Introduction
Currently, gastric cancer (GC) is one of the most
common cancer types and the leading cause of cancer-associated
mortality worldwide (1–6). According to the 2020 global cancer
statistics, GC) remained a significant global health burden in
2020, causing 10.89 million new cancer cases and 7.69 million
deaths worldwide. This positioned it as the fifth most common
malignancy and the fourth leading cause of cancer-related mortality
(7). In the early stages,
GC-associated symptoms are unclear or absent. In the majority of
cases, it has progressed to a stage that is not amenable to radical
surgery at the time of diagnosis. In most cancer databases, GC is
labeled stomach adenocarcinoma (STAD) because, in 95% of GC cases,
this is the predominant histological subtype of gastrointestinal
malignancy (8). To date,
therapeutic outcomes for STAD remain limited, with a 5-year overall
survival (OS) rate <10% in patients with advanced STAD (9). Recurrence is common even after
resection (10). Therefore, the
identification of new biomarkers is key for guiding systemic
treatment strategies. This highlights the urgency of developing an
accurate prognostic model and novel therapeutic targets for
patients with STAD.
Ferroptosis is a type of iron-dependent programmed
cell death that is distinct from apoptosis, necrosis and autophagy.
The primary mechanism is the catalysis of lipid peroxidation of
unsaturated fatty acids highly expressed on the cell membrane in
the presence of divalent iron or ester oxygenase, which induces
cell death. This results in a decrease in the regulation of the
antioxidant system (glutathione system) (11–17).
It is hypothesized that ferroptosis is associated with the
occurrence and development of tumors, such as liver and gastric
cancer (12,18–21).
The association between ferroptosis and GC has also been
increasingly recognized (22–36),
and ferroptosis is hypothesized to serve a vital role in GC
development and progression (37,38).
Thus, targeting ferroptosis may be a potential therapeutic strategy
for patients with GC.
Ferroptosis is a double-edged sword in
gastrointestinal disease, since the inhibition of ferroptosis
relieves the symptoms of intestinal injury, and the induction of
ferroptosis via pharmacological agonists or bioactive compounds
inhibits the proliferation of GC (32). Ferroptosis inducers affect different
steps in ferroptosis to regulate GC proliferation, invasion and
metastasis, although development of drug resistance in GC cells
poses a hurdle (23). However,
limitations exist in previous studies (23,31,39–41).
Firstly, the efficacy of the prognostic models for GC based on
ferroptosis-related genes is poor, as demonstrated by the fact that
the log-rank P-values in the prognostic models are often not low
enough (P>0.01), the hazard ratios of high- vs. low-risk are not
high enough and the optimal area under the curve (AUC) values of
the receiver operating characteristic (ROC) curves are often ~0.7
(39) . Secondly, considering the
dual nature of ferroptosis, its role in GC progression remains
controversial, and needs to be clarified with more evidence.
Thirdly, the roles of numerous important drivers and suppressors
[solute Carrier Family 7 Member 11 (SLC7A11)/Glutathione Peroxidase
4 (GPX4)] of ferroptosis in GC progression remain unclear. Finally,
novel therapeutic strategies based on targeting ferroptosis,
including potential small molecule drugs and microRNAs (miRNAs or
miRs) as molecular drugs to regulate gene expression, remain to be
explored.
The present study aimed to identify key
ferroptosis-related genes associated with the development of GC to
construct prognostic models and identify new molecular mechanisms
and potential treatment strategies. The present findings may serve
as a reference for future studies on the mechanism underlying
ferroptosis and treatment of GC.
Materials and methods
Key gene screening and survival
prediction via the Cox regression model
Ferroptosis-related genes were collected from FerrDb
(zhounan.org/ferrdb/current/), which is an open-source, open
access, manually curated and continuously updated database
(Fig. 1). There were 12 ferroptosis
markers, 369 ferroptosis driver items and 348 ferroptosis
suppressor (or inhibitor) items. Following deduplication, 484
ferroptosis-related genes remained. Subsequently, using the Gene
Expression Profiling Interactive Analysis 2
database(gepia.cancer-pku.cn/), the 500 top OS- and
progression-free survival (PFS)-associated genes were identified,
and 910 survival-related genes were identified following
deduplication. In addition, there were 4,640 differentially
expressed genes (DEGs) between STAD and adjacent normal tissue. The
intersection of these three sets was analyzed, and three common
elements were found in addition to 124 STAD-ferroptosis genes and
13 survival-ferroptosis genes. These 140 genes were used to
establish the prognostic models.
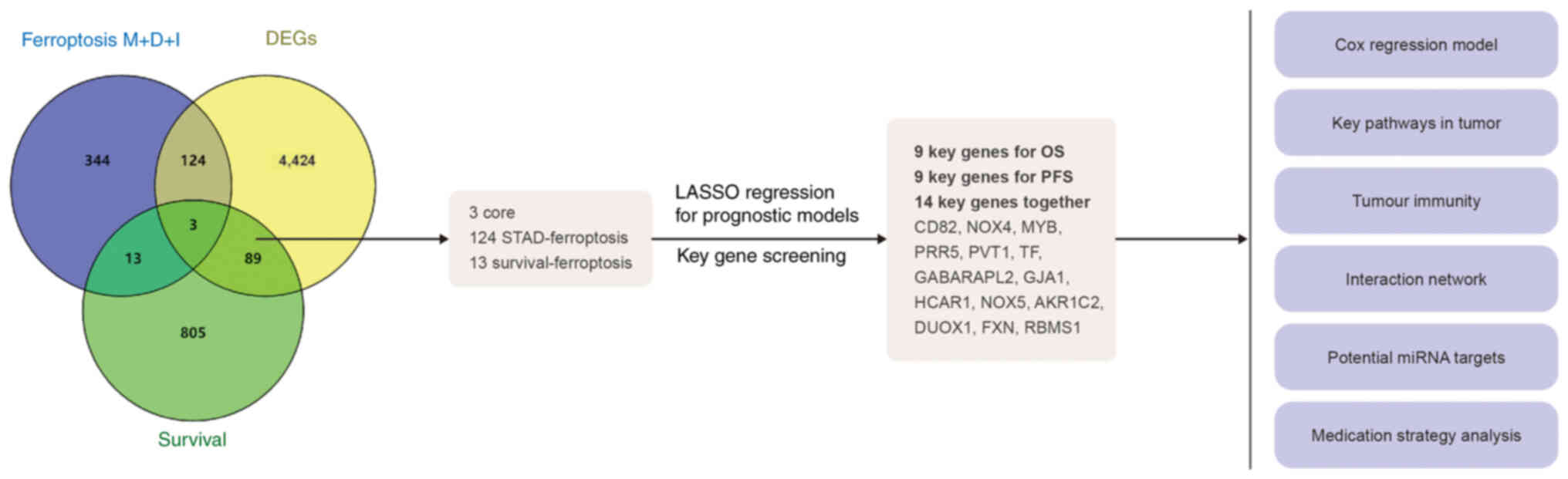 | Figure 1.Flow chart of the present study. A
total of 140 key ferroptosis-related genes were obtained, and 14
key genes were screened by LASSO and prognostic models for the
following analyses: i) Optimized Cox regression model, ii) key
pathway analysis in tumors, iii) immunity analysis and iv)
identification of potential miRNAs and drug targeting key
carcinogenic ferroptosis genes in STAD. M, marker; D, driver; I,
inhibitor; LASSO, least absolute shrinkage and selection operator
regression; miRNA, microRNA; STAD, stomach adenocarcinoma; DEG,
differentially expressed gene; OS, overall survival; PFS,
progression-free survival; NOX, NADPH oxidase; MYB, MYB
proto-oncogene, transcription factor; PRR5, proline-rich protein 5;
PVT1, Pvt1 oncogene; TF, transferrin; GABARAPL2, GABA type A
receptor-associated protein-like 2; GJA1, gap junction protein α1;
HCAR1, hydroxycarboxylic acid receptor 1; AKR1C2, aldo-keto
reductase family 1 member C2; DUOX1, dual oxidase 1; FXN, frataxin;
RBMS1, RNA binding motif single stranded interacting protein 1. |
For OS and PFS prediction, two multivariate Cox
regression models were established based on gastric cancer (STAD)
samples from The Cancer Genome Atlas (TCGA; Project ID: TCGA-STAD;
Data Portal: http://portal.gdc.cancer.gov/), as described in the
original pan-cancer analysis by TCGA Research Network (42). Least absolute shrinkage and
selection operator (LASSO) regression algorithm was used for
feature selection using 10-fold cross-validation and nine features
were selected. The equations of the Cox regression models were
calculated as risk scores, and patients were divided into high- and
low-risk groups using the median value as the cutoff (1.85). The
log-rank test was used to compare differences in survival between
the high- and low-risk groups. For the Kaplan-Meier curves, the
P-values and hazard rations (HRs) with 95% confidence intervals
(CIs) were generated using log-rank tests. ROC curves with AUC
values were constructed to assess the efficacy of the Cox
regression models. The key features (genes) of the models were
collected, and aggregated into the key gene set. As the performance
of OS was still not satisfactory (as the aim was to reduce the
likelihood ratio test and the log-rank P-value to
<1×10−12), another Cox regression model was
constructed after the key genes were obtained using the Tumor
Immune Estimation Resource (TIMER) database
(timer2.compbio.cn/timer1/). The model incorporated demographic and
clinical features(age, tumor stage), and removed certain
unimportant gene expression features(MYB, PRP5) to ensure that the
log-rank P-value was <1×10−12.
Roles of key genes in STAD
For each key ferroptosis gene in STAD, the
expression between tumor and adjacent normal tissue was compared.
The expression trends from stages I to IV were acquired via the
Gene Set Cancer Analysis (GSCA) online tool
(guolab.wchscu.cn/GSCA/#/). Using the same tool (based on the TCGA
datasets), the association between gene expression and different
types of survival [disease-specific survival (DSS), OS and PFS]
were presented in a bubble plot (where red indicates increased
risk). Additionally, the association between key gene expression
levels and the activation/inhibition of important pathways in
cancer development were using the GSCA tool.
Key-gene interaction networks
Using the GeneMANIA online tool (genemania.org/),
the interaction networks between 14 key genes were explored by
focusing on genetic and physical protein interactions and
co-expression.
Tumor immunity analysis based on key
ferroptosis genes in STAD
Using the TIMER database, the immune characteristics
of the cells were evaluated. Correlation between key ferroptosis
genes in STAD and tumor-infiltrating immune cells was determined
using Pearson's coefficient. The present study focused only on
immune cells with expression of key ferroptosis genes >0.3
(P<0.001).
Potential miRNAs that target key
carcinogenic ferroptosis genes in STAD
Among the 14 key ferroptosis genes, the carcinogenic
genes in STAD were selected according to the following criteria: i)
Coefficient in the prognostic equation should be >0.05 (for
either PFS or OS) and ii) expression tendency should be in
increasing order from stage I to IV or significant risk factors for
OS and PFS (univariate analysis). Overall, seven genes were
regarded as key carcinogenic ferroptosis genes in STAD. Using the
miRWalk database(mirwalk.umm.uni-heidelberg.de/), the miRNAs
targeting these seven genes were downloaded. The miRNAs with the
most matching pairs (base-pairing sequences with the seven genes)
and the greatest number of target genes were considered to have a
potential therapeutic value.
