Introduction
Colorectal cancer (CRC) is a leading cause of
cancer-related deaths worldwide, with metastatic CRC (mCRC) posing
therapeutic challenges (1).
Treatment strategies often rely on targeted therapies guided by
biomarkers such as rat sarcoma virus (RAS) and rapidly
accelerated fibrosarcoma (RAF) mutations. These mutations
influence key biological processes, including angiogenesis, cell
proliferation, and apoptosis, and serve as critical prognostic
markers (2–13).
KRAS is a small GTP-binding protein that
plays a critical role in transmitting growth signals downstream
from the epidermal growth factor receptor (EGFR). KRAS gene
mutations are present in approximately half of CRC cases.
Approximately 90% of these mutations occurring in KRAS exon
2 (codon 12, 13) (1). KRAS
exon 2 mutations are the most common predictor of resistance to the
anti-EGFR drugs, cetuximab and panitumumab, in patients with mCRC,
given that KRAS mutations reduce the intrinsic GTPase
activity of Ras, causing it to remain in its active, GTP-bound
state. Thus, despite EGFR inhibition, downstream proliferative
signaling persists, leading to resistance (3–7,9).
Subsequently, reports from the European consortium indicated that
other KRAS mutations, including KRAS exons 3 (codon
59, 61) and 4 (codon 117, 146), and NRAS mutations,
including NRAS exons 2 (codon 12, 13), 3 (codon 59, 61), and
4 (codon 117, 146), were also associated with resistance to
anti-EGFR antibody therapy (14–21).
These findings led to a consensus that all RAS mutations can
predict resistance to anti-EGFR antibodies (8,10,11).
However, KRAS exons 3 and 4, and NRAS mutations are
relatively rare, and prior studies have examined only a limited
number of cases. Additionally, in real-world clinical practice,
mCRC with RAS mutations is commonly treated with
chemotherapy regimens that include angiogenesis inhibitors, not
anti-EGFR monoclonal antibodies. However, it remains unclear
whether the prognosis in patients with KRAS exons 3 and 4,
or NRAS mutations is comparable to that in patients with
KRAS exon 2 mutations in this treatment setting.
Historically, only KRAS exon 2 mutations were
detectable and therefore recognized as the primary RAS
mutations associated with resistance to anti-EGFR antibody therapy.
With advances in sequencing technologies, additional mutations in
KRAS non-exon 2 regions of KRAS as well as
NRAS mutations became identifiable. As a result, the
definition of anti-EGFR resistance biomarkers expanded from
KRAS exon 2 mutations alone to encompass the entire spectrum
of RAS mutations. However, despite this unified classification, the
biological characteristics, treatment responses, and prognostic
implications of KRAS exon 2 mutations vs. KRAS
non-exon 2 mutations remain insufficiently understood. Currently,
patients with any RAS mutation are broadly categorized into
a single group, although this approach may overlook meaningful
heterogeneity within the RAS-mutated population.
Against the background this study aimed to focus on
the relatively rare KRAS exons 3 and 4, and NRAS
mutations to evaluate their impact on prognosis, clinical
characteristics, and efficacy of angiogenesis inhibitors The study
also sought to determine whether these impacts are comparable to
those observed with KRAS exon 2 mutations. These mutations
are categorized as minor RAS mutations in this study.
Patients and methods
Patient selection and
characteristics
This retrospective cohort study was conducted at
Osaka International Cancer Institute. Patients who underwent tissue
RAS testing between August 2018 and December 2023 were
included in this analysis. Tumor tissue samples for RAS testing
were obtained as part of routine clinical care, and no additional
samples were collected specifically for this study. As this was a
retrospective observational study, no formal sample size
calculation was performed. Instead, all eligible patients treated
during the study period were included to enhance the
representativeness and generalizability of the findings. Tumor
tissue samples were analyzed to determine RAS mutation
status using the MEBGEN RASKET™-B kit. Information regarding RAS
mutation subtypes was extracted from existing medical records.
Patients with histologically confirmed colorectal
adenocarcinoma and documented RAS mutations were included.
Patients who had not received systemic chemotherapy for recurrent
or metastatic CRC were excluded. For patients who experienced
recurrence during or within 6 months of completing adjuvant
chemotherapy, the start date of adjuvant chemotherapy was
considered the initiation point for all statistical analyses,
rather than the initiation point for mCRC treatment.
Demographic and clinical data, including age, sex,
Eastern Cooperative Oncology Group (ECOG) performance status,
location of the primary tumor, pathological differentiation,
microsatellite instability status, metastatic disease
characteristics, first-line chemotherapy regimen, and the best
efficacy of first-line chemotherapy, were extracted from electronic
medical records. Extracted metastatic characteristics included the
number of metastatic organs and the presence or absence of liver
and lung metastases. Tumor location was categorized both as
right-sided (from the cecum to the transverse colon) or left-sided
(from the splenic flexure to the rectum) and as colonic (from the
cecum to the sigmoid colon) or rectal (rectum). RAS mutation
subtypes were recorded and categorized into two groups: KRAS
exon 2 mutations (codons 12 and 13) and minor RAS mutations
(KRAS exons 3 and 4, and NRAS mutations). These
categorizations allowed for further subgroup analyses based on
clinical and molecular characteristics. We report the efficacy
results using a data cutoff of December 2024.
RAS mutation analysis
Tumor tissues were obtained from primary or
metastatic sites and preserved as formalin-fixed paraffin-embedded
(FFPE) specimens. DNA was extracted from the FFPE blocks, and
RAS mutation testing was performed using the MEBGEN
RASKET™-B kit (22,23).
