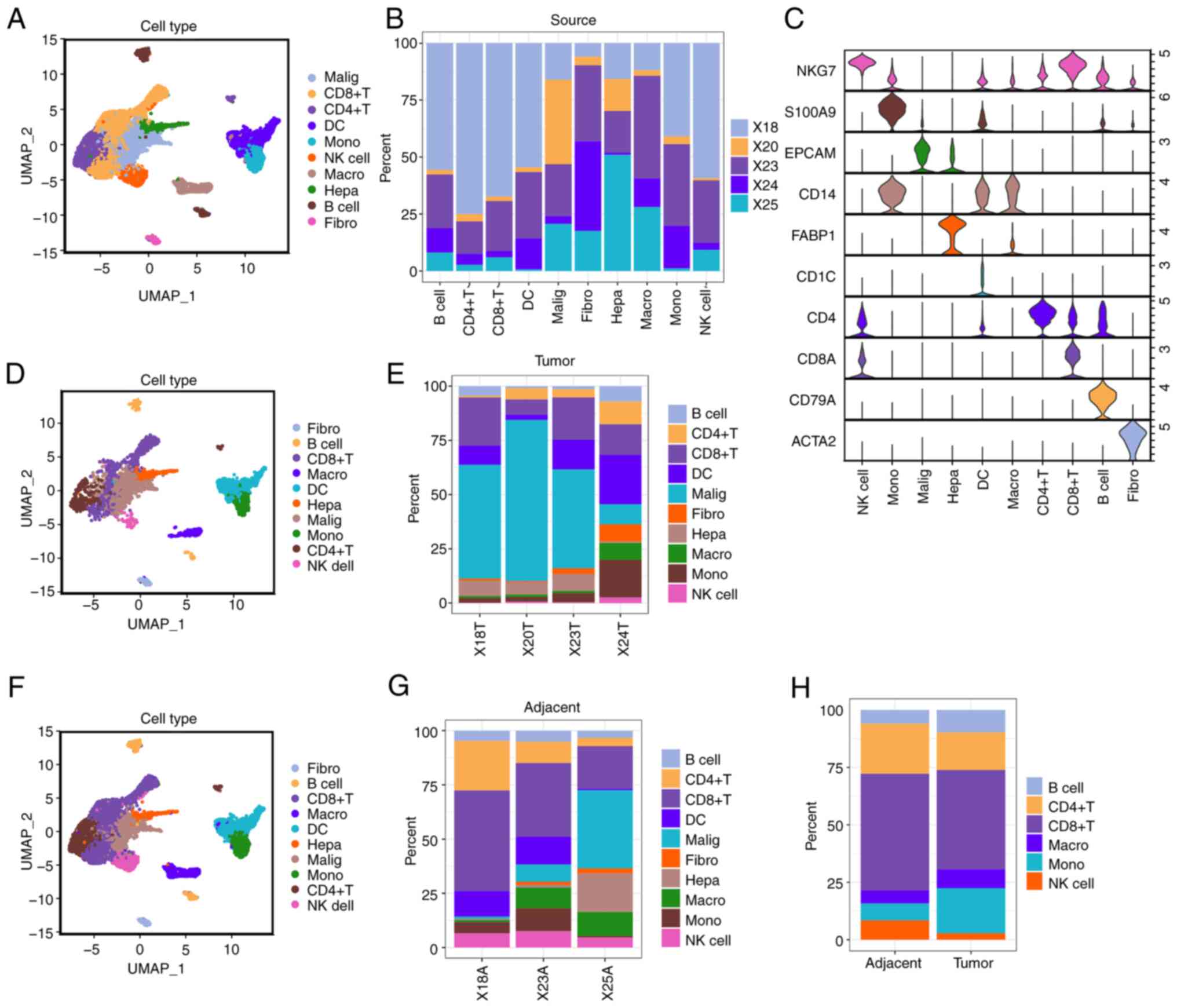
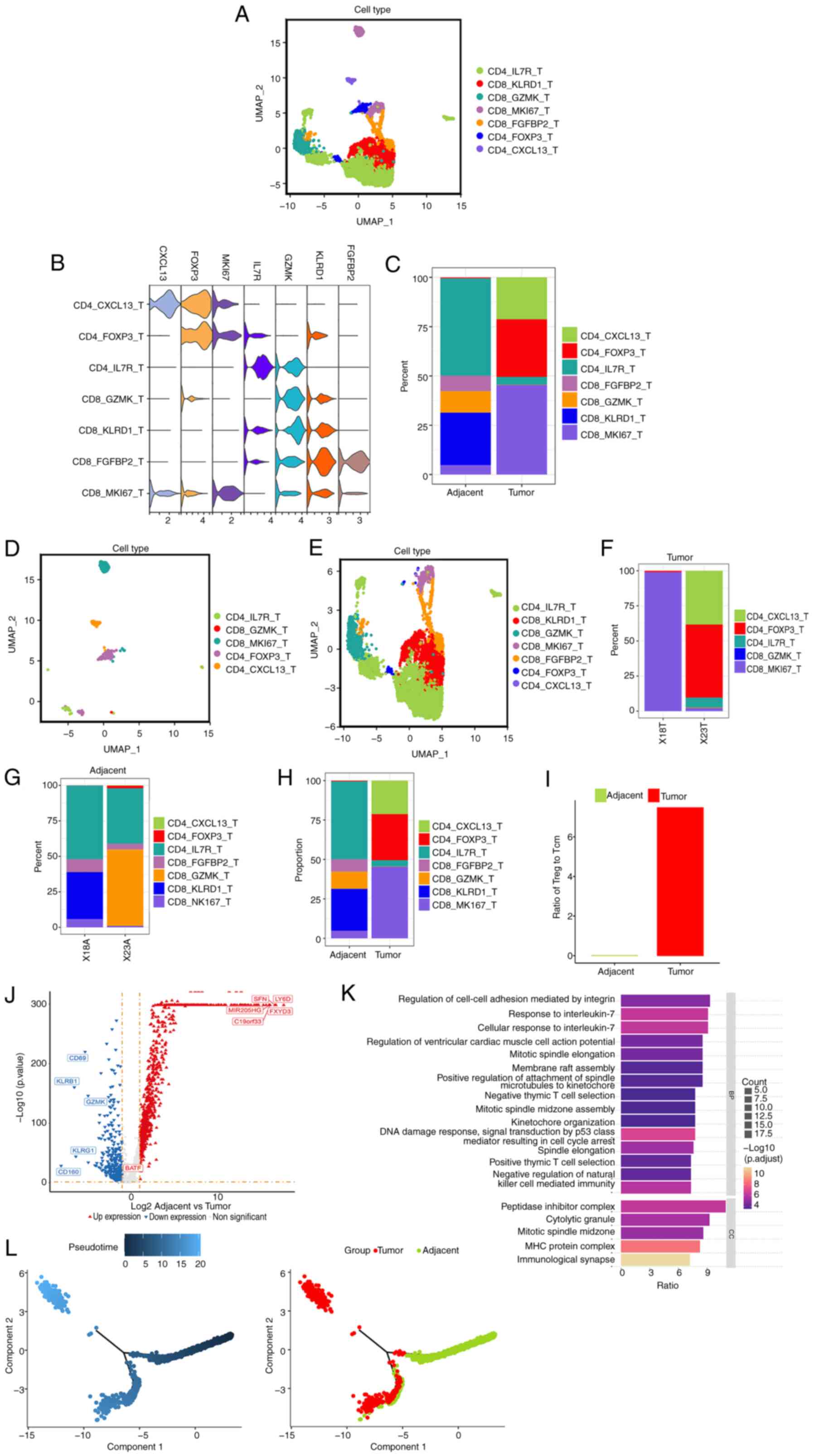
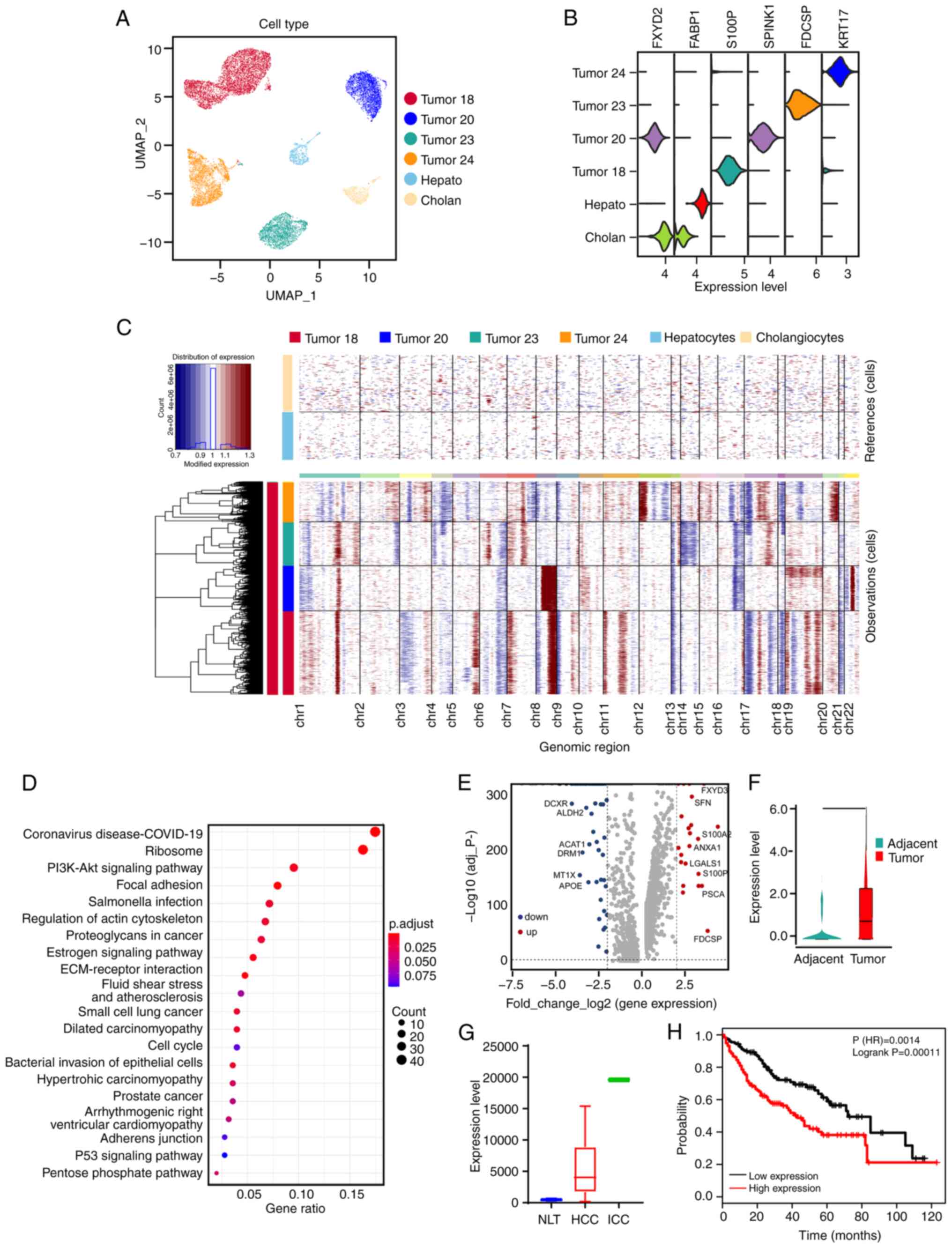
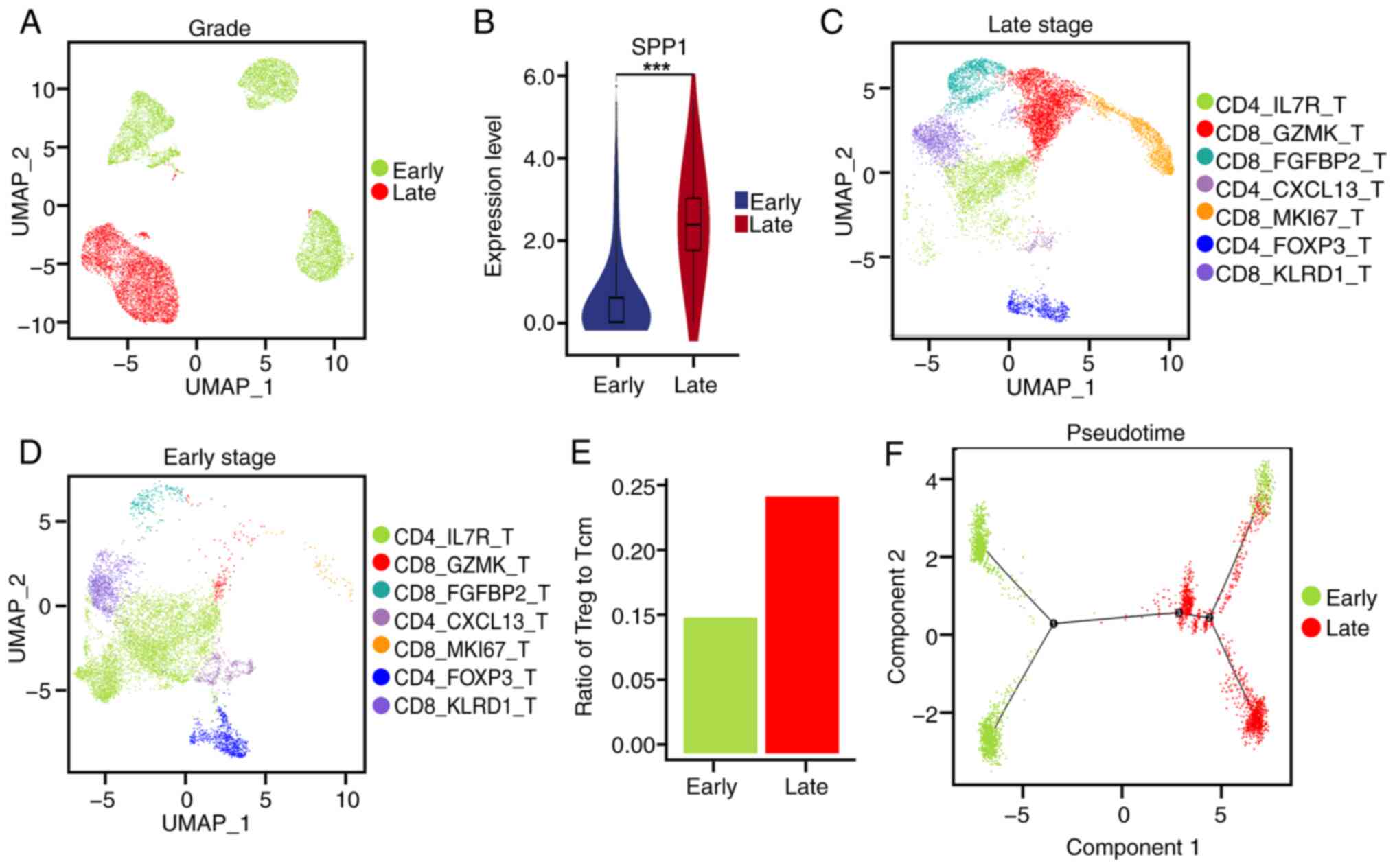
|
1
|
Zhang M, Yang H, Wan L, Wang Z, Wang H, Ge
C, Liu Y, Hao Y, Zhang D, Shi G, et al: Single-cell transcriptomic
architecture and intercellular crosstalk of human intrahepatic
cholangiocarcinoma. J Hepatol. 73:1118–1130. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Loeuillard E, Yang J, Buckarma E, Wang J,
Liu Y, Conboy C, Pavelko KD, Li Y, O'Brien D, Wang C, et al:
Targeting tumor-associated macrophages and granulocytic
myeloid-derived suppressor cells augments PD-1 blockade in
cholangiocarcinoma. J Clin Invest. 130:5380–5396. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yuan H, Lin Z, Liu Y, Jiang Y, Liu K, Tu
