Introduction
Globally, lung cancer remains a leading cause of
cancer mortality, with 2.2 million new cases and 1.8 million deaths
annually (1). Prognosis is notably
poor, demonstrating a 5-year survival rate of 10–20% globally and
<7% for metastatic disease, largely due to late-stage diagnosis
(2–4). Among its subtypes, lung adenocarcinoma
(LUAD) is the most frequently diagnosed form of lung cancer.
Despite previous advancements in the treatment of various cancers,
the 5-year survival rate for lung cancer remains low (5). Consequently, identifying novel
predictive gene signatures is crucial for improving prognosis and
developing targeted therapeutic strategies for patients with
LUAD.
The transient receptor potential (TRP) channels
constitute a superfamily of non-selective cation channels that
serve a pivotal role in calcium homeostasis and calcium-mediated
signal transduction (6,7). Calcium-dependent signaling pathways
are critical regulators of tumor cell survival, proliferation,
invasiveness and therapeutic resistance, underscoring the
importance of TRP channels as key modulators of carcinogenesis and
tumor progression (8,9). Accumulating evidence suggests that
tumors can markedly influence the expression and activation of TRP
channels. Notably, specific subfamilies of TRP channels, including
TRPV, TRPM and TRPC, have been strongly implicated in the
initiation and progression of various cancers (10). For instance, TRPV4 has been shown to
induce apoptosis in human lung cancer cells via the p38 MAPK
pathway, highlighting its potential as a therapeutic target in lung
cancer (11). Despite these
advances, comprehensive bioinformatics analyses exploring the role
of TRP channel-related genes in LUAD remain limited.
TRP channels are involved in both tumorigenesis and
antitumor processes. However, their specific functions in LUAD
remain poorly understood. To address this gap, a systematic
investigation into the expression levels of TRP channel-related
genes was performed in normal lung tissues compared with LUAD
tissues. The present study aimed to explore the prognostic value of
these genes and elucidate their potential connections with focal
cell death and the tumor immune microenvironment.
However, the interaction between TRP channel
dysregulation and immune cell infiltration in LUAD, a hallmark of
tumor aggressiveness, remains uncharacterized. Therefore, the
present study aimed to investigate the relationship between this
TRP channel-related signature and the tumor immune microenvironment
in LUAD, and to explore its potential clinical utility for
therapeutic stratification. We employed comprehensive bioinformatic
analyses combined with experimental validation to assess immune
cell infiltration patterns associated with the signature and
evaluate its implications for patient prognosis and treatment
response.
Materials and methods
Datasets
RNA sequencing (RNA-seq) data from 535 patients with
LUAD and 59 normal human lung tissues were obtained from The Cancer
Genome Atlas (TCGA) database (TCGA-BRCA) (https://portal.gdc.cancer.gov/repository). The data
were downloaded as raw counts and normalized to transcripts per
million using the TCGAbiolinks R package (version 2.25.9) (12). Quality control steps included: i)
Sample filtering, where patients with incomplete survival
information or missing clinical annotations (such as tumor stage
and age) were excluded; ii) batch effect correction where the
ComBat algorithm from the sva R package (version 3.42.0) (13) was applied to adjust for batch
effects across sequencing batches; and iii) gene expression
filtering where genes with low expression (counts <10 in >90%
of samples) were removed, retaining 19,856 protein-coding genes for
downstream analysis.
For external validation, RNA-seq data (GSE50081) and
clinical metadata from 127 patients with LUAD were retrieved from
the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) using the GEOquery
R package (version 2.62.2) (14).
Raw microarray data were normalized using robust multi-array
average preprocessing using the limma R package (version 3.50.3)
(15). Probes were annotated to
gene symbols using the platform-specific annotation file (GPL570,
Affymetrix Human Genome U133 Plus 2.0 Array).
Identification of differentially
expressed TRP channel-associated genes
A total of 43 TRP channel-associated genes were
identified using the following steps: i) GeneCards Database
(https://www.genecards.org/): Genes were
retrieved using the keyword ‘TRP channel’ and filtered by a
relevance score ≥4.0 (top 20% high-confidence associations; score
range, 0–10); i) Online Mendelian Inheritance in Man database
(OMIM; http://www.omim.org/): Genes were
selected based on experimental evidence (such as functional studies
and cancer-related phenotypes) supporting direct roles in TRP
channel activity or carcinogenesis; and iii) functional validation:
Only genes encoding TRP channel proteins or directly regulating
their activity (such as calcium transport and channel gating) were
retained and indirect regulators were excluded. A full list of the
43 genes, including relevance scores, functional annotations and
supporting references, is provided in Table SI.
Consensus clustering
Consensus clustering was performed using the k-means
method to identify distinct TRP channel-related gene expression
patterns. The optimal number of clusters and their stability were
determined using the ConsensusClusterPlus R package (16). Clustering was repeated 1,000 times
to ensure robustness.
Development and validation of the TRP
channel-associated gene prognostic model
Differentially expressed genes (DEGs) between TRP
channel-associated patterns were identified in TCGA cohort using an
adjusted P-value of <0.05 and an absolute log2 fold change
(log2FC) >1.5. The prognostic significance of TRP
channel-associated genes was evaluated using Cox regression
analysis. The least absolute shrinkage and selection operator
(LASSO) method was applied to select and shrink variables. The risk
score for each patient was calculated using the following formula:
Risk score=(X1 × Y1) + (X2 × Y2) + (X3 × Y3) + (X4 × Y4), where X
represents the regression coefficients and Y denotes the gene
expression levels. Patients were stratified into high- and low-risk
groups based on the median risk score.
Principal component analysis (PCA) and t-distributed
stochastic neighbor embedding (t-SNE) were performed using the
Rtsne (v0.17) (17) and ggplot2
(v3.5.1) (18) R packages,
respectively, to visualize gene expression patterns (19). Time-dependent receiver operating
characteristic (ROC) curve analysis was performed using the
survival (v3.6–4) (20), timeROC
(v0.4) (21) and survminer (v0.4.9)
(22) R packages. The prognostic
model was further validated using an independent LUAD cohort from
the GEO database (GSE50081). Univariate and multivariate Cox
regression analyses were performed to assess the predictive
significance of the gene signature.
The prognostic utility of the risk model was further
assessed using the IMvigor210 cohort, which included transcriptomic
profiles and clinical outcomes of 348 patients with urothelial
carcinoma receiving anti-PD-L1 therapy. Participants were
stratified into four response categories: Complete response (CR),
partial response (PR), stable disease (SD) and progressive disease
(PD).
Functional enrichment analysis of DEGs
between high- and low-risk groups
Functional enrichment analysis, including Gene
Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analyses, was performed using the clusterProfiler R package
(v4.0.5) (23). Single-sample gene
set enrichment analysis (ssGSEA) was performed using the GSVA
package (v1.52.0) (24) to evaluate
immune cell infiltration scores and the activity of immune-related
pathways.
To investigate the association between signature
genes (ANLN, CREG2, RHOF, CDCP1) and immune cell infiltration in
LUAD, the TIMER database (http://timer.cistrome.org/) was used. Spearman
correlation analysis was performed to evaluate the relationships
between gene expression levels and immune cell infiltration
abundances, including CD8+ T cells, macrophages and
other immune subsets.
Drug sensitivity prediction using the
prophet algorithm
Drug response data (IC50 values) for LUAD
cell lines were obtained from the publicly available Genomics of
Drug Sensitivity in Cancer (GDSC) database (https://www.cancerrxgene.org/). The Prophet algorithm
(25), a machine learning-based
tool, was employed to model the relationship between the expression
levels of the four-gene TRP channel-related signature and drug
sensitivity. . Briefly, the input data of the gene expression
profiles of ANLN, CREG2, RHOF and CDCP1 from TCGA-LUAD and GEO
cohorts were normalized and integrated with GDSC-derived
IC50 values. Prophet used a generalized additive model
to regress gene expression against log-transformed IC50
values for model training, accounting for non-linear relationships
and interaction effects. The model was internally validated using
10-fold cross-validation within TCGA cohort. Predictive accuracy
was assessed using Pearson correlation between predicted and
observed IC50 values.
Cell culture
The LUAD cell line H1299 (Shaanxi Fuheng
Biotechnology Co., Ltd.), and normal bronchial epithelial cells,
BEAS-2B (BeNa Culture Collection), were used in the present study.
H1299 cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (HyClone™; Cytiva), while BEAS-2B cells
were maintained in Bronchial Epithelial Cell Growth Medium (BEGM;
Lonza Group Ltd.) containing the manufacturer's recommended
supplements (BEGM BulletKit; Lonza). Both cell lines were incubated
at 37°C in a humidified 5% CO2 atmosphere and passaged
using 0.25% trypsin-EDTA (Gibco; Thermo Fisher Scientific, Inc.).
Mycoplasma contamination was routinely excluded using the
MycoAlert™ Detection Kit (Lonza Group Ltd.).
Reverse transcription
(RT)-quantitative (q)PCR
For RT-qPCR validation, total RNA was extracted from
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and RNA purity was verified using a NanoDrop
2000 spectrophotometer (Thermo Fisher Scientific, Inc.). Reverse
transcription was performed with 1 µg RNA using the PrimeScript™ RT
Reagent Kit (Takara Bio, Inc.), followed by qPCR amplification
using a QuantStudio 5 System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with TB Green® Premix Ex Taq™ II
(Takara Bio, Inc.). The thermal profile included an initial
denaturation at 95°C for 30 sec, 40 cycles at 95°C for 5 sec and
60°C for 34 sec and melt curve analysis. Gene expression levels
were normalized to β-actin using the 2−ΔΔCq method
(26), with triplicate technical
replicates (27). Primer sequences
were as follows: β-actin-forward (F) 5′-GCACCACACCTTCTACAATGAGC-3′,
β-actin-reverse (R) 5′-GGATAGCACAGCCTGGATAGCAAC-3′, ANLN-F
5′-ACTCAGTCACTTCCAGTAACAG-3′, ANLN-R 5′-GCTAGATTTCGTCATTTCGCAT-3′,
CREG2-F 5′-ATGAAGAACCCCATGGCCTC-3′, CREG2-R,
5′-AAAACATGGCTTGCTTGGCA-3′, RHOF-F, 5′-CTCCTTCCCCGAGCACTACG-3′,
RHOF-R 5′-TAGCAGATGAGCACGAGGTG-3′, CDCP1-F
5′-CAACATCAATACTGAGATGCCG-3′, CDCP1-R
5′-GTAGCAGATGCCCATATACCAT-3′.
Statistical analysis
All statistical analyses were performed using R
software (version 4.1.2; R Foundation for Statistical Computing).
Data are presented as mean ± SEM. Comparisons between normal and
tumor groups were performed using an unpaired two-tailed Student's
t-test (n=3 independent experiments). P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification of TRP-related DEGs in
LUAD
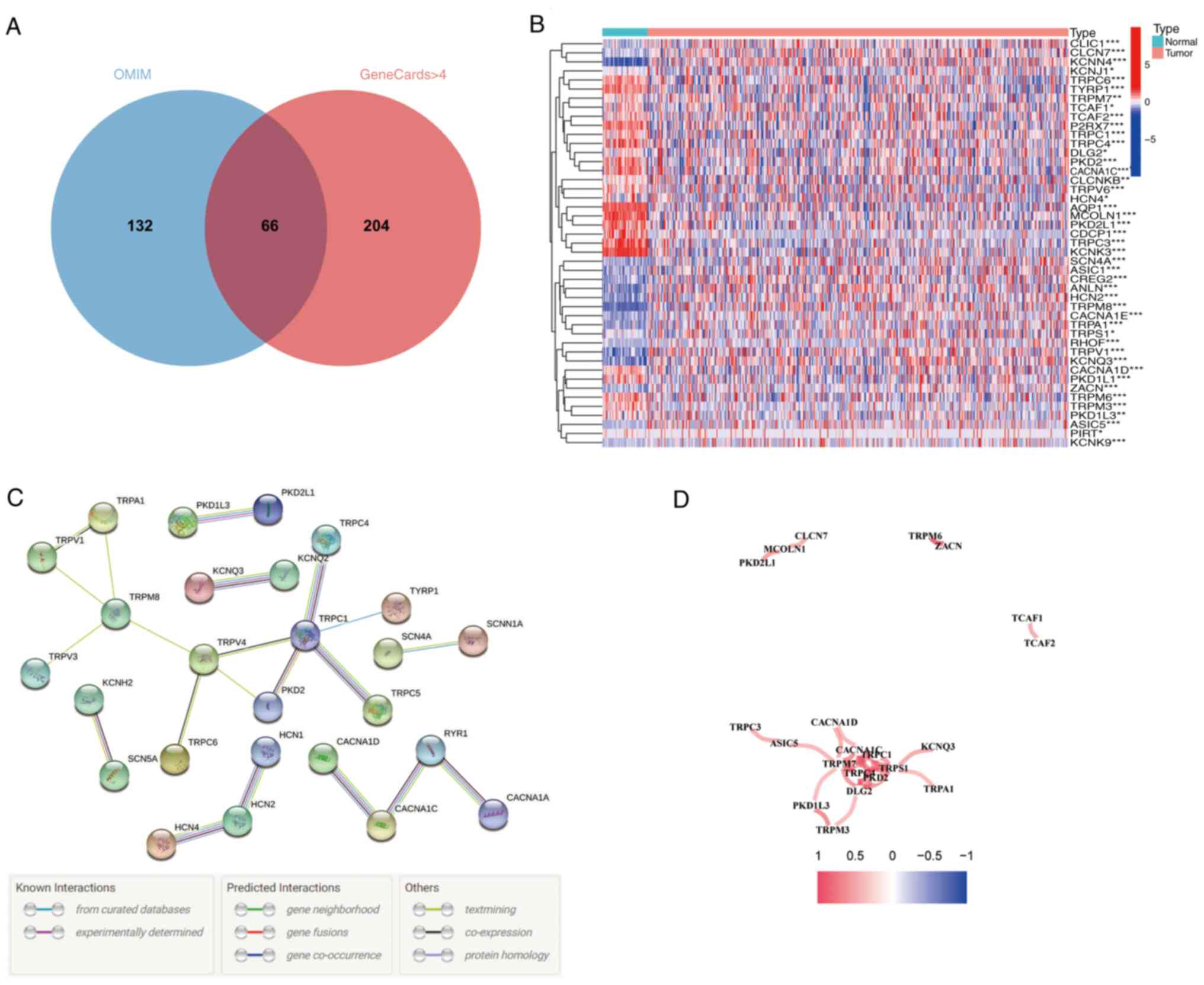
A Venn diagram revealed that 66 TRP channel-related
genes were identified as the intersection between the GeneCards and
OMIM databases (Fig. 1A).
Expression levels of these 66 genes were compared between 59 normal
lung tissues and 535 LUAD tissues from TCGA database, leading to
the identification of 43 DEGs (Fig.
1B). To further explore the interactions among these TRP
channel-related genes, a protein-protein interaction (PPI) network
was constructed (Fig. 1C), and the
correlation network of these genes was visualized (Fig. 1D).
Subgroups of LUAD identified by TRP
channel-encoding genes
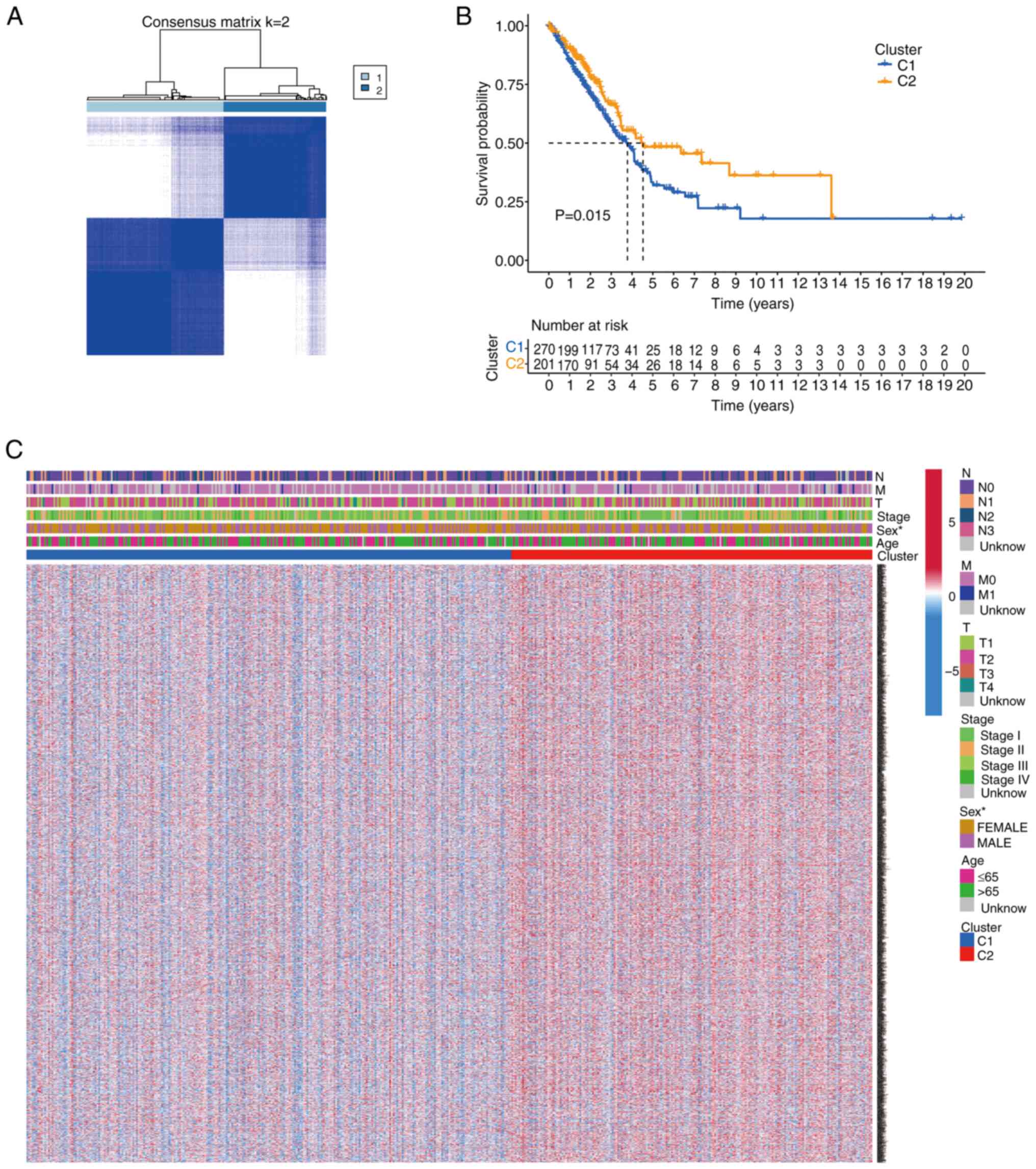
To investigate the relationship between the 43 TRP
channel-encoding DEGs and LUAD subtypes, consensus clustering
analysis was performed on 535 patients with LUAD from TCGA cohort.
The optimal number of clusters (k=2) was determined based on
clustering stability (Fig. 2A).
Patients in Cluster 2 exhibited significantly shorter overall
survival (OS) compared with those in Cluster 1 (P=0.015; Fig. 2B). A heatmap displaying gene
expression profiles and clinical parameters (such as age, stage and
sex) revealed that sex was a distinguishing factor between the two
clusters (Fig. 2C).
Prognostic model construction and
validation
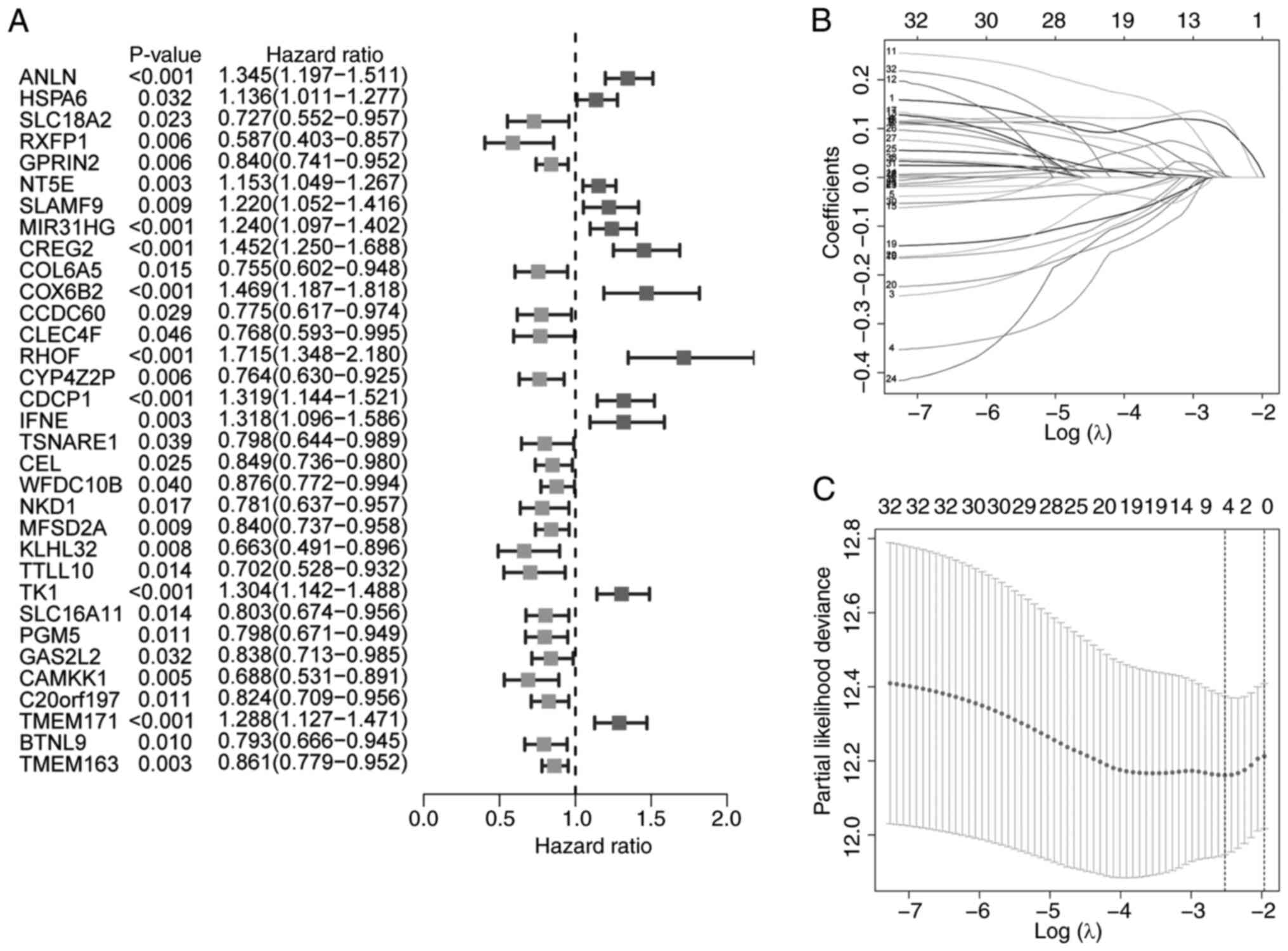
Univariate Cox regression analysis was used to
identify survival-associated genes (Fig. 3A). A four-gene TRP channel-related
signature was developed using LASSO Cox regression analysis, with
the optimal λ value selected (Fig. 3B
and C). The risk score was calculated as follows: Risk
score=(0.102 × ANLN expression) + (0.114 × CREG2 expression) +
(0.001 × RHOF expression) + (0.004 × CDCP1 expression).
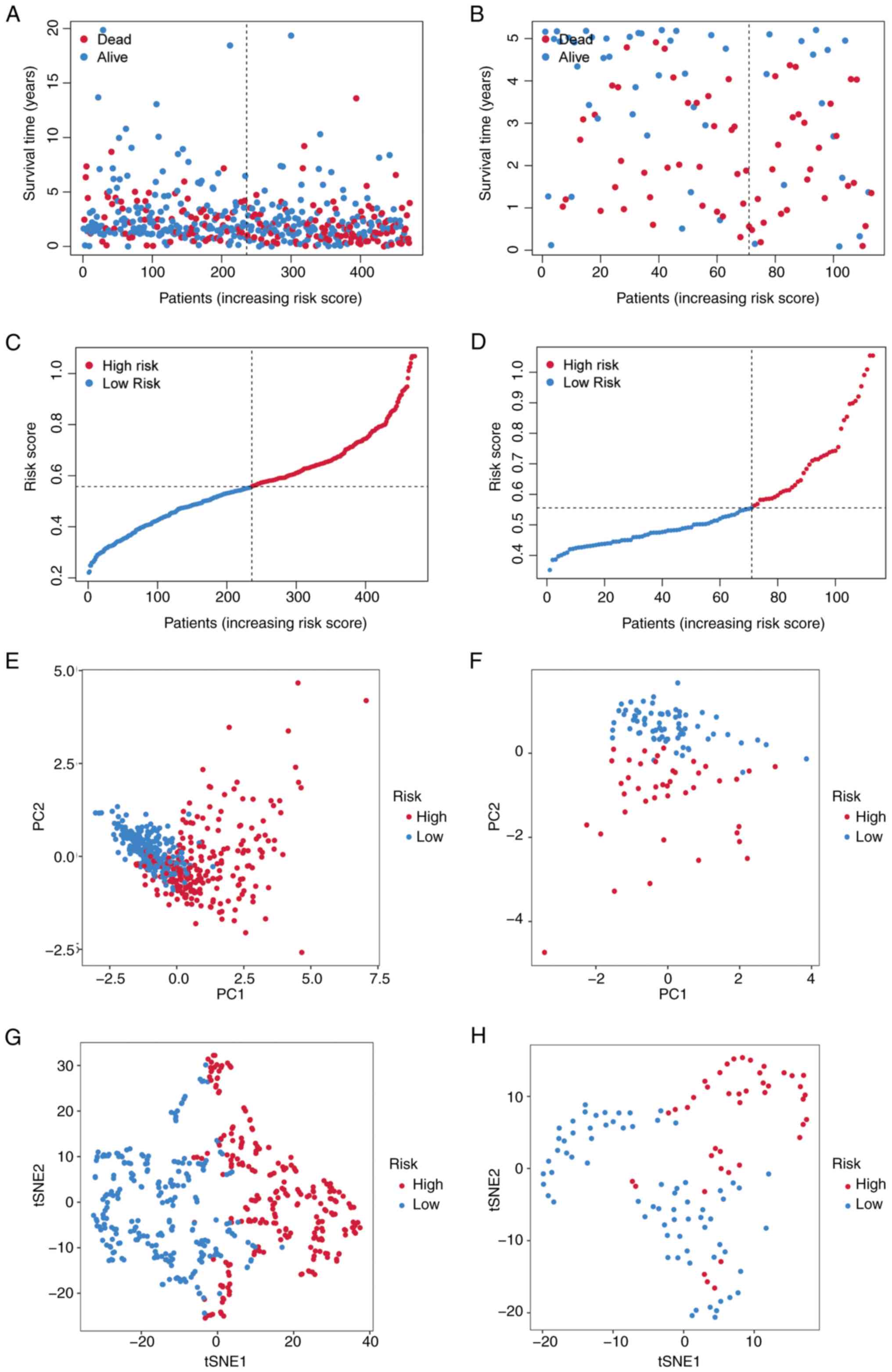
Patients were stratified into high- and low-risk
groups based on the median risk score. The risk stratification
model effectively segregated patients into high- and low-risk
prognostic groups, with higher risk scores predominantly assigning
patients to the high-risk category and lower scores to the low-risk
category (Fig. 4A and C). PCA and
t-SNE demonstrated distinct clustering patterns between the risk
groups (Fig. 4E and G).
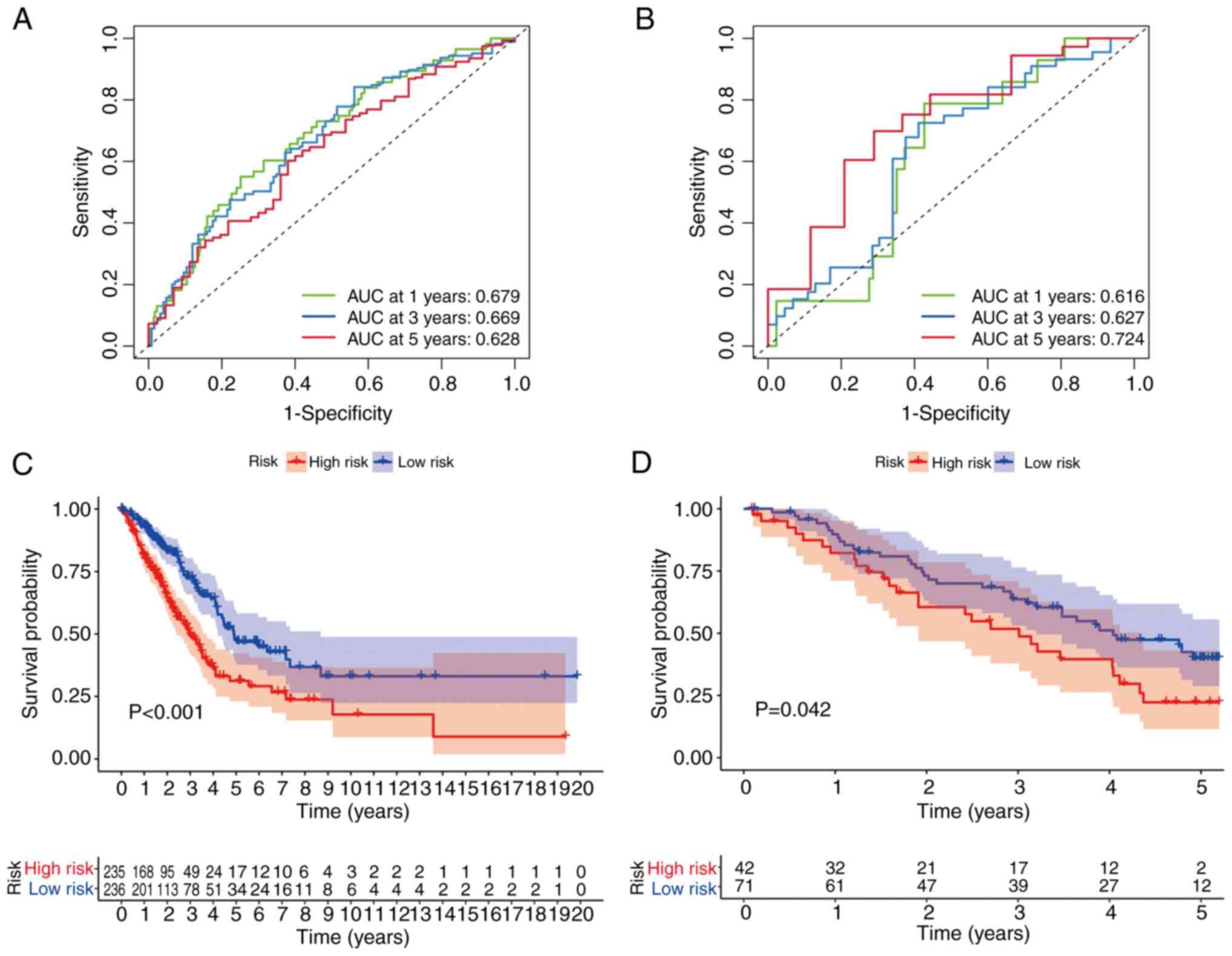
Time-dependent ROC analysis revealed that the 1-, 3- and 5-year
area under the curve (AUC) values for the risk score were 0.679,
0.669 and 0.628, respectively (Fig.
5A). High-risk patients exhibited significantly shorter
survival times compared with low-risk patients (P<0.001;
Fig. 5C).
Validation of the four-gene TRP
channel-related signature in the GEO cohort
The prognostic model was validated using an
independent LUAD cohort from the GEO database. Patients were
stratified into high- and low-risk groups based on the median risk
score derived from TCGA cohort. The risk stratification model
demonstrated discriminative capacity for patient survival outcomes
(Fig. 4B and D). PCA and t-SNE
analyses further supported the distinct clustering of risk groups
(Fig. 4F and H). Time-dependent ROC
analysis showed AUC values of 0.616, 0.627 and 0.724 for 1-, 3- and
5-year survival, respectively (Fig.
5B). High-risk patients had significantly shorter survival
times compared with low-risk patients (P<0.042; Fig. 5D).
Independent prognostic value of the
four-gene TRP channel-related signature
The risk score independently predicted poor survival
in both TCGA [hazard ratio (HR)=10.1; 95% confidence interval (CI),
4.6–22.3] and GEO cohorts (HR=7.5; 95% CI, 1.8–32.2; univariate
Cox), which remained significant after multivariate adjustment
(TCGA, HR=6.3; 95% CI, 2.7–15.1; GEO, HR=8.2; 95% CI, 1.7–39.4)
(Fig. 6A-D). A heatmap of
clinicopathological characteristics and gene expression profiles
further illustrated the differences between high- and low-risk
subgroups (Fig. 6E). High-risk
patients in the IMvigor210 cohort exhibited significantly worse
overall survival compared with the low-risk group (HR=1.8; 95% CI,
1.2–2.7; P=0.038; Fig. 6F),
demonstrating the prognostic value of the TRP channel-related
signature in an immunotherapy context. Patients achieving objective
responses (CR/PR) had significantly lower risk scores compared with
non-responders (SD/PD; P=0.021; Fig.
6G), suggesting that the signature predicts not only survival
but also therapeutic efficacy. ROC analysis demonstrated moderate
predictive accuracy for immunotherapy response (AUC=0.592; 95% CI,
0.516–0.667; Fig. 6H), highlighting
the potential of the risk score as a standalone biomarker despite
multifactorial resistance mechanisms.
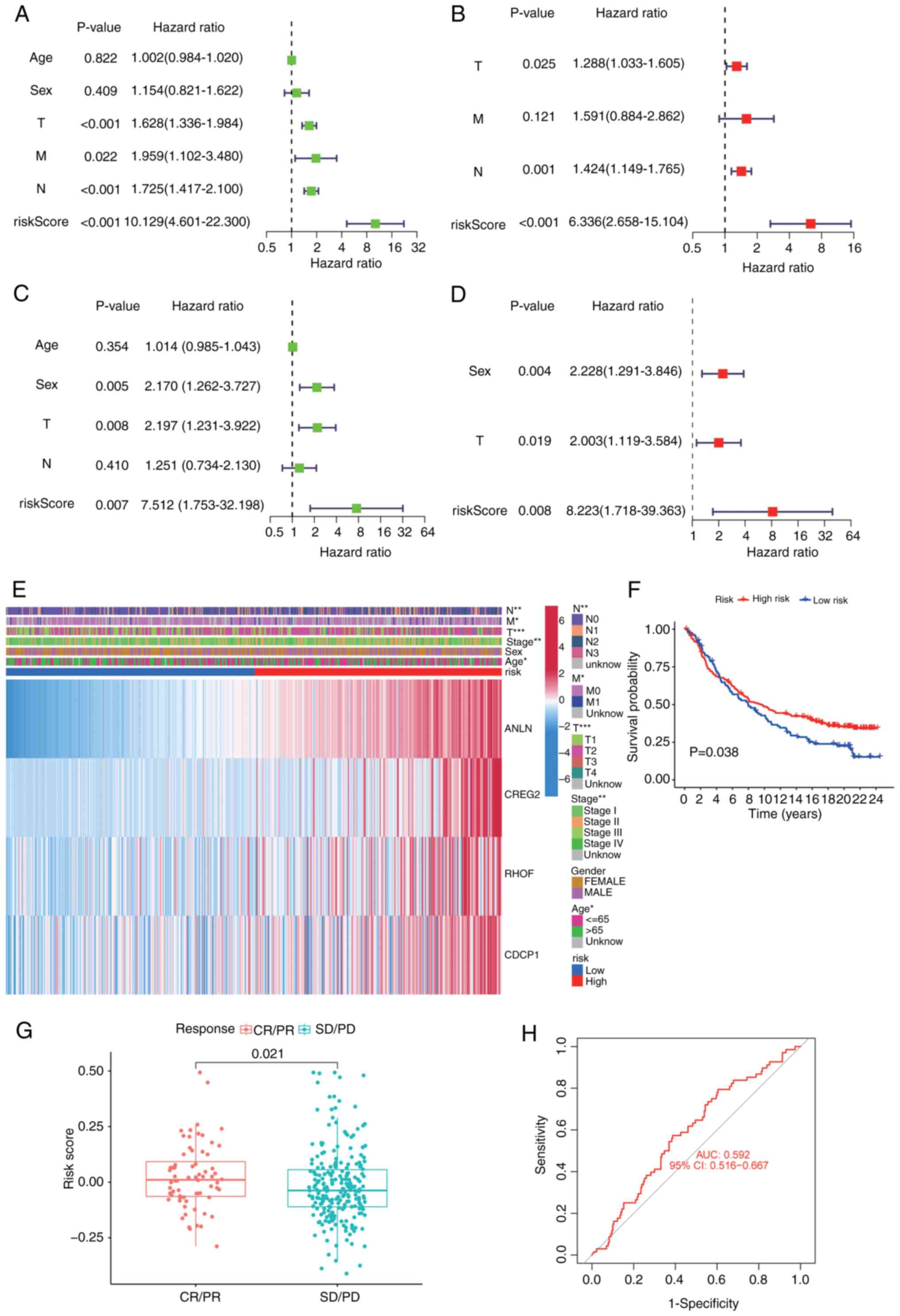 | Figure 6.Univariate and multivariate Cox
regression analyses for the risk score. (A) TCGA univariate
independent prognostic analysis results. (B) TCGA multifactor
independent prognostic analysis results. (C) GEO univariate
independent prognostic analysis results. (D) GEO multifactor
independent prognostic analysis results. (E) Heatmap depicting the
clinicopathological characteristics and gene expression variations
between the high-risk and low-risk groups. (F) Kaplan-Meier
survival curves for high- and low-risk groups in the IMvigor210
cohort (P=0.038). (G) Risk score distribution in IMvigor210
patients stratified by immunotherapy response (CR/PR vs. SD/PD).
(H) Receiver operating characteristic curve evaluating the risk
score's ability to predict immunotherapy response (AUC=0.592).
*P<0.05; **P<0.01; ***P<0.001. TCGA, The Cancer Genome
Atlas; GEO, gene expression omnibus; CR, complete response; PR,
partial response; SD, stable disease; PD, progressive disease; CI,
confidence interval; ANLN, anillin; CREG2, cellular repressor of
E1A stimulated genes 2; RHOF, Ras homolog family member F,
filopodia associated; CDCP1, CUB domain containing protein 1; T,
tumor stage; N, lymph node stage; M, metastasis stage. |
To facilitate clinical translation, a nomogram was
constructed incorporating the risk score, age, TNM stage and sex
(Fig. S1A). Each variable was
assigned a weighted point contribution, with the risk score showing
the strongest prognostic impact. Calibration plots demonstrated
notable concordance between predicted and observed survival rates
at 1, 3 and 5 years (Fig.
S1B).
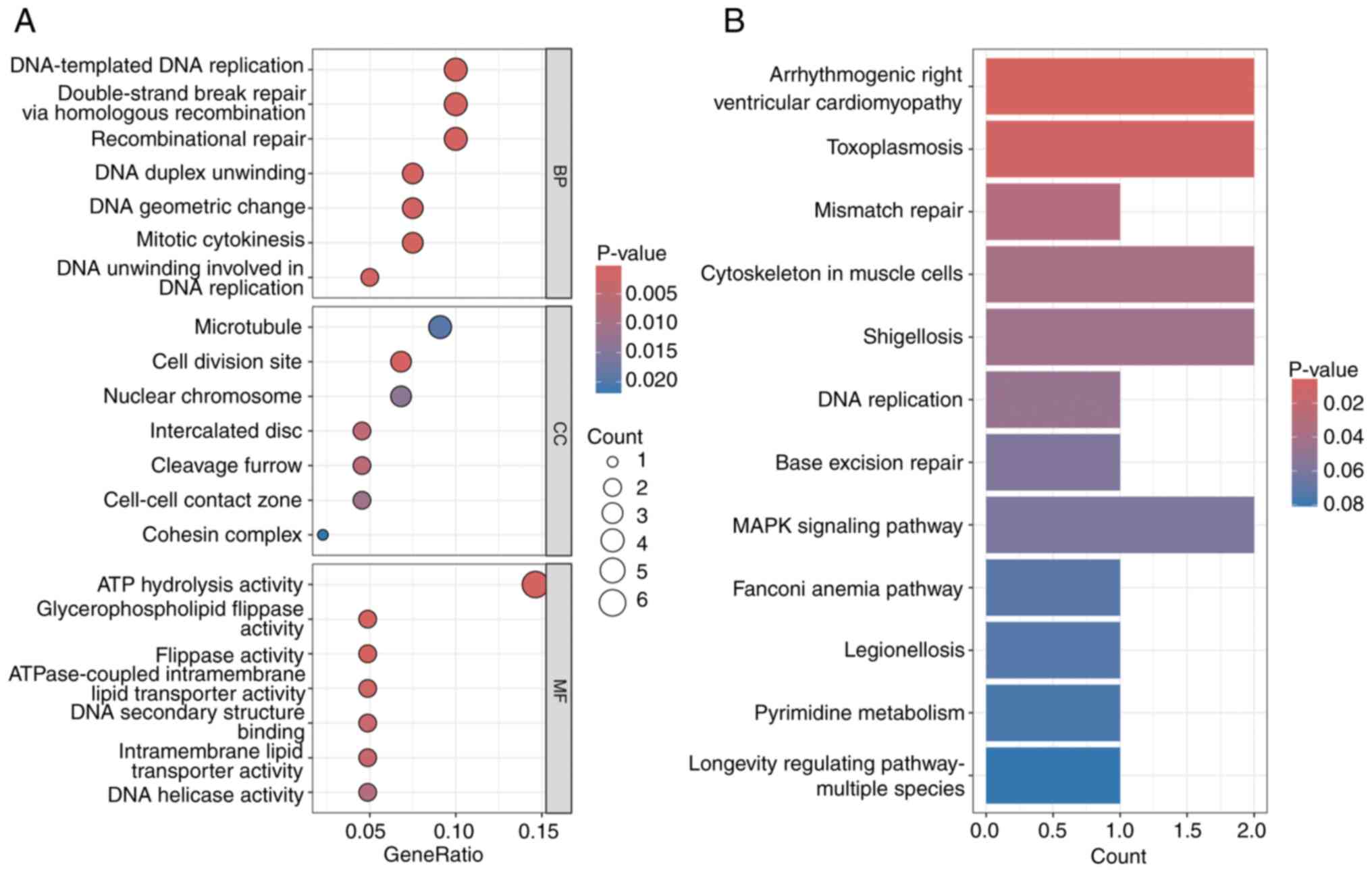
Functional analysis of DEGs between
high- and low-risk groups
Functional enrichment analysis of DEGs (log2FC
>1.5; false discovery rate <0.05) revealed significant
associations with cellular processes, environmental information
processing, genetic information processing and human diseases in
KEGG analysis (Fig. 7B). GO
analysis highlighted pathways associated with mitotic cytokinesis
and protein signal transduction (Fig.
7A).
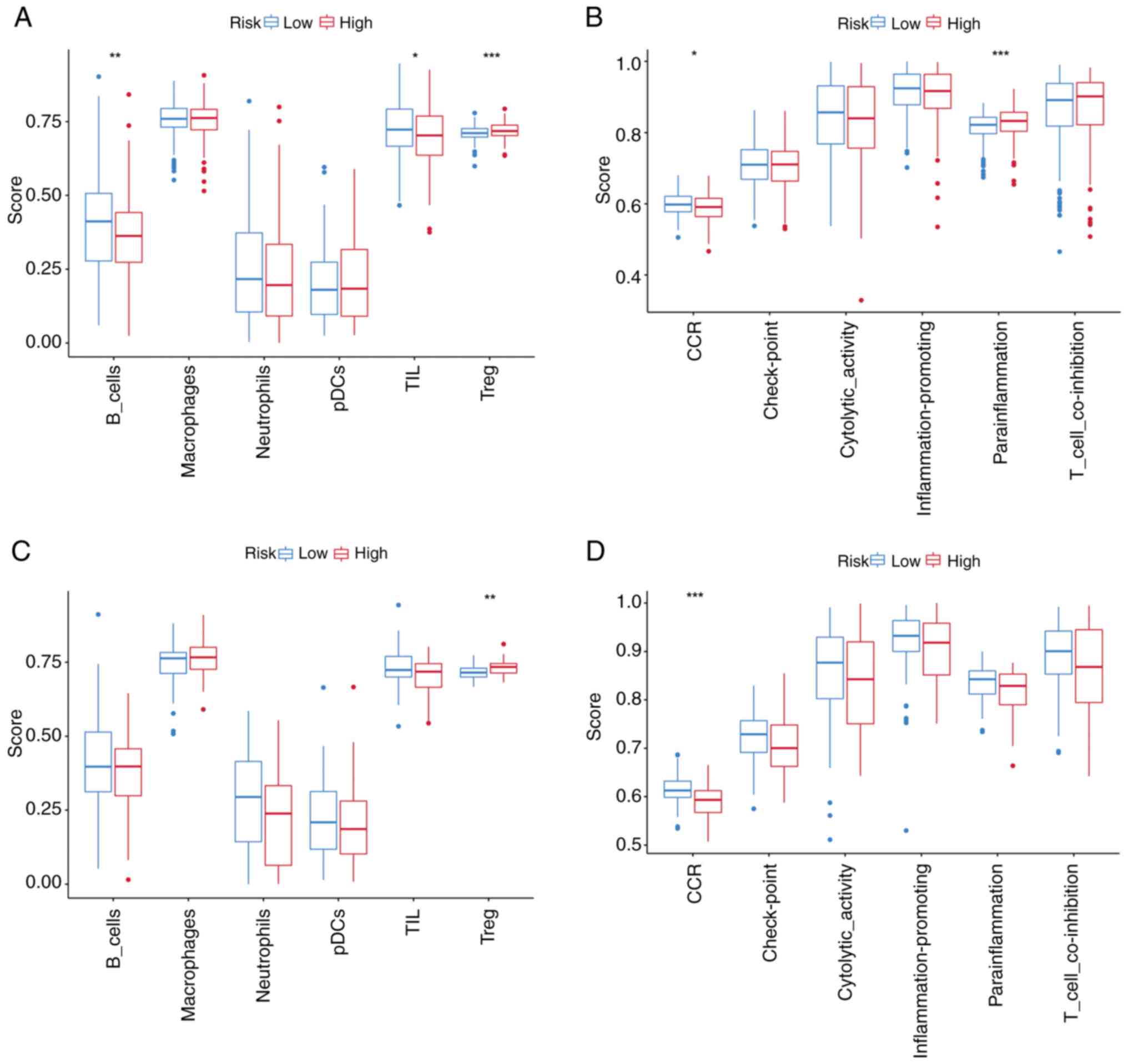
Immune microenvironment and
therapeutic implications
ssGSEA revealed elevated immune cell infiltration,
including higher levels of B cells and tumor-infiltrating
lymphocytes (TILs), in the low-risk subgroup compared with the
high-risk subgroup within TCGA cohort (Fig. 8A). Functional immune profiling
further demonstrated enhanced activity of chemokine-mediated
signaling (‘CCR’) and ‘parainflammation’ pathways in the low-risk
group (Fig. 8B), with consistent
trends observed in the GEO cohort (Fig.
8C and D). To investigate the role of TRP channel-related genes
in immune regulation, their associations with immune cell
infiltration were analyzed using the TIMER database. Notably, the
TRP channel-related signature exhibited a significant negative
correlation with B cell infiltration (P<0.05), suggesting
potential suppression of B cell recruitment via specific regulatory
pathways. Collectively, these findings underscore the dual role of
TRP channel-related signatures in shaping both pro-inflammatory and
immunosuppressive microenvironments (Fig. S2).
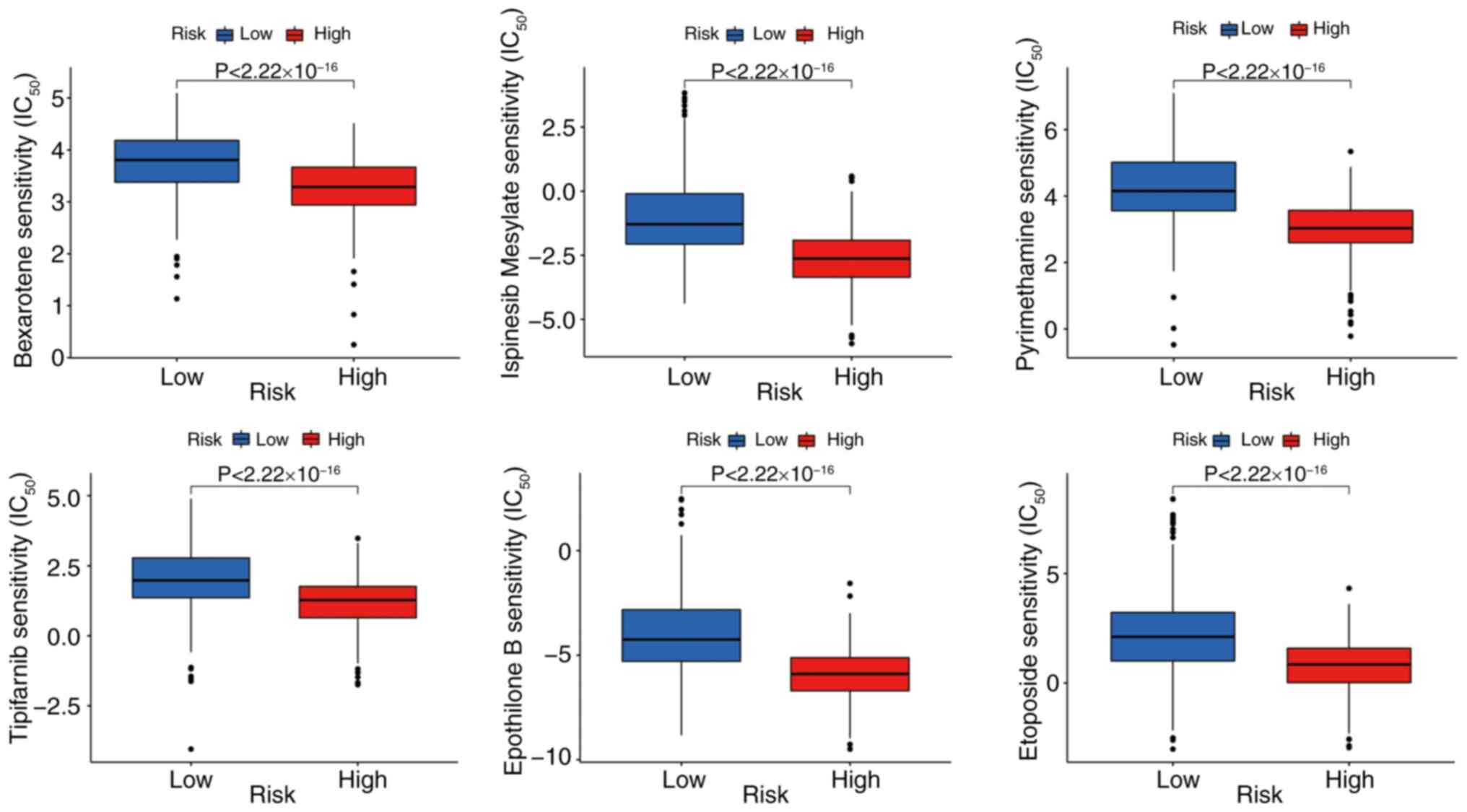
Response to treatment in high- and
low-risk groups
The Prophet algorithm was used to predict
chemotherapy response based on the IC50. A total of
eight small-molecule compounds, including bexarotene, ispinesib,
pyrimethamine, tipifarnib, etoposide, paclitaxel, ruxolitinib and
vinorelbine, showed significant differences in sensitivity between
high- and low-risk groups (Fig.
9).
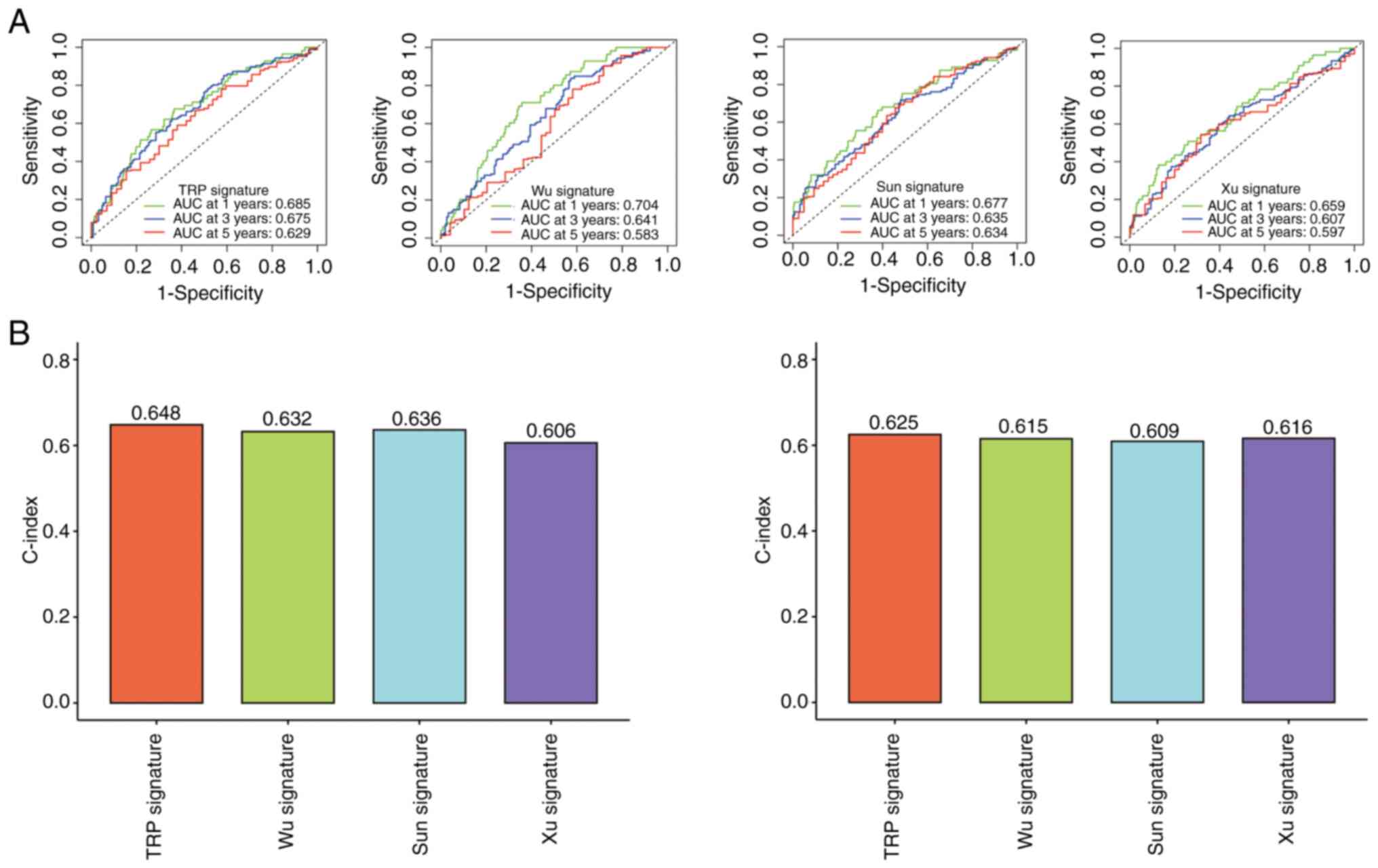
The ROC curve and concordance (C)-index were used to
compare the models. The prognostic model was compared with three
previously published LUAD signatures (28–30).
The AUC and C-index values of the present model were superior to
those of the other models (Fig. 10A
and B).
Validation of the four-gene TRP
channel-related signature expression in LUAD cells
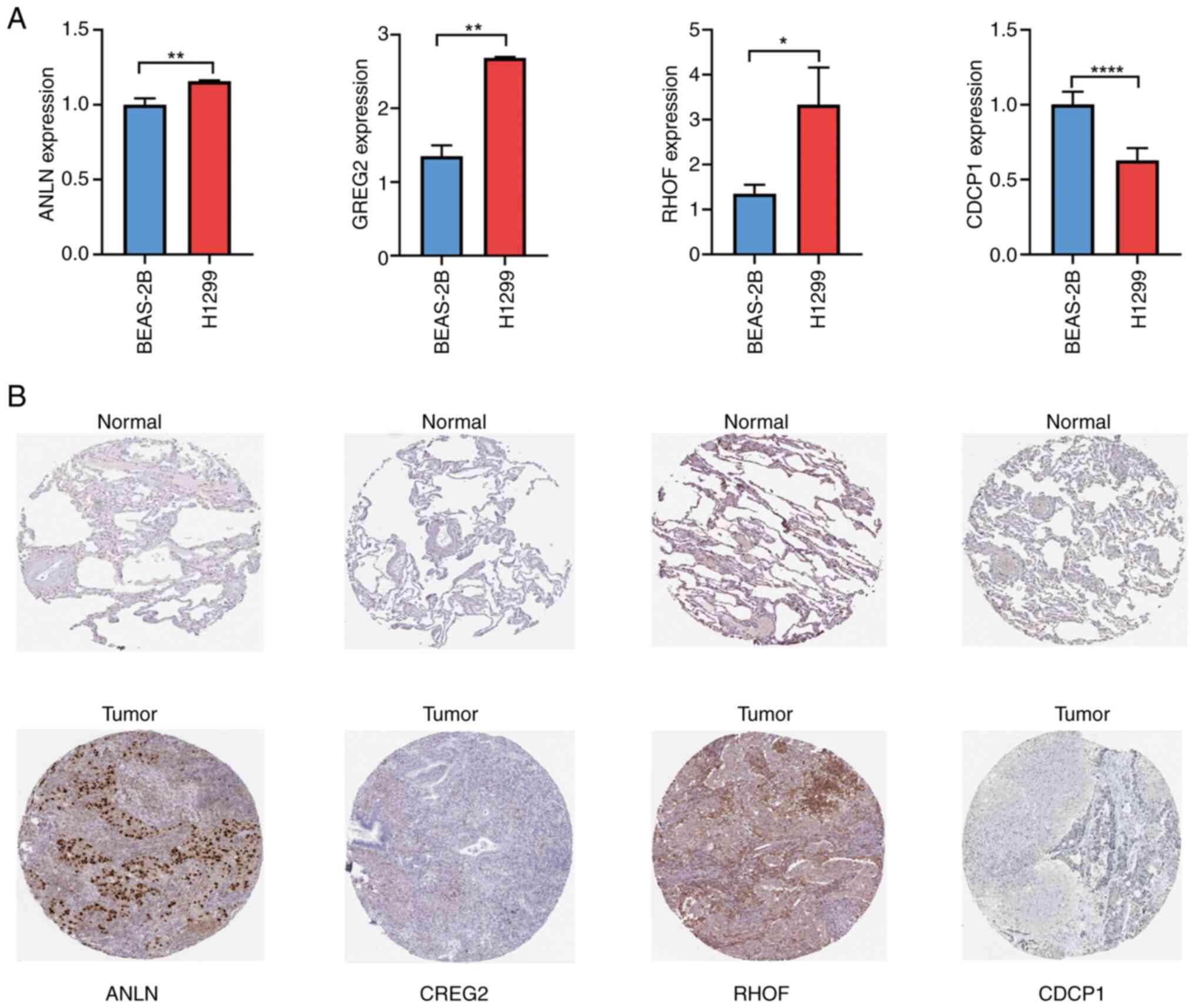
RT-qPCR analysis demonstrated that the mRNA
expression levels of ANLN, CREG2 and RHOF were significantly higher
in the LUAD cell line (H1299) compared with the normal lung cell
line (BEAS-2B), while CDCP1 expression was lower (Fig. 11A). Immunohistochemical analysis
further validated these findings at the protein level (Fig. 11B), consistent with the mRNA
expression results. To further validate the expression trends,
RNA-seq data from the Cancer Cell Line Encyclopedia (CCLE) were
analyzed. Among 30 cancer cell lines, ANLN, CREG2 and RHOF genes
exhibited significantly higher expression levels in LUAD cells,
while the CDCP1 gene displayed a low expression pattern (Fig. S3). This pan-cell line consistency
reinforces the reliability of the signature.
Discussion
The present study aimed to investigate the
expression levels of TRP channel-related genes in LUAD, their
prognostic significance and their role in the tumor
microenvironment. Compared with normal tissues, LUAD tissues
exhibited significantly elevated expression of ANLN, CREG2 and
RHOF, while CDCP1 expression was markedly reduced. Using consensus
clustering based on the expression profiles of 43 TRP
channel-associated DEGs, two distinct LUAD subgroups were
identified: Cluster 1 and Cluster 2. Patients in Cluster 2 were
associated with early tumor stage and grade, while Cluster 1
patients exhibited a lower probability of survival.
A prognostic risk score model was developed for the
TCGA-LUAD cohort, incorporating four TRP channel-related genes
(ANLN, CREG2, RHOF and CDCP1). This model effectively stratified
patients with LUAD into low- and high-risk groups, with low-risk
patients demonstrating markedly improved survival outcomes. The
prognostic utility of this signature was further validated in an
independent GEO-LUAD cohort. Notably, the risk score increased
progressively with tumor progression, and both univariate and
multivariate Cox regression analyses confirmed the four-gene
signature as an independent prognostic factor.
Among the four genes, ANLN, a scaffold protein
critical for cytokinesis, is overexpressed in LUAD and promotes
tumor proliferation through PI3K/Akt activation (31,32).
RHOF, a Rho GTPase regulating cytoskeletal dynamics and
epithelial-mesenchymal transition (EMT), was also identified as a
key player (33). Both genes were
upregulated in high-risk patients. Consistent with these findings,
pathway analysis revealed significant enrichment of PI3K-Akt
signaling and cell cycle pathways, thereby supporting their
mechanistic roles in mitotic dysregulation and tumor
progression.
CREG2, a secreted glycoprotein involved in
pluripotent stem cell differentiation (34), has not been previously studied in
LUAD. CREG2 regulates angiogenesis and immune responses by
modulating VEGF and TGF-β pathways (35). High CREG2 expression was protective
in the present model. KEGG analysis highlighted suppressed
angiogenesis pathways (such as VEGF signaling) in high-risk
patients, consistent with the role of CREG2 in maintaining vascular
homeostasis. This implies that CREG2 downregulation may impair
antitumor immunity and promote hypoxia-driven progression. CDCP1 is
a transmembrane protein that promotes metastasis in multiple
cancers by activating Wnt/β-catenin signaling and EMT (36). In the model, low CDCP1 expression
was associated with poor prognosis, aligning with its reported role
in suppressing LUAD metastasis (37,38).
Pathway analysis revealed significant downregulation of Wnt
signaling in high-risk patients, suggesting that CDCP1 loss may
drive aggressive phenotypes via Wnt pathway inactivation. These
findings underscore the importance of TRP channel-related genes in
cancer biology.
TRP channels are critical regulators of
intracellular calcium homeostasis, which is essential for immune
cell activation and cytokine production (39). The ssGSEA results revealed
significantly higher infiltration of B cells and TILs in the
low-risk group. This aligns with previous studies showing that TRP
channel-mediated calcium influx promotes T-cell receptor signaling
and B-cell differentiation. For instance, TRPV1 activation enhances
cytotoxic T-cell activity by upregulating IFN-γ secretion (40), while TRPM2 inhibition suppresses
regulatory T-cell function, thereby reducing immune suppression
(41). The upregulation of CDCP1 (a
negative regulator of TRPC6) in high-risk patients may impair
calcium-dependent T-cell activation, contributing to the observed
immunosuppressive microenvironment. The IMvigor210 analysis extends
the findings beyond chemotherapy, implicating TRP channel
dysregulation in immunotherapy resistance. The association between
low-risk scores and improved PD-L1 response may stem from enhanced
immune infiltration, a hypothesis supported by elevated
CD8+ T-cell levels and IFN-γ signaling in low-risk
patients.
The low-risk group exhibited stronger activity in
CCR pathways. TRP channels, particularly TRPC and TRPV subfamilies,
regulate chemokine receptor expression and immune cell chemotaxis
(42). For example, TRPV4
activation in endothelial cells facilitates leukocyte
transmigration by upregulating C-X-C motif chemokine (CXC) ligand
12/CXC receptor type 4 signaling (43). The downregulation of RHOF (a Rho
GTPase associated with TRPM7) in low-risk patients may enhance
dendritic cell migration via cytoskeletal remodeling, promoting
antigen presentation and TIL recruitment (44).
Parainflammation, a state of chronic, low-grade
inflammation, was more pronounced in the low-risk group compared
with the high-risk group. TRP channels such as TRPA1 and TRPV1 are
known to mediate neurogenic inflammation by releasing
pro-inflammatory neuropeptides (45). In LUAD, sustained TRP channel
activation may drive parainflammation, paradoxically favoring
immune surveillance by recruiting natural killer cells and M1
macrophages (46). Conversely,
high-risk patients showed reduced parainflammation, potentially due
to ANLN overexpression, which has been associated with
immunosuppressive cytokine secretion and macrophage polarization
toward the M2 phenotype.
Drug susceptibility analysis suggested that
high-risk patients may exhibit favorable responses to specific
chemotherapy agents, such as bexarotene, ispinesib and paclitaxel.
These findings highlight the potential of TRP channel-related genes
as predictive biomarkers for therapeutic efficacy in patients with
LUAD.
Previous studies have advanced the understanding of
TRP-related biology in LUAD, including a 12-gene immune-focused
model (47), a five-gene tryptophan
metabolism signature (48) and an
eight-gene cytokine-centric model (49). While these studies established the
prognostic value of TRP-associated pathways, the present study
provides critical distinctions. Firstly, a minimalist four-gene
signature focused on TRP channel-encoding genes was developed,
avoiding overfitting while achieving superior accuracy (AUC,
0.679–0.724). Secondly, the signature introduces novel biomarkers
(CREG2, CDCP1) not previously associated with LUAD, expanding the
mechanistic scope of TRP biology to include angiogenesis and
metastasis suppression. Thirdly, the model's utility in predicting
anti-PD-L1 response was validated using the IMvigor210 cohort,
bridging TRP dysregulation to immunotherapy resistance, a finding
absent in prior studies. Finally, the integration of multi-omics
validation (CCLE and cell lines) and clinical translation tools
(nomogram) addresses limitations of earlier bioinformatics-centric
approaches, offering a robust framework for personalized
prognosis.
Despite these insights, the present study has
several limitations. First, the sample size and training cohort for
the prognostic model were relatively modest, which may affect the
generalizability of the findings. Second, additional clinical data
are needed to further validate the nomogram and enhance its
reliability. Third, while RT-qPCR validation in H1299 cells and
cross-dataset consistency (TCGA, Human Protein Atlas and single
cell RNA-seq) support the biological plausibility of the signature,
the lack of functional evidence in genetically heterogeneous LUAD
models warrants investigation. Future work will prioritize
mechanistic studies using patient-derived organoids and isogenic
cell line panels to address tumor heterogeneity. Fourth, while the
prognostic model was validated in both TCGA and GEO cohorts,
potential batch effects arising from differences in sequencing
platforms, sample collection protocols and normalization methods
between these datasets may influence the generalizability of the
signature. Fifth, Drug sensitivity (IC50) models rely on
cell line data (GDSC), which may not fully reflect in vivo
tumor complexity; therefore, there is need for clinical validation.
Sixth, the lack of validation using tissues from a real-life
patient cohort limits the clinical applicability of the present
findings. While computational validation through public datasets
(TCGA and GEO) provides preliminary support, direct experimental
confirmation in prospectively collected clinical specimens is
essential to strengthen translational relevance. While the
IMvigor210 cohort validates the prognostic utility of the signature
in an immunotherapy context, future studies should prioritize
LUAD-specific immunotherapy cohorts to confirm translational
applicability.
In conclusion, the present study addresses critical
gaps in TRP-related LUAD research by delivering a concise,
experimentally validated prognostic tool with novel insights into
immune modulation and therapeutic response. By focusing on TRP
channel-encoding genes, uncovering B-cell interactions and
validating immunotherapy utility, the field can be expanded beyond
prior metabolic or immune-centric models. These contributions
underscore the necessity of the present work and its potential to
refine clinical decision-making in LUAD management.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by ‘14th Five-Year Plan’ Key
Discipline-Nantong Medical Innovation Team (Oncology; grant no.
102).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
DY and SH conceived the project. YC analyzed the
data. XF contributed towards the interpretation of the data. All
authors wrote, read and approved the final version of the
manuscript. DY and SH confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
LUAD
|
lung adenocarcinoma
|
|
PFS
|
progression-free survival
|
|
OS
|
overall survival
|
|
CI
|
confidence intervals
|
|
RFS
|
relapse free survival
|
|
HR
|
hazard ratio
|
|
TCGA
|
The Cancer Genome Atlas
|
References
|
1
|
Siegel RL, Kratzer TB, Giaquinto AN, Sung
H and Jemal A: Cancer statistics, 2025. CA Cancer J Clin. 75:10–45.
2025.PubMed/NCBI
|
|
2
|
Yang C, Liu Y, Huang Z, Liu S, Zhang X,
Liu Q and Dai J: Recent advances and challenges of cellular
immunotherapies in lung cancer treatment. Exp Hematol Oncol.
14:942025. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
No authors listed. Lung Cancer. Am Fam
Physician. 105:Online2022.
|
|
4
|
Zhang Y, Luo G, Etxeberria J and Hao Y:
Global patterns and trends in lung cancer incidence: A
population-based study. J Thorac Oncol. 16:933–944. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000–14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rodrigues T, Sieglitz F and Bernardes GJ:
Natural product modulators of transient receptor potential (TRP)
channels as potential anti-cancer agents. Chem Soc Rev.
45:6130–6137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Samanta A, Hughes TET and Moiseenkova-Bell
VY: Transient receptor potential (TRP) channels. Subcell Biochem.
87:141–165. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pan F, Wang K, Zheng M, Ren Y, Hao W and
Yan J: A TRP family based signature for prognosis prediction in
head and neck squamous cell carcinoma. J Oncol. 2022:87576562022.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ouadid-Ahidouch H, Dhennin-Duthille I,
Gautier M, Sevestre H and Ahidouch A: TRP channels: Diagnostic
markers and therapeutic targets for breast cancer? Trends Mol Med.
19:117–124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Büch TRH, Büch EAM, Boekhoff I, Steinritz
D and Aigner A: Role of chemosensory TRP channels in lung cancer.
Pharmaceuticals (Basel). 11:902018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Y, Wang J and Liu X: TRPV4 induces
apoptosis via p38 MAPK in human lung cancer cells. Braz J Med Biol
Res. 54:e108672021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mounir M, Lucchetta M, Silva TC, Olsen C,
Bontempi G, Chen X, Noushmehr H, Colaprico A and Papaleo E: New
functionalities in the TCGAbiolinks package for the study and
integration of cancer data from GDC and GTEx. PLoS Comput Biol.
15:e10067012019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen H, Su X, Li Y, Dang C and Luo Z:
Identification of metabolic reprogramming-related genes as
potential diagnostic biomarkers for diabetic nephropathy based on
bioinformatics. Diabetol Metab Syndr. 16:2872024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tu X, Huang H, Xu S, Li C and Luo S:
Single-cell transcriptomics reveals immune infiltrate in sepsis.
Front Pharmacol. 14:11331452023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wilkerson MD and Hayes DN:
ConsensusClusterPlus: A class discovery tool with confidence
assessments and item tracking. Bioinformatics. 26:1572–1573. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tong M, Tu Q, Wang L, Chen H, Wan X and Xu
Z: Joint analysis of single-cell RNA sequencing and bulk
transcriptome reveals the heterogeneity of the urea cycle of
astrocytes in glioblastoma. Neurobiol Dis. 208:1068352025.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
O'Connell TM: Pathway volcano: An
interactive tool for pathway guided visualization of differential
expression data. Bioinformatics. 41:btaf3672025. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Melit Devassy B, George S and Nussbaum P:
Unsupervised clustering of hyperspectral paper data using t-SNE. J
Imaging. 6:292020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang S, Huang G, Li X, Zhang Z, Peng K,
Zhu L, Zhang C and Niu TT: Global, regional and national
retinoblastoma burden in children under 10 years of age from 1990
to 2021: Trend analysis based on the global burden of disease study
2021. PLoS One. 20:e03278322025. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li H, Li G, Gao X, Chen C, Cui Z, Cao X
and Su J: Development of a reliable risk prognostic model for lung
adenocarcinoma based on the genes related to endotheliocyte
senescence. Sci Rep. 15:126042025. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Chen W, Zhou J, Chen J, Cao G,
Huang C, Lu X, Chen X, Luo R, Huang H, et al: Association between
early antibiotic treatment after admission and mortality of
acute-on-chronic liver failure patients with bacterial infection: A
multicenter retrospective study. Virulence. 16:25097572025.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li-Fei M, Ren SM, Wang J, Zhao WJ, Chen J
and Hu WT: Risk scoring model for lung adenocarcinoma based on
PD-L1 related signature reveals prognostic predictability and
correlation with tumor immune microenvironment genes was
constructed. Front Immunol. 16:16019822025. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
SCOT-HEART Investigators, . Newby DE,
Adamson PD, Berry C, Boon NA, Dweck MR, Flather M, Forbes J, Hunter
A, Lewis S, et al: Coronary CT angiography and 5-year risk of
myocardial infarction. N Engl J Med. 379:924–933. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang N and Liu D: Identification and
validation a necroptosis-related prognostic signature and
associated regulatory axis in stomach adenocarcinoma. Onco Targets
Ther. 14:5373–5383. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun S, Wang Y, Li M and Wu J:
Identification of TRP-related subtypes, development of a prognostic
model, and characterization of tumor microenvironment infiltration
in lung adenocarcinoma. Front Mol Biosci. 9:8613802022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu J, Li L, Zhang H, Zhao Y, Zhang H, Wu S
and Xu B: A risk model developed based on tumor microenvironment
predicts overall survival and associates with tumor immunity of
patients with lung adenocarcinoma. Oncogene. 40:4413–4424. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu Q and Chen Y: An aging-related gene
signature-based model for risk stratification and prognosis
prediction in lung adenocarcinoma. Front Cell Dev Biol.
9:6853792021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Suzuki C, Daigo Y, Ishikawa N, Kato T,
Hayama S, Ito T, Tsuchiya E and Nakamura Y: ANLN plays a critical
role in human lung carcinogenesis through the activation of RHOA
and by involvement in the phosphoinositide 3-kinase/AKT pathway.
Cancer Res. 65:11314–11325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu J, Zheng H, Yuan S, Zhou B, Zhao W, Pan
Y and Qi D: Overexpression of ANLN in lung adenocarcinoma is
associated with metastasis. Thorac Cancer. 10:1702–1709. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li S, Liu Y, Bai Y, Chen M, Cheng D, Wu M
and Xia J: Ras homolog family member F, filopodia associated
promotes hepatocellular carcinoma metastasis by altering the
metabolic status of cancer cells through RAB3D. Hepatology.
73:2361–2379. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kunita R, Otomo A and Ikeda JE:
Identification and characterization of novel members of the CREG
family, putative secreted glycoproteins expressed specifically in
brain. Genomics. 80:456–460. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Weivoda MM, Chew CK, Monroe DG, Farr JN,
Atkinson EJ, Geske JR, Eckhardt B, Thicke B, Ruan M, Tweed AJ, et
al: Identification of osteoclast-osteoblast coupling factors in
humans reveals links between bone and energy metabolism. Nat
Commun. 11:872020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He Y, Davies CM, Harrington BS, Hellmers
L, Sheng Y, Broomfield A, McGann T, Bastick K, Zhong L, Wu A, et
al: CDCP1 enhances Wnt signaling in colorectal cancer promoting
nuclear localization of β-catenin and E-cadherin. Oncogene.
39:219–233. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Khan T, Kryza T, Lyons NJ, He Y and Hooper
JD: The CDCP1 signaling hub: A target for cancer detection and
therapeutic intervention. Cancer Res. 81:2259–2269. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Alajati A, D'Ambrosio M, Troiani M, Mosole
S, Pellegrini L, Chen J, Revandkar A, Bolis M, Theurillat JP,
Guccini I, et al: CDCP1 overexpression drives prostate cancer
progression and can be targeted in vivo. J Clin Invest.
130:2435–2450. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hwang SM, Lee JY, Park CK and Kim YH: The
role of TRP channels and PMCA in brain disorders: Intracellular
calcium and pH homeostasis. Front Cell Dev Biol. 9:5843882021.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Balood M, Ahmadi M, Eichwald T, Ahmadi A,
Majdoubi A, Roversi K, Roversi K, Lucido CT, Restaino AC, Huang S,
et al: Nociceptor neurons affect cancer immunosurveillance. Nature.
611:405–412. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lory NC, Nawrocki M, Corazza M, Schmid J,
Schumacher V, Bedke T, Menzel S, Koch-Nolte F, Guse AH, Huber S and
Mittrücker HW: TRPM2 is not required for T-Cell activation and
differentiation. Front Immunol. 12:7789162022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kozai D, Ogawa N and Mori Y: Redox
regulation of transient receptor potential channels. Antioxid Redox
Signal. 21:971–986. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sullivan JM, Bagnell AM, Alevy J, Avila
EM, Mihaljević L, Saavedra-Rivera PC, Kong L, Huh JS, McCray BA,
Aisenberg WH, et al: Gain-of-function mutations of TRPV4 acting in
endothelial cells drive blood-CNS barrier breakdown and motor
neuron degeneration in mice. Sci Transl Med. 16:eadk13582024.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tian X, Nanding K, Dai X, Wang Q, Wang J
and Morigen Fan L: Pattern recognition receptor mediated innate
immune response requires a Rif-dependent pathway. J Autoimmun.
134:1029752023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liang Q, Wang JW, Bai YR, Li RL, Wu CJ and
Peng W: Targeting TRPV1 and TRPA1: A feasible strategy for natural
herbal medicines to combat postoperative ileus. Pharmacol Res.
196:1069232023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Datta A, Lee JH, Flandrin O, Horneman H,
Lee J, Metruccio MME, Bautista D, Evans DJ and Fleiszig SMJ: TRPA1
and TPRV1 ion channels are required for contact lens-induced
corneal parainflammation and can modulate levels of resident
corneal immune cells. Invest Ophthalmol Vis Sci. 64:212023.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
He M, Wu G, Wang Z, Ren K, Yang Z and Xue
Q: Development and validation of a TRP-related gene signature for
overall survival prediction in lung adenocarcinoma. Front Genet.
13:9056502022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Z, Zhang J, Zuo C, Chen H, Wang L,
Xie Y, Ma H, Min S, Wang X and Lian C: Identification and
validation of tryptophan-related gene signatures to predict
prognosis and immunotherapy response in lung adenocarcinoma reveals
a critical role for PTTG1. Front Immunol. 15:13864272024.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Guo Y and Liu N: Systematic analysis and
identification of molecular subtypes of TRP-related genes and
prognosis prediction in lung adenocarcinoma. J Oncol.
2022:53882832022. View Article : Google Scholar : PubMed/NCBI
|