Potential drugs based on key
ferroptosis genes in STAD
The GSCA tool was used to identify potential drugs
for treating STAD. Genomics of Drug Sensitivity in Cancer (GDSC)
and Cancer Therapeutics Response Portal (CTRP) databases
(cancerrxgene.org/; portals.broadinstitute.org/ctrp/) were used to
obtain the half-maximal inhibitory concentration (IC50)
values of the drugs. The drugs were ranked by the integrated level
of the correlation coefficient and false discovery rate values, and
the top 30 ranked drugs were shown in bubble plots. In addition,
theoretically, if IC50 is significantly negatively
correlated with a greater number of carcinogenic mRNAs, this
suggests a greater likelihood that the drug will exert an anti-STAD
effect.
Human tissue collection
The tumors and surrounding normal tissue (distance,
3–5 cm from the tumor tissue) of 20 patients (12 males and 8
females, aged 18–60 years) with GC who underwent surgical resection
in The First Affiliated Hospital of Kunming Medical University
(Kunming, China) from April to July 2024 were collected. The
inclusion criteria were as follows: i) Patients diagnosed with GC
by pathological examination; ii) aged ≥18 years and ≤60 years; iii)
received tumor resection surgery; iv)did not receive preoperative
chemotherapy, radiotherapy, biotherapy or traditional Chinese
medicine; and v) followed the normal follow-up requirements. In
addition, the exclusion criteria were as follows: i) Patients with
other primary types of cancer; ii) with needle or blood phobia; and
iii) pregnant women or lactating mothers. The present study was
approved by The First Affiliated Hospital of Kunming Medical
University Ethics Committee (approval no. 2024 lunshen L no. 78).
All samples were obtained with written informed consent obtained
from patients, and the study adhered to the ethical principles of
the Declaration of Helsinki.
Cell culture and transfection
The GC cell line AGS was purchased from American
Type Culture Collection and cultured in RPMI-1640 medium (Procell)
supplemented with 10% fetal bovine serum (Procell) and 1%
penicillin/streptomycin in 5% CO2 in a humidified
atmosphere at 37°C. Following 24 h culture, the Homo sapiens
(hsa)-miR-501-5p inhibitor (delivered as siRNA) and hsa-miR-484
mimic were transfected separately, along with their respective
negative controls, using Lipofectamine 3000 (Invitrogen, Thermo
Fisher Scientific), and incubated. The concentrations used were 50
nM for miR-484 mimic and 100 nM for miR-501-5p siRNA. Transfection
was carried out at 37°C for 6 h. Cells were harvested 48 h
post-transfection for subsequent assays. hsa-miR-484 mimic,
hsa-miR-501-5p inhibitor (siRNA format) and non-targeting scrambled
negative controls were designed and synthesized by Shanghai
GeneChem Co., Ltd. The sequences were as follows: miR-484 mimic:
Forward, 5′-UCAGGCUCAGUCCCCUCCCGAU-3′ and
R:5′-CGGGAGGGGACUGAGCCUGAGC-3′ and mimic negative controls:
Forward, 5′-UUGUACUACACAAAAGUACUG-3′ and reverse,
5′-GUACUUUUGUGUAGUACAAGC-3′; miR-501-5p siRNA:
F:5′-UCUCACCCAGGGACAAAGGA-3′ and R:5′-UCCUUUGUCCCUGGGUGAGA-3′ and
siRNA negative controls: F:5′-UUCUCCGAACGUGUCACGUUU-3′ and
R:5′-ACGUGACACGUUCGGAGAAUU-3′.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® was used to isolate RNA from GC
cell lines and tissue. According to the manufacturer's
instructions, cDNA was synthesized from RNA using a PrimeScript™ RT
reagent kit (Takara). TB Green® Premix Ex Taq™ II FAST
qPCR kit (Takara) was used for qPCR. qPCR was performed using the
following thermocycling conditions: Initial denaturation at 95°C
for 3 min, followed by 45 cycles of 95°C for 5 sec and 60°C for 30
sec. The amplification of DNA was performed with TB
Green® Premix Ex Taq (Takara). CFX96™ Real-Time System
was used to evaluate the amplification of each gene. The primer
sequences were as follows: hsa-miR-501-5p: forward:
5′-AUCCUUUGUCCCUGGGUGAGA-3′ and reverse: 5′-GTGCAGGGTCCGAGGT-3′;
hsa-miR-484: Forward: 5′-UCAGGCUCAGUCCCCUCCCGAU-3′ and reverse:
5′-GTGCAGGGTCCGAGGT-3′ and U6 sense: Forward:
5′-CTCGCTTCGGCAGCACA-3′ and reverse: 5′-AACGCTTCACGAATTTGCGT-3′.
The miRNA primers were provided by Tiangen Biotech Co., Ltd. U6 was
used as an endogenous control. The results were determined using
the 2−ΔΔCq method (43).
All the experiments were repeated three times.
Cell viability assay
GC AGS cells were treated with DMSO,
(5Z)-7-oxozeanol (0, 1, 2, 4 µmol/l), selumetinib (0, 12.5, 25, 50
µmol/l), RDEA119 (0, 5, 50, 100 µmol/l), AZ628 (0, 0.5, 1, 1.5
µmol/l), dabrafenib(0, 0.25, 0.5, 1 µmol/l) or trametinib (0, 0.5,
5, 50 µmol/l) (all MedChemExpress) for 0, 24, 48 or 72 h, 37°C.
Following digestion with trypsin, the total number of cells was
counted on a cell counting plate. RPMI-1640 (Procell) containing
5,000 AGS cells was added to each well of a 96-well plate. Cell
Counting Kit-8 assay (Beyotime Institute of Biotechnology) was used
to observe the proliferative capacity (incubation for 2 h).
Absorbance values of the cells at 450 nm were determined. All the
experiments were repeated three times.
Western blotting
Cell Lysis buffer (Cell Signaling Technology)
containing PMSF (1 mM, Beyotime Institute of Biotechnology) was
used to lyse AGS cells. The BCA method was used to determine the
protein concentration. After that, the protein samples (20 µg/lane)
were separated using 8–12% sodium dodecyl sulfate-polyacrylamide
gel electrophoresis gels, and transferred to polyvinylidene
difluoride membranes (MilliporeSigma), which were incubated for 2 h
in 5% skimmed milk at 25°C. Subsequently, primary antibodies were
added overnight at 4°C. Following 2 h incubation with the secondary
antibody at 25°C, the membranes were washed three times with
TBS-Tween 20 (0.1%), and ECL (cat. no. WBULS0100; Millipore) was
added to the membrane, which was placed on a GelDoc imaging system.
ImageJ software (v1.54f, http://imagej.net/ij/) was used to analyze the optical
density. The antibodies were as follows: Anti-aldo-keto reductase
family 1 member C2 (AKR1C2; 1:200; cat. no. 13035; Cell Signaling
Technology, Inc.), anti-transferrin (TF; 1:500; cat no. 17435-1-AP;
Proteintech Group, Inc.), anti-NADPH oxidase (NOX) 4 (1:500; cat.
no. 14347-1-AP; Proteintech Group, Inc.), anti-RNA binding motif
single stranded interacting protein 1 (RBMS1; 1:400; cat. no.
ab150353; Abcam), anti-β-actin (1:2,000; cat. no. 66009-1-Ig;
Proteintech Group, Inc.) and HRP-conjugated goat anti-mouse IgG
(1:1,000; cat. no. D110087; Sangon Biotech Co., Ltd.). β-actin was
used as a loading control for normalization. All the experiments
were repeated three times.
Statistical analysis
GraphPad Prism 8.0 (GraphPad; Dotmatics) or SPSS
26.0 (IBM Corp.) statistical software were used to analyze the
data. Kolmogorov-Smirnov test was used to assess normal
distribution. Data conforming to a normal distribution are
presented as the mean ± standard deviation. Paired samples were
compared with paired Student's t-test. Comparisons of >2 groups
were made with one-way ANOVA followed by Dunnett's test. P<0.05
was considered to indicate a statistically significant difference.
Each experiment was independently repeated three times.
Results
Key ferroptosis genes in STAD and
prognostic models
The intersection analysis of 484 ferroptosis- and
910 survival-related genes and 4,640 DEGs in STAD revealed that 140
genes were notable ferroptosis genes in STAD. Using the 140 key
genes, the LASSO regression algorithm was used for feature
selection, and two Cox regression models for OS and FPS were
generated.
For OS prediction (Fig.
2), the regression model was as follows: Risk score=(−0.0065) ×
CD82 + (0.08) × NOX4 + (−0.0219) × MYB proto-oncogene,
transcription factor (MYB) + (−6×10−4) × proline rich
protein 5 (PRR5) + (−0.026) × Pvt1 oncogene (PVT1) + (0.1876) ×
GABA type A receptor associated protein like 2 (GABARAPL2) +
(0.0711) × gap junction protein α1 (GJA1) + (0.0086) ×
hydroxycarboxylic acid receptor 1 (HCAR1) + (0.5351) × NOX5. This
model contained nine features (Fig.
2A-C), among which, NOX4, NOX5, GABARAPL2 and GJA1 were the
most important risk factors. It had satisfactory efficacy in terms
of 5-6-year survival (AUC >0.72; Fig. 2D). The HR of the high-risk group was
2.66 (Fig. 2D). The additional OS
prediction model was constructed using seven genes(CD82, NOX4,
PVT1, GABARAPL2, TF, HCAR1and NOX5) along with several clinical
features (age, tumor stage; Fig.
2E), and validated its performance using standard Kaplan-Meier
curves and ROC curves (Fig. 2F-G).
Kaplan-Meier curves stratified patients into prognostically
distinct risk groups (log-rank P<0.0001), with the low-risk
group exhibiting a median survival time (6.02 years) four times
longer than that of the high-risk group (1.51 years). All high-risk
patients died within 5 years, whereas low-risk patients
demonstrated sustained survival with cases remaining alive at the
10-year mark (Fig. 2F). ROC
analysis confirmed the model's robust discriminative ability from 1
to 9 years (all AUC >0.70), with peak predictive performance at
6 years (AUC=0.787, 95% CI: 0.620–0.954). Strong predictive
validity was also maintained at 1 (AUC=0.713) and 2 years
(AUC=0.751), highlighting its utility for both short- and long-term
survival prediction (Fig. 2G).