M, Yao N, Qu C and Hong J: Intrahepatic cholangiocarcinoma induced
M2-polarized tumor-associated macrophages facilitate tumor growth
and invasiveness. Cancer Cell Int. 20:5862020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y,
Guo X, Kang B, Hu R, Huang JY, Zhang Q, et al: Landscape of
infiltrating T cells in liver cancer revealed by single-cell
sequencing. Cell. 169:1342–1356.e16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim RD, Chung V, Alese OB, El-Rayes BF, Li
D, Al-Toubah TE, Schell MJ, Zhou JM, Mahipal A, Kim BH and Kim DW:
A Phase 2 multi-institutional study of nivolumab for patients with
advanced refractory biliary tract cancer. JAMA Oncol. 6:888–894.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song G, Shi Y, Zhang M, Goswami S, Afridi
S, Meng L, Ma J, Chen Y, Lin Y, Zhang J, et al: Global immune
characterization of HBV/HCV-related hepatocellular carcinoma
identifies macrophage and T-cell subsets associated with disease
progression. Cell Discov. 6:902020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
D'Avola D, Villacorta-Martin C,
Martins-Filho SN, Craig A, Labgaa I, von Felden J, Kimaada A,
Bonaccorso A, Tabrizian P, Hartmann BM, et al: High-density single
cell mRNA sequencing to characterize circulating tumor cells in
hepatocellular carcinoma. Sci Rep. 8:115702018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chai X, Wang J, Li H, Gao C, Li S, Wei C,
Huang J, Tian Y, Yuan J, Lu J, et al: Intratumor microbiome
features reveal antitumor potentials of intrahepatic