Assays with the RASKET-B kit were performed according to the
manufacturer's protocol. Briefly, this multiplex PCR-based assay
was specifically designed to detect mutations in KRAS and
NRAS genes across exons 2, 3, and 4. The mutations included
those in KRAS codons 12 (G12S, G12C, G12R, G12D, G12V, and
G12A), 13 (G13S, G13C, G13R, G13D, G13V, and G13A), KRAS
codon 59 (A59T and A59G), 61 (Q61K, Q61E, Q61L, Q61P, Q61R, and
Q61H), 117 (K117N), and 146 (A146T, A146P, and A146V), as well as
NRAS codons 12 (G12S, G12C, G12R, G12D, G12V, and G12A), 13
(G13S, G13C, G13R, G13D, G13V, and G13A), 59 (A59T and A59G), 61
(Q61K, Q61E, Q61L, Q61P, Q61R, and Q61H), 117 (K117N), and 146
(A146T, A146P, and A146V). All procedures were conducted according
to the manufacturer's protocol, ensuring accurate mutation
identification with high sensitivity and specificity.
Assessment and statistical
analysis
The primary outcome of the study was overall
survival (OS), defined as the time from treatment initiation to
death from any cause or the last follow-up date. The study included
a comparison of OS between patients with KRAS exon 2
mutations and those with minor RAS mutations to assess
differences in clinical outcomes. Patients who were alive at the
end of the study period were censored at their most recent
follow-up date (December 2024). Secondary outcomes included the
prevalence of tumor location (analyzed as right-sided vs.
left-sided and colon vs. rectum) by RAS mutation subtype,
descriptive statistics of clinical characteristics, the
relationship between metastatic disease features and survival
outcomes, and progression-free survival (PFS) analysis for patients
treated with bevacizumab in the first-line setting.
Survival analyses were conducted using the
Kaplan-Meier method to generate survival curves, with differences
between groups evaluated using the log-rank test. Cox proportional
hazards regression models were used to estimate hazard ratios (HRs)
and 95% confidence intervals (CIs) for survival outcomes.
χ2 tests or Fisher's exact test were applied to
categorical variables, such as tumor location or the presence of
liver and lung metastases, depending on the expected cell counts.
P<0.05 was considered to indicate a statistically significant
difference. Subgroup analyses were performed to explore the impact
of RAS mutation subtypes, metastatic organ involvement, and
other clinical factors on survival outcomes. Statistical analyses
were conducted using EZR Version 1.63 (Saitama Medical Center,
Jichi Medical University, Japan). Results were summarized as means
with standard deviations, or medians with interquartile ranges, as
appropriate.
Ethical considerations
This study, conducted following The Declaration of
Helsinki, was approved by the institutional review board of Osaka
International Cancer Institute (approval no. IRB 24122) (24). Given the retrospective nature of our
study, which utilized anonymized patient data, the requirement for
informed consent was waived.
Results
Patient characteristics and
frequencies of RAS mutation subtypes
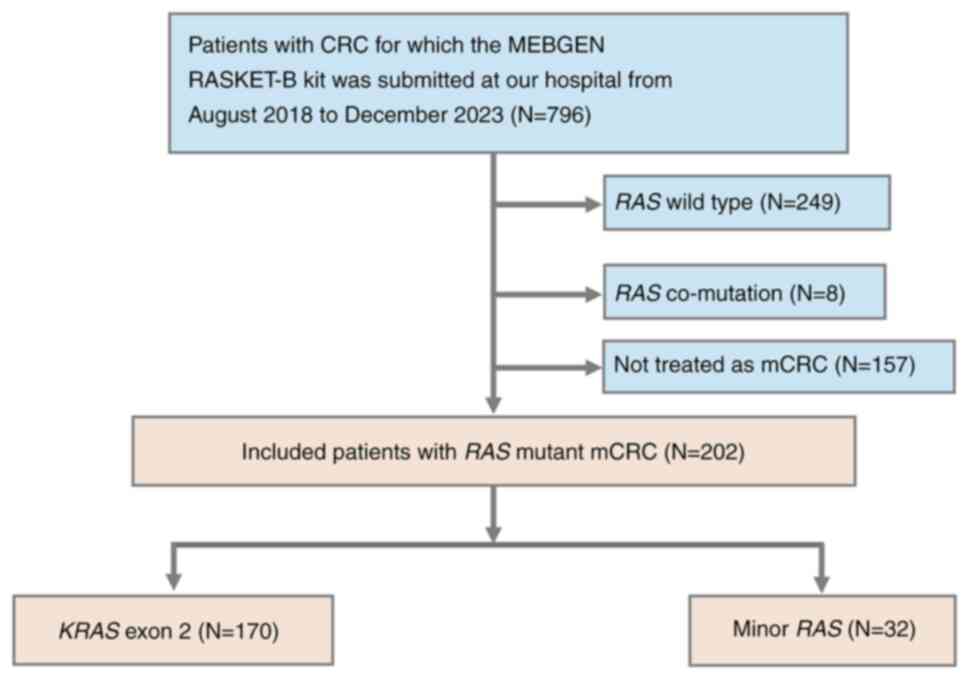
A total of 796 patients with CRC submitted the
MEBGEN RASKET™-B kit at our hospital between August 2018
and December 2023. All samples were available for analysis. Of
these, 429 patients with RAS wild-type mutations were
excluded. The remaining 367 patients with RAS mutations
included 202 individuals who had received systemic chemotherapy for
advanced or recurrent CRC. Among these, 170 patients (84%) had
KRAS exon 2 mutations, whereas 32 (16%) exhibited minor
RAS mutations (Fig. 1).
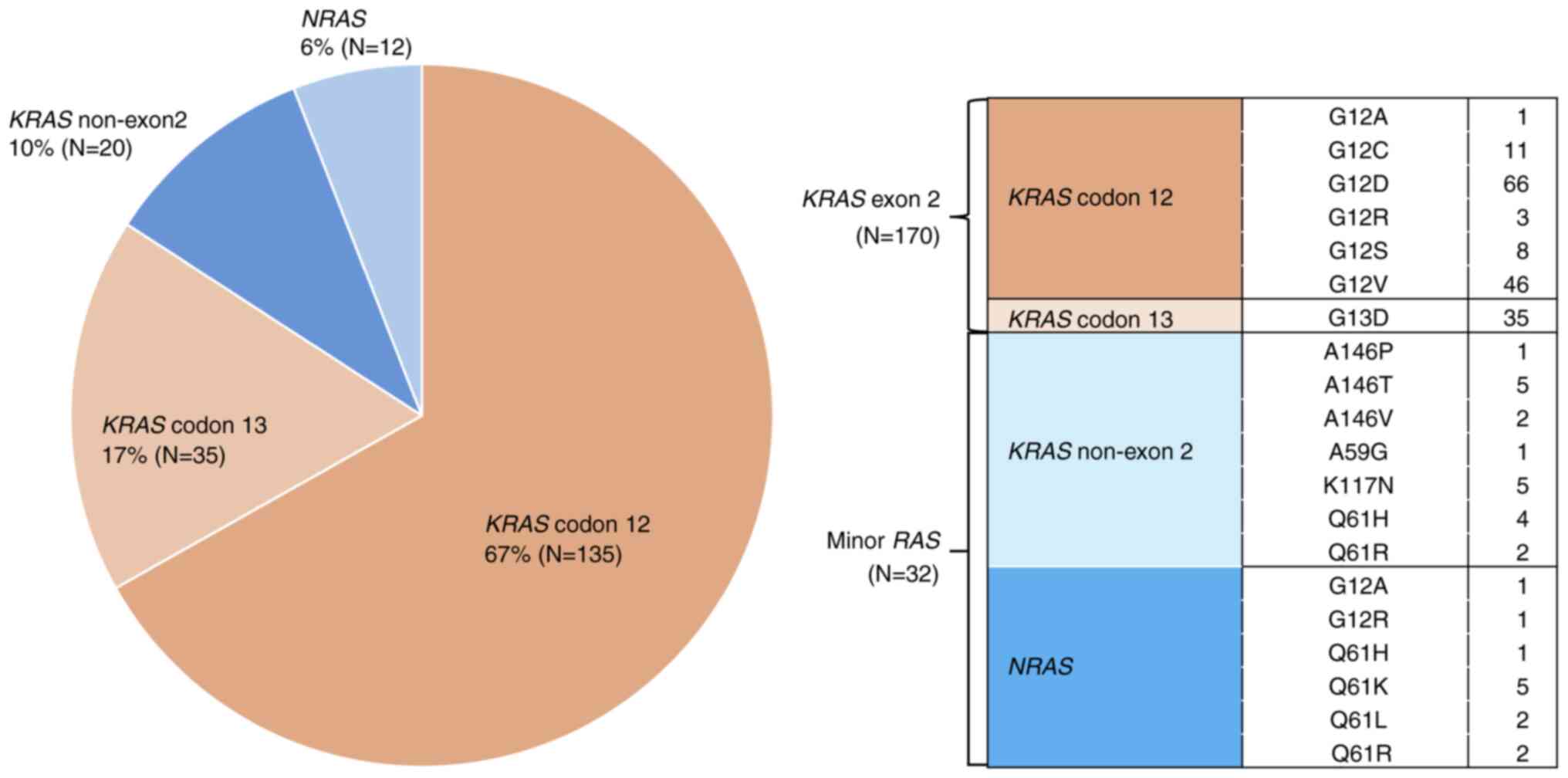
Analysis of RAS mutation subtypes revealed that KRAS
codon 12 mutations were the most frequent, accounting for 67% of
cases (n=135), followed by KRAS codon 13 (17%, n=35) and
non-exon 2 mutations (10%, n=20), and NRAS mutations (6%,
n=12) (Fig. 2). These findings are
consistent with those from prior studies, highlighting the
predominance of KRAS exon 2 mutations in mCRC and the
relatively rare occurrence of minor RAS mutations (1).
Clinicopathological characteristics of
patients with RAS Mutation subtypes
The demographic and clinicopathological
characteristics of those patients are shown in Table I. Correlation between RAS
mutation status and age, sex, ECOG PS, primary site of disease,
tumor sidedness, pathological differentiation, microsatellite
instability status, previous surgery, previous adjuvant
chemotherapy, number of metastatic sites, liver metastasis, lung
metastasis was evaluated. Minor RAS mutations were more
common in left-sided colorectal cancer (81%); however, no
significant differences in background were observed between the two
groups in any category.
 | Table I.Clinical characteristics of patients
with RAS mutant metastatic colorectal cancer (N=202). |
Table I.
Clinical characteristics of patients
with RAS mutant metastatic colorectal cancer (N=202).
| Characteristic | KRAS exon 2
mutation (N=170) | Minor RAS
mutation (N=32) | P-value |
|---|
| Median age, years
(range) | 66 (26–90) | 67 (44–84) |
|
| Sex,
female/male | 82/88 (48/52) | 17/15 (53/47) | 0.70 |
| ECOG PS,
0/1/≥2 | 116/45/9
(68/26/5) | 18/11/3
(56/34/9) | 0.31 |
| Sidedness,
left/right | 111/59 (65/35) | 26/6 (81/19) | 0.10 |
| Differentiation,
tub1/tub2/por/sig/muc/pap | 65/68/9/0/21/7 | 13/11/3/1/4/0 |
|
|
|
(38/40/5/0/12/4) |
(41/34/9/3/13/0) | 0.30 |
| Microsatellite
instability, MSS/MSI-H/unknown | 133/1/36
(78/1/21) | 28/1/3
(88/3/9) | 0.12 |
| Stage
IV/recurrence | 91/79 (54/46) | 14/18 (44/56) | 0.34 |
| Previous surgery,
yes/no | 115/55 (68/32) | 18/14 (56/44) | 0.23 |
| Previous adjuvant
chemotherapy, yes/no | 45/125 (26/74) | 5/27 (16/84) | 0.27 |
| Number of sites of
metastasis, 0/1/2/≥3 | 23/81/50/16
(14/48/29/9) | 3/16/11/2
(9/50/34/6) | 0.87 |
| Liver metastasis,
yes/no | 86/84 (51/49) | 13/19 (41/59) | 0.34 |
| Lung metastasis,
yes/no | 68/102 (40/60) | 15/17 (47/53) | 0.56 |
OS in patients with RAS mutation
subtypes
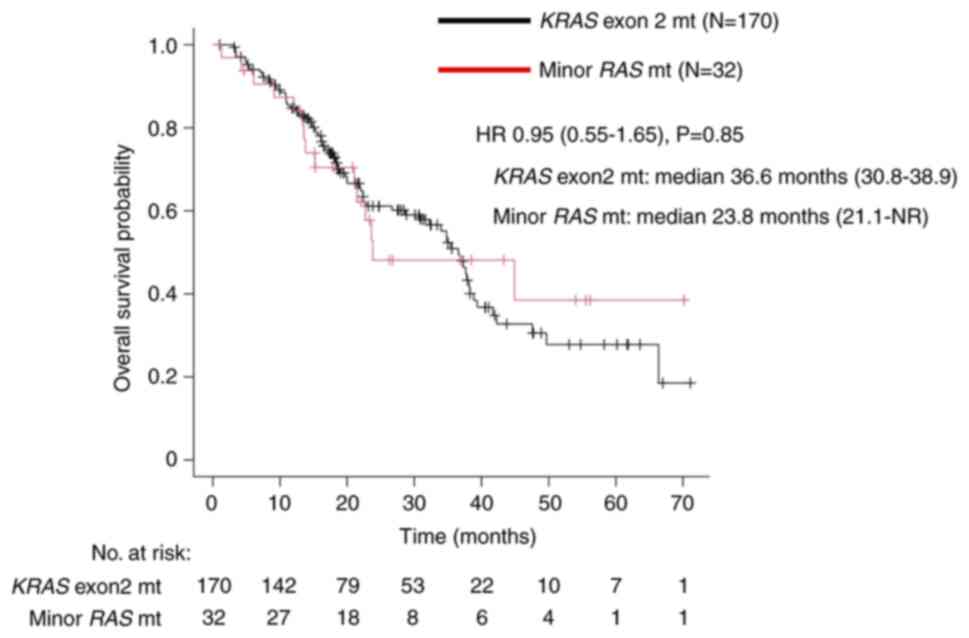
The median OS in the KRAS exon 2 mutation
group was 36.6 months (95% CI: 30.8–38.9), whereas that in the
minor RAS mutation group was 23.8 months (95% CI: 21.1-not
reached) (Fig. 3). The HR for OS
between the groups was 0.95 (95% CI: 0.55–1.65, P=0.85), suggesting
that the impact of KRAS exon 2 mutation and minor RAS
mutation on prognosis is almost comparable.
OS and PFS in patients receiving
first-line bevacizumab-containing therapy
Among 202 patients with RAS mutations, 156
received first-line treatment with bevacizumab-containing
chemotherapy. The cohort included 134 patients with KRAS
exon 2 mutations and 22 with minor RAS mutations. We
investigated the relationship between RAS mutation subtype
rate and age, sex, ECOG PS, tumor sidedness, number of metastatic
sites, liver metastasis, lung metastasis, first-line systemic
chemotherapy regimen, and the best efficacy of first-line
chemotherapy (Table II). Liver
metastases were significantly less common in the minor RAS
group, reported in only 32% of cases compared to 56% in the
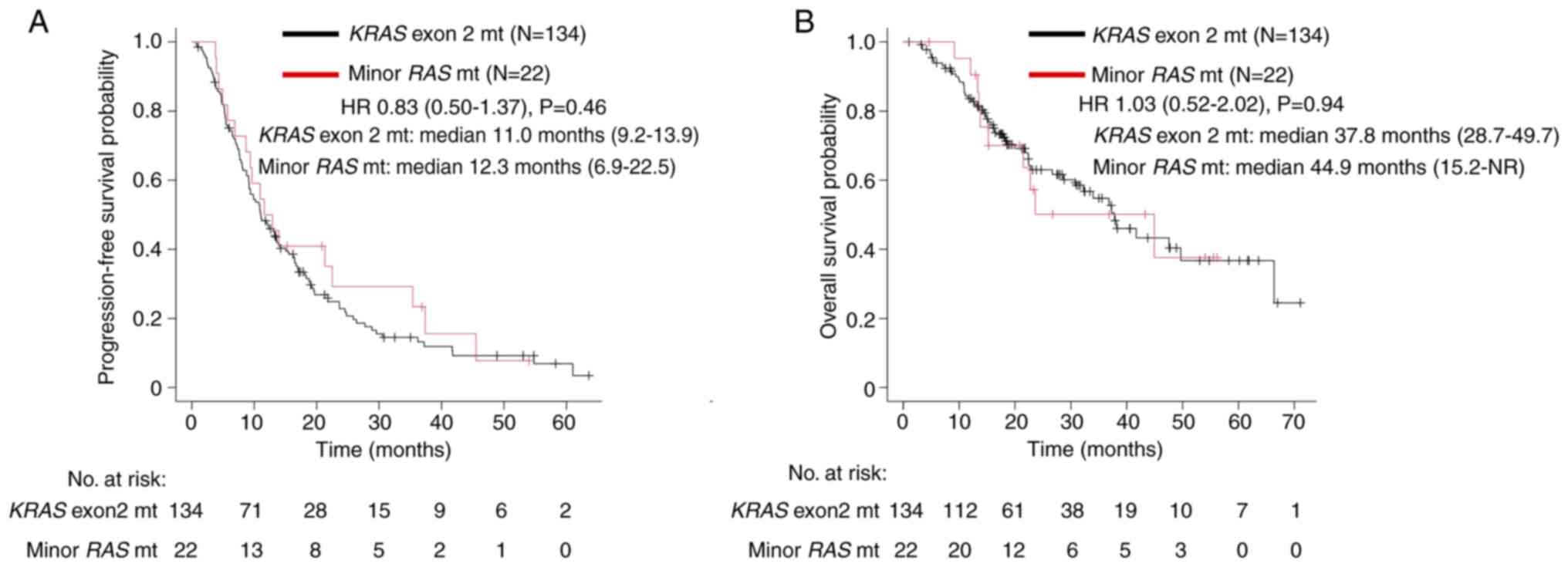
KRAS exon 2 group (P=0.04). A trend toward improved OS (37.8
vs. 44.9 months, P=0.94) and PFS (11.0 vs. 12.3 months, P=0.46) was
not noted between the two groups (Fig.
4).
 | Table II.Clinical characteristics of patients
treated with bevacizumab at first line with RAS mutant
metastatic colorectal cancer (N=156). |
Table II.
Clinical characteristics of patients
treated with bevacizumab at first line with RAS mutant
metastatic colorectal cancer (N=156).
| Characteristic | KRAS exon2
mutation (N=134) | Minor RAS
mutation (N=22) | P-value |
|---|
| Median age, years
(range) | 67 (37–90) | 66 (45–79) |
|
| Sex,
female/male | 65/69 (49/51) | 12/10 (55/45) | 0.65 |
| ECOG PS,
0/1/≥2 | 90/38/6
(67/28/4) | 15/6/1
(68/27/5) | >0.99 |
| Sidedness,
Left/right | 82/52 (61/39) | 18/4 (82/ 18) | 0.09 |
| Number of sites of
metastasis, 0/1/2/≥3 | 5/72/43/14
(4/54/32/10) | 2/12/7/1
(9/55/32/5) | 0.59 |
| Liver metastasis,
yes/no | 75/59 (56/44) | 7/15 (32/68) | 0.04 |
| Lung metastasis,
yes/no | 58/76 (43/57) | 12/10 (55/45) | 0.36 |
| First line regimen,
triplet/doublet/other | 6/108/20
(4/81/15) | 1/16/5
(5/73/23) | 0.58 |
The efficacy of the first-line chemotherapy was
evaluated in 117 patients (100 with KRAS exon 2 mutations
and 17 with minor RAS mutations) with target lesions.
Complete response (CR), partial response (PR), stable disease (SD),
and progressive disease (PD) were observed in 1, 48, 39, and 12%,
respectively, for the KRAS exon 2 mutation group, and 6, 47,
41, and 6%, respectively, for minor RAS mutation group
(Table III). The overall response
rate (ORR) was 49% in patients with KRAS exon 2 mutations
and 53% in those with minor RAS mutations (P=0.80).
 | Table III.ORR in patients treated with
bevacizumab with different subtypes of RAS mutations
(N=117). |
Table III.
ORR in patients treated with
bevacizumab with different subtypes of RAS mutations
(N=117).
| Response | KRAS exon 2
mutation (N=100) | Minor RAS
mutation (N=17) | P-value |
|---|
| CR | 1 (1) | 1 (6) |
|
| PR | 48 (48) | 8 (47) |
|
| SD | 39 (39) | 7 (41) |
|
| PD | 12 (12) | 1 (6) |
|
| ORR | 49 (49) | 9 (53) | 0.80 |
Discussion
Minor RAS mutations are rare and not fully
understood. Therefore, our study provides insights into the
distinct clinical and prognostic characteristics of patients with
mCRC and minor RAS mutations by comparing them with those
with KRAS exon 2 mutations. Based on our real-world clinical
experience, our initial hypothesis posited that minor RAS
mutations would not confer a worse prognosis than KRAS exon
2 mutations.
This study investigated the frequency and subtypes
of RAS mutations (KRAS exon 2 and minor RAS
mutations) and evaluated the prognostic outcomes and the impact of
first-line treatment with anti-angiogenic agents in both groups.
Our data demonstrated that the median OS and PFS were comparable
between the two groups. Consistent with our initial hypothesis,
minor RAS and KRAS exon 2 mutations were suggested to
have similar effects on prognosis and treatment outcomes.
Accordingly, we believe that current treatment strategies for mCRC
with RAS mutations remain appropriate and should not be
altered based on the RAS mutation subtype. While our results
align with this view, previous studies have reported conflicting
findings. Some have indicated poorer prognoses in patients with
minor RAS mutations compared to those with KRAS exon
2 mutations, whereas others have suggested that NRAS
mutations may be associated with better outcomes (25–27).
Interestingly, despite previous conflicting reports, the relatively
favorable survival observed in the minor RAS group may be partially
attributable to the lower incidence of liver metastases.
Nonetheless, as no current evidence supports a causal relationship
between minor RAS mutations and reduced liver involvement, further
studies are warranted to elucidate the biological basis of this
observation. In addition to metastatic patterns, molecular
differences may also contribute to prognostic variability. For
example, Takane et al demonstrated that CRC with NRAS
mutations is associated with a distinct DNA methylation epigenotype
(LME) compared to CRC with KRAS mutations (26), which could partly explain the
differences in prognosis. Furthermore, Ogura et al reported
that NRAS mutations were more prevalent in distal colon
cancers compared to KRAS mutations, potentially contributing
to a more favorable prognosis (27). These conflicting findings highlight
the need for larger studies that can analyze NRAS and
KRAS non-exon 2 mutations separately. Moreover, the small
number of patients with NRAS or KRAS non-exon 2
mutations in most studies may limit statistical power and
contribute to the inconsistent findings. This study did not assess
the pathogenicity of individual minor RAS mutations, and such
analysis was beyond the scope of this retrospective
investigation.
KRAS mutations were initially recognized
solely as biomarkers for resistance to anti-EGFR antibody
therapies. However, recent advancements have led to the FDA
approval of targeted therapies such as adagrasib plus cetuximab and
sotorasib plus panitumumab for KRAS G12C mutations,
expanding treatment options (28,29).
Additionally, new drugs targeting KRAS G12D and KRAS
G12V mutations are currently under development, with promising
potential for clinical application. Moreover, several
pan-RAS inhibitors are being developed, which may eventually
address minor RAS mutations (30). Since minor RAS mutations are
rare driver mutations, conducting randomized controlled trials in
CRC is challenging. Therefore, the data from our study on the
efficacy and survival outcomes of conventional chemotherapy in
patients with minor RAS mutations could serve as valuable
historical control data for future therapeutic developments. These
findings may provide a foundational dataset for evaluating the
efficacy of novel treatment strategies.
We also evaluated the clinical characteristics of
patients in both groups. During the study period, the MEBGEN
RASKET™-B kit was used to analyze 796 cases of CRC, of
which 429 were tumors with wild-type RAS. Approximately half
of the cases had RAS mutations, consistent with previous
reports. Among these, KRAS exon 2 mutations accounted for
84% (67% KRAS codon 12 mutations and 17% KRAS codon
13 mutations), whereas minor RAS mutations accounted for 16%
(10% KRAS non-exon 2 mutations and 6% NRAS
mutations). KRAS exon 2 mutations are reportedly present in
approximately 35–40% of CRC cases (30–35% KRAS codon 12
mutations and 4–8% KRAS codon 13 mutations), whereas minor
RAS mutations are observed in about 10–15% (3–6% of
mutations in KRAS exons 3 and 4, NRAS exons 2 and 3,
and less than 1% in NRAS exon 4) (17,31).
Considering that approximately half of CRC cases harbor RAS
mutations, our findings regarding the proportion of each RAS
mutation type within the RAS-mutant group were generally
consistent with previously reported findings.
An investigation into the distribution of
clinicopathological characteristics and RAS mutation
subtypes revealed no statistically significant findings; however,
the proportion of left-sided lesions in minor RAS mutations
was 81%, indicating a high tendency. Consistent with prior studies,
our study identified NRAS mutations most frequently in
rectal and sigmoid colon cancers (18). Reports have suggested that CIMP-high
is associated with a continuous increase in frequency from the
rectum to the ascending colon. Conversely, KRAS codon 61 and
146 mutations had higher frequencies in cecal cancers, with a
higher prevalence of CpG island methylator phenotype (CIMP)-low,
compared to KRAS wild-type cases (32,33).
These discrepancies may be due to small sample sizes in various
studies. Notably, a few Japanese studies have reported a higher
frequency of KRAS non-exon 2 mutations in left-sided CRC
that aligns with our findings (25). Despite this predominance, no
associations were identified with microsatellite instability
(MSI)-H status or pathological differentiation. The biological
mechanisms underlying the correlation between minor RAS mutations
and left-sided tumors need to be further explored.
As a single-center study with an adequate number of
cases, this research represents a significant contribution to
understanding the characteristics of minor RAS mutations.
Future studies with larger sample sizes, combined with data from
the recently introduced OncoGuide™ EpiLight™
methylation detection kit in Japan, may help establish the
statistical significance of the continuous model for mutation
distribution (34–36).
In conclusion, this study elucidates the prognostic
impact, clinical characteristics, and effects of anti-angiogenic
therapy in patients with minor RAS and KRAS exon 2
mutations. Consistent with previously reported findings from Japan,
minor RAS mutations were more common in left-sided
colorectal cancer. The association of minor RAS mutations
with left-sided tumors warrants further investigation into
underlying biological mechanisms. The prognostic impact of minor
RAS mutations appeared to be comparable to that of
KRAS exon 2 mutations, suggesting that current treatment
strategies may remain unchanged for RAS-mutant CRC. However,
further studies should validate these findings. The results
highlight the necessity of therapeutic advancements targeting all
RAS mutations to improve the prognosis of mCRC with
RAS mutations.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
TK and YK conceived and designed the study. YK
performed data acquisition. TK and YK confirm the authenticity of
all the raw data. Data analysis was performed by TK and YK. All
authors including DS, TO, YA, RM, MK, TS, TO, MN, MI, YK, JN, NS,
TY, MT and MY contributed to the interpretation of clinical data.
Statistical analysis was conducted by YK. The manuscript was
prepared and edited by TK, MT and YK. All authors reviewed the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study, conducted following The Declaration of
Helsinki, was approved by the institutional review board of Osaka
International Cancer Institute (approval no. IRB 24122). Given the
retrospective nature of our study, which utilized anonymized
patient data, the requirement for informed consent was waived.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, AI tools
(ChatGPT) were used to improve the readability and language of the
manuscript, and subsequently, the authors revised and edited the
content produced by the AI tools as necessary, taking full
responsibility for the ultimate content of the present
manuscript.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
mCRC
|
metastatic CRC
|
|
RAS
|
rat sarcoma virus
|
|
RAF
|
rapidly accelerated fibrosarcoma
|
|
EGFR
|
epidermal growth factor receptor
|
|
ECOG
|
Eastern Cooperative Oncology
Group
|
|
HR
|
hazard ratio
|
|
CI
|
confidence interval
|
|
OS
|
overall survival
|
|
PFS
|
progression-free survival
|
|
CR
|
complete response
|
|
PR
|
partial response
|
|
SD
|
stable disease
|
|
PD
|
progressive disease
|
|
ORR
|
overall response rate
|
|
MSI
|
microsatellite instability
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Amado RG, Wolf M, Peeters M, Van Cutsem E,
Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et
al: Wild-type KRAS is required for panitumumab efficacy in patients
with metastatic colorectal cancer. J Clin Oncol. 41:3278–3286.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bokemeyer C, Bondarenko I, Hartmann JT, de
Braud F, Schuch G, Zubel A, Celik I, Schlichting M and Koralewski
P: Efficacy according to biomarker status of cetuximab plus
FOLFOX-4 as first-line treatment for metastatic colorectal cancer:
The OPUS study. Ann Oncol. 22:1535–1546. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dahabreh IJ, Terasawa T, Castaldi PJ and
Trikalinos TA: Systematic review: Anti-epidermal growth factor
receptor treatment effect modification by KRAS mutations in
advanced colorectal cancer. Ann Intern Med. 154:37–49. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Douillard JY, Siena S, Cassidy J,
Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham
D, Jassem J, et al: Randomized, phase III trial of panitumumab with
infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4)
versus FOLFOX4 alone as first-line treatment in patients with
previously untreated metastatic colorectal cancer: The PRIME study.
J Clin Oncol. 28:4697–4705. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Karapetis CS, Khambata-Ford S, Jonker DJ,
O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD,
Robitaille S, et al: K-ras mutations and benefit from cetuximab in
advanced colorectal cancer. N Engl J Med. 359:1757–1765. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lièvre A, Bachet JB, Le Corre D, Boige V,
Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, et al:
KRAS mutation status is predictive of response to cetuximab therapy
in colorectal cancer. Cancer Res. 66:3992–3995. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schirripa M, Cohen SA, Battaglin F and
Lenz HJ: Biomarker-driven and molecular targeted therapies for
colorectal cancers. Semin Oncol. 45:124–132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Van Cutsem E, Köhne CH, Hitre E, Zaluski
J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G,
et al: Cetuximab and chemotherapy as initial treatment for
metastatic colorectal cancer. N Engl J Med. 360:1408–1417. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Venook AP, Niedzwiecki D, Lenz HJ,
Innocenti F, Fruth B, Meyerhardt JA, Schrag D, Greene C, O'Neil BH,
Atkins JN, et al: Effect of first-line chemotherapy combined with
cetuximab or bevacizumab on overall survival in patients with KRAS
wild-type advanced or metastatic colorectal cancer: A randomized
clinical trial. JAMA. 317:2392–2401. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Watanabe J, Muro K, Shitara K, Yamazaki K,
Shiozawa M, Ohori H, Takashima A, Yokota M, Makiyama A, Akazawa N,
et al: Panitumumab vs bevacizumab added to standard first-line
chemotherapy and overall survival among patients with RAS
wild-type, left-sided metastatic colorectal cancer: A randomized
clinical trial. JAMA. 329:1271–1282. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Heinemann V, von Weikersthal LF, Decker T,
Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmüller
C, Kahl C, Seipelt G, et al: FOLFIRI plus cetuximab versus FOLFIRI
plus bevacizumab as first-line treatment for patients with
metastatic colorectal cancer (FIRE-3): A randomised, open-label,
phase 3 trial. Lancet Oncol. 15:1065–1075. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jones JC, Renfro LA, Al-Shamsi HO, Schrock
AB, Rankin A, Zhang BY, Kasi PM, Voss JS, Leal AD, Sun J, et al:
Non-V600 BRAF mutations define a clinically distinct
molecular subtype of metastatic colorectal cancer. J Clin Oncol.
35:2624–2630. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bokemeyer C, Köhne CH, Ciardiello F, Lenz
HJ, Heinemann V, Klinkhardt U, Beier F, Duecker K, van Krieken JH
and Tejpar S: FOLFOX4 plus cetuximab treatment and RAS mutations in
colorectal cancer. Eur J Cancer. 51:1243–1252. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
De Roock W, Claes B, Bernasconi D, De
Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V,
Papamichael D, Laurent-Puig P, et al: Effects of KRAS, BRAF, NRAS,
and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy
in chemotherapy-refractory metastatic colorectal cancer: A
retrospective consortium analysis. Lancet Oncol. 11:753–762. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Douillard JY, Oliner KS, Siena S,
Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham
D, Jassem J, et al: Panitumumab-FOLFOX4 treatment and RAS mutations
in colorectal cancer. N Engl J Med. 369:1023–1034. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kawazoe A, Shitara K, Fukuoka S, Kuboki Y,
Bando H, Okamoto W, Kojima T, Fuse N, Yamanaka T, Doi T, et al: A
retrospective observational study of clinicopathological features
of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with
metastatic colorectal cancer. BMC Cancer. 15:2582015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maughan TS, Adams RA, Smith CG, Meade AM,
Seymour MT, Wilson RH, Idziaszczyk S, Harris R, Fisher D, Kenny SL,
et al: Addition of cetuximab to oxaliplatin-based first-line
combination chemotherapy for treatment of advanced colorectal
cancer: Results of the randomised phase 3 MRC COIN trial. Lancet.
377:2103–2114. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peeters M, Oliner KS, Price TJ, Cervantes
A, Sobrero AF, Ducreux M, Hotko Y, André T, Chan E, Lordick F, et
al: Analysis of KRAS/NRAS mutations in a phase III study of
panitumumab with FOLFIRI compared with FOLFIRI alone as second-line
treatment for metastatic colorectal cancer. Clin Cancer Res.
21:5469–5479. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sorich MJ, Wiese MD, Rowland A,
Kichenadasse G, McKinnon RA and Karapetis CS: Extended RAS
mutations and anti-EGFR monoclonal antibody survival benefit in
metastatic colorectal cancer: A meta-analysis of randomized,
controlled trials. Ann Oncol. 26:13–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Van Cutsem E, Lenz HJ, Köhne CH, Heinemann
V, Tejpar S, Melezínek I, Beier F, Stroh C, Rougier P, van Krieken
JH and Ciardiello F: Fluorouracil, leucovorin, and irinotecan plus
cetuximab treatment and RAS mutations in colorectal cancer. J Clin
Oncol. 33:692–700. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Taniguchi H, Okamoto W, Muro K, Akagi K,
Hara H, Nishina T, Kajiwara T, Denda T, Hironaka S, Kudo T, et al:
Clinical validation of newly developed multiplex kit using Luminex
xMAP technology for detecting simultaneous RAS and BRAF mutations
in colorectal cancer: Results of the RASKET-B study. Neoplasia.
20:1219–1226. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoshino T, Muro K, Yamaguchi K, Nishina T,
Denda T, Kudo T, Okamoto W, Taniguchi H, Akagi K, Kajiwara T, et
al: Clinical validation of a multiplex kit for RAS mutations in
colorectal cancer: Results of the RASKET (RAS KEy testing)
prospective, multicenter study. EBioMedicine. 2:317–323. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
World Medical Association, . World medical
association declaration of Helsinki. Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ikoma T, Shimokawa M, Kotaka M, Matsumoto
T, Nagai H, Boku S, Shibata N, Yasui H and Satake H: Clinical and
prognostic features of patients with detailed RAS/BRAF-mutant
colorectal cancer in Japan. BMC Cancer. 21:5182021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takane K, Akagi K, Fukuyo M, Yagi K,
Takayama T and Kaneda A: DNA methylation epigenotype and clinical
features of NRAS-mutation(+) colorectal cancer. Cancer Med.
6:1023–1035. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ogura T, Kakuta M, Yatsuoka T, Nishimura
Y, Sakamoto H, Yamaguchi K, Tanabe M, Tanaka Y and Akagi K:
Clinicopathological characteristics and prognostic impact of
colorectal cancers with NRAS mutations. Oncol Rep. 32:50–56. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yaeger R, Weiss J, Pelster MS, Spira AI,
Barve M, Ou SI, Leal TA, Bekaii-Saab TS, Paweletz CP, Heavey GA, et
al: Adagrasib with or without cetuximab in colorectal cancer with
mutated KRAS G12C. N Engl J Med. 388:44–54. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fakih MG, Salvatore L, Esaki T, Modest DP,
Lopez-Bravo DP, Taieb J, Karamouzis MV, Ruiz-Garcia E, Kim TW,
Kuboki Y, et al: Sotorasib plus panitumumab in refractory
colorectal cancer with mutated KRAS G12C. N Engl J Med.
389:2125–2139. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Holderfield M, Lee BJ, Jiang J, Tomlinson
A, Seamon KJ, Mira A, Patrucco E, Goodhart G, Dilly J, Gindin Y, et
al: Concurrent inhibition of oncogenic and wild-type RAS-GTP for
cancer therapy. Nature. 629:919–926. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Watanabe T, Yoshino T, Uetake H, Yamazaki
K, Ishiguro M, Kurokawa T, Saijo N, Ohashi Y and Sugihara K: KRAS
mutational status in Japanese patients with colorectal cancer:
Results from a nationwide, multicenter, cross-sectional study. Jpn
J Clin Oncol. 43:706–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA,
Masugi Y, Shi Y, Song M, da Silva A, Gu M, et al: Fusobacterium
nucleatum in colorectal carcinoma tissue according to tumor
location. Clin Transl Gastroenterol. 7:e2002016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Imamura Y, Lochhead P, Yamauchi M, Kuchiba
A, Qian ZR, Liao X, Nishihara R, Jung S, Wu K, Nosho K, et al:
Analyses of clinicopathological, molecular, and prognostic
associations of KRAS codon 61 and codon 146 mutations in colorectal
cancer: Cohort study and literature review. Mol Cancer. 13:1352014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ouchi K, Takahashi S, Yamada Y, Tsuji S,
Tatsuno K, Takahashi H, Takahashi N, Takahashi M, Shimodaira H,
Aburatani H and Ishioka C: DNA methylation status as a biomarker of
anti-epidermal growth factor receptor treatment for metastatic
colorectal cancer. Cancer Sci. 106:1722–1729. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamauchi M, Lochhead P, Morikawa T,
Huttenhower C, Chan AT, Giovannucci E, Fuchs C and Ogino S:
Colorectal cancer: A tale of two sides or a continuum? Gut.
61:794–797. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yamauchi M, Morikawa T, Kuchiba A, Imamura
Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower
C, et al: Assessment of colorectal cancer molecular features along
bowel subsites challenges the conception of distinct dichotomy of
proximal versus distal colorectum. Gut. 61:847–854. 2012.
View Article : Google Scholar : PubMed/NCBI
|