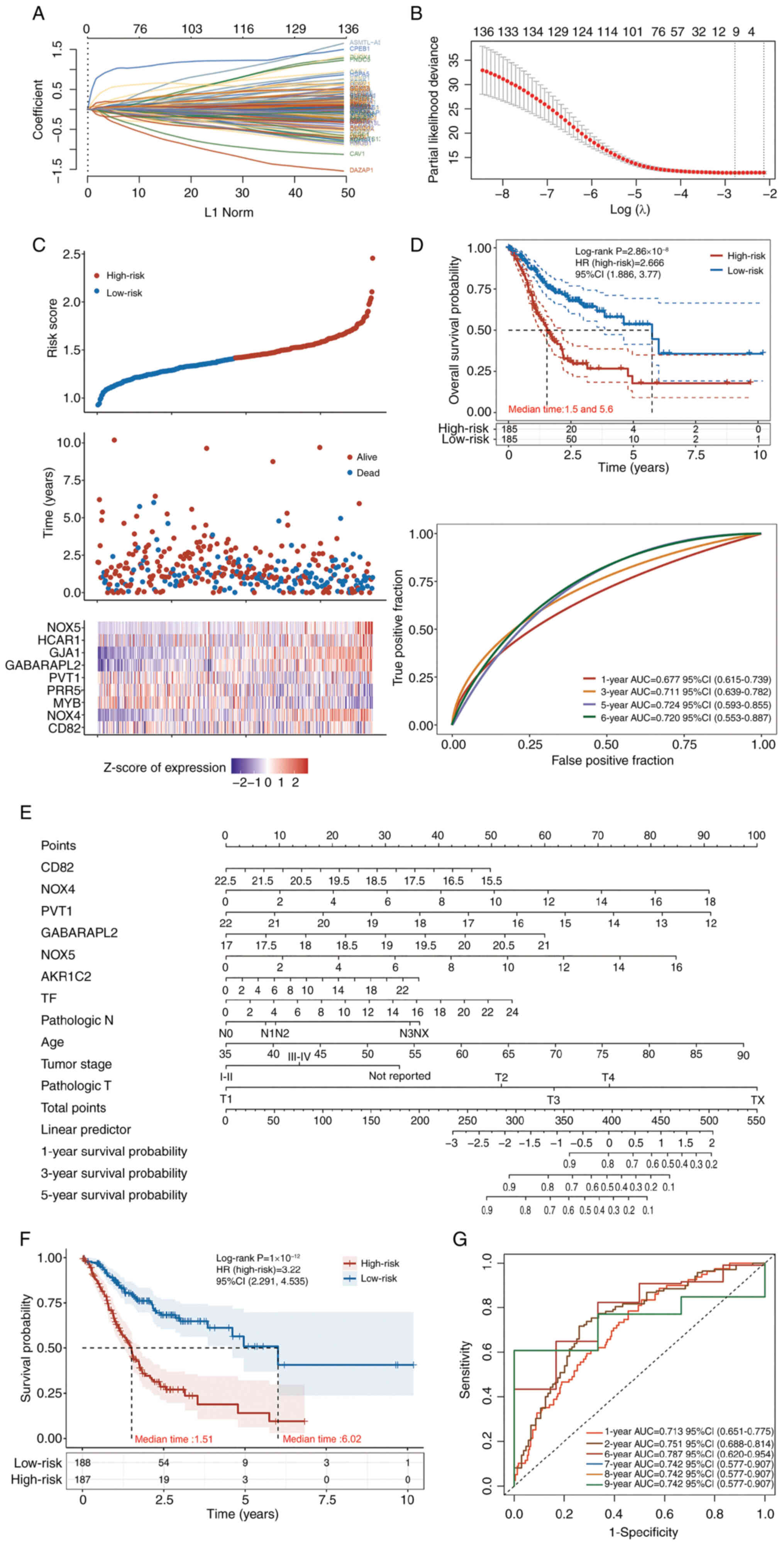 | Figure 2.LASSO analysis and Cox regression
models for OS prediction. (A) Coefficients of the selected features
are shown by the λ parameter, where the horizontal coordinate
represents the value of the independent variable λ and the vertical
coordinate represents the coefficients of the independent
variables. (B) Partial likelihood deviance plotted against log(λ)
values calculated through the LASSO Cox regression model. (C)
Association between expression of nine key genes in patients with
gastric adenocarcinoma from The Cancer Genome Atlas database with
patient risk scores and survival times. (D) Median OS time of the
low- and high-risk groups, and receiver operating characteristic
curve of the Cox regression model for the prediction of OS. (E-G)
Cox regression models constructed with the key genes from TIMER
database. (E) Nomogram of the constructed COX regression model,
visually presenting the model; (F) KM curve of the model; (G) ROC
curve of the model. LASSO, least absolute shrinkage and selection
operator; OS, overall survival; HR, hazard ratio; AUC, area under
the curve; CI, confidence interval; NOX, NADPH oxidase; MYB, MYB
proto-oncogene, transcription factor; PRR5, proline rich protein 5;
PVT1, Pvt1 oncogene; GABARAPL2, GABA type A receptor associated
protein like 2; GJA1, gap junction protein α1; HCAR1,
hydroxycarboxylic acid receptor 1. |
For PFS prediction (Fig.
3), the regression model was as follows: Risk score=(−0.0907) ×
AKR1C2 + (0.0074) × dual oxidase 1 + (−0.0384) × MYB + (−0.0483) ×
frataxin (FXN) + (0.118) × RBMS1 + (0.0132) × GABARAPL2 + (0.0883)
× TF + (0.066) × HCAR1 + (0.7408) × NOX5. This model also had nine
features (Fig. 3A-C), among which,
AKR1C2, RBMS1, NOX5, GABARAPL2, TF and HCAR1 were the most
important risk factors. It had good performance in the evaluation
of 6-9-year PFS; AUC of the 6-year PFS was >0.8 (Fig. 3D). The HR of the high-risk group was
2.827 (Fig. 2D).
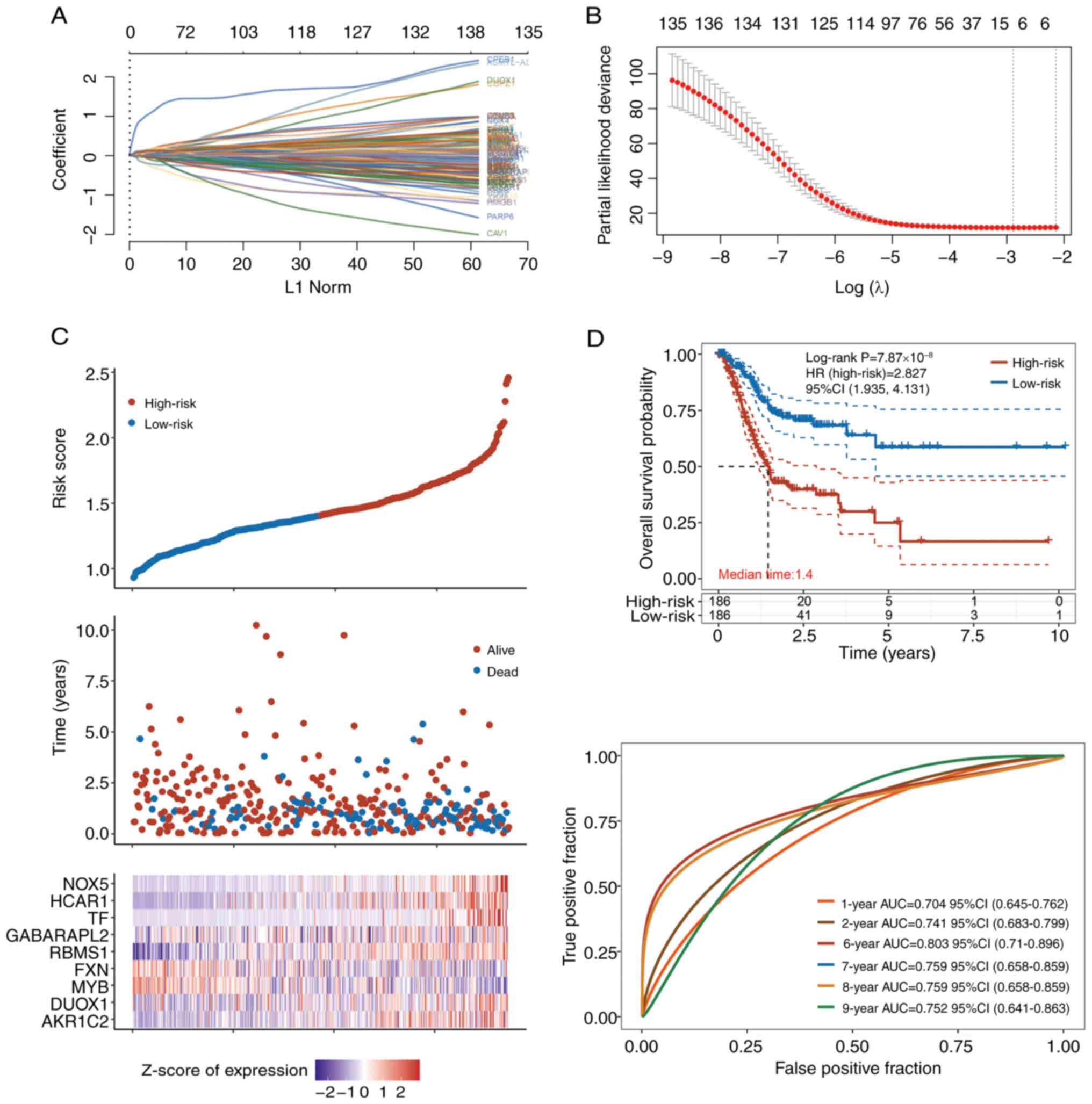 | Figure 3.LASSO analysis and Cox regression
models for PFS prediction. (A) Coefficients of the selected
features are shown by the λ parameter, with the horizontal
coordinate representing the value of the independent variable λ and
the vertical coordinate representing the coefficients of the
independent variables. (B) Partial likelihood deviance plotted
against log(λ) values calculated via the LASSO Cox regression
model. (C) Associations between risk scores and survival times of
patients with expressing nine key genes in patients with stomach
adenocarcinoma from The Cancer Genome Atlas database. (D) Median
PFS of the low- and high-risk groups, and receiver operating
characteristic curve of the Cox regression model for the prediction
of PFS. LASSO, least absolute shrinkage and selection operator;
PFS, progression-free survival; HR, hazard ratio; AUC, area under
the curve; CI, confidence interval; NOX, NADPH oxidase; MYB, MYB
proto-oncogene, transcription factor; GABARAPL2, GABA type A
receptor associated protein like 2; HCAR1, hydroxycarboxylic acid
receptor 1; TF, transferrin; AKR1C2, aldo-keto reductase family 1
member C2; DUOX1, dual oxidase 1; FXN, frataxin; RBMS1, RNA binding
motif single stranded interacting protein 1. |
Together, there were 14 unique key ferroptosis genes
identified in STAD (the combination of the nine features in the OS
model and the nine features in the PFS model), among which, NOX4,
NOX5, AKR1C2, RBMS1, GABARAPL2, GJA1, TF and HCAR1 may be the most
important carcinogenic genes (risk genes).
Roles of key genes in STAD
Among the 14 key genes in STAD, MYB, NOX4, PVT1 and
PRR5 exhibited increased expression in STAD tumors compared with
normal tissue (Fig. 4A). NOX4,
AKR1C2, GJA1, GABARAPL2, HCAR1, RBMS1 and TF expression tended to
increase from stages I to IV (Fig.
4B). TF, RBMS1, NOX5, NOX4, HCAR1, GJA1 and GABARAPL2 were
significant risk factors for OS and PFS (Fig. 4C). Moreover, NOX5 was a significant
risk factor not only for OS and PFS, but also for disease-free
interval and DSS. The associations between key genes and important
cancer pathways are shown in Fig.
4D. These genes suppress apoptosis and cell cycle progression
but promote epithelial-mesenchymal transition (EMT) and activate
protein kinase B and receptor tyrosine kinase (44–49).
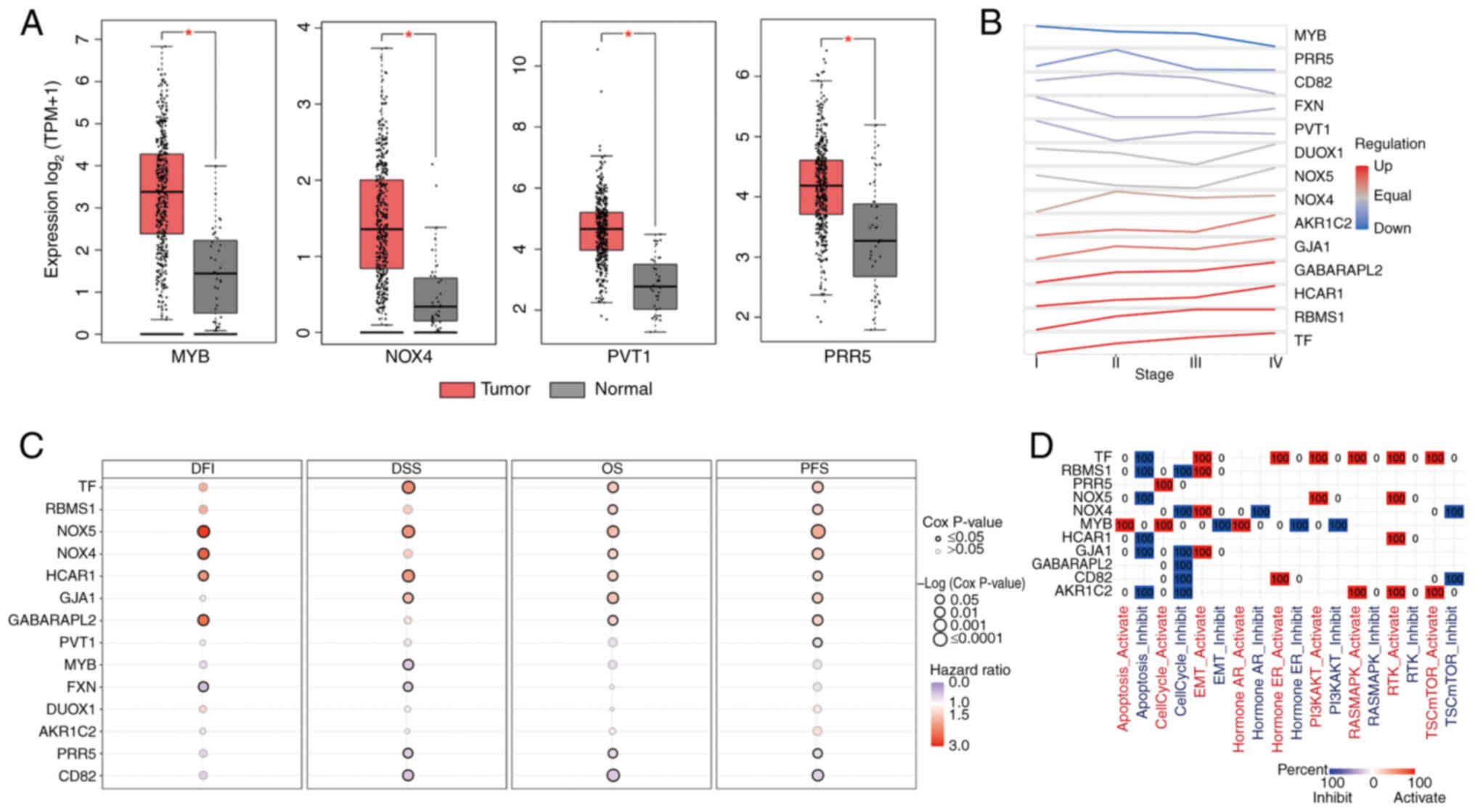 | Figure 4.Roles of key ferroptosis genes in
STAD. (A) Among the 14 key genes in STAD, MYB, NOX4, PVT1 and PRR5
expression was increased in STAD tumors compared with normal
tissue. *P<0.05. (B) Expression trend of each key gene from
stages I to IV. (C) Risk genes for DFI, DSS, OS and PFS among the
14 key genes. (D) Association between key genes and cancer
pathways. STAD, stomach adenocarcinoma; DFI, disease-free interval;
DSS, disease-specific survival; OS, overall survival; PFS,
progression-free survival; NOX, NADPH oxidase; MYB, MYB
proto-oncogene, transcription factor; PRR5, proline-rich protein 5;
PVT1, Pvt1 oncogene; TF, transferrin; GABARAPL2, GABA type A
receptor associated protein like 2; GJA1, gap junction protein α1;
HCAR1, hydroxycarboxylic acid receptor 1; AKR1C2, aldo-keto
reductase family 1 member C2; DUOX1, dual oxidase 1; FXN, frataxin;
RBMS1, RNA binding motif single stranded interacting protein 1;
TPM, transcripts per million; A, activation; I, inhibition. |
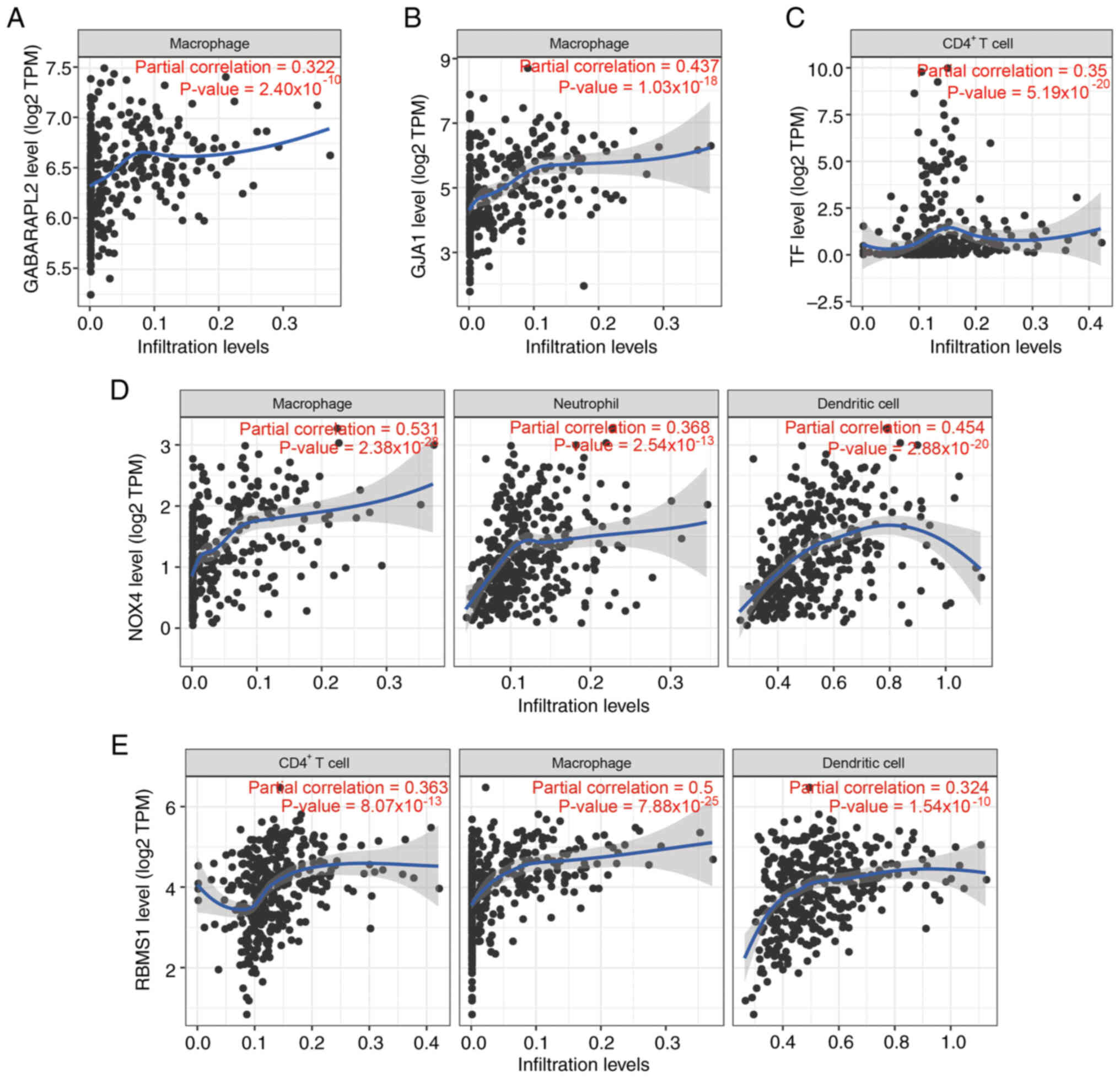
Tumor immunity associated with key
ferroptosis genes in STAD
The present study focused on key ferroptosis genes
correlated (r>0.3; P<0.001) with immune cells in STAD tumors.
GABARAPL2 and GJA1 expression was positively associated with
macrophages (Fig. 5A and B). TF
expression was positively associated with CD4+ T cells
(Fig. 5C). The NOX4 expression
levels were positively correlated with the levels of macrophages,
neutrophils and dendritic cells (Fig.
5D). RBMS1 expression was positively correlated with
CD4+ T cells, macrophages and dendritic cells (Fig. 5E). These results suggest these key
ferroptosis genes may impact the immune microenvironment.
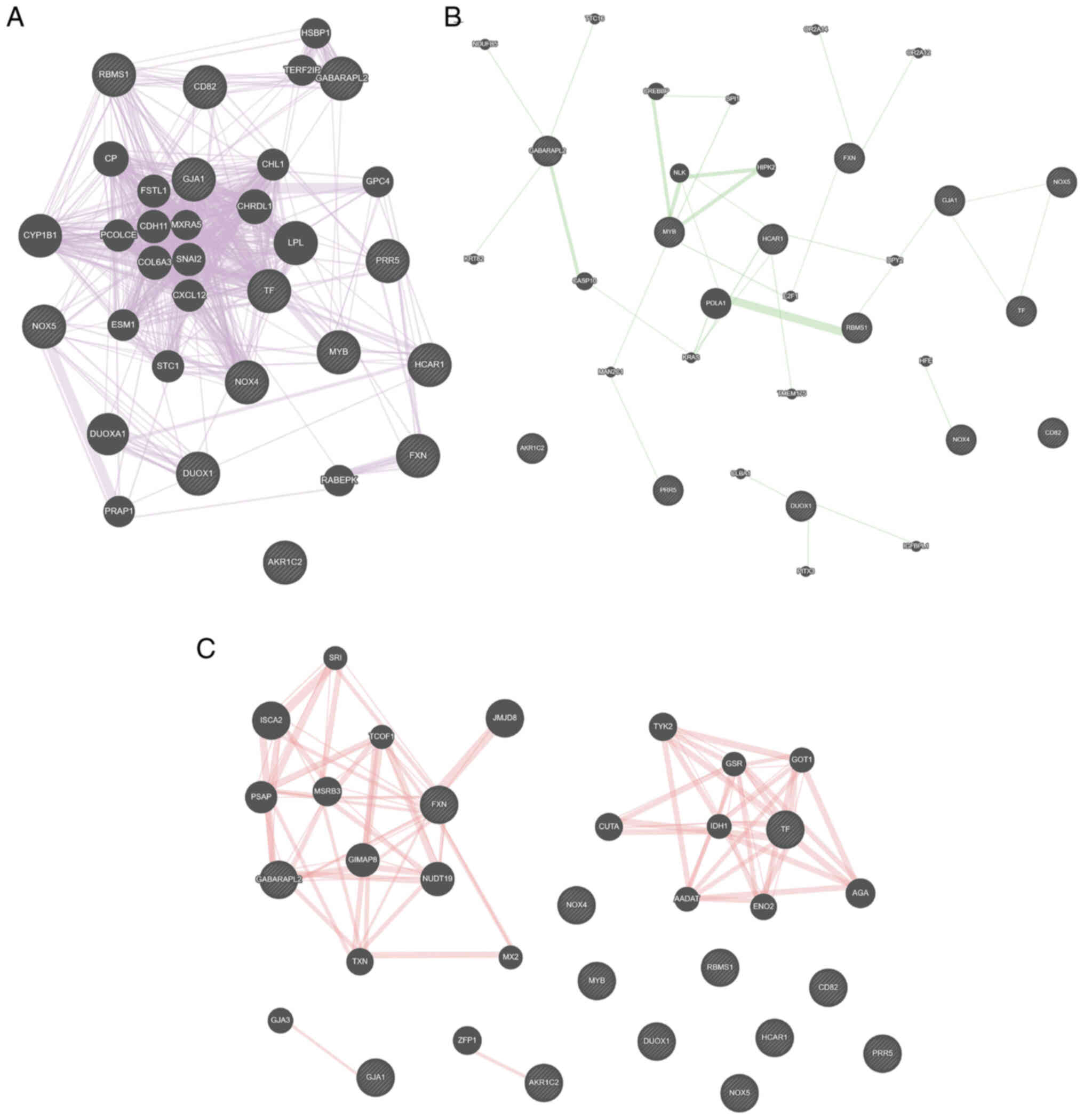
Key gene interaction networks
Using the GeneMANIA online tool, GJA1, NOX4, NOX5
and TF hub genes were identified in a co-expression network of 14
key genes (Fig. 6A). RBMS1, GJA1,
NOX5 and TF were key nodes in the genetic interaction network
(Fig. 6B), while FXN and GABARAPL2
were important nodes in the physical interaction network (Fig. 6C).
Potential miRNAs that target key
carcinogenic ferroptosis genes in STAD
Key genes were selected based on significant
prognostic weights in the Cox model (absolute value of coefficients
>0.05), positive correlation between gene expression and tumor
staging, or significant association with poor survival in
univariate analysis (P<0.05). As a result, seven genes were
regarded as key carcinogenic ferroptosis genes in STAD among the 14
key ferroptosis genes. Using the miRWalk database, the miRNAs
targeting these seven genes were downloaded. hsa-miR-505-5p and
hsa-miR-6795-5p presented the most targets (12 and 11 targets,
respectively; Fig. 7A). In
addition, the top miRNAs with paired key genes (six genes each)
were hsa-miR-6795-5p, hsa-miR-6758-5p, hsa-miR-501-5p and
hsa-miR-484 (Fig. 7B). Therefore,
these miRNAs (hsa-miR-6795-5p, hsa-miR-6758-5p, hsa-miR-501-5p,
hsa-miR-505-5p and hsa-miR-484) may have potential therapeutic
value for STAD; targets of these miRNAs are presented in Fig. 7C.
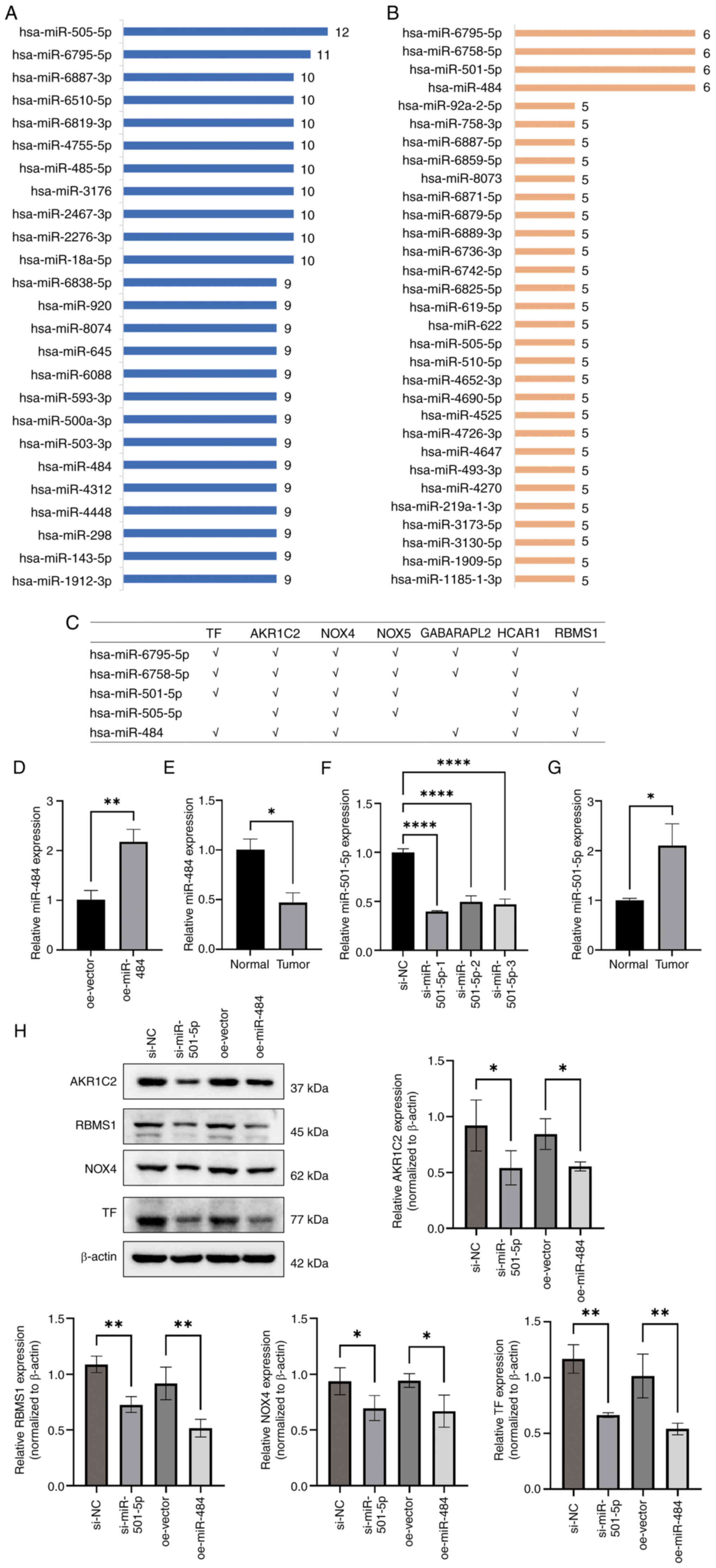 | Figure 7.Potential miRNAs that target key
carcinogenic ferroptosis genes in stomach adenocarcinoma. (A) Top
miRNAs with the most matching pairs to key genes. (B) Top miRNAs
with the most targeted genes. (C) A total of five miRNAs
(hsa-miR-6795-5p, hsa-miR-6758-5p, hsa-miR-501-5p, hsa-miR-505-5p
and hsa-miR-484) targeted the majority of genes (six targets each).
Relative mRNA expression of miR-484 in (D) GC cells with miR-484
overexpression and (E) tumor and surrounding normal tissue.
Relative mRNA expression of miR-501-5p in (F) GC cell line treated
with siRNAs and (G) tumor and surrounding normal tissue. (H) A
total of four risk genes for GC (AKR1C2, RBMS1, NADPH oxidase 4 and
TF) were detected via western blotting. *P<0.05; **P<0.01;
****P<0.001. miRNA/miR, microRNA; hsa, Homo sapiens; GC,
gastric cancer; siRNA, small interfering RNA; nc, negative control;
oe, overexpression; GABARAPL2, GABA type A receptor-associated
protein-like 2; TF, transferrin; AKR1C2, aldo-keto reductase family
1 member C2; NOX, NADPH oxidase; HCAR1, hydroxycarboxylic acid
receptor 1; RBMS1, RNA binding motif single stranded interacting
protein 1. |
Through target gene prediction and pathway
association analysis, target genes of miR-501-5P and miR-484
involved in core ferroptosis-related biological processes such as
lipid peroxidation, iron ion metabolism and oxidative stress were
identified (50–52). By contrast, the other candidates
lacked direct literature support or had unknown functional
relevance and require further investigation. miR-501-5p and miR-484
expression was evaluated in GC and surrounding normal tissue.
miR-484 expression was decreased, while miR-501-5P expression was
increased, in cancer tissues compared with normal tissue (Fig. 7E and G). miR-484 was overexpressed
and miR-501-5p was knocked down in AGS cells (Fig. 7D and F). Overexpression of miR-484
and the knockdown of miR-501-5p significantly decreased the
expression of the GC-associated high-risk genes AKR1C2, RBMS1, NOX4
and TF (Fig. 7H).
Potential drugs based on key
ferroptosis genes in STAD
Using the GDSC and CTRP databases, potential drugs
were identified on the basis of the seven key ferroptosis-related
carcinogenic genes. The top 30 ranked drugs in the GDSC and CTRP
databases are shown in Fig. 8A and
B. The present study focused on drugs with multiple targets
(which may have greater therapeutic value). In the CTRP database,
four drugs had the most targets (targeting three genes):
PHA-793887, SNS-032, CR-1-31B and saracatinib (Fig. 8C). According to the GDSC database
(Fig. 8D), six drugs targeted four
genes simultaneously: (5Z)-7-Oxozeaenol, selumetinib, RDEA119,
AZ628, dabrafenib and trametinib. Together, these drugs may be
candidate ferroptosis-related medications for the treatment of
STAD. In AGS cells, all six drugs significantly induced GC cell
death to varying degrees (Fig.
8E-J), and significantly reduced the expression of the
high-risk genes (Fig. 8K).
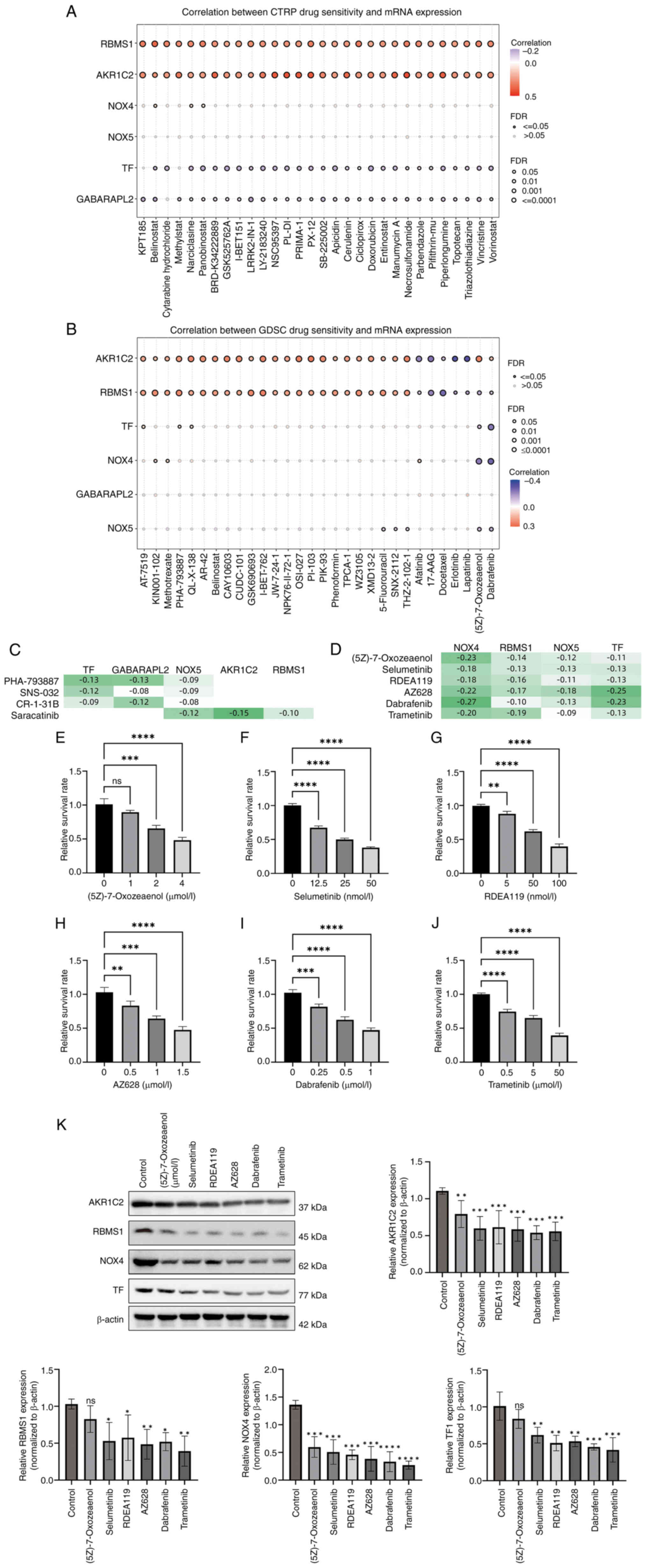 | Figure 8.Potential drugs based on key
ferroptosis genes in stomach adenocarcinoma. Top 30 ranked drugs in
(A) GDSC and (B) CTRP database based on the seven key carcinogenic
genes. (C) In the CTRP database, the following four drugs had the
most targets (each targeting three genes): PHA-793887, SNS-032,
CR-1-31B and saracatinib. (D) In the GDSC database, six drugs
targeted four genes simultaneously, namely (5Z)-7-oxozeaenol,
selumetinib, RDEA119, AZ628, dabrafenib and trametinib. Drug
toxicity of (E) (5Z)-7-oxozeaenol, (F) selumetinib, (G) RDEA119,
(H) AZ628, (I) dabrafenib and (J) trametinib towards gastric cancer
cells. (K) Expression of four risk genes (AKR1C2, RBMS1, NOX4 and
TF) was downregulated in gastric cancer cells treated with the
aforementioned drugs. *P<0.05; **P<0.01; ***P<0.005;
****P<0.001. GDSC, Genomics of Drug Sensitivity in Cancer; CTRP,
Cancer Therapeutics Response Portal; AKR1C2, aldo-keto reductase
family 1 member C2; RBMS1, RNA-binding motif single stranded
interacting protein 1; NOX, NADPH oxidase; TF, transferrin; FDR,
false discovery rate; ns, not significant. |
Discussion
The present study investigated the key
ferroptosis-associated genes involved in STAD development. A total
of 14 key genes was identified, including seven carcinogenic genes
that promote STAD development and are risk factors for survival.
For OS and PFS prediction, two models were constructed; by
combining age and stage information, another powerful OS model was
generated. GABARAPL2, GJA1, NOX4 and RBMS1 may impact the immune
microenvironment. Moreover, five miRNAs (hsa-miR-6795-5p,
hsa-miR-6758-5p, hsa-miR-501-5p, hsa-miR-505-5p and hsa-miR-484)
with potential therapeutic value for STAD were identified via the
targeting of carcinogenic genes. Finally, it was hypothesized that
the following drugs may be effective in treating STAD:
(5Z)-7-Oxozeaenol, selumetinib, RDEA119, AZ628, dabrafenib and
trametinib.
Unlike previous studies focusing on individual
ferroptosis regulators (such as GPX4 or SLC7A11), the present work
identified a novel seven-gene risk signature that predicts GC
prognosis (53–55). This multi-gene approach provides a
more comprehensive framework for targeting ferroptosis in
heterogeneous tumors. The present results align with previous
report linking ferroptosis resistance to GC metastasis but
identified RBMS1 as a dual regulator of iron metabolism and EMT
(44,56).
In the current optimized Cox hazard model, the
log-rank P-value of OS was 2.55×10−14. To the best of
our knowledge, this value is the lowest among all currently
available models. Previous studies have established prognostic
models with log-rank P-values (high- vs. low-risk) ranging from
0.0001 to 0.0100 (1,57–61).
Moreover, the AUC of the ROC curve for the prediction of PFS was
>0.8, which is markedly improved compared with that reported in
the majority of previous studies (1,61,62). A
previous study explored novel immune ferroptosis-related genes
associated with clinical and prognostic features in patients with
GC (38). However, performance of
the model was not satisfactory (P=0.046 in the test cohort; the
best AUC value was ~0.7). In summary, the present prognostic model
constructed using ferroptosis genes is one of the best performing
models for GC prognosis.
On the basis of the coefficients of the prognostic
model, the interaction networks and association with tumor
immunity, it was found that NOX5, NOX4 and GABARAPL2 served more
prominent cancer-promoting roles compared with the other key genes.
NOX5 is a strong reactive oxygen species producer. It mediates the
crosstalk between tumor cells and cancer-associated fibroblasts by
regulating the cytokine network (63). To the best of our knowledge, only
one study has investigated the genetic alteration and mRNA
expression of the NOX family in patients with GC (63), the aforementioned bioinformatics
study demonstrated decreased NOX5 expression in GC; to the best of
our knowledge, however, no study has elucidated the role of NOX5 in
GC. The present study highlights the prognostic and carcinogenic
role of NOX5 in GC and hence expands the range of GC targets. As
shown in a previous study (46),
NOX4 expression was also increased in GC compared with normal
tissues, which is in line with the present findings. Additionally,
the aforementioned study reported NOX4 is a potential prognostic
marker in GC and implicate that the use of NOX inhibitor targeting
NOX4 and DUOX1 may be an effective strategy for GC therapy
(46). NOX4 was a valid biomarker
for STAD (64). Similarly, a
previous study involving LASSO analysis used NOX4 for the
construction of a hypoxia-related gene prognostic model for GC
(65). However, none of the
aforementioned studies revealed the cancer-promoting mechanism of
NOX4. By contrast, the present study suggested that NOX4 and NOX5
(two important ferroptosis drivers) may drive GC progression by
promoting ferroptosis.
GABARAPL2 is a mitophagy-related gene (66–69)
and a ferroptosis driver. A previous study established and
validated a nomogram model based on GABARAPL2 and cell division
cycle 37, HSP90 cochaperone (CDC37) (49). GABARAPL2 and CDC37 display different
immune infiltration states and are prognostic biomarkers and
candidate therapeutic targets of GC (49). Another study revealed different
expression patterns of GABARAPL2 in GC and normal tissues, but it
was not a powerful independent prognostic factor (70). The present study revealed that
ferroptosis drivers, but not inhibitors, serve primarily
pro-carcinogenic roles, suggesting that the inhibition of
ferroptosis is a potential strategy to combat GC progression.
The present study identified miRNAs
(hsa-miR-6795-5p, hsa-miR-6758-5p, hsa-miR-501-5p, hsa-miR-505-5p
and hsa-miR-484) with potential therapeutic implications. A
previous study revealed that the upregulation of miR-501-5p
activates the Wnt/β-catenin signaling pathway and enhances the stem
cell-like phenotype in GC (71),
and another study revealed that miRNA-501-5p promotes cell
proliferation and migration in GC by downregulating
lysophosphatidic acid receptor 1 (72). The aforementioned conclusions
contradict that of the present study; thus, whether hsa-miR-501-5p
exerts an anticancer effect remains to be determined. The
expression of miR-484 is downregulated in GC (73). In 2020, an in vitro study
revealed that miR-484 suppresses the proliferation, migration and
invasion, and induces the apoptosis of GC cells (74). Moreover, downregulation of miR-484
is associated with poor prognosis and GC progression (75). The aforementioned studies are
consistent with the present hypothesis that miR-484 may serve as a
potential molecular agent against GC progression. The present study
experimentally validated miR-484 and miR-501-5p due to their
established roles in ferroptosis-associated processes. miR-484 was
downregulated in GC tissues. Through its seed sequence 5′-UCAGG-3′,
miR-484 targets the 3′-UTRs of NOX4, TF, and RBMS1, inducing mRNA
degradation and reducing target protein expression by 60–70%.
Consequently, it blocks NADPH oxidase activity, iron ion uptake,
and reverses epithelial-mesenchymal transition, synergistically
inhibiting ferroptosis, immune evasion, and metastasis (44,76).
Regarding miR-501-5p, experimental inhibition of this miRNA
unexpectedly reduced the expression of target genes AKR1C2 and
GABARAPL2. This paradoxical effect may stem from: (1) indirect regulatory networks (e.g.,
miR-501-5p suppresses tumor suppressors LPAR1; inhibiting
miR-501-5p elevates LPAR1 expression, indirectly reducing AKR1C2)
(72); (2) ceRNA competitive mechanisms (77); and (3) miRNA concentration-dependent effects.
These findings reveal its dual role in directly targeting oncogenes
while indirectly maintaining oncogenic networks. The other miRNAs
(miR-6795-5p, miR-6758-5p and hsa-miR-505-5p) represent novel
candidates with potential therapeutic value but require functional
characterization in future.
A panel of drugs (5Z-7-oxozeaenol, selumetinib,
RDEA119, AZ628, dabrafenib and trametinib) for the treatment of
STAD was investigated in the present study. 5Z-7-oxozeaenol is a
selective TGFβ-activated kinase 1 inhibitor (78). A previous study reported that
5Z-7-oxozeaenol increases the expression levels of cytosolic
cytochrome c and cleaved caspase 3 and apoptosis rate in GC
cells (79). Despite the different
mechanisms, the results are consistent with the conclusions of the
present study, and support the potential of 5Z-7-oxozeaenol as a
candidate anti-GC drug. The MEK inhibitor selumetinib is a potent,
orally active inhibitor of the MAPK/ERK pathway, and in
vitro experiments suggested that selumetinib should be
validated in prospective clinical trials (80). Moreover, the VIKTORY trial was
designed to classify patients with metastatic GC on the basis of
clinical sequencing and focused on eight biomarker groups, among
which, selumetinib was evaluated with or without chemotherapy
(81). However, the effectiveness
of selumetinib in the clinic is not yet clear. Similar to
selumetinib, RDEA119 is a MEK inhibitor that exhibits an anticancer
effect on multiple cancer cells, including GC cells (82). The potential for improving tumor
microenvironment of AZ628 has been proposed in another
bioinformatics study, but no validation was performed (83). Dabrafenib is a BRAF inhibitor and
its action on GC may increase susceptibility to immunotherapy
(84). Recently, the US Food and
Drug Administration accelerated the approval of darafenib in
combination with trametinib for the treatment of unresectable or
metastatic solid tumors (including GC) with the BRAFV600E mutation,
and this combination has been recommended in the latest National
Comprehensive Cancer Network guidelines for GC (85). Trametinib is a MEK inhibitor used to
inhibit the growth of GC (86–88).
In addition, the combination of dabrafenib and trametinib may be
promising in the clinical treatment of GC.
The present study has limitations. Regarding
bioinformatics methods, LASSO regression performance is greatly
influenced by data quality. The present study uses three databases,
and the sample size was limited. In the future, it is necessary to
expand the database samples and optimize the data quality to obtain
more accurate results. LASSO regression selects the most important
variables, but the explanatory power of these variables may be low.
Therefore, it may be necessary to combine other analytical methods
to improve the explanatory power of LASSO regression in the future.
Although numerous key genes, miRNAs and inhibitors have been
screened, the interaction between them and the regulatory mechanism
of iron-mediated cell death remains unclear. The present results
should be verified through basic experiments and clinical research.
While weighted correlation network analysis and LASSO regression
robustly identify gene signatures, these approaches may overlook
non-linear gene interactions. Additionally, bulk RNA-sequencing
data cannot resolve cell type-specific ferroptosis mechanisms,
necessitating future single-cell analyses. Prioritized targets
(such as NOX4) should be validated using NOX4-knockout AGS cell
lines and patient-derived xenograft models treated with ferroptosis
inducers erastin/artesunate, alongside RNA interference-mediated
gene silencing.
In summary, 14 key ferroptosis-related genes,
including seven carcinogenic genes, that promoted STAD development
and are risk factors for survival were identified in the present
study. For OS and PFS prediction, two models were constructed, and
five miRNAs (hsa-miR-6795-5p, hsa-miR-6758-5p, hsa-miR-501-5p,
hsa-miR-505-5p and hsa-miR-484) with potential therapeutic value
for STAD were identified through the targeting of carcinogenic
genes. The present results revealed that (5Z)-7-oxozeaenol,
selumetinib, RDEA119, AZ628, dabrafenib and trametinib may be
effective in treating STAD.
Acknowledgements
Not applicable.
Funding
The present study was supported by Kunming Medical University
First Affiliated Hospital Science and Technology Talent Training
Program (Leading Talent; grant no. L-2019024), Sub-project of
Yunnan Clinical Medical Research Center (grant no. 202102AA100062),
Scientific Research Fund Project of Yunnan Education Department
(grant no. 2024Y223), Kunming Medical University graduate Student
Innovation Fund (grant no. 2024S045), Yunnan Fundamental Research
Projects (grant no. 202401AT070170) and First-Class Discipline Team
of Kunming Medical University (grant no. 2024XKTDYS02).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
HW conceived and designed the study, analyzed data
and wrote the manuscript. HC conceived and designed the study,
interpreted data and edited the manuscript. JJL and DZ performed
experiments and analyzed data. DW analyzed data. MSH and MWL
interpreted data and constructed figures. SYH interpreted data. LQM
conceived and designed the study. HW and LQM confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Ethics approval was obtained from the ethics
committee of the First Affiliated Hospital of Kunming Medical
University (approval no. 2024 lunshen L No.78). The procedures used
in the present study adhere to the principles of CFDA/GCP and the
Declaration of Helsinki. All participants provided voluntary
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chang J, Wu H, Wu J, Liu M, Zhang W, Hu Y,
Zhang X, Xu J, Li L, Yu P and Zhu J: Constructing a novel
mitochondrial-related gene signature for evaluating the tumor
immune microenvironment and predicting survival in stomach
adenocarcinoma. J Transl Med. 21:1912023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jin W, Ou K, Li Y, Liu W and Zhao M:
Metabolism-related long noncoding RNA in the stomach cancer
associated with 11 AMMLs predictive nomograms for OS in STAD. Front
Genet. 14:11271322023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang J, Liu D, Wang Q and Xie Y:
Identification of basement membrane-related signatures in gastric
cancer. Diagnostics (Basel). 13:18442023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu B, Fu L, Guo X, Hu H, Li Y, Shi Y,
Zhang Y, Han S, Lv C and Tian Y: Multiomics profiling and digital
image analysis reveal the potential prognostic and
immunotherapeutic properties of CD93 in stomach adenocarcinoma.
Front Immunol. 14:9848162023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao W, Lin J, Cheng S, Li H, Shu Y and Xu
C: Comprehensive analysis of COMMD10 as a novel prognostic
biomarker for gastric cancer. PeerJ. 11:e146452023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao Z, Mak TK, Shi Y, Li K, Huo M and
Zhang C: Integrative analysis of cancer-associated fibroblast
signature in gastric cancer. Heliyon. 9:e192172023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
8
|
Ajani JA, Lee J, Sano T, Janjigian YY, Fan
D and Song S: Gastric adenocarcinoma. Nat Rev Dis Primers.
3:170362017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takahari D: Second-line chemotherapy for
patients with advanced gastric cancer. Gastric Cancer. 20:395–406.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Johnston FM and Beckman M: Updates on
management of gastric cancer. Curr Oncol Rep. 21:672019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hirschhorn T and Stockwell BR: The
development of the concept of ferroptosis. Free Radic Biol Med.
133:130–143. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao W, Wang X, Zhou Y, Wang X and Yu Y:
Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor
immunotherapy. Signal Transduct Target Ther. 7:1962022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang D, Minikes AM and Jiang X:
Ferroptosis at the intersection of lipid metabolism and cellular
signaling. Mol Cell. 82:2215–2227. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu J, Kang R and Tang D: Signaling
pathways and defense mechanisms of ferroptosis. FEBS J.
289:7038–7050. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu P, Wang W, Li Z, Li Y, Yu X, Tu J and
Zhang Z: Ferroptosis: A new regulatory mechanism in osteoporosis.
Oxid Med Cell Longev. 2022:26344312022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang Y, Wang Y, Guo L, Gao W, Tang TL and
Yan M: Interaction between macrophages and ferroptosis. Cell Death
Dis. 13:3552022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao L, Zhou X, Xie F and Zhang L, Yan H,
Huang J, Zhang C, Zhou F, Chen J and Zhang L: Ferroptosis in cancer
and cancer immunotherapy. Cancer Commun (Lond). 42:88–116. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gong C, Ji Q, Wu M, Tu Z, Lei K, Luo M,
Liu J, Lin L, Li K, Li J, et al: Ferroptosis in tumor immunity and
therapy. J Cell Mol Med. 26:5565–5579. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Liu J, Xu Y, Wu R, Chen X, Song X,
Zeh H, Kang R, Klionsky DJ, Wang X and Tang D: Tumor heterogeneity
in autophagy-dependent ferroptosis. Autophagy. 17:3361–3374. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liao P, Wang W, Wang W, Kryczek I, Li X,
Bian Y, Sell A, Wei S, Grove S, Johnson JK, et al: CD8(+) T cells
and fatty acids orchestrate tumor ferroptosis and immunity via
ACSL4. Cancer Cell. 40:365–378.e6. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo T, Wang Y and Wang J: Ferroptosis
assassinates tumor. J Nanobiotechnology. 20:4672022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu D, Wang C, Yu L and Yu R: Induction of
ferroptosis by ATF3 elevation alleviates cisplatin resistance in
gastric cancer by restraining Nrf2/Keap1/xCT signaling. Cell Mol
Biol Lett. 26:262021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gu R, Xia Y, Li P, Zou D, Lu K, Ren L,
Zhang H and Sun Z: Ferroptosis and its Role in Gastric Cancer.
Front Cell Dev Biol. 10:8603442022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang G, Xiang Z, Wu H, He Q, Dou R, Lin
Z, Yang C, Huang S, Song J, Di Z, et al: The lncRNA
BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer
peritoneal metastasis by regulating VDAC3 ubiquitination. Int J
Biol Sci. 18:1415–1433. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li D, Wang Y, Dong C, Chen T, Dong A, Ren
J, Li W, Shu G, Yang J, Shen W, et al: CST1 inhibits ferroptosis
and promotes gastric cancer metastasis by regulating GPX4 protein
stability via OTUB1. Oncogene. 42:83–98. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin Z, Song J, Gao Y, Huang S, Dou R,
Zhong P, Huang G, Han L, Zheng J, Zhang X, et al: Hypoxia-induced
HIF-1α/lncRNA-PMAN inhibits ferroptosis by promoting the
cytoplasmic translocation of ELAVL1 in peritoneal dissemination
from gastric cancer. Redox Biol. 52:1023122022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Song Z, Liu Y, Ma X, Wang W, Ke Y,
Xu Y, Yu D and Liu H: Identification of ferroptosis as a novel
mechanism for antitumor activity of natural product derivative a2
in gastric cancer. Acta Pharm Sin B. 11:1513–1525. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma M, Kong P, Huang Y, Wang J, Liu X, Hu
Y, Chen X, Du C and Yang H: Activation of MAT2A-ACSL3 pathway
protects cells from ferroptosis in gastric cancer. Free Radic Biol
Med. 181:288–299. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ouyang S, Li H, Lou L, Huang Q, Zhang Z,
Mo J, Li M, Lu J, Zhu K, Chu Y, et al: Inhibition of
STAT3-ferroptosis negative regulatory axis suppresses tumor growth
and alleviates chemoresistance in gastric cancer. Redox Biol.
52:1023172022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Song S, Wen F, Gu S, Gu P, Huang W, Ruan
S, Chen X, Zhou J, Li Y, Liu J and Shu P: Network pharmacology
study and experimental validation of Yiqi Huayu decoction inducing
ferroptosis in gastric cancer. Front Oncol. 12:8200592022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Zheng L, Shang W, Yang Z, Li T,
Liu F, Shao W, Lv L, Chai L, Qu L, et al: Wnt/beta-catenin
signaling confers ferroptosis resistance by targeting GPX4 in
gastric cancer. Cell Death Differ. 29:2190–2202. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu C, Liu Z and Xiao J: Ferroptosis: A
double-edged sword in gastrointestinal disease. Int J Mol Sci.
22:124032021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu X and Li Y, Wu Y, Wang M, Lu Y, Fang Z,
Wang H and Li Y: Increased ATF2 expression predicts poor prognosis
and inhibits sorafenib-induced ferroptosis in gastric cancer. Redox
Biol. 59:1025642023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang H, Hu Y, Weng M, Liu X, Wan P, Hu Y,
Ma M, Zhang Y, Xia H and Lv K: Hypoxia inducible lncRNA-CBSLR
modulates ferroptosis through m6A-YTHDF2-dependent modulation of
CBS in gastric cancer. J Adv Res. 37:91–106. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang Z, Zou S, Zhang Y, Zhang J, Zhang P,
Xiao L, Xie Y, Meng M, Feng J, Kang L, et al: ACTL6A protects
gastric cancer cells against ferroptosis through induction of
glutathione synthesis. Nat Commun. 14:41932023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang H, Wang M, He Y, Deng T, Liu R, Wang
W, Zhu K, Bai M, Ning T, Yang H, et al: Chemotoxicity-induced
exosomal lncFERO regulates ferroptosis and stemness in gastric
cancer stem cells. Cell Death Dis. 12:11162021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee JY, Nam M, Son HY, Hyun K, Jang SY,
Kim JW, Kim MW, Jung Y, Jang E, Yoon SJ, et al: Polyunsaturated
fatty acid biosynthesis pathway determines ferroptosis sensitivity
in gastric cancer. Proc Natl Acad Sci USA. 117:32433–32442. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xiao C, Dong T, Yang L, Jin L, Lin W,
Zhang F, Han Y and Huang Z: Identification of novel immune
ferropotosis-related genes associated with clinical and prognostic
features in gastric cancer. Front Oncol. 12:9043042022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Deng H, Lin Y, Gan F, Li B, Mou Z, Qin X,
He X and Meng Y: Prognostic model and immune infiltration of
ferroptosis subcluster-related modular genes in gastric cancer. J
Oncol. 2022:58135222022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cheng X, Dai E, Wu J, Flores NM, Chu Y,
Wang R, Dang M, Xu Z, Han G, Liu Y, et al: Atlas of metastatic
gastric cancer links ferroptosis to disease progression and
immunotherapy response. Gastroenterology. 167:1345–1357. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yue Z, Yuan Y, Zhou Q, Sheng J and Xin L:
Ferroptosis and its current progress in gastric cancer. Front Cell
Dev Biol. 12:12893352024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cancer Genome Atlas Research Network, .
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu M, Li H, Zhang H, Zhou H, Jiao T, Feng
M, Na F, Sun M, Zhao M, Xue L and Xu L: RBMS1 promotes gastric
cancer metastasis through autocrine IL-6/JAK2/STAT3 signaling. Cell
Death Dis. 13:2872022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lodhi MS, Khan MT, Bukhari SMH, Sabir SH,
Samra ZQ, Butt H and Akram MS: Probing transferrin receptor
overexpression in gastric cancer mice models. ACS Omega.
6:29893–29904. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
You X, Ma M, Hou G, Hu Y and Shi X: Gene
expression and prognosis of NOX family members in gastric cancer.
Onco Targets Ther. 11:3065–3074. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang H, Zhan J, Zhou J, Liu L, He Y, Le
Y, Liu W, Zhou L, Liu Y and Xiang X: Identification of HCAR1 as a
ferroptosis-related biomarker of gastric cancer based on a novel
ferroptosis-related prognostic model and in vitro experiments.
Carcinogenesis. 7:bgaf0302025. View Article : Google Scholar
|
|
48
|
Pourjamal N, Shirkoohi R, Rohani E and
Hashemi M: The expression analysis of MEST1 and GJA1 genes in
gastric cancer in association with clinicopathological
characteristics. Int J Hematol Oncol Stem Cell Res. 18:83–91.
2024.PubMed/NCBI
|
|
49
|
Wang Z, Chen C, Ai J, Shu J, Ding Y, Wang
W, Gao Y, Jia Y and Qin Y: Identifying mitophagy-related genes as
prognostic biomarkers and therapeutic targets of gastric carcinoma
by integrated analysis of single-cell and bulk-RNA sequencing data.
Comput Biol Med. 163:1072272023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huang DH, Wang GY, Zhang JW, Li Y, Zeng XC
and Jiang N: MiR-501-5p regulates CYLD expression and promotes cell
proliferation in human hepatocellular carcinoma. Jpn J Clin Oncol.
45:738–744. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gu Y, Wu S, Fan J, Meng Z, Gao G, Liu T,
Wang Q, Xia H, Wang X and Wu K: CYLD regulates cell ferroptosis
through Hippo/YAP signaling in prostate cancer progression. Cell
Death Dis. 15:792024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Huang M, Cheng S, Li Z, Chen J, Wang C, Li
J and Zheng H: Preconditioning exercise inhibits neuron ferroptosis
and ameliorates brain ischemia damage by skeletal muscle-derived
exosomes via regulating miR-484/ACSL4 axis. Antioxid Redox Signal.
41:769–792. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang J, Tian T, Li X, Yan Y, Zhao D, Ji
S, Ni J, Zhang J, Liu K, Qing H and Quan Z: p53 inhibits OTUD5
transcription to promote GPX4 degradation and induce ferroptosis in
gastric cancer. Clin Transl Med. 15:e702712025. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lu D, Yuan L, Wang Z, Xu D, Meng F, Jia S,
Li Y, Li W and Nan Y: Dioscin induces ferroptosis to suppress the
metastasis of gastric cancer through the SLC7A11/GPX4 axis. Free
Radic Res. 59:426–441. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang H, Li C, Meng S and Kuang YT: The
LINC01094/miR-545-3p/SLC7A11 signaling axis promotes the
development of gastric cancer by regulating cell growth and
ferroptosis. Biochem Genet. https://doi.org/10.1007/s10528-024-10959-3
|
|
56
|
Xu Y, Hao J, Chen Q, Qin Y, Qin H, Ren S,
Sun C, Zhu Y, Shao B, Zhang J and Wang H: Inhibition of the
RBMS1/PRNP axis improves ferroptosis resistance-mediated
oxaliplatin chemoresistance in colorectal cancer. Mol Carcinog.
63:224–237. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yu R, Li Z, Zhang C, Song H, Deng M, Sun
L, Xu L, Che X, Hu X, Qu X, et al: Elevated limb-bud and heart
development (LBH) expression indicates poor prognosis and promotes
gastric cancer cell proliferation and invasion by upregulating
Integrin/FAK/Akt pathway. PeerJ. 7:e68852019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Berthelet E, Pickles T, Lee KW, Liu M and
Truong PT; Prostate Cancer Outcomes Initiative, : Long-term
androgen deprivation therapy improves survival in prostate cancer
patients presenting with prostate-specific antigen levels >20
ng/ml. Int J Radiat Oncol Biol Phys. 63:781–787. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Huang R, Lu TL and Zhou R: Identification
and immune landscape analysis of fatty acid metabolism genes
related subtypes of gastric cancer. Sci Rep. 13:204432023.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yuan Q, Deng D, Pan C, Ren J, Wei T, Wu Z,
Zhang B, Li S, Yin P and Shang D: Integration of transcriptomics,
proteomics, and metabolomics data to reveal HER2-associated
metabolic heterogeneity in gastric cancer with response to
immunotherapy and neoadjuvant chemotherapy. Front Immunol.
13:9511372022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang X, Zhang W, Guo Y, Zhang Y, Bai X and
Xie Y: Identification of critical prognosis signature associated
with lymph node metastasis of stomach adenocarcinomas. World J Surg
Oncol. 21:612023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Feng A, He L, Chen T and Xu M: A novel
cuproptosis-related lncRNA nomogram to improve the prognosis
prediction of gastric cancer. Front Oncol. 12:9579662022.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen J, Wang Y, Zhang W, Zhao D, Zhang L,
Zhang J, Fan J and Zhan Q: NOX5 mediates the crosstalk between
tumor cells and cancer-associated fibroblasts by regulating
cytokine network. Clin Transl Med. 11:e4722021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xiao R, Wang S, Guo J, Liu S, Ding A, Wang
G, Li W, Zhang Y, Bian X, Zhao S and Qiu W: Ferroptosis-related
gene NOX4, CHAC1 and HIF1A are valid biomarkers for stomach
adenocarcinoma. J Cell Mol Med. 26:1183–1193. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Guo J, Xing W, Liu W, Liu J, Zhang J and
Pang Z: Prognostic value and risk model construction of hypoxic
stress-related features in predicting gastric cancer. Am J Transl
Res. 14:8599–8610. 2022.PubMed/NCBI
|
|
66
|
Chan JCY and Gorski SM: Unlocking the gate
to GABARAPL2. Biol Futur. 73:157–169. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Polletta L, Vernucci E, Carnevale I,
Arcangeli T, Rotili D, Palmerio S, Steegborn C, Nowak T,
Schutkowski M, Pellegrini L, et al: SIRT5 regulation of
ammonia-induced autophagy and mitophagy. Autophagy. 11:253–270.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Scicluna K, Dewson G, Czabotar PE and
Birkinshaw RW: A new crystal form of GABARAPL2. Acta Crystallogr F
Struct Biol Commun. 77:140–147. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang Z, Gu H, Li Q, Zheng J, Cao S, Weng
C and Jia H: GABARAPL2 is critical for growth restriction of
Toxoplasma gondii in HeLa cells treated with gamma
interferon. Infect Immun. 88:e00054–e00020. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wang M, Jing J, Li H, Liu J, Yuan Y and
Sun L: The expression characteristics and prognostic roles of
autophagy-related genes in gastric cancer. PeerJ. 9:e108142021.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fan D, Ren B, Yang X, Liu J and Zhang Z:
Upregulation of miR-501-5p activates the wnt/β-catenin signaling
pathway and enhances stem cell-like phenotype in gastric cancer. J
Exp Clin Cancer Res. 35:1772016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ma X, Feng J, Lu M, Tang W, Han J, Luo X,
Zhao Q and Yang L: microRNA-501-5p promotes cell proliferation and
migration in gastric cancer by downregulating LPAR1. J Cell
Biochem. 121:1911–1922. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zare A, Ahadi A, Larki P, Omrani MD, Zali
MR, Alamdari NM and Ghaedi H: The clinical significance of miR-335,
miR-124, miR-218 and miR-484 downregulation in gastric cancer. Mol
Biol Rep. 45:1587–1595. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu J and Li SM: MiR-484 suppressed
proliferation, migration, invasion and induced apoptosis of gastric
cancer by targeting CCL-18. Int J Exp Pathol. 101:203–214. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li Y, Liu Y, Yao J, LI R and Fan X:
Downregulation of miR-484 is associated with poor prognosis and
tumor progression of gastric cancer. Diagn Pathol. 15:252020.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang L and Gong W: NOX4 regulates gastric
cancer cell invasion and proliferation by increasing ferroptosis
sensitivity through regulating ROS. Int Immunopharmacol.
132:1120522024. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhang D, Yan H, Li H, Hao S, Zhuang Z, Liu
M, Sun Q, Yang Y, Zhou M, Li K and Hang C: TGFβ-activated kinase 1
(TAK1) inhibition by 5Z-7-Oxozeaenol attenuates early brain injury
after experimental subarachnoid Hemorrhage. J Biol Chem.
290:19900–19909. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yang Y, Qiu Y, Tang M, Wu Z, Hu W and Chen
C: Expression and function of transforming growth
factor-β-activated protein kinase 1 in gastric cancer. Mol Med Rep.
16:3103–3110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ahn S, Brant R, Sharpe A, Dry JR, Hodgson
DR, Kilgour E, Kim K, Kim ST, Park SH, Kang WK, et al: Correlation
between MEK signature and Ras gene alteration in advanced gastric
cancer. Oncotarget. 8:107492–107499. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Lee J, Kim ST, Kim K, Lee H, Kozarewa I,
Mortimer PGS, Odegaard JI, Harrington EA, Lee J, Lee T, et al:
Tumor genomic profiling guides patients with metastatic gastric
cancer to targeted treatment: The VIKTORY umbrella trial. Cancer
Discov. 9:1388–1405. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Liu Z and Xing M: Induction of
sodium/iodide symporter (NIS) expression and radioiodine uptake in
nonthyroid cancer cells. PLoS One. 7:e317292012. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wu X, Zhou F, Cheng B, Tong G, Chen M, He
L, Li Z, Yu S, Wang S and Lin L: Immune activity score to assess
the prognosis, immunotherapy and chemotherapy response in gastric
cancer and experimental validation. PeerJ. 11:e163172023.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Liu J, Zhang B, Zhang Y, Zhao H, Chen X,
Zhong L and Shang D: Oxidative stress and autophagy-mediated immune
patterns and tumor microenvironment infiltration characterization
in gastric cancer. Aging (Albany NY). 15:12513–12536. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ajani JA, D'Amico TA, Bentrem DJ, Corvera
CU, Das P, Enzinger PC, Enzler T, Gerdes H, Gibson MK, Grierson P,
et al: Gastric cancer, version 2.2025, NCCN clinical practice
guidelines in oncology. J Natl Compr Canc Netw. 23:169–191. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Dai C, Shen L, Jin W, Lv B, Liu P, Wang X,
Yin Y, Fu Y, Liang L, Ma Z, et al: Physapubescin B enhances the
sensitivity of gastric cancer cells to trametinib by inhibiting the
STAT3 signaling pathway. Toxicol Appl Pharmacol. 408:1152732020.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Liu H, Yao Y, Zhang J and Li J: MEK
inhibition overcomes everolimus resistance in gastric cancer.
Cancer Chemother Pharmacol. 85:1079–1087. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang Z, Chen Y, Li X, Zhang Y, Zhao X,
Zhou H, Lu X, Zhao L, Yuan Q, Shi Y, et al: Tegaserod maleate
suppresses the growth of gastric cancer in vivo and in vitro by
targeting MEK1/2. Cancers (Basel). 14:35922022. View Article : Google Scholar : PubMed/NCBI
|