cholangiocarcinoma. Gut Microbes. 15:21562552023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jono S, Peinado C and Giachelli CM:
Phosphorylation of osteopontin is required for inhibition of
vascular smooth muscle cell calcification. J Biol Chem.
275:20197–1203. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Christensen B, Kazanecki CC, Petersen TE,
Rittling SR, Denhardt DT and Sørensen ES: Cell type-specific
post-translational modifications of mouse osteopontin are
associated with different adhesive properties. J Biol Chem.
282:19463–19472. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Oyama M, Kariya Y, Kariya Y, Matsumoto K,
Kanno M, Yamaguchi Y and Hashimoto Y: Biological role of
site-specific O-glycosylation in cell adhesion activity and
phosphorylation of osteopontin. Biochem J. 475:1583–1595. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song G, Shi Y, Meng L, Ma J, Huang S,
Zhang J, Wu Y, Li J, Lin Y, Yang S, et al: Single-cell
transcriptomic analysis suggests two molecularly subtypes of
intrahepatic cholangiocarcinoma. Nat Commun. 13:16422022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma L, Wang L, Khatib SA, Chang CW,
Heinrich S, Dominguez DA, Forgues M, Candia J, Hernandez MO, Kelly
M, et al: Single-cell atlas of tumor cell evolution in response to
therapy in hepatocellular carcinoma and intrahepatic
cholangiocarcinoma. J Hepatol. 75:1397–1408. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Paulis YW, Huijbers EJ, van der Schaft DW,
Soetekouw PM, Pauwels P, Tjan-Heijnen VC and Griffioen AW: CD44
enhances tumor aggressiveness by promoting tumor cell plasticity.
Oncotarget. 6:19634–19646. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Katagiri YU, Sleeman J, Fujii H, Herrlich
P, Hotta H, Tanaka K, Chikuma S, Yagita H, Okumura K, Murakami M,
et al: CD44 variants but not CD44s cooperate with beta1-containing
integrins to permit cells to bind to osteopontin independently of
arginine-glycine-aspartic acid, thereby stimulating cell motility
and chemotaxis. Cancer Res. 59:219–226. 1999.PubMed/NCBI
|
|
16
|
Rao G, Wang H, Li B, Huang L, Xue D, Wang
X, Jin H, Wang J, Zhu Y, Lu Y, et al: Reciprocal interactions
between tumor-associated macrophages and CD44-positive cancer cells
via osteopontin/CD44 promote tumorigenicity in colorectal cancer.
Clin Cancer Res. 19:785–797. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pietras A, Katz AM, Ekström EJ, Wee B,
Halliday JJ, Pitter KL, Werbeck JL, Amankulor NM, Huse JT and
Holland EC: Osteopontin-CD44 signaling in the glioma perivascular
niche enhances cancer stem cell phenotypes and promotes aggressive
tumor growth. Cell Stem Cell. 14:357–369. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Klement JD, Paschall AV, Redd PS, Ibrahim
ML, Lu C, Yang D, Celis E, Abrams SI, Ozato K and Liu K: An
osteopontin/CD44 immune checkpoint controls CD8+ T cell activation
and tumor immune evasion. J Clin Invest. 128:5549–5560. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hornburg M, Desbois M, Lu S, Guan Y, Lo
AA, Kaufman S, Elrod A, Lotstein A, DesRochers TM, Munoz-Rodriguez
JL, et al: Single-cell dissection of cellular components and
interactions shaping the tumor immune phenotypes in ovarian cancer.
Cancer Cell. 39:928–944.e6. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lun ATL, McCarthy DJ and Marioni JC: A
step-by-step workflow for low-level analysis of single-cell RNA-seq
data with bioconductor. F1000Res. 5:21222016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Korsunsky I, Millard N, Fan J, Slowikowski
K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR and Raychaudhuri
S: Fast, sensitive and accurate integration of single-cell data
with Harmony. Nat Methods. 16:1289–1296. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hao Q, Li J, Zhang Q, Xu F, Xie B, Lu H,
Wu X and Zhou X: Single-cell transcriptomes reveal heterogeneity of
high-grade serous ovarian carcinoma. Clin Transl Med. 11:e5002021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao J, Spielmann M, Qiu X, Huang X,
Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers
FJ, et al: The single-cell transcriptional landscape of mammalian
organogenesis. Nature. 566:496–502. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sergushichev AA: An algorithm for fast
preranked gene set enrichment analysis using cumulative statistic
calculation. bioRxiv. 0600122016.
|
|
25
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aibar S, González-Blas CB, Moerman T,
Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC,
Geurts P, Aerts J, et al: SCENIC: Single-cell regulatory network
inference and clustering. Nat Methods. 14:1083–1086. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bao MM, Kennedy JM, Dolinger MT, Dunkin D,
Lai J and Dubinsky MC: Cytomegalovirus colitis in a patient with
severe treatment refractory ulcerative colitis. Crohns Colitis 360.
6:otae0142024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Menyhárt O, Nagy Á and Győrffy B:
Determining consistent prognostic biomarkers of overall survival
and vascular invasion in hepatocellular carcinoma. R Soc Open Sci.
5:1810062018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Traag VA, Waltman L and van Eck NJ: From
louvain to leiden: Guaranteeing well-connected communities. Sci
Rep. 9:52332019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao
R, Modak M, Carotta S, Haslinger C, Kind D, et al: Landscape and
dynamics of single immune cells in hepatocellular carcinoma. Cell.
179:829–845.e20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jia C, Ma Y, Wang M, Liu W, Tang F and
Chen J: Evidence of omics, immune infiltration, and
pharmacogenomics for BATF in a pan-cancer cohort. Front Mol Biosci.
9:8447212022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lauer S and Gresham D: An evolving view of
copy number variants. Curr Genet. 65:1287–1295. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sodek J, Chen J, Nagata T, Kasugai S,
Todescan R Jr, Li IW and Kim RH: Regulation of osteopontin
expression in osteoblasts. Ann N Y Acad Sci. 760:223–241. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sato Y, Harada K, Sasaki M and Nakanuma Y:
Clinicopathological significance of S100 protein expression in
cholangiocarcinoma. J Gastroenterol Hepatol. 28:1422–1429. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang Z, Jin Q, Hu W, Dai L, Xue Z, Man D,
Zhou L, Xie H, Wu J and Zheng S: 14-3-3σ downregulation suppresses
ICC metastasis via impairing migration, invasion, and anoikis
resistance of ICC cells. Cancer Biomark. 19:313–325. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kumar MP, Du J, Lagoudas G, Jiao Y, Sawyer
A, Drummond DC, Lauffenburger DA and Raue A: Analysis of
single-cell RNA-Seq identifies cell-cell communication associated
with tumor characteristics. Cell Rep. 25:1458–1468.e4. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Richardson JR, Schöllhorn A, Gouttefangeas
C and Schuhmacher J: CD4+ T cells: Multitasking cells in the duty
of cancer immunotherapy. Cancers (Basel). 13:5962021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim HJ and Cantor H: CD4 T-cell subsets
and tumor immunity: The helpful and the not-so-helpful. Cancer
Immunol Res. 2:91–98. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tay RE, Richardson EK and Toh HC:
Revisiting the role of CD4+ T cells in cancer
immunotherapy-new insights into old paradigms. Cancer Gene Ther.
28:5–17. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Aizarani N, Saviano A, Sagar, Mailly L,
Durand S, Herman JS, Pessaux P, Baumert TF and Grün D: A human
liver cell atlas reveals heterogeneity and epithelial progenitors.
Nature. 572:199–204. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kang K, Wang X, Meng C, He L, Sang X,
Zheng Y and Xu H: The application of single-cell sequencing
technology in the diagnosis and treatment of hepatocellular
carcinoma. Ann Transl Med. 7:7902019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shroff RT and Bachini M: Treatment options
for biliary tract cancer: Unmet needs, new targets and
opportunities from both physicians' and patients' perspectives.
Future Oncol. 20:1435–1450. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vincelette ND, Yu X, Kuykendall AT, Moon
J, Su S, Cheng CH, Sammut R, Razabdouski TN, Nguyen HV, Eksioglu
EA, et al: Trisomy 8 defines a distinct subtype of
myeloproliferative neoplasms driven by the MYC-alarmin axis. Blood
Cancer Discov. 5:276–297. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Popek-Marciniec S, Styk W,
Wojcierowska-Litwin M, Chocholska S, Szudy-Szczyrek A,
Samardakiewicz M, Swiderska-Kolacz G, Czerwik-Marcinkowska J and
Zmorzynski S: Association of chromosome 17 aneuploidy, TP53
deletion, expression and Its rs1042522 variant with multiple
myeloma risk and response to thalidomide/bortezomib treatment.
Cancers (Basel). 15:47472023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hallek M and Al-Sawaf O: Chronic
lymphocytic leukemia: 2022 Update on diagnostic and therapeutic
procedures. Am J Hematol. 96:1679–1705. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang F, Luo M, Qu H and Cheng Y: BAP1
promotes viability and migration of ECA109 cells through
KLF5/CyclinD1/FGF-BP1. FEBS Open Bio. 11:1497–1503. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang Y, Du W, Chen Z and Xiang C:
Upregulation of PD-L1 by SPP1 mediates macrophage polarization and
facilitates immune escape in lung adenocarcinoma. Exp Cell Res.
359:449–457. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cai X, Zhang H and Li T: The role of SPP1
as a prognostic biomarker and therapeutic target in head and neck
squamous cell carcinoma. Int J Oral Maxillofac Surg. 51:732–741.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Song Z, Chen W, Athavale D, Ge X, Desert
R, Das S, Han H and Nieto N: Osteopontin takes center stage in
chronic liver disease. Hepatology. 73:1594–1608. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Morse C, Tabib T, Sembrat J, Buschur KL,
Bittar HT, Valenzi E, Jiang Y, Kass DJ, Gibson K, Chen W, et al:
Proliferating SPP1/MERTK-expressing macrophages in idiopathic
pulmonary fibrosis. Eur Respir J. 54:18024412019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu L, Zhang R, Deng J, Dai X, Zhu X, Fu
Q, Zhang H, Tong Z, Zhao P, Fang W, et al: Construction of TME and
Identification of crosstalk between malignant cells and macrophages
by SPP1 in hepatocellular carcinoma. Cancer Immunol Immunother.
71:121–136. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Storrs EP, Chati P, Usmani A, Sloan I,
Krasnick BA, Babbra R, Harris PK, Sachs CM, Qaium F, Chatterjee D,
et al: High-dimensional deconstruction of pancreatic cancer
identifies tumor microenvironmental and developmental stemness
features that predict survival. NPJ Precis Oncol. 7:1052023.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nallasamy P, Nimmakayala RK, Karmakar S,
Leon F, Seshacharyulu P, Lakshmanan I, Rachagani S, Mallya K, Zhang
C, Ly QP, et al: Pancreatic tumor microenvironment factor promotes
cancer stemness via SPP1-CD44 axis. Gastroenterology.
161:1998–2013.e7. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zheng Y, Zhao L, Xiong Z, Huang C, Yong Q,
Fang D, Fu Y, Gu S, Chen C, Li J, et al: Ursolic acid targets
secreted phosphoprotein 1 to regulate Th17 cells against metabolic
dysfunction-associated steatotic liver disease. Clin Mol Hepatol.
30:449–467. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shevde LA and Samant RS: Role of
osteopontin in the pathophysiology of cancer. Matrix Biol.
37:131–141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yan Z, Hu X, Tang B and Deng F: Role of
osteopontin in cancer development and treatment. Heliyon.
9:e210552023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhao H, Chen Q, Alam A, Cui J, Suen KC,
Soo AP, Eguchi S, Gu J and Ma D: The role of osteopontin in the
progression of solid organ tumour. Cell Death Dis. 9:3562018.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kawamura K, Iyonaga K, Ichiyasu H, Nagano
J, Suga M and Sasaki Y: Differentiation, maturation, and survival
of dendritic cells by osteopontin regulation. Clin Diagn Lab
Immunol. 12:206–212. 2005.PubMed/NCBI
|
|
60
|
Cui G, Chen J, He J, Lu C, Wei Y, Wang L,
Xu X, Li L, Uede T and Diao H: Osteopontin promotes dendritic cell
maturation and function in response to HBV antigens. Drug Des Devel
Ther. 9:3003–3016. 2015.PubMed/NCBI
|
|
61
|
Wen Y, Feng D, Wu H, Liu W, Li H, Wang F,
Xia Q, Gao WQ and Kong X: Defective initiation of liver
regeneration in osteopontin-deficient mice after partial
hepatectomy due to insufficient activation of IL-6/Stat3 pathway.
Int J Biol Sci. 11:1236–1247. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu Y, Zhang L, Ju X, Wang S and Qie J:
Single-cell transcriptomic analysis reveals macrophage-tumor
crosstalk in hepatocellular carcinoma. Front Immunol.
13:9553902022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
He C, Sheng L, Pan D, Jiang S, Ding L, Ma
X, Liu Y and Jia D: Single-cell transcriptomic analysis revealed a
critical role of SPP1/CD44-mediated crosstalk between macrophages
and cancer cells in glioma. Front Cell Dev Biol. 9:7793192021.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cheng M, Liang G, Yin Z, Lin X, Sun Q and
Liu Y: Immunosuppressive role of SPP1-CD44 in the tumor
microenvironment of intrahepatic cholangiocarcinoma assessed by
single-cell RNA sequencing. J Cancer Res Clin Oncol. 149:5497–5512.
2023. View Article : Google Scholar : PubMed/NCBI
|