ovarian
cancer but also presents circLARP4 as a potential therapeutic
target. Further research has focused on circ-itchy E3 ubiquitin
protein ligase (ITCH), another circRNA with notable antitumor
properties in ovarian cancer. Overexpression of circITCH has been
shown to significantly suppress cell proliferation and to promote
apoptosis through direct interaction with miR-10a, by acting as a
ceRNA and regulating the circITCH/miR-145/RASA1 axis. These
mechanisms have been shown to effectively inhibit ovarian cancer
cell proliferation both in vitro and in vivo
(87). The dual functionality of
circITCH underscores its potential as a versatile therapeutic
agent. The emerging understanding of these circRNA-mediated tumor
suppression mechanisms demonstrates novel possibilities for ovarian
cancer treatments. By targeting specific circRNAs or their
downstream pathways, it may be possible to develop more effective
and less toxic therapeutic strategies (88). Moreover, the tissue-specific
expression patterns of circRNAs offer the potential for highly
targeted therapies with reduced off-target effects. This
characteristic could be particularly beneficial in developing
personalized treatment approaches for patients with ovarian cancer
(2). As research in this field
progresses, it is essential to further investigate the safety and
efficacy of circRNA-based therapies. Future studies should focus on
optimizing delivery methods, understanding potential side effects
and exploring combination therapies with existing treatment
modalities (89). The therapeutic
value of circRNAs in ovarian cancer is increasingly evident. These
molecules offer promising targets for developing novel treatment
strategies that could potentially improve outcomes for patients
with this type of malignancy.
Potential of circRNA as a vaccine
candidate in ovarian cancer
circRNAs have emerged as promising candidates for
cancer vaccines, with potential applications extending to ovarian
cancer. Their unique stability and capacity for durable protein
expression make them particularly suitable for vaccine development,
offering potential options in the field of hard-to-treat
malignancies. Li et al (90)
demonstrated the efficacy of circRNA cancer vaccines in driving
immunity against challenging types of cancer. This previous study
established a novel circRNA vaccine platform by encapsulating
antigen-coding circRNA in lipid nanoparticles (LNPs). This
innovative approach triggered robust innate and adaptive immune
activation, showing superior antitumor efficacy in multiple mouse
tumor models. Furthermore, the circRNA-LNP vaccine exhibited higher
stability and initiated more durable protein expression than its
linear counterpart in vitro; in vivo, the circRNA-LNP
vaccine elicited marked innate immune responses and potent
antigen-specific T-cell responses, comparable to modified mRNA-LNP
vaccines. While specific studies on circRNA vaccines for ovarian
cancer are currently limited, the potential is considerable.
Certain circRNAs, such as circ-Wolf-Hirschhorn syndrome candidate 1
(WHSC1), have been identified as being highly expressed in ovarian
cancer tissues, and serve crucial roles in cancer cell
proliferation, apoptosis, migration and invasion (91). These circRNAs could potentially
serve as targets for vaccine development. The ability of circRNAs
to regulate key cancer-related proteins, such as MUC1 and hTERT in
the case of circWHSC1, provides a molecular basis for their
potential as vaccine candidates. However, it is important to note
that the application of circRNA vaccines in ovarian cancer is still
in its early stages. Further research is needed to identify ovarian
cancer-specific circRNAs suitable for vaccine development and to
optimize circRNA-LNP formulations for maximum efficacy in ovarian
cancer models. In addition, the safety and immunogenicity of
circRNA vaccines should be evaluated in preclinical ovarian cancer
studies, and potential combinations with existing immunotherapies
or targeted treatments should be investigated. As research
progresses, the unique properties of circRNAs, such as their
resistance to exonuclease degradation and ability to be translated
into proteins without 5′ capping or polyadenylation, may prove
advantageous in developing effective and long-lasting vaccines
against ovarian cancer (20). While
specific applications in ovarian cancer require further
investigation, the initial results of circRNA vaccines in other
cancer models are promising and could potentially revolutionize
immunotherapy strategies for ovarian cancer in the future (92).
circRNA and endometrial cancer
Endometrial cancer is a significant health concern,
ranking as the second most prevalent gynecological malignancy in
China and the foremost in developed nations. In 2022, China
reported ~84,520 new cases of endometrial cancer (93). Various risk factors have been
associated with this type of cancer, including obesity, diabetes,
polycystic ovary syndrome, infertility, early menarche and late
menopause (94). Current screening
methods for endometrial cancer primarily rely on transvaginal
ultrasound, hysteroscopy and endometrial biopsy (95). However, these techniques have the
following limitations: Transvaginal ultrasound results can be
subjective, whereas hysteroscopy and endometrial biopsy are
invasive procedures, restricting their widespread use in clinical
screening (96). The standard
treatment approach for endometrial cancer typically involves
surgery followed by chemotherapy, often using a combination of
carboplatin and PTX (97). Despite
these interventions, challenges persist, including poor prognosis
and drug resistance in some cases (98). Given these challenges, there is a
pressing need to explore novel diagnostic and therapeutic
strategies to enhance survival rates for patients with endometrial
cancer. circRNAs have emerged as promising candidates in this
regard. These unique RNA molecules, characterized by their
covalently closed loop structure, have shown potential in both the
diagnosis and treatment of various types of cancer, including
endometrial cancer (99). A
previous study demonstrated that circRNAs serve key roles in
endometrial cancer pathogenesis, influencing cell proliferation,
invasion and metastasis (100).
Moreover, certain circRNAs have been identified as potential
biomarkers for early detection and prognostic prediction in
endometrial cancer (101). The
exploration of circRNA-based therapeutic approaches, such as
circRNA-mediated gene therapy or circRNA-targeted drug delivery
systems, represents a notable option in endometrial cancer research
(102).
Mechanistic studies of circRNA in
endometrial cancer
A previous study has provided valuable insights into
the biogenesis mechanisms of circRNAs, which can be categorized
into three main models: The intron pairing model, the exon skipping
model and the RBP model. The intron pairing model posits that
complementary sequences within introns form stable secondary
structures, such as hairpins, which facilitate back-splicing and
the formation of circRNAs (103).
In endometrial cancer, the intron pairing model may explain the
high expression levels of specific circRNAs, such as circTNFRSF21,
which is significantly upregulated and competes with miR-1227 to
activate the MAPK13/ATF2 signaling pathway, promoting cancer cell
proliferation and cell cycle progression (104). Similarly, overexpression of
circWHSC1, driven by intron pairing, enhances cell proliferation,
migration and invasion while inhibiting apoptosis (105). The exon skipping model suggests
that certain exons are skipped during the normal linear splicing
process, leading to the formation of circRNAs (106). In endometrial cancer,
hsa_circ_0061140 is formed through exon skipping and interacts with
miR-149-5p to upregulate STAT3, thereby promoting cancer
progression and migration (107).
The RBP model involves the direct modulation of back-splicing by
specific RBPs, such as Quaking and Muscleblind. In endometrial
cancer, the abnormal expression of these proteins may contribute to
the biogenesis of circRNAs like circWHSC1 and hsa_circ_0061140,
influencing cancer development (3).
Diagnostic value of circRNA in
endometrial cancer
Previous studies have highlighted the potential of
circRNAs as diagnostic biomarkers in endometrial cancer. These
investigations have demonstrated distinctive circRNA expression
patterns in cancer tissues, offering novel potential for early
detection and prognostic prediction (102,108). In 2019, Ye et al (104) used sequencing techniques to
compare circRNA expression profiles in stage III endometrial cancer
tissues with adjacent non-cancerous endometrial tissues;
significant differential expression of circRNAs between these
tissue types was demonstrated. Through bioinformatics analysis, it
was identified that the
hsa_circ_0039659/hsa-miR-542-3p/hsa-let-7c-5p pathway may be a
critical predictor for stage III endometrial cancer. This discovery
not only enhances the understanding of the molecular mechanisms
underlying endometrial cancer progression but also presents a
potential diagnostic tool for advanced-stage disease. Further
supporting the diagnostic value of circRNAs, Chen et al
(109) noted that, while no
significant difference in the number of linear RNA transcripts was
detected between endometrial cancer tissues and healthy tissues, a
marked reduction in circRNA levels was observed within cancer
tissues. The comprehensive analysis identified ~120 differentially
expressed circRNAs, which may contribute to the susceptibility of
endometrial tissues to carcinogenesis. This distinct circRNA
signature in cancer tissues underscores the potential of these
molecules as specific and sensitive biomarkers for endometrial
cancer. These findings collectively suggest that circRNAs hold
considerable promise as diagnostic and prognostic biomarkers for
endometrial cancer. The unique expression patterns of circRNAs in
cancer tissues, coupled with their stability and tissue-specific
expression, make them attractive candidates for non-invasive
diagnostic tests. Moreover, the identification of specific
circRNA-miRNA-mRNA pathways, such as the
hsa_circ_0039659/hsa-miR-542-3p/hsa-let-7c-5p axis, provides
possibilities for targeted therapies and personalized medicine
approaches. Future research should focus on validating these
findings in larger, diverse patient cohorts and exploring the
potential of circRNA-based liquid biopsies for the early detection
of endometrial cancer. Additionally, investigating the functional
roles of these differentially expressed circRNAs may provide deeper
insights into the pathogenesis of endometrial cancer and identify
new therapeutic targets.
Therapeutic value of circRNA in
endometrial cancer
Advances in circRNA research have demonstrated
promising therapeutic potential in endometrial cancer. A notable
discovery in this field is the identification of hsa_circ_0001860,
a novel circRNA associated with medroxyprogesterone acetate (MPA)
resistance in patients with endometrial cancer. This circRNA has
been shown to be negatively correlated with histological grade and
lymph node metastasis, suggesting its potential as a biomarker for
disease progression and treatment response (110). Further investigation into the
molecular mechanisms of hsa_circ_0001860 has reported a role in
regulating SMAD7 expression levels and activating the SMAD7/EMT
signaling pathway. Notably, hsa_circ_0001860 has been shown to
enhance cancer cell sensitivity to MPA by targeting miR-520h.
Experimental studies have demonstrated that overexpression of
hsa_circ_0001860, coupled with miR-520h knockdown, can
significantly increase cellular sensitivity to MPA treatment. These
findings highlight the potential of hsa_circ_0001860 as a
therapeutic target for MPA-resistant endometrial cancer (110). By modulating hsa_circ_0001860, it
may be possible to overcome drug resistance and improve treatment
outcomes in patients with advanced or recurrent endometrial cancer.
This approach aligns with the growing interest in circRNAs as both
diagnostic biomarkers and therapeutic targets in various types of
cancer, including endometrial cancer (111). The discovery of hsa_circ_0001860
and its role in MPA resistance provides novel options for
personalized medicine in endometrial cancer treatment. Future
research should focus on developing strategies to manipulate this
circRNA in vivo and to assess its efficacy in clinical
settings. Additionally, exploring the interactions between
hsa_circ_0001860 and other molecular pathways involved in
endometrial cancer progression could provide a more comprehensive
understanding of its therapeutic potential. As knowledge regarding
circRNA biology is improved, it is likely that more circRNAs with
therapeutic value in endometrial cancer will be identified. This
emerging field offers possibilities for improving endometrial
cancer diagnosis, prognosis and treatment, potentially leading to
more effective and targeted therapies for patients with this
malignancy.
Potential as a vaccine candidate in
endometrial cancer
circRNAs have emerged as promising molecules in
cancer research, particularly for gynecological malignancies such
as endometrial cancer. Initially recognized for their potential as
biomarkers and therapeutic targets, recent advances have
highlighted circRNAs as novel vaccine candidates (112). The unique properties of circRNAs,
including high stability and resistance to exonuclease-mediated
degradation, make them attractive for vaccine development (113). The covalently closed structure
circRNAs enables efficient protein expression and potentially
allows for rolling circle translation. When engineered and
encapsulated in LNPs, circRNA vaccines have demonstrated the
ability to elicit robust innate and adaptive immune responses,
showing superior antitumor efficacy in preclinical models (114). Research has indicated that
circRNA-LNP complexes can induce potent antigen-specific T-cell
responses, which are crucial for combating solid tumors such as
endometrial cancer (115). The
circRNA vaccine platform offers an innovative approach for
developing cancer RNA vaccines that may be particularly applicable
to endometrial cancer, which is often considered challenging to
treat. The potential of circRNA vaccines extends beyond
monotherapy; a previous study suggested that circRNA-based
approaches may serve as both primary and adjuvant therapies,
potentially synergizing with existing immunotherapies to combat
obesity-related endometrial cancer (116). For example, a circRNA OVA-luc-LNP
vaccine has exhibited efficacy in suppressing immune-exclusive
tumor progression, inducing regression in immune-desert tumors and
preventing metastasis (90).
Compared with traditional mRNA vaccines, circRNA vaccines
demonstrate superior stability, prolonged protein expression and
the ability to initiate robust immune responses without nucleotide
modifications (117). These
characteristics position circRNA vaccines as a promising
alternative in cancer immunotherapy and have potential to improve
endometrial cancer treatment and outcomes in the future. The
ability of circRNA vaccines to collaborate with adoptive cell
transfer therapy and to suppress late-stage immune-exclusive tumor
progression by enhancing T-cell receptor T-cell therapy (TCR-T)
cell persistence demonstrates their potential as a cancer therapy
(90). Further investigation into
circRNA vaccines for endometrial cancer is warranted, as it could
lead to novel therapies. The unique properties of circRNAs,
combined with their potential for enhanced translation efficiency
through engineering, position them as a promising option in
effective cancer vaccine development.
circRNA and cervical cancer
Cervical cancer remains a notable health issue for
women worldwide, ranking as the second most common malignancy after
breast cancer. It is also associated with a high mortality rate,
being a leading cause of global cancer-related mortality.
Persistent infection with HPV is the primary driver of cervical
cancer progression. In China, cervical cancer mortality is slightly
higher in rural areas than in urban regions, with midwestern areas
exhibiting mortality rates about twice those in eastern regions
(118). Additionally, recent
trends have indicated a decreasing average age of onset for
cervical cancer, suggesting a rise in incidence among younger women
(119,120). Current treatments for cervical
cancer typically involve a combination of surgery, chemotherapy and
radiotherapy. However, poor prognosis and low survival rates due to
distant metastasis and lymphatic spread continue to challenge
patient outcomes (121). Thus,
there is an urgent need to identify new therapeutic approaches and
early diagnostic biomarkers to improve survival rates. circRNAs
have garnered significant attention in cancer research due to their
unique properties, including high stability and resistance to
exonuclease-mediated degradation (122). These characteristics make circRNAs
promising candidates for developing novel therapeutic strategies
and diagnostic tools for cervical cancer (123). Studies have suggested that
circRNAs can serve as effective biomarkers for early detection and
may offer new opportunities for targeted therapies (5,124).
For example, some circRNAs (including circ_0010235 and circCCDC66)
have been shown to interact with miRNAs and proteins involved in
cancer pathways, influencing tumor growth and metastasis (125). The potential application of
circRNA-based therapies could lead to more effective treatment
options for cervical cancer, addressing the current limitations of
conventional therapies. Research into the role of circRNAs in
cervical cancer is still in its early stages but holds promise.
Advances in circRNA studies may identify breakthrough therapies,
improving prognosis and survival rates for patients with cervical
cancer (Table II).
 | Table II.Summary of potential circRNA
biomarkers and therapeutic targets in cervical cancer. |
Table II.
Summary of potential circRNA
biomarkers and therapeutic targets in cervical cancer.
| circRNA | Expression
change | Potential as
biomarker | Potential as a
therapeutic target | Related
mechanism | (Refs.) |
|---|
|
hsa_circ_0000745 | Upregulated | Diagnostic
marker | Inhibition of tumor
progression | Regulates
miR-3178/E2F3 signaling pathway | (49) |
| circMYBL2 | Upregulated | Early diagnostic
marker | Inhibition of tumor
growth | Regulates FUS/EGFR
signaling pathway | (132) |
|
hsa_circ_0023404 | Upregulated | High (negatively
correlated with overall survival) | High | Promotes cancer
development by sponging miR-136, leading to increased expression of
TFCP2 and subsequent activation of the YAP pathway | (126) |
| circ_0005576 | Upregulated | High | High | Promotes cell
proliferation and metastasis by sponging miR-153-3p and promoting
kinesin family member 20A expression | (128) |
| circFOXO3a | Downregulated in
serum | High (lower levels
associated with lymph node metastasis and deeper stromal
invasion) | Potential (complex
role) | Lower serum levels
associated with higher overall survival and recurrence-free
survival | (130) |
| circATP8A2 | Upregulated | High (associated
with advanced FIGO stage and myometrial invasion depth) | Potential | Upregulation
closely related to disease severity and progression | (131) |
Mechanistic studies of circRNA in
cervical cancer
Previous research has unveiled significant roles of
circRNAs in the pathogenesis and progression of cervical cancer,
offering novel insights into potential diagnostic and therapeutic
approaches (47). Notably, survival
analyses on hsa_circ_0023404 demonstrated an inverse correlation
between hsa_circ_0023404 expression and overall patient survival,
with higher expression levels associated with poorer outcomes.
Mechanistically, hsa_circ_0023404 functions as a miRNA sponge,
specifically targeting miR-136. This interaction led to increased
expression levels of the transcription factor CP2 and subsequent
activation of the YAP pathway, ultimately driving cancer
progression (126). Another
notable circRNA, circ_0005576, has been shown to be markedly
upregulated in cervical cancer tissues. Localized in the cytoplasm,
this circRNA serves a key role in cell proliferation and metastasis
(127). Functional studies have
demonstrated that knockdown of circ_0005576 can inhibit these
malignant behaviors, while its overexpression may enhance them. The
underlying mechanism involves circ_0005576 sponging miR-153-3p,
thereby promoting the expression of KIF20A, a key player in cancer
cell division and movement (128).
Given that HPV infection is the primary etiological factor in
cervical cancer, exploring the interplay between HPV and circRNAs
presents a promising avenue for future research. Understanding
these relationships could potentially identify novel therapeutic
targets and strategies for cervical cancer treatment (129). These mechanistic studies highlight
the complex regulatory networks involving circRNAs in cervical
cancer and underscore their potential as both biomarkers and
therapeutic targets. Further investigation into the functional
roles of circRNAs and their interactions with other molecular
players in cervical cancer pathogenesis is warranted to fully
harness their clinical potential.
Diagnostic value of circRNA in
cervical cancer
circRNAs have emerged as promising diagnostic and
prognostic biomarkers in cervical cancer due to their stability and
tissue-specific expression patterns. A previous study highlighted
the potential of specific circRNAs in predicting disease
progression and patient outcomes; Tang et al (130) demonstrated that lower serum levels
of circFoxO3a are associated with positive lymph node metastasis
and increased depth of stromal invasion in patients with cervical
cancer. Notably, patients with lower circFOXO3a serum levels
exhibited higher overall survival and relapse-free survival rates,
suggesting its complex role in cancer progression and its potential
as a prognostic marker. Further emphasizing the diagnostic
potential of circRNAs, Ding and Zhang (131) identified circ-ATPase phospholipid
transporting 8A2 (ATP8A2) as being significantly upregulated in
both cervical cancer tissues and cell lines. This finding indicated
that elevated levels of circATP8A2 may be strongly associated with
advanced International Federation of Gynecology and Obstetrics
staging and the degree of myometrial invasion. This association
underscores the potential of circATP8A2 as a biomarker for disease
severity and progression. These studies collectively demonstrate
the promising role of circRNAs as diagnostic and prognostic
biomarkers in cervical cancer. The differential expression of
circRNAs such as circFOXO3a and circATP8A2 in relation to
clinicopathological features offers new avenues for non-invasive
diagnosis and personalized treatment strategies. However, further
large-scale clinical studies are required to validate these
findings and to establish standardized protocols for circRNA-based
diagnostics in cervical cancer management.
Therapeutic value of circRNA in
cervical cancer
Previous research has unveiled the potential of
circRNAs as therapeutic targets in cervical cancer, particularly in
addressing drug resistance. In a novel study from 2021, Dong et
al (132) provided information
on the role of circMYBL2 in PTX resistance, a common challenge in
cervical cancer treatment, demonstrating that circMYBL2 may serve a
key role in modulating the sensitivity of cervical cancer cells to
PTX. Overexpression of circ-MYB proto-oncogene like 2 (MYBL2) was
found to significantly reduce the growth-inhibitory effects of PTX
on cancer cells; by contrast, the knockdown of circMYBL2 enhanced
the efficacy of PTX, demonstrating its potential as a therapeutic
target. The mechanism underlying this effect was shown to involve
the regulation of miR-665; the study showed that overexpression or
inhibition of miR-665 could reverse the effects of circMYBL2 on PTX
sensitivity. These findings suggest that circMYBL2 functions as a
positive regulator of drug resistance and malignant behavior in
cervical cancer cells through its interaction with miR-665. This
discovery provides novel options for developing targeted therapies
for patients with PTX-resistant cervical cancer. By manipulating
circMYBL2 levels or disrupting its interaction with miR-665, it may
be possible to re-sensitize resistant tumors to PTX treatment. The
identification of circMYBL2 as a key player in drug resistance
highlights the broader potential of circRNAs as therapeutic targets
in cervical cancer. This research not only provides insights into
the mechanisms of drug resistance but also offers a promising
strategy for overcoming this notable clinical challenge. Future
studies may focus on developing circRNA-based therapeutics or using
circMYBL2 as a biomarker for predicting PTX response in patients
with cervical cancer (133).
Several other circRNAs, such as circEIF4G2 (134) and circ_0006528 (135), have also been implicated in
cervical cancer progression and drug resistance, further
highlighting the therapeutic potential of circRNAs in this
malignancy.
Potential as a vaccine candidate in
cervical cancer
Advances in circRNA research have demonstrated
potential for their application as cancer vaccines, including for
cervical cancer. This approach leverages the unique properties of
circRNAs to stimulate robust immune responses against malignancies.
For example, Niu et al (136) demonstrated the ability of a
circRNA-based cancer vaccine to induce potent innate and adaptive
immune responses in preclinical models, showcasing its therapeutic
potential against types of difficult-to-treat cancer. For instance,
combining circRNA-LNP vaccines with adoptive T cell transfer (OT-I
cells) achieved complete tumor regression in all treated mice with
advanced B16-OVA melanoma, while monotherapy (circRNA vaccine
alone) resulted in 60% survival. This combination strategy extended
survival beyond 60 days in 100% of mice, compared with rapid
mortality in control groups. The potential mechanism is illustrated
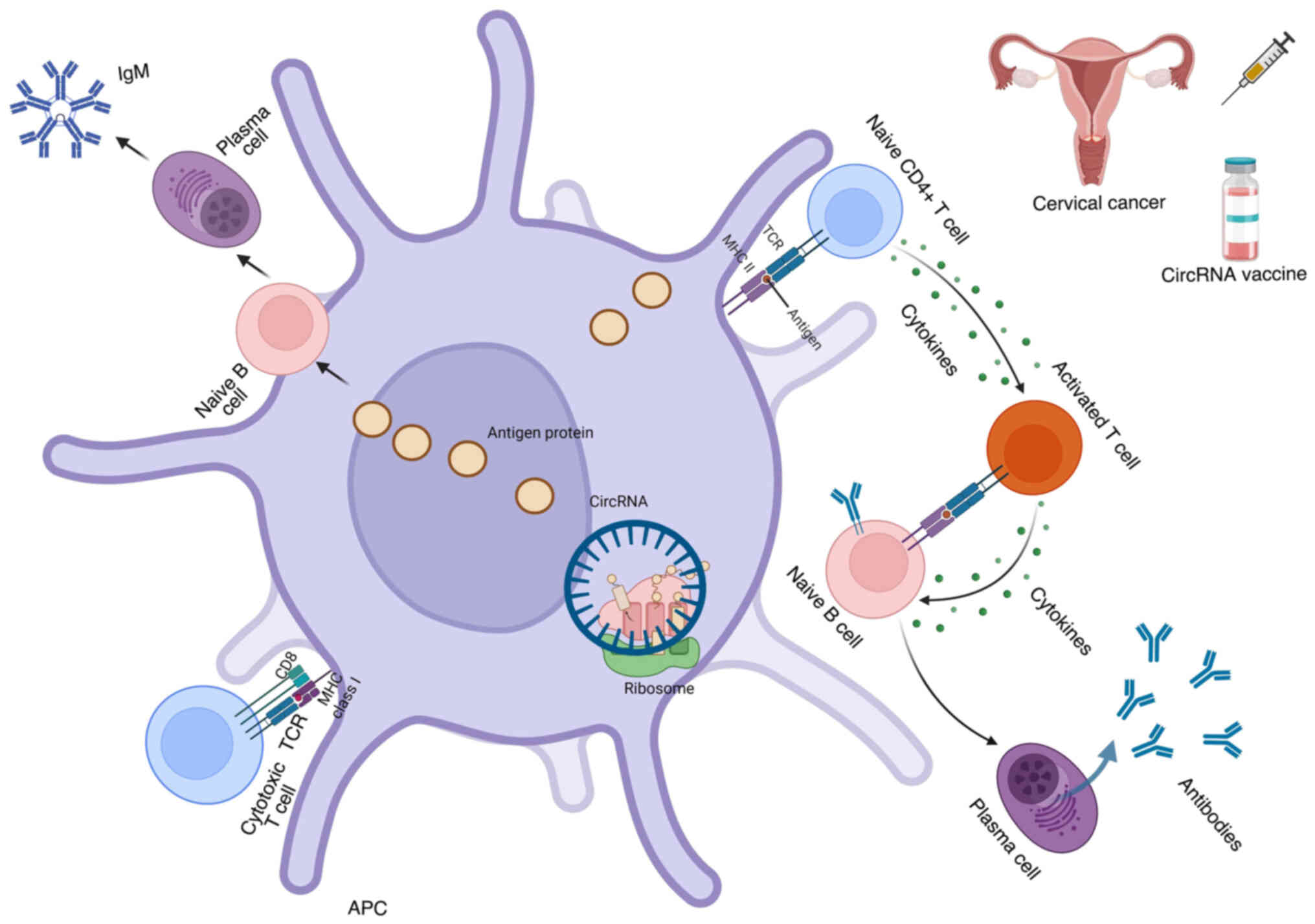
in Fig. 1.
While direct studies on circRNA-based vaccines for
cervical cancer are limited, research on the role of circRNAs in
cervical cancer progression provides valuable insights into their
potential as vaccine candidates. For example, Ou et al
(137) reported that circAMOTL1
can promote cervical cancer growth by acting as a sponge for
miR-485-5p, leading to increased AMOTL1 levels. This finding
highlights the possibility of targeting specific circRNAs involved
in cancer progression for vaccine development. Furthermore, Wang
et al (138) emphasized the
significance of genome-wide perturbations in A-to-I RNA editing in
dysregulating circRNAs and promoting cervical cancer development.
This previous study identified three RNA editing sites as potential
biomarkers or therapeutic targets, underscoring the therapeutic
potential of focusing on RNA editing dysregulation in cervical
cancer treatment. The unique stability and tissue-specific
expression patterns of circRNAs make them attractive candidates for
vaccine development. By designing circRNA vaccines that encode
specific tumor antigens or target circRNAs involved in cervical
cancer progression, researchers may be able to elicit more targeted
and effective immune responses against this disease. While these
findings are promising, it is important to note that the
development of circRNA-based vaccines for cervical cancer is still
in its early stages. Further research is needed to fully elucidate
the mechanisms by which circRNA vaccines stimulate antitumor
immunity, and to optimize their delivery and efficacy in clinical
settings. Nonetheless, the potential of circRNAs as vaccine
candidates offers a potential new option for cervical cancer
immunotherapy, which may lead to more effective and personalized
treatment strategies in the future.
Challenges and limitations of
circRNA-based therapies for gynecological cancer
CircRNAs hold potential as therapeutic tools;
however, efficiently and safely translating them into effective
treatments requires addressing several critical challenges. Despite
the significant potential of circRNAs, efficient and safe delivery
remains a major challenge. Current strategies for delivering
circRNAs include lipid nanoparticles, viral vectors and exosomes.
Each of these delivery systems has its limitations. For instance,
viral vectors, while highly efficient, can trigger immune responses
or insertional mutagenesis (139).
Lipid nanoparticles, on the other hand, require further
optimization for stability and tissue specificity (140). Additionally, ensuring that
circRNAs are delivered to tumor sites while minimizing their
accumulation in non-targeted tissues is a pressing issue (141).
The ability of circRNAs to regulate gene expression
by interacting with miRNAs and RBPs can also lead to non-specific
targeting effects, causing off-target effects. circRNAs may
simultaneously regulate multiple miRNAs, thereby affecting multiple
signaling pathways and leading to unintended biological
consequences (124). Therefore, in
developing circRNA therapies, extensive in vivo studies and
high-throughput screening are necessary to ensure the specificity
of their targeting effects while avoiding interference with normal
cellular functions (142).
The clinical translation of circRNA therapies also
depends on strict regulatory requirements. As an emerging
biotherapeutic tool, circRNAs must meet the standards of
international drug regulatory agencies such as the U.S. Food and
Drug Administration. Research on the long-term safety,
immunogenicity and metabolic pathways of circRNA drugs is still in
its early stages. Before circRNA therapies can be used clinically,
uniform quality control standards must be established, and rigorous
preclinical and clinical trials must be conducted to ensure their
safety and efficacy (143).
Conclusion
The emerging field of circRNA research offers
promising options for advancing the understanding and management of
gynecological malignancies. The present review has highlighted the
diverse roles of circRNAs in ovarian, endometrial and cervical
cancer, from their involvement in tumor progression to their
potential as diagnostic and prognostic biomarkers. The unique
properties of circRNAs, including their stability, tissue-specific
expression and diverse regulatory functions, position them as
valuable tools in gynecological oncology. The roles of circRNAs as
miRNA sponges, protein scaffolds and even potential peptide
encoders underscore their complex involvement in cancer
pathogenesis. While notable progress has been made in elucidating
the functions of circRNAs in gynecological cancer, much remains to
be explored. Future research should focus on validating
circRNA-based biomarkers, investigating tissue-specific and
cancer-specific circRNA-mediated transcriptional regulation,
exploring the therapeutic potential of targeting circRNAs and
elucidating the functional significance of circRNA-encoded peptides
in gynecological cancer. As the understanding of circRNAs in
gynecological malignancies improves, the development of novel
diagnostic tools, prognostic markers and targeted therapies is
anticipated. These advances hold the potential to markedly improve
patient outcomes and to reshape the landscape of gynecological
cancer management.
Acknowledgements
Not applicable.
Funding
The present study was provided financial support from the
following projects: the scholarship under the China Scholarship
Council (grant no. 202308210316), Liaoning Province Science and
Technology Program Joint Program Fund Project (grant no.
2023-MSLH-059), Postgraduate Education Teaching Research and Reform
Project of Jinzhou Medical University (grant no. YJ2023-018), Jie
Bang Gua Shuai Project of Science & Technology Department of
Liaoning Province (grant no. 2022JH1/10800070) and Basic Scientific
Research Project of Colleges and Universities of Education
Department of Liaoning Province (key project) (grant no.
1821240403).
Availability of data and materials
Not applicable.
Author's contributions
YL was responsible for writing the manuscript and
investigation of the subject, such as conducting literature reviews
to understand the current state of research. HA edited,
conceptualized and supervised the present study. Data
authentication is not applicable. All authors have read and
approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Conn VM, Chinnaiyan AM and Conn SJ:
Circular RNA in cancer. Nat Rev Cancer. 24:597–613. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qin M, Zhang C and Li Y: Circular RNAs in
gynecologic cancers: Mechanisms and implications for chemotherapy
resistance. Front Pharmacol. 14:11947192023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang J, Luo Z, Zheng Y, Duan M, Qiu Z and
Huang C: CircRNA as an Achilles heel of cancer: Characterization,
biomarker and therapeutic modalities. J Transl Med. 22:7522024.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zong ZH, Du YP, Guan X, Chen S and Zhao Y:
CircWHSC1 promotes ovarian cancer progression by regulating MUC1
and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer
Res. 38:4372019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Y, Yang Z, Zeng M, Wang Y, Chen X, Li
S, Zhao X and Sun Y: Circular RNA differential expression profiles
and bioinformatics analysis of hsa_circRNA_079422 in human
endometrial carcinoma. J Obstet Gynaecol. 43:22288942023.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Heidari-Ezzati S, Moeinian P,
Ahmadian-Nejad B, Maghbbouli F, Abbasi S, Zahedi M, Afkhami H,
Shadab A and Sajedi N: The role of long non-coding RNAs and
circular RNAs in cervical cancer: Modulating miRNA function. Front
Cell Dev Biol. 12:13087302024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lei B, Tian Z, Fan W and Ni B: Circular
RNA: A novel biomarker and therapeutic target for human cancers.
Int J Med Sci. 16:292–301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He AT, Liu J, Li F and Yang BB: Targeting
circular RNAs as a therapeutic approach: Current strategies and
challenges. Signal Transduct Target Ther. 6:1852021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Misir S, Wu N and Yang BB: Specific
expression and functions of circular RNAs. Cell Death Differ.
29:481–491. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nisar S, Bhat AA, Singh M, Karedath T,
Rizwan A, Hashem S, Bagga P, Reddy R, Jamal F, Uddin S, et al:
Insights into the role of CircRNAs: Biogenesis, characterization,
functional, and clinical impact in human malignancies. Front Cell
Dev Biol. 9:6172812021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Papadopoulos N and Trifylli EM: Role of
exosomal circular RNAs as microRNA sponges and potential targeting
for suppressing hepatocellular carcinoma growth and progression.
World J Gastroenterol. 30:994–998. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Panda AC: Circular RNAs Act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hentze MW and Preiss T: Circular RNAs:
Splicing's enigma variations. EMBO J. 32:923–925. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 Is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen YG, Chen R, Ahmad S, Verma R, Kasturi
SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, et al:
N6-methyladenosine modification controls circular RNA immunity. Mol
Cell. 76:96–109.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Mo Y, Gong Z, Yang X, Yang M,
Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al: Circular RNAs in
human cancer. Mol Cancer. 16:252017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tashakori N, Mikhailova MV, Mohammedali
ZA, Mahdi MS, Ali Al-Nuaimi AM, Radi UK, Alfaraj AM and Kiasari BA:
Circular RNAs as a novel molecular mechanism in diagnosis,
prognosis, therapeutic target, and inhibiting chemoresistance in
breast cancer. Pathol Res Pract. 263:1555692024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Holdt LM, Kohlmaier A and Teupser D:
Circular RNAs as therapeutic agents and targets. Front Physiol.
9:12622018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ragan C, Goodall GJ, Shirokikh NE and
Preiss T: Insights into the biogenesis and potential functions of
exonic circular RNA. Sci Rep. 9:20482019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang T, Zhang X and Zheng B:
Identification of intronic lariat-derived circular RNAs in
arabidopsis by RNA deep sequencing. Methods Mol Biol. 2362:93–100.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhong Y, Yang Y, Wang X, Ren B, Wang X,
Shan G and Chen L: Systematic identification and characterization
of exon-intron circRNAs. Genome Res. 34:376–393. 2024.PubMed/NCBI
|
|
27
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo Z, Cao Q, Zhao Z and Song C:
Biogenesis, features, functions, and disease relationships of a
specific circular RNA: CDR1as. Aging Dis. 11:1009–1020. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu J, Li Q, Wu Z, Xu W and Jiang R:
Circular RNA-mediated miRNA sponge & RNA binding protein in
biological modulation of breast cancer. Noncoding RNA Res.
9:262–276. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pisignano G, Michael DC, Visal TH, Pirlog
R, Ladomery M and Calin GA: Going circular: History, present, and
future of circRNAs in cancer. Oncogene. 42:2783–2800. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao X, Zhong Y, Wang X, Shen J and An W:
Advances in circular RNA and its applications. Int J Med Sci.
19:975–985. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li L, Wei C, Xie Y, Su Y, Liu C, Qiu G,
Liu W, Liang Y, Zhao X, Huang D and Wu D: Expanded insights into
the mechanisms of RNA-binding protein regulation of circRNA
generation and function in cancer biology and therapy. Gen Dis.
1013832024.
|
|
35
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nature reviews.
Nat Rev Mol Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang A, Zheng H, Wu Z, Chen M and Huang
Y: Circular RNA-protein interactions: functions, mechanisms, and
identification. Theranostics. 10:3503–3517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen L and Shan G: CircRNA in cancer:
Fundamental mechanism and clinical potential. Cancer Lett.
505:49–57. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hwang HJ and Kim YK: Molecular mechanisms
of circular RNA translation. Exp Mol Med. 56:1272–1280. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang X, Mei J, Wang H, Gu D, Ding J and
Liu C: The emerging roles of circular RNAs in ovarian cancer.
Cancer Cell Int. 20:2652020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gao S, Zhao T, Meng F, Luo Y, Li Y and
Wang Y: Circular RNAs in endometrial carcinoma (Review). Oncol Rep.
48:2122022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ye C, Liu Z, Xie Q, Tang Y, Zeng J, Feng
Z, Liu J and Xie H: Adeno-associated virus mediated artificial
circular RNA for triggering cancer immunotherapy to treat prostate
cancer. Front Oncol. 15:14435712025. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lv H, Wei X and Lin Y: Immunoregulatory
role of exosomal CircRNAs in the tumor microenvironment. Front
Oncol. 15:14537862025. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jarlstad Olesen MT and S Kristensen L:
Circular RNAs as microRNA sponges: Evidence and controversies.
Essays Biochem. 65:685–696. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Piwecka M, Glažar P, Hernandez-Miranda LR,
Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda
Jara CA, Fenske P, et al: Loss of a mammalian circular RNA locus
causes miRNA deregulation and affects brain function. Science.
357:eaam85262017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Heydarnia E, Dorostgou Z, Hedayati N,
Mousavi V, Yahyazadeh S, Alimohammadi M, Gheibi M, Heidari P, Igder
S, Mafi A and Vakili O: Circular RNAs and cervical cancer: Friends
or foes? A landscape on circRNA-mediated regulation of key
signaling pathways involved in the onset and progression of
HPV-related cervical neoplasms. Cell Commun Signal. 22:1072024.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ghafouri-Fard S, Khoshbakht T, Hussen BM,
Sarfaraz S, Taheri M and Ayatollahi SA: Circ_CDR1as: A circular RNA
with roles in the carcinogenesis. Pathol Res Pract. 236:1539682022.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jiao J, Zhang T, Jiao X, Huang T, Zhao L,
Ma D and Cui B: hsa_circ_0000745 promotes cervical cancer by
increasing cell proliferation, migration, and invasion. J Cell
Physiol. 235:1287–1295. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang Y, Yin L and Sun X: CircRNA
hsa_circ_0002577 accelerates endometrial cancer progression through
activating IGF1R/PI3K/Akt pathway. J Exp Clin Cancer Res.
39:1692020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ma Y, Zheng L, Gao Y, Zhang W, Zhang Q and
Xu Y: A comprehensive overview of circRNAs: Emerging biomarkers and
potential therapeutics in gynecological cancers. Front Cell Dev
Biol. 9:7095122021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rao D, Yu C, Sheng J, Lv E and Huang W:
The emerging roles of circFOXO3 in cancer. Front Cell Dev Biol.
9:6594172021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang L, Chen J and Lu C: Circular RNA
Foxo3 enhances progression of ovarian carcinoma cells. Aging.
13:22432–22443. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R,
Thorne RF, Zhang XD, Hu W and Wu M: CircACC1 regulates assembly and
activation of AMPK complex under metabolic stress. Cell Metab.
30:157–173.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhou M, Yang Z, Wang D, Chen P and Zhang
Y: The circular RNA circZFR phosphorylates Rb promoting cervical
cancer progression by regulating the SSBP1/CDK2/cyclin E1 complex.
J Exp Clin Cancer Res. 40:482021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Guan X, Zong ZH, Liu Y, Chen S, Wang LL
and Zhao Y: circPUM1 promotes tumorigenesis and progression of
ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther
Nucleic Acids. 18:882–892. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ and
Xu RH: Circular RNA: Metabolism, functions and interactions with
proteins. Mol Cancer. 19:1722020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xue C, Li G, Lu J and Li L: Crosstalk
between circRNAs and the PI3K/AKT signaling pathway in cancer
progression. Signal Transduct Target Ther. 6:4002021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou
X, Xie X and Tang H: circFBXW7 inhibits malignant progression by
sponging miR-197-3p and encoding a 185-aa protein in
triple-negative breast cancer. Mol Ther Nucleic Acids. 18:88–98.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhao J, Lee EE, Kim J, Yang R, Chamseddin
B, Ni C, Gusho E, Xie Y, Chiang CM, Buszczak M, et al: Transforming
activity of an oncoprotein-encoding circular RNA from human
papillomavirus. Nat Commun. 10:23002019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ju X, Tang Y, Qu R and Hao S: The emerging
role of circ-SHPRH in cancer. Onco Targets Ther. 14:4177–4188.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang S, Cheng C, Lin Z, Xiao L, Su X,
Zheng L, Mu Y, Liao M, Ouyang R, Li W, et al: The global burden and
associated factors of ovarian cancer in 1990–2019: Findings from
the global burden of disease study 2019. BMC Public Health.
22:14552022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Bowtell DD, Böhm S, Ahmed AA, Aspuria PJ,
Bast RC Jr, Beral V, Berek JS, Birrer MJ, Blagden S, Bookman MA, et
al: Rethinking ovarian cancer II: Reducing mortality from
high-grade serous ovarian cancer. Nat Rev Cancer. 15:668–679. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian cancer statistics, 2018. CA Cancer J Clin. 68:284–296.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Colombo N, Ledermann JA and Lorusso D:
Newly diagnosed and relapsed epithelial ovarian cancer: ESMO
clinical practice guideline for diagnosis, treatment and follow-up.
Ann Oncol. 34:127–144. 2023. View Article : Google Scholar
|
|
69
|
Tendulkar S and Dodamani S:
Chemoresistance in ovarian cancer: Prospects for new drugs.
Anticancer Agents Med Chem. 21:668–678. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Xing Y, Liang X, Lv X, Cheng Y, Du J, Liu
C and Yang Y: New insights into the role of circular RNAs in
ovarian cancer. Pathol Res Pract. 238:1540732022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song
X, Gorshkov K, Sun Q, Lin H, Zheng X, et al: CircRNA-SORE mediates
sorafenib resistance in hepatocellular carcinoma by stabilizing
YBX1. Signal Transduct Target Ther. 5:2982020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Luo Y, Fu Y, Huang R, Gao M, Liu F, Gui R
and Nie X: CircRNA_101505 sensitizes hepatocellular carcinoma cells
to cisplatin by sponging miR-103 and promotes oxidored-nitro
domain-containing protein 1 expression. Cell Death Discov.
5:1212019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang H, Zhang Y, Miao H, Xu T, Nie X and
Cheng W: CircRAD23B promotes proliferation and carboplatin
resistance in ovarian cancer cell lines and organoids. Cancer Cell
Int. 24:422024. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Gan X, Zhu H, Jiang X, Obiegbusi SC, Yong
M, Long X and Hu J: CircMUC16 promotes autophagy of epithelial
ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer.
19:452020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li QH, Liu Y, Chen S, Zong ZH, Du YP,
Sheng XJ and Zhao Y: circ-CSPP1 promotes proliferation, invasion
and migration of ovarian cancer cells by acting as a miR-1236-3p
sponge. Biomed Pharmacother. 114:1088322019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Foruzandeh Z, Zeinali-Sehrig F, Nejati K,
Rahmanpour D, Pashazadeh F, Seif F and Alivand MR: CircRNAs as
potent biomarkers in ovarian cancer: A systematic scoping review.
Cell Mol Biol Lett. 26:412021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881.e13. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ahmed I, Karedath T, Andrews SS, Al-Azwani
IK, Mohamoud YA, Querleu D, Rafii A and Malek JA: Altered
expression pattern of circular RNAs in primary and metastatic sites
of epithelial ovarian carcinoma. Oncotarget. 7:36366–36381. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Pei C, Wang H, Shi C, Zhang C and Wang M:
CircRNA hsa_circ_0013958 may contribute to the development of
ovarian cancer by affecting epithelial-mesenchymal transition and
apoptotic signaling pathways. J Clin Lab Anal. 34:e232922020.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Sun Y, Chen X, Shi Y, Teng F, Dai C, Ge L,
Xu J and Jia X: hsa_circ_0020093 suppresses ovarian cancer
progression by modulating LRPPRC activity and miR-107/LATS2
signaling. Biol Direct. 19:692024. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang N, Jin Y, Hu Q, Cheng S, Wang C,
Yang Z and Wang Y: Circular RNA hsa_circ_0078607 suppresses ovarian
cancer progression by regulating miR-518a-5p/Fas signaling pathway.
J Ovarian Res. 13:642020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Shabaninejad Z, Vafadar A, Movahedpour A,
Ghasemi Y, Namdar A, Fathizadeh H, Pourhanifeh MH, Savardashtaki A
and Mirzaei H: Circular RNAs in cancer: New insights into functions
and implications in ovarian cancer. J Ovarian Res. 12:842019.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Malviya A and Bhuyan R: Circular RNAs in
cancer: Roles, mechanisms, and therapeutic potential across
colorectal, gastric, liver, and lung carcinomas. Discov Oncol.
16:52025. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhu Y, Liang L, Zhao Y, Li J, Zeng J, Yuan
Y, Li N and Wu L: CircNUP50 is a novel therapeutic target that
promotes cisplatin resistance in ovarian cancer by modulating p53
ubiquitination. J Nanobiotechnology. 22:352024. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Wang Y, Chen X and Yang Y:
CircRNA-regulated glucose metabolism in ovarian cancer: An emerging
landscape for therapeutic intervention. Clin Transl Oncol.
26:584–596. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lu H, Guo Q, Mao G, Zhu J and Li F:
CircLARP4 suppresses cell proliferation, invasion and glycolysis
and promotes apoptosis in non-small cell lung cancer by targeting
miR-135b. Onco Targets Ther. 13:3717–3728. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Luo L, Gao YQ and Sun XF: Circular RNA
ITCH suppresses proliferation and promotes apoptosis in human
epithelial ovarian cancer cells by sponging miR-10a-α. Eur Rev Med
Pharmacol Sci. 22:8119–8126. 2018.PubMed/NCBI
|
|
88
|
Verduci L, Tarcitano E, Strano S, Yarden Y
and Blandino G: CircRNAs: Role in human diseases and potential use
as biomarkers. Cell Death Dis. 12:4682021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Arnaiz E, Sole C, Manterola L,
Iparraguirre L, Otaegui D and Lawrie CH: CircRNAs and cancer:
Biomarkers and master regulators. Semin Cancer Biol. 58:90–99.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li H, Peng K, Yang K, Ma W, Qi S, Yu X, He
J, Lin X and Yu G: Circular RNA cancer vaccines drive immunity in
hard-to-treat malignancies. Theranostics. 12:6422–6436. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang X, Yuan Y, Wang X, Wang H, Zhang L
and He J: CircWHSC1 (CircNSD2): A novel circular RNA in multiple
cancers. Clin Med Insights Oncol. 18:117955492412547812024.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Du Y, Zuber PK, Xiao H, Li X, Gordiyenko Y
and Ramakrishnan V: Efficient circular RNA synthesis for potent
rolling circle translation. Nat Biomed Eng. Dec 13–2024.(Epub ahead
of print). View Article : Google Scholar
|
|
93
|
Li J, Li X, Quan C, Li X, Wan C and Wu X:
Genomic profile of Chinese patients with endometrial carcinoma. BMC
Cancer. 23:8882023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Morice P, Leary A, Creutzberg C,
Abu-Rustum N and Darai E: Endometrial cancer. Lancet.
387:1094–1108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Du Y, Xu Y, Qin Z, Sun L, Chen Y, Han L
and Zheng A: The oncology safety of diagnostic hysteroscopy in
early-stage endometrial cancer: A systematic review and
meta-analysis. Front Oncol. 11:7427612021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Clarke MA, Long BJ, Del Mar Morillo A,
Arbyn M, Bakkum-Gamez JN and Wentzensen N: Association of
endometrial cancer risk with postmenopausal bleeding in women: A
systematic review and meta-analysis. JAMA Intern Med.
178:1210–1222. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Colombo N, Creutzberg C, Amant F, Bosse T,
González-Martín A, Ledermann J, Marth C, Nout R, Querleu D, Mirza
MR, et al: ESMO-ESGO-ESTRO consensus conference on endometrial
cancer: Diagnosis, treatment and follow-up. Ann Oncol. 27:16–41.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Kok PS, Antill YC, Scott CL and Lee CK:
The impact of single agent PD-1 or PD-L1 inhibition on advanced
endometrial cancers: Meta-analysis. ESMO Open. 7:1006352022.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Tao M, Zheng M, Xu Y, Ma S, Zhang W and Ju
S: CircRNAs and their regulatory roles in cancers. Mol Med.
27:942021. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Xu H, Gong Z, Shen Y, Fang Y and Zhong S:
Circular RNA expression in extracellular vesicles isolated from
serum of patients with endometrial cancer. Epigenomics. 10:187–197.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhang W, Xu C, Yang Z, Zhou J, Peng W,
Zhang X, Li H, Qu S and Tao K: Circular RNAs in tumor immunity and
immunotherapy. Mol Cancer. 23:1712024. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Włodarczyk K, Kuryło W, Pawłowska-Łachut
A, Skiba W, Suszczyk D, Pieniądz P, Majewska M, Boniewska-Bernacka
E and Wertel I: circRNAs in endometrial cancer-a promising
biomarker: State of the art. Int J Mol Sci. 25:63872024. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Das A, Sinha T, Shyamal S and Panda AC:
Emerging role of circular RNA-protein interactions. Noncoding RNA.
7:482021.PubMed/NCBI
|
|
104
|
Ye F, Tang QL, Ma F, Cai L, Chen M, Ran
XX, Wang XY and Jiang XF: Analysis of the circular RNA
transcriptome in the grade 3 endometrial cancer. Cancer Manag Res.
11:6215–6227. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Liu Y, Chen S, Zong ZH, Guan X and Zhao Y:
CircRNA WHSC1 targets the miR-646/NPM1 pathway to promote the
development of endometrial cancer. J Cell Mol Med. 24:6898–6907.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Mumtaz PT, Taban Q, Dar MA, Mir S, Haq ZU,
Zargar SM, Shah RA and Ahmad SM: Deep insights in circular RNAs:
From biogenesis to therapeutics. Biol Proced Online. 22:102020.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Liu Y, Chang Y and Cai Y: Hsa_circ_0061140
promotes endometrial carcinoma progression via regulating
miR-149-5p/STAT3. Gene. 745:1446252020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Shi R, Zhao R, Shen Y, Wei S, Zhang T,
Zhang J, Shu W, Cheng S, Teng H and Wang H: IGF2BP2-modified
circular RNA circCHD7 promotes endometrial cancer progression via
stabilizing PDGFRB and activating JAK/STAT signaling pathway.
Cancer Gene Ther. 31:1221–1236. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Chen BJ, Byrne FL, Takenaka K, Modesitt
SC, Olzomer EM, Mills JD, Farrell R, Hoehn KL and Janitz M:
Analysis of the circular RNA transcriptome in endometrial cancer.
Oncotarget. 9:5786–5796. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yuan S, Zheng P, Sun X, Zeng J, Cao W, Gao
W, Wang Y and Wang L: Hsa_Circ_0001860 promotes Smad7 to enhance
MPA resistance in endometrial cancer via miR-520h. Front Cell Dev
Biol. 9:7381892021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang
Y, Li X, Wu Z, Yang D, Zhou Y, et al: Circular RNAs in cancer:
Emerging functions in hallmarks, stemness, resistance and roles as
potential biomarkers. Mol Cancer. 18:902019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Ren Y, Manoharan T, Liu B, Cheng CZM, En
Siew B, Cheong WK, Lee KY, Tan IJ, Lieske B, Tan KK and Chia G:
Circular RNA as a source of neoantigens for cancer vaccines. J
Immunother Cancer. 12:e0084022024. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Xie J, Ye F, Deng X, Tang Y, Liang JY,
Huang X, Sun Y, Tang H, Lei J, Zheng S and Zou Y: Circular RNA: A
promising new star of vaccine. J Transl Int Med. 11:372–381. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Liu X, Zhang Y, Zhou S, Dain L, Mei L and
Zhu G: Circular RNA: An emerging frontier in RNA therapeutic
targets, RNA therapeutics, and mRNA vaccines. J Control Release.
348:84–94. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Li W, Liu JQ, Chen M, Xu J and Zhu D:
Circular RNA in cancer development and immune regulation. J Cell
Mol Med. 26:1785–1798. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Takenaka K, Olzomer EM, Hoehn KL,
Curry-Hyde A, Jun Chen B, Farrell R, Byrne FL and Janitz M:
Investigation of circular RNA transcriptome in obesity-related
endometrial cancer. Gene. 855:1471252023. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Su P and Zhang L, Zhou F and Zhang L:
Circular RNA vaccine, a novel mRNA vaccine design strategy for
SARS-CoV-2 and variants. MedComm (2020). 3:e1532022. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Xiaohui C: Analysis cervical cancer among
urban and rural residents in Xiangyang, Hubei, 2017–2019. Prog Med
Sci. 9:1–5. 2025.
|
|
119
|
Agarwal R, King JB, Gopalani SV and
Senkomago V: Cervical cancer incidence and trends among women aged
15–29 years by county-level economic status and rurality-United
States, 2007–2020. Cancer Epidemiol. 94:1027302025. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Tanaka S, Palmer M and Katanoda K: Trends
in cervical cancer incidence and mortality of young and middle
adults in Japan. Cancer Sci. 113:1801–1807. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Liu X, Meng Q, Wang W, Zhou Z, Zhang F and
Hu K: Predictors of distant metastasis in patients with cervical
cancer treated with definitive radiotherapy. J Cancer.
10:3967–3974. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ogasawara A and Hasegawa K: Recent
advances in immunotherapy for cervical cancer. Int J Clin Oncol.
30:434–448. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Wan J, Wang Z, Wang L, Wu L, Zhang C, Zhou
M, Fu ZF and Zhao L: Circular RNA vaccines with long-term lymph
node-targeting delivery stability after lyophilization induce
potent and persistent immune responses. mBio. 15:e01775232024.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Saleem A, Khan MU, Zahid T, Khurram I,
Ghani MU, Ullah I, Munir R, Calina D and Sharifi-Rad J: Biological
role and regulation of circular RNA as an emerging biomarker and
potential therapeutic target for cancer. Mol Biol Rep. 51:2962024.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Ma Y, Wang T, Zhang X, Wang P and Long F:
The role of circular RNAs in regulating resistance to cancer
immunotherapy: Mechanisms and implications. Cell Death Dis.
15:3122024. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Zhang J, Zhao X, Zhang J, Zheng X and Li
F: Circular RNA hsa_circ_0023404 exerts an oncogenic role in
cervical cancer through regulating miR-136/TFCP2/YAP pathway.
Biochem Biophys Res Commun. 501:428–433. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Shi Y, He R, Yang Y, He Y, Shao K, Zhan L
and Wei B: Circular RNAs: Novel biomarkers for cervical, ovarian
and endometrial cancer (Review). Oncol Rep. 44:1787–1798.
2020.PubMed/NCBI
|
|
128
|
Ma H, Tian T, Liu X, Xia M, Chen C, Mai L,
Xie S and Yu L: Upregulated circ_0005576 facilitates cervical
cancer progression via the miR-153/KIF20A axis. Biomed
Pharmacother. 118:1093112019. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Hsu CY, Faisal A, Jumaa SS, Gilmanova NS,
Ubaid M, Athab AH, Mirzaei R and Karampoor S: Exploring the impact
of circRNAs on cancer glycolysis: Insights into tumor progression
and therapeutic strategies. Noncoding RNA Res. 9:970–994. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Tang C, Xie Y, Yu T, Liu N, Wang Z,
Woolsey RJ, Tang Y, Zhang X, Qin W, Zhang Y, et al:
m6A-dependent biogenesis of circular RNAs in male germ
cells. Cell Res. 30:211–228. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Ding L and Zhang H: Circ-ATP8A2 promotes
cell proliferation and invasion as a ceRNA to target EGFR by
sponging miR-433 in cervical cancer. Gene. 705:103–108. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Dong M, Li P, Xie Y, Wang Z and Wang R:
CircMYBL2 regulates the resistance of cervical cancer cells to
paclitaxel via miR-665-dependent regulation of EGFR. Drug Dev Res.
82:1193–1205. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Zhou M, Liu L, Wang J and Liu W: The role
of long noncoding RNAs in therapeutic resistance in cervical
cancer. Front Cell Dev Biol. 10:10609092022. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Mao Y, Zhang L and Li Y: circEIF4G2
modulates the malignant features of cervical cancer via the
miR-218/HOXA1 pathway. Mol Med Rep. 19:3714–3722. 2019.PubMed/NCBI
|
|
135
|
Liu G, Zhang Z, Song Q, Guo Y, Bao P and
Shui H: Circ_0006528 contributes to paclitaxel resistance of breast
cancer cells by regulating miR-1299/CDK8 axis. Onco Targets Ther.
13:9497–9511. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Niu D, Wu Y and Lian J: Circular RNA
vaccine in disease prevention and treatment. Signal Transduct
Target Ther. 8:3412023. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Ou R, Lv J, Zhang Q, Lin F, Zhu L, Huang
F, Li X, Li T, Zhao L, Ren Y and Xu Y: circAMOTL1 motivates AMOTL1
expression to facilitate cervical cancer growth. Mol Ther Nucleic
Acids. 19:50–60. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Wang Y, Xu X, Yu S, Jeong KJ, Zhou Z, Han
L, Tsang YH, Li J, Chen H, Mangala LS, et al: Systematic
characterization of A-to-I RNA editing hotspots in microRNAs across
human cancers. Genome Res. 27:1112–1125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Liu Y, Ou Y and Hou L: Advances in
RNA-based therapeutics: Challenges and innovations in RNA delivery
systems. Curr Issues Mol Biol. 47:222024. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Xu F, Xiao Q, Du WW, Wang S and Yang BB:
CircRNA: Functions, applications and prospects. Biomolecules.
14:15032024. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Zhang F, Jiang J, Qian H, Yan Y and Xu W:
Exosomal circRNA: Emerging insights into cancer progression and
clinical application potential. J Hematol Oncol. 16:672023.
View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Feng XY, Zhu SX, Pu KJ, Huang HJ, Chen YQ
and Wang WT: New insight into circRNAs: Characterization,
strategies, and biomedical applications. Exp Hematol Oncol.
12:912023. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
O'Leary E, Jiang Y, Kristensen LS, Hansen
TB and Kjems J: The therapeutic potential of circular RNAs. Nat Rev
Genet. 26:230–244. 2025. View Article : Google Scholar : PubMed/NCBI
|