Introduction
Lactylation, a novel post-translational modification
(PTM) characterized by the covalent attachment of lactate to lysine
residues, has emerged as a critical regulatory mechanism in cancer
biology (1). Initially identified
as a metabolic byproduct of anaerobic glycolysis, lactate is now
recognized as a dynamic signaling molecule that orchestrates
cellular adaptation through lactylation. This modification, first
reported on histones in 2019, bridges metabolic reprogramming and
epigenetic regulation, influencing key oncogenic processes such as
proliferation, metastasis, immune evasion and resistance to therapy
(2). Within the tumor
microenvironment (TME), lactate accumulation drives lactylation of
histones and non-histone proteins, modulating interactions among
cancer cells, cancer-associated fibroblasts (CAFs),
tumor-associated macrophages (TAMs) and immune cells (3). Notably, lactylation reshapes chromatin
structure, alters enzymatic activities in glycolysis and the
tricarboxylic acid cycle, and sustains immunosuppressive niches by
polarizing TAMs towards an M2 phenotype and impairing cytotoxic
T-cell function (4). Furthermore,
lactylation contributes to genomic instability, angiogenesis and
phenotypic plasticity through mechanisms involving DNA repair
modulation and metabolic-epigenetic crosstalk. Emerging therapeutic
strategies targeting lactylation-associated enzymes [such as
lactate dehydrogenase A (LDHA) and monocarboxylate transporter
(MCT1/4)] or disrupting lactate-driven feedback loops hold promise
for overcoming chemoresistance and enhancing the efficacy of
immunotherapy (5). The present
review summarizes the current insights into the role of lactylation
across the hallmarks of cancer, highlighting its potential as a
diagnostic biomarker and therapeutic target in precision
oncology.
Lactate and lactylation
Initially regarded as a metabolic byproduct of
anaerobic glycolysis, lactate is now recognized as more than its
historical perception as mere waste. Emerging evidence highlights
its critical roles in energy metabolism (6), signaling regulation (7,8) and
cellular homeostasis (9).
Lactylation, a PTM involving covalent attachment of lactate to
lysine residues, mirrors acetylation in mechanism and importance.
First identified on histones by Zhang et al (2) in 2019, lactylation has since been
detected on non-histone proteins, establishing a
metabolic-epigenetic link. This modification, driven by glycolytic
end-product lactate, is mechanistically associated with cancer
progression, inflammatory responses and stem cell regulation
(1). Previous studies underscore
the profound impact of lactylation on the hallmarks of cancer,
including proliferation, metastasis and drug resistance, through
modulation of metabolic enzyme activity (10).
Lactylation-related proteins
Histone lactylation and non-histone
lactylation
PTMs involve the covalent addition of chemical
groups to amino acid residues, altering protein function. Common
PTMs, including phosphorylation, acetylation and ubiquitination
regulate chromatin dynamics and gene expression (11). Histones, comprising core (H2A, H2B,
H3 and H4) and linker (H1 and H5) subtypes, undergo PTM, and these
PTMs are involved in epigenetic regulation (12). Lactylation modification modulates
chromatin structure, DNA-histone interactions and transcriptional
activity. Hypoxia-induced pyruvate accumulation drives lactylation,
particularly at histone 3 lysine 18 (H3K18), H4K5 and H4K18
residues, while H4K5 lactylation (H4K5la) is associated with a poor
prognosis in neuroblastoma (13,14).
In neuroblastoma studies, elevated lactylation levels of the 5th
lysine residue of histone H4 (H4K5la) were found to be notably
associated with a poor clinical prognosis in patients (15).
Non-histone lactylation occurs across organelles
(nucleus, mitochondria and lysosomes) and regulates several
processes, including glycolysis, inflammation and oxidative stress
(16–18). By altering protein conformation,
charge and stability, lactylation impacts enzyme activity and
molecular interactions. For example, in hepatocellular carcinoma
(HCC), adenylate kinase 2 lactylation at K28 suppresses
mitochondrial function, driving metastasis (19). In lung squamous cell carcinoma,
glucose transporter 1-driven lactate accumulation induces
lactylation of non-histone proteins [such as
N6-adenosine-methyltransferase 70 kDa subunit (METTL3)], thereby
enhancing the immunosuppressive tumor microenvironment, reducing
the response to immunotherapy and promoting tumor progression
(20). Similarly, in pancreatic
ductal adenocarcinoma (PDAC), lactylation of the ENSA protein
inhibits the phosphatase PP2A, activating the STAT3/CCL2 axis. This
axis recruits tumor-associated TAMs and impairs cytotoxic T cell
activity (21).
Non-histone protein lysine lactylation (Kla), an
emerging PTM, has gained notable attention in oncology research.
However, elucidating its mechanisms and achieving clinical
translation present substantial challenges. Although enzymes such
as p300, alanyl-tRNA synthetase (AARS1) and lysine
acetyltransferase 8 (KAT8) have been reported as potential
lactyltransferases, most possess acyltransferase activity for
acetylation and crotonylation (Kcr). For example, KAT8 catalyzes
eukaryotic translation elongation factor 1a2 K408 lactylation to
promote protein synthesis in colorectal cancer (CRC). However, its
affinity for lactoyl-CoA (Km ≈28 µM) is markedly lower compared
with for acetyl-CoA (Km ≈12 µM), raising questions about its
preferential catalysis of lactylation under high-lactate conditions
(22). Non-enzymatic lactylation
mediated by lactoylglutathione has been reported in liver and
breast cancer; this type of modification is not enzyme-regulated,
making it difficult to target via catalytic enzyme inhibition, and
its physiological importance remains unclear. Functional outcomes
of lactylation also exhibit tissue-specific discrepancies. For
example, p53 K120 lactylation suppresses its tumor-suppressive
function in gastric cancer (23)
yet stabilizes the protein and enhances DNA repair in glioma
(24). Such context-dependent
effects underscore that lactylation functions are highly dependent
on the microenvironment, suggesting that single mechanistic models
can oversimplify the actual complexity.
Regulatory machinery of protein
lactylation
Lactylation, a dynamic PTM, is regulated by three
classes of proteins: Writers (lactyltransferases), erasers
(delactylases) and readers (lactylation-recognition factors).
Collectively, these components orchestrate lactate modification on
histones and non-histone proteins by mediating its addition,
removal and functional interpretation, thereby modulating gene
expression and cellular physiology (Fig. 1). Writers catalyze the addition of a
lactate group to lysine residues: p300, a histone acetyltransferase
(HAT) with dual roles in acetylation and lactylation, was first
identified as a lactyltransferase by Zhang et al (2), linking it to pathologies including
pulmonary fibrosis (25), several
types of cancer (26) and
Parkinson's disease (27). Lysine
acetyltransferase 7 (HBO1), a MYST-family acyltransferase, mediates
several histone modifications such as acetylation, Kcr and
lactylation (28). HBO1
preferentially targets H3K9la, and elevated H3K9la levels in
cervical cancer are strongly associated with HBO1 overexpression
(5), driven by its high affinity
for lactyl-CoA to regulate oncogenic genes. Erasers remove lactate
modifications to reset histone states: Histone deacetylase (HDAC)
1–3 and sirtuin (SIRT) 1–3 function as zinc- and
NAD+-dependent deacetylases with broad delactylase
activity, cleaving both ε-N-L-lactylated and D-lactylated residues
(29). SIRT2 specifically removes
H3K18la/H4K8la to suppress neuroblastoma proliferation and
migration (30), while SIRT3
inhibits HCC by regulating cyclin E2 lactylation levels (31). Readers transduce lactylation
signals: SWI/SNF related BAF chromatin remodeling complex subunit
ATPase 4 (Brg1), a chromatin remodeler, recognizes H3K18la at
promoters of pluripotency genes to facilitate transcriptional
reprogramming (32).
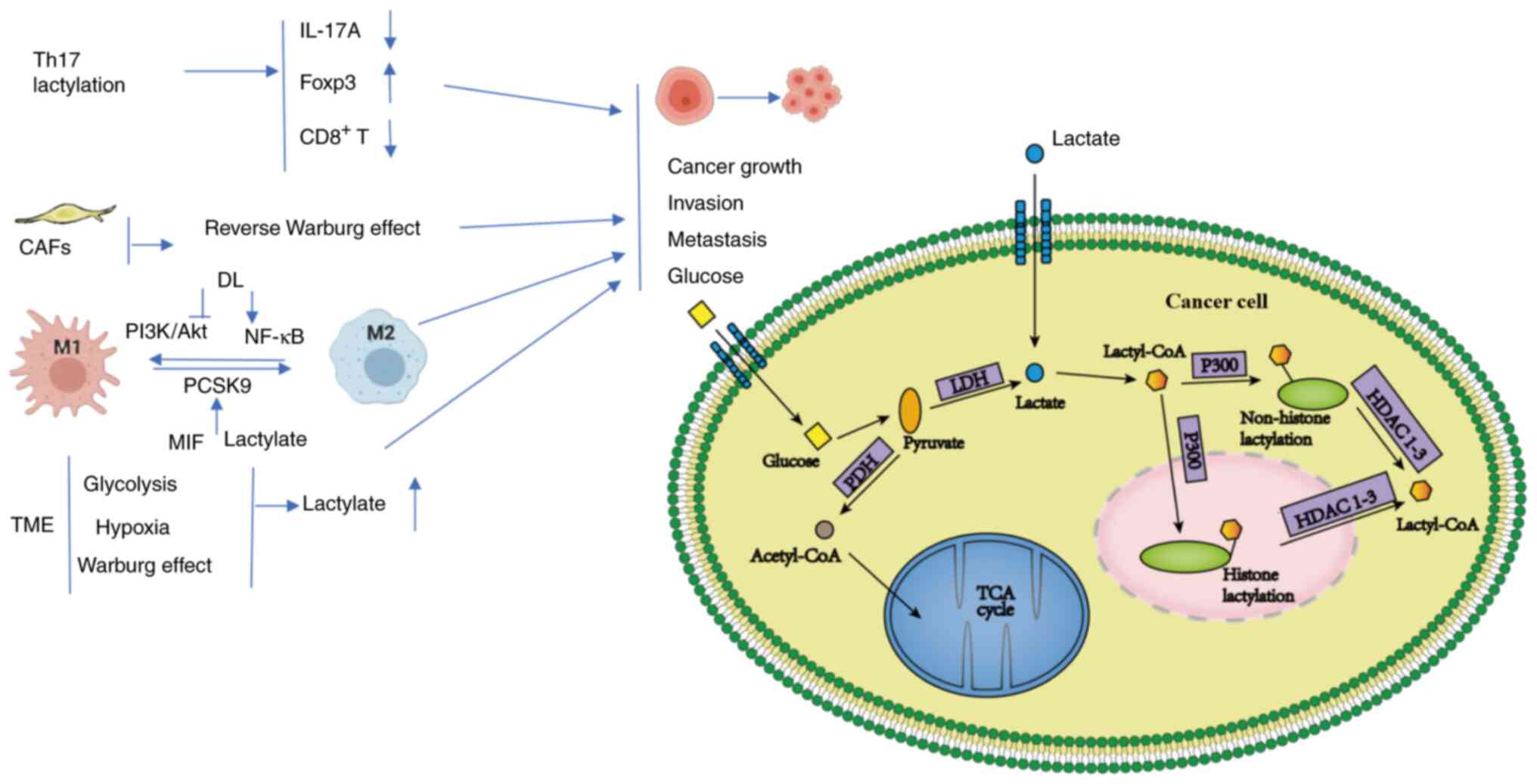 | Figure 1.In the TME, lactate polarizes TAMs to
the M2 phenotype through histone lactylation. Lactate-treated Th17
cells markedly reduce IL-17A and upregulate Foxp3. The high-energy
metabolites (such as lactate and pyruvate) produced by CAFs via
aerobic glycolysis are translocated into adjacent epithelial cancer
cells. DL can promote the transformation of M2 to M1 macrophages by
regulating the lactylation of macrophages, specifically through the
inhibition of the PI3K/Akt pathway and the activation of the NF-κB
pathway. PCSK9 affect the polarization of TAMs in colon cancer by
regulating MIF and lactate levels. Higher glycolysis, hypoxia and
the Warburg effect elevate lactate levels and promote tumor growth,
evasion and metastasis. In cancer cells, lactate produced from
glucose and exogenous lactate is further metabolized to produce
lactyl-CoA, which can enter and exit the nucleus. p300, as a writer
of lactylations, can add lactyl groups to lysine residues of
histones or non-histone proteins to form lactylations, while
erasers HDAC1-3 remove lactate modification motifs from histones or
non-histone proteins and restore them to their original state. MIF,
macrophage migration inhibitory factor; DL, D-lactic acid; TCA
cycle, tricarboxylic acid cycle; TME, tumor microenvironment; PDH,
pyruvate dehydrogenase; LDH, lactate dehydrogenase; PCSK9,
proprotein convertase subtilisin/kexin type 9; CAFs,
cancer-associated fibroblasts; TAMs, tumor-associated macrophages;
HDAC, histone deacetylase. |
Interaction of lactylation with other
PTMs
Lysine lactylation, as a novel metabolism-dependent
PTM, engages in a complex and dynamic interplay with other PTMs.
This network orchestrates tumor progression through mechanisms
including competitive occupancy of modification sites, shared
catalytic enzymes, cascading signaling and epigenetic
reprogramming. Kla exhibits substrate competition with lysine
acetylation (Kac) and Kcr, a relationship governed by fluctuating
metabolite concentrations and shared enzymatic machinery. HATs
p300/CREB-binding protein (CBP) and HBO1 possess dual
lactyltransferase/acetyltransferase activity and can catalyze
multiple acylation types. For example, HBO1 preferentially
catalyzes either lactylation (H3K9la) or acetylation (H3K9ac) at
histone H3K9, with the type of modification dictated by the
intracellular lactate-to-acetyl-CoA ratio (33,34).
Similarly, de-modifying enzymes HDAC1-3 and SIRT1-3 exhibit both
deacetylase and delactylase activities; SIRT2, for example, removes
H3K18la and H4K8la modifications (33). Lactate and acetyl-CoA compete for
the same lysine residues. In the acidic TME, lactate accumulation
drives H3K18la to replace H3K18ac, activating immunosuppressive
genes such as programmed death-ligand 1 (PD-L1). Conversely, under
glucose-replete conditions, acetylation predominates, promoting
oncogene transcription (35,36).
Kcr, another prominent Kac, serves key roles in
diseases such as liver cancer (37)
and glioblastoma (GBM) stem cells (38) by modulating histone structure and
function. A previous study suggested potential synergistic roles
for Kla and Kcr in processes such as neural differentiation and
cell proliferation (39). However,
due to its greater hydrophobicity, Kcr can antagonize Kla
modifications in open chromatin regions; for example, H3K56la
suppresses Kcr-mediated activation of inflammatory genes (40). Kla and phosphorylation mutually
amplify oncogenic signals through positive feedback loops. Histone
lactylation (such as H3K18la) activates genes encoding cell cycle
kinases (for example TTK protein kinase and BUB1 mitotic checkpoint
serine/threonine kinase B). Their protein products phosphorylate
LDHA at Tyr239, enhancing LDHA activity and lactate production,
promoting further lactylation (14). Similarly, AMPK phosphorylation of
p300 at Ser89 enhances its lactyltransferase activity, establishing
a metabolic stress-p300 lactylation amplification loop (34). Furthermore, transcription factor EB
(TFEB) lactylation at K91 impedes binding via the action of the E3
ubiquitin ligase WW domain containing E3 ubiquitin protein ligase
2, reducing TFEB ubiquitination and degradation. This stabilization
enhances autophagy and lysosomal biogenesis, supporting tumor
survival (41).
Kla is intricately integrated within the intricate
dynamic PTM network and is particularly interconnected with other
types of lysine acylation (such as Kac and Kcr) via shared core
enzymatic machinery (such as p300/CBP and HBO1 with dual
lactyl/acyl transferase activity; HDACs/SIRTs with dual
deacetylase/delactylase activity). This interdependence underpins
the ‘metabolite-concentration-dependent substrate competition’
hypothesis: Fluctuations in metabolite abundance (such as lactate
and acetyl-CoA) competitively regulate the balance between
different types of acylation, with cascading effects, and pyruvate
dehydrogenase complex component X acetylation inhibits lactate
production, subsequently downregulating H3K56 lactylation (29,39).
However, the downstream mechanistic effects of these interactions
remain largely unknown. Critical unresolved questions include: Do
Kla and Kac/Kcr directly compete for occupancy at identical lysine
residues? How do the distinct chemical properties of the modifying
groups (lactyl, acetyl and the more hydrophobic crotonyl) lead to
the recruitment of specific reader proteins and functional
divergence (such as the Brg1-specific recognition of H3K18la)? How
are these interactions tissue- or tumor-type dependent?
For the emerging Kcr, evidence for functional
synergy or antagonism with Kla in cancer is limited, and the
differential impact of their chemical properties on chromatin
remodeling and signal transduction is poorly understood. The
hijacking of shared regulatory factors by drug-induced
modifications (such as lysine isonicotinylation) highlights the
sensitivity and complexity of the PTM network, yet the pathological
importance of endogenous analogues remains unknown.
Current research often remains descriptive or
focused on isolated pathways. Notable limitations include
insufficient investigation of spatiotemporal dynamics, lack of
rigorous causal validation and inadequate critical assessment of
model systems and technological constraints. These gaps hinder a
comprehensive understanding of the precise role of this
sophisticated metabolic-epigenetic coupling network in
tumorigenesis. Future research should employ tools such as
site-specific mutagenesis, time-resolved omics and metabolic
gradient models to evaluate the kinetics of inter-modification
competition, identify specific effector molecules and assess the
consequences of network perturbation. Overcoming the current
fragmented understanding is essential to providing a solid
theoretical foundation for the development of precision anticancer
strategies targeting key nodes within the PTM network.
Lactylation and the TME
Under aerobic conditions, cancer cells often
metabolize glucose to lactate rather than producing carbon dioxide
and water via mitochondrial oxidative phosphorylation (OXPHOS).
This phenomenon, known as the Warburg effect (42,43),
leads to increased intracellular and extracellular lactate
concentrations. As a hallmark of cancer metabolism, the Warburg
effect enables cancer cells to meet their high ATP demands by
upregulating glycolysis even when oxygen is sufficient. In the TME,
lactate regulates interactions between cells, CAFs, TAMs and
tumor-infiltrating lymphocytes (TILs), thereby establishing a
microenvironment favorable for tumor cell growth and immune evasion
(3).
TAMs, a key immune cell component in the TME, serve
a crucial role in carcinogenesis and progression. Based on their
functions, TAMs are classified into the pro-inflammatory,
anticancer M1 type and the anti-inflammatory, pro-cancer M2 type.
Lactate can promote the polarization of TAMs towards an
immunosuppressive M2 phenotype through histone lactylation when
lactate levels are increased (2). T
cells serve a central role in antitumor immunity while also being
key components of the TME. Lactylation of moesin at K72 enhances
the suppressive function of regulatory T cells (Tregs).
Concurrently, extracellular lactate upregulates programmed cell
death protein 1 (PD-1) expression via G protein-coupled receptor 81
(GPR81) signaling, inducing exhaustion in CD8+ T cells
(44). Furthermore, lactate
promotes the expansion of PD-L1+ neutrophils through the
MCT1/NF-κB axis, ultimately compromising antitumor immunity
(45).
The formation of the TME is primarily based on
glycolysis, hypoxia and the Warburg effect, which collectively
influence tumor cell growth, invasion and metastasis. In the TME,
CAFs, rather than cancer cells themselves, utilize aerobic
glycolysis, a phenomenon known as the reverse Warburg effect
(46). The high-energy metabolites
(such as lactate and pyruvate) produced by CAFs through aerobic
glycolysis are transported into adjacent epithelial cancer cells,
where they undergo aerobic mitochondrial metabolism. This metabolic
pattern increases ATP production in cancer cells, thereby promoting
cancer growth and metastasis (47).
In summary, the metabolic characteristics of TME,
notably the Warburg effect and reverse Warburg effect, drive
substantial lactate accumulation. Lactylation serves as a pivotal
metabolic-epigenetic coupling mechanism that extensively regulates
the functional states and interactions of a range of cell types in
the TME, including cancer cells, CAFs, immune cells and other
stromal components. Collectively, these lactate-driven
modifications foster an immunosuppressive and pro-tumorigenic
microenvironment that supports tumor growth, invasion, metastasis
and immune evasion. This understanding provides a crucial
foundation for deciphering tumor progression and developing novel
therapeutic strategies targeting the TME.
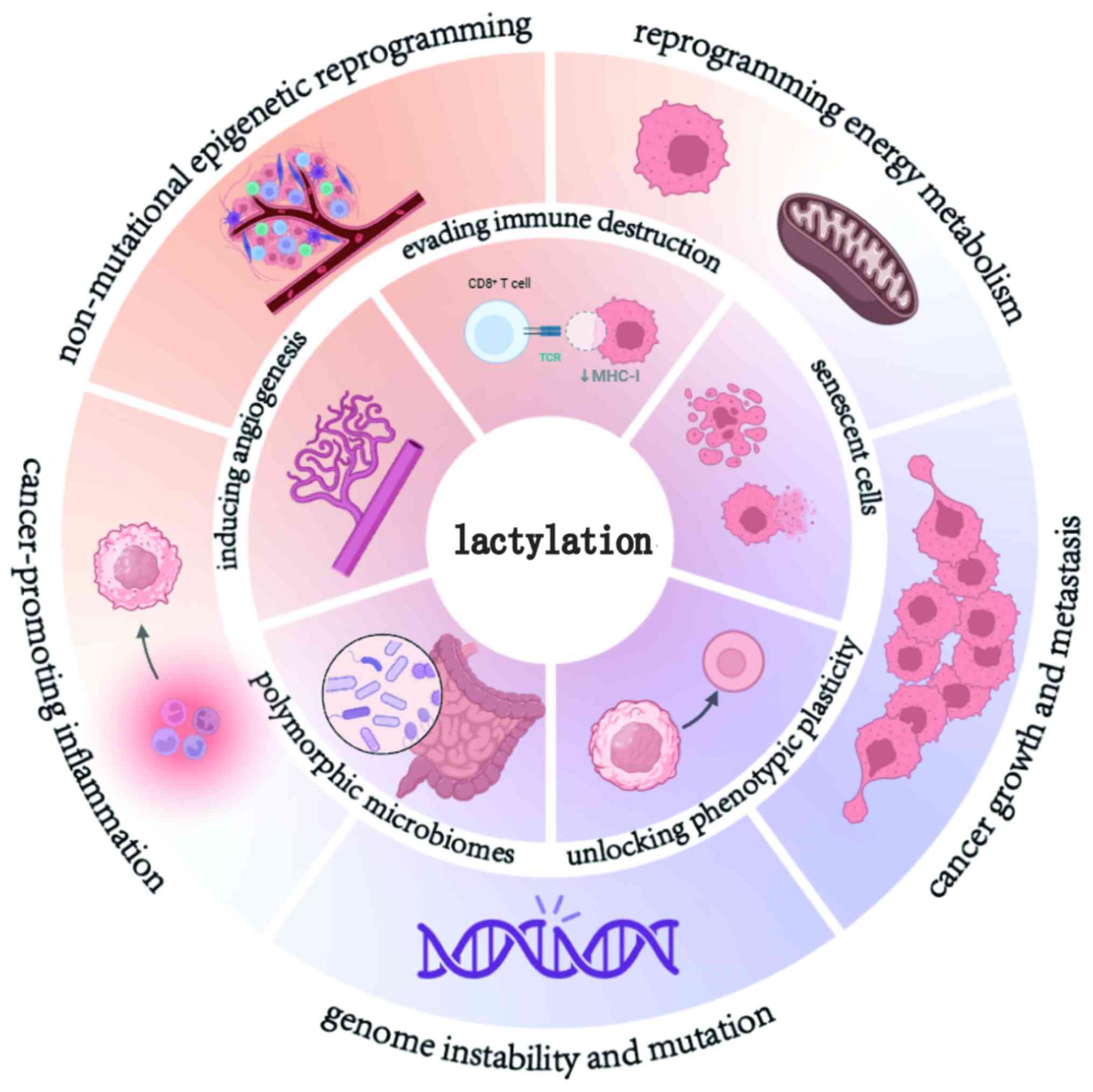
Lactylation and the hallmarks of cancer
The hallmarks of cancer are pivotal characteristics
employed in cancer research to distinguish tumor cells from normal
cells. Since their conception, the hallmarks have expanded from the
initial six to 14 (48,49), including sustaining proliferative
signaling, evading growth suppressors, resisting cell death,
enabling replicative immortality, inducing angiogenesis, activating
invasion and metastasis, genome instability and mutation, promoting
inflammation, reprogramming energy metabolism, evading immune
destruction, unlocking phenotypic plasticity, non-mutational
epigenetic reprogramming, polymorphic microbiomes and senescent
cells. Enabling replicative immortality and activating invasion and
metastasis are closely associated with cancer growth and
metastasis. These 14 hallmarks are collectively summarized in
Fig. 2. The hallmarks of cancer
provide a systematic framework for understanding tumor biology.
Lactylation serves as a central node (Fig. 3), exerting a pivotal role across the
10 established cancer hallmarks via a metabolic-epigenetic
bidirectional regulatory axis.
Metabolic reprogramming-driven
malignant proliferation and metastasis
Lactylation serves as both a core effector and
driver of metabolic reprogramming. By establishing self-reinforcing
positive feedback loops, it not only directly couples glycolysis
with epigenetic reprogramming but also maintains cancer stem cell
properties and promotes angiogenesis. This multifaceted mechanism
drives tumor growth, invasion and metastasis across multiple
dimensions.
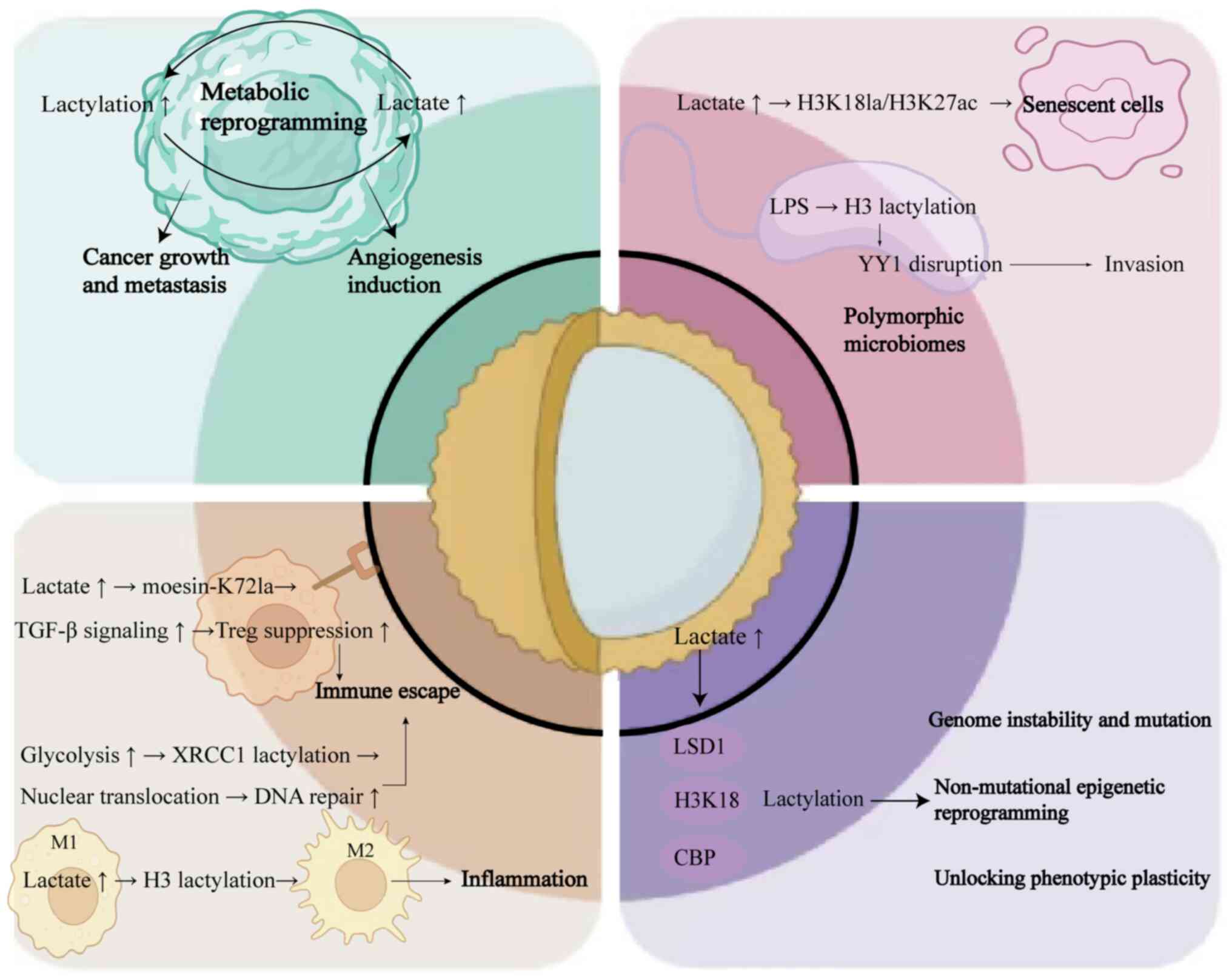
Lactylation and metabolic
reprogramming
Lactylation directly couples glycolysis with
epigenetic reprogramming, forming self-reinforcing pro-tumorigenic
circuits. Histone lactylation serves as a critical link between
metabolic reprogramming and transcriptomic dysregulation in cancer
cells (4). In malignant cells,
metabolic alterations induce fluctuations in lactate levels, which
manifest as changes in histone lactylation. Conversely, shifts in
the histone lactylation landscape reshape the transcriptomic
profile to accommodate the metabolic reprogramming demands within
the TME. This establishes a metabolic reprogramming-lactylation
positive feedback loop that accelerates tumor progression. For
example, in clear cell renal cell carcinoma (ccRCC), lactylation
levels are markedly elevated compared with normal kidney tissue
(50). This increase is
particularly pronounced upon loss of the key tumor suppressor von
Hippel-Lindau (VHL) protein. VHL deficiency leads to
hypoxia-inducible factor (HIF) accumulation, which directly
promotes lactate production via glycolysis. Furthermore,
lactylation enhances the transcriptional activity of
platelet-derived growth factor receptor β (PDGFRβ). Activation of
the PDGFRβ signaling pathway subsequently elevates lactylation
levels, establishing a self-reinforcing circuit that drives ccRCC
progression (50). Similarly, in
pancreatic cancer, lactate from TAMs induces lactylation of
nucleolar and spindle-associated protein 1 (NUSAP1), upregulating
its expression. NUSAP1 forms a transcriptional complex with c-Myc
and HIF-1α. These transcription factors bind to the LDHA promoter,
enhancing LDHA expression, thereby promoting glycolysis and
increasing lactate production. This creates another positive
feedback loop (51). Collectively,
these metabolic reprogramming-lactylation positive feedback
circuits serve a pivotal role in driving tumor growth and
metastasis.
Lactylation in tumor growth and
metastasis
Tumor growth and metastasis are governed by multiple
factors, including cancer stemness and tumor angiogenesis. Cancer
stemness refers to the capacity of tumor cells for self-renewal and
differentiation into a range of cell types, which is essential for
tumor progression and dissemination. Activation of the NF-κB
signaling pathway induces histone H3 lactylation, increasing
expression of the long non-coding RNA LINC01127. LINC01127 sustains
GBM stemness through the MAP4K4/JNK/NF-κB axis (52). Furthermore, Feng et al
(53) demonstrated that liver
cancer stem cells (LCSCs) exhibit elevated glycolytic flux, lactate
accumulation and lactylation levels compared with HCC cells.
Notably, histone H3K56la is closely associated with the
tumorigenicity and stemness maintenance of LCSCs. High glycolytic
flux provides the lactate substrate driving H3K9la/H3K56la
modifications, while lactylation, in-turn, upregulates glycolytic
enzyme expression. This establishes an LCSC-specific
metabolic-epigenetic memory that perpetuates a self-sustaining
loop.
Lactylation and inducing
angiogenesis
As the circulatory system is a major route of
metastasis, tumor angiogenesis is critically important for both
tumor metastasis and growth. Lactate, functions as a potent
pro-angiogenic factor and promotes angiogenesis through multiple
mechanisms (54–56). Lactate activates the GPR81 receptor
on endothelial cells, stimulating angiogenesis and subsequently
promoting tumor cell proliferation and survival (55). Through HIF-dependent pathways,
lactate, transported via MCT1, inhibits prolyl hydroxylases,
thereby stabilizing HIF-1α. This stabilization enhances VEGF
expression and angiogenesis. In HIF-independent pathways, lactate
promotes angiogenesis via ROS generation and NF-κB/IL-8 signaling
(54,55). Mounting evidence has established
lactylation as a critical regulator of tumor angiogenesis. For
example, Luo et al (57)
demonstrated that lactylation of HIF1α at K672 promotes
angiogenesis in prostate cancer by enhancing transcription of the
cell migration-inducing and hyaluronan-binding protein gene.
Furthermore, as previously discussed, NUSAP1 forms a
transcriptional complex with c-Myc and HIF-1α that binds to the
LDHA promoter, augmenting LDHA expression to drive angiogenesis and
metastasis in pancreatic ductal adenocarcinoma (51).
Microbiota, senescence and the
lactylation epigenetic regulatory axis
Lactylation and polymorphic microbiomes
Emerging evidence has demonstrated that gut
microbiota exerts autonomous regulatory effects on oncogenesis and
tumor evolution (49). Notably,
intestinal microbial diversity and compositional dynamics markedly
modulate cancer initiation, malignant transformation and
therapeutic responsiveness (58,59).
Mechanistically, microbial-epigenetic crosstalk may involve
lactylation-mediated pathways.
In pathogen-induced epigenetic remodeling,
enteropathogens (such as Salmonella) induce marked
alterations in long non-coding RNA networks within colonic
epithelia. Wang et al (60)
revealed that lipopolysaccharide (LPS)-mediated histone lactylation
at the LINC00152 promoter disrupts YY1 transcription factor
binding, facilitating CRC invasion and metastatic dissemination. In
bacterial metabolic adaptation, quantitative lactylome profiling of
Escherichia coli identified 1,047 modification sites, with
NAD-dependent lysine deacetylase-mediated delactylation of pyruvate
kinase I K382 serving as a critical regulator of bacterial
glycolysis. This post-translational control enhances pyruvate
kinase activity, potentiating bacterial glycolytic flux and
proliferation (61).
Lactylation and senescent cells
Cellular senescence is characterized by irreversible
cell cycle exit accompanied by distinct morphological and metabolic
alterations, including activation of the senescence-associated
secretory phenotype (SASP). The SASP entails secretion of bioactive
mediators (chemokines, cytokines and proteases) that exert
paracrine oncogenic effects within TME, driving neoplastic
proliferation, apoptotic resistance, angiogenesis, metastatic
dissemination and immune suppression (49). Pioneering work by Maekawa and
Yamanaka (62) identified GLIS
family zinc finger 1 (GLIS1) as a pluripotency-associated
transcription factor capable of reprogramming both normal and
senescent cells. During early reprogramming, GLIS1 directly binds
glycolytic gene loci to activate their transcription while
repressing somatic gene chromatin, inducing metabolic rewiring from
OXPHOS to dominance of glycolysis with concomitant lipogenesis.
This glycolytic surge increases intracellular acetyl-CoA and
lactate pools, fueling secondary epigenetic modifications including
H3K27 acetylation (H3K27ac) and H3K18 lactylation (H3K18la) at loci
associated with pluripotency. These chromatin accessibility changes
facilitate transcriptional reprogramming through structural
relaxation (63). Notably, Rong
et al (64) revealed that
GLIS1 overexpression in exhausted HCC-infiltrating CD8+
T cells promotes T cell dysfunction via a serum/glucocorticoid
regulated kinase 1-STAT3-PD-1 axis activation, mechanistically
linking metabolic control to immune evasion.
Immune microenvironment remodeling and
immune evasion
Within the TME, lactate modulates interactions among
a range of cell types, including tumor cells, CAFs, TAMs and TILs,
to shape an immunosuppressive milieu conducive to tumor growth and
immune evasion. This process is fundamentally associated with key
hallmarks of cancer: Tumor-promoting inflammation, inducing
angiogenesis and evading immune destruction.
Lactylation and tumor-promoting
inflammation
Lactate-mediated polarization of TAMs toward the M2
phenotype constitutes a central mechanism in its promotion of
tumor-associated inflammation. Beyond histone lactylation, multiple
specific pathways have been elucidated. Wang et al (65) demonstrated that PCSK9 inhibition
reduced M2 polarization by modulating MIF and lactate levels. Han
et al (66) reported that
D-lactate promotes M2-to-M1 transition through PI3K/AKT pathway
suppression and NF-κB pathway activation. Liu et al
(67) established that lactate
stabilizes HIF-2α via mTORC1-dependent inhibition of the
TFEB/ATP6V0d2 axis, enhancing expression of M2-associated genes
(such as VEGF). Zhang et al (8) identified lactate binding to
mitochondrial antiviral-signaling protein as an inhibitor of
retinoic acid-inducible gene I signaling, indirectly facilitating
M2 polarization. Lactate-induced histone lactylation functions as a
‘lactate clock’, enabling macrophages to acquire LPS tolerance and
sustain M2-related gene expression (2,68).
Collectively, these mechanisms program TAMs to exert
immunosuppressive, pro-angiogenic and tumor-promoting
functions.
Lactylation and evading immune
destruction
Evading immune destruction, a defining hallmark of
cancer (10), is garnering
increasing attention. Lactate within the TME serves a pivotal role
in tumor immune evasion. It not only drives the polarization of T
cells and macrophages toward immunosuppressive phenotypes, such as
promoting Treg differentiation and M2-type TAM formation, but also
enhances the functionality of these immunosuppressive cells through
multiple mechanisms, thereby facilitating tumor cell escape from
immune surveillance (69).
Lactate amplifies immunosuppression by expanding the
population and augmenting the activity of myeloid-derived
suppressor cells (MDSCs), resulting in impaired cytotoxic T cell
and natural killer cell function (70). Furthermore, lactylation modulates
DNA repair capacity, indirectly promoting immune evasion. Li et
al (71) demonstrated that
glycolytic reprogramming enhanced X-ray repair cross-complementing
protein 1 (XRCC1) lactylation at K247, promoting its nuclear
translocation and DNA repair function, ultimately conferring
resistance to radiotherapy. Similarly, Chen et al (72) found that nibrin (NBS1) lactylation
markedly enhanced DNA repair efficiency. This enhanced genomic
maintenance capability indirectly supports immune escape.
In summary, lactylation orchestrates immune evasion
by constructing a multi-tiered immunosuppressive network through
reprogramming TAM functionality, suppressing effector T cell
activity, activating inhibitory immune cells (Tregs, MDSCs and
PD-L1+ neutrophils), and bolstering tumor cell intrinsic
resistance (via enhanced DNA repair). This highlights lactylation
as a central metabolic-epigenetic mechanism underpinning tumor
immune evasion, providing a compelling rationale for therapeutic
strategies targeting lactylation to enhance immunotherapy
efficacy.
Tumor phenotypic plasticity
Lactylation serves as a central metabolic-epigenetic
coupling hub that simultaneously drives genomic instability,
non-mutational epigenetic reprogramming and phenotypic plasticity.
This dynamic adaptive capacity enables tumor cells to overcome
differentiation barriers and acquire hallmark malignant progression
capabilities under microenvironmental stress.
Lactylation in genome instability and
mutation
Genomic mutations fundamentally involve alterations
in DNA sequences, including nucleotide
additions/substitutions/deletions, single/double-strand breaks and
structural rearrangements such as translocations, duplications or
deletions.
Chen et al (73) demonstrated that MRE11 homolog,
double strand break repair nuclease (MRE11), a core nuclease in DNA
repair, undergoes robust lactylation catalyzed by the
acetyltransferase CBP. Lactylation at MRE11-K673 enhances MRE11-DNA
binding, thereby promoting DNA end resection and homologous
recombination repair. Epigenetic alterations notably impact genomic
stability through two primary mechanisms. DNA methylation
dysregulation is where aberrant hyper/hypomethylation at regulatory
regions can mimic mutational effects to drive carcinogenesis.
Additionally, histone modification-mediated chromatin remodeling is
where an altered chromatin architecture may induce chromosomal
rearrangements and instability, disrupting cell cycle progression
and checkpoint control. For example, p53 (the guardian of the
genome) maintains genomic integrity. Zong et al (74) identified AARS1 as a lactate sensor
and lactyltransferase that mediates global lysine lactylation in
tumor cells, targeting key proteins including p53. Beyond proteins,
lactylation may potentially occur on DNA/RNA sequences, analogous
to documented mRNA acetylation (75).
Lactylation and non-mutational
epigenetic reprogramming
Epigenetic reprogramming, alterations in gene
expression without changes in DNA sequence, serves fundamental
roles in development, differentiation and tissue homeostasis
(49). Within the TME, aberrant
conditions such as hypoxia and nutrient deprivation drive extensive
epigenetic modifications that enable cancer cells to acquire
hallmark malignant capabilities. Hypoxia-induced lactate
accumulation suppresses ten-eleven translocation demethylase
activity by reducing α-ketoglutarate levels (76). This leads to CpG island
hypermethylation and subsequent silencing of tumor suppressor gene
promoters (such as CDK inhibitor 2A), a mutation-independent
mechanism enriched in gliomas and CRC (63,77).
In IDH-mutant gliomas, lactylation-mediated metabolic dysregulation
further induces aberrant methylation of CCCTC-binding factor
insulators, reducing oncogene expression (such as PDGFRA) to drive
tumorigenesis. Stromal cells within the TME, including CAFs, innate
immune cells and endothelial cells, undergo non-genetic epigenetic
reprogramming upon recruitment to solid tumors, enhancing their
pro-tumorigenic functions. Lactate additionally facilitates cancer
cell migration by modulating integrin binding to extracellular
matrix components (78).
Furthermore, CAFs alter cancer cell NAD+/NADH ratios via
lactate shuttling, activating SIRT1-dependent PPARG coactivator 1a
to increase mitochondrial biogenesis and activity, thereby
remodeling cancer cell metabolism (79).
Lactylation and unlocking phenotypic
plasticity
The relationship between lactylation and tumor
phenotypic plasticity, an emerging cancer hallmark enabling
malignant progression through evasion of terminal differentiation,
remains incompletely elucidated (49). Phenotypic plasticity subverts normal
developmental processes wherein terminally differentiated cells
exit the cell cycle irreversibly, instead promoting
dedifferentiation (progenitor-like reversion), differentiation
blockade (progenitor-stage arrest) or trans-differentiation
(heterologous trait acquisition). Mechanistic evidence reveals
context-dependent functions. In BRAF inhibitor-resistant melanoma,
lactate accumulation induces lysine-specific histone demethylase 1A
(LSD1)-K503 lactylation, impeding tripartite motif-containing
protein 21-mediated degradation to stabilize LSD1-FosL1 complexes,
which suppress transferrin receptor transcription and confer
ferroptosis resistance, and this is reversed by LSD1 inhibitors
(80). GBM utilizes GTP
cyclohydrolase I-synthesized lactoyl-CoA to cooperate with p300,
driving H3K18la-mediated growth/differentiation factor 15
upregulation, and this sustains cancer stemness and radioresistance
(81).
Age-related macular degeneration models further
demonstrate that histone lactylation and alkB homolog 3,
a-ketoglutarate dependent dioxygenase-driven glycolysis form a
feedforward loop accelerating mesenchymal transition and fibrosis
(82). Beyond oncology, lactylation
remodels chromatin accessibility during embryonic stem cell
differentiation toward extraembryonic endoderm and coordinates
mesenchymal-epithelial transition in somatic reprogramming.
Notably, tissue-specific functional paradoxes exist; loss of
α-major histocompatibility complex-K1897 lactylation exacerbates
heart failure in cardiomyocytes (83), while neuronal arrestin b1-K195
lactylation induces S100A9-mediated mitochondrial apoptosis
(84).
These opposing outcomes underscore the
microenvironmental context, including lactate gradients and
substrate specificity, as determinants of the functional
directionality of lactylation. The spatiotemporal dynamics of
lactylation constitute a master regulator of phenotypic plasticity.
Future research should integrate high-resolution spatiotemporal
mapping (single-cell lactylome profiling across TME niches),
multi-omics dynamic modeling (lactylation-metabolism-oscillation
coupling) and context-specific delivery tools (precision
interventions) to elucidate the global roles of lactylation in TME
heterogeneity, cell fate decisions and therapeutic resistance,
providing novel paradigms to overcome plasticity-driven treatment
failure and metastasis.
Table I
systematically summarizes the lactylation sites, regulatory
mechanisms and clinical implications across key cancer hallmarks,
providing a comprehensive overview of its multifaceted roles.
 | Table I.Lactylation hallmarks in cancer. |
Table I.
Lactylation hallmarks in cancer.
| Cancer
hallmark | Lactylation
site | Tumor type | Key
enzymes/regulators |
Pathway/mechanism | Clinical
function/impact | (Refs.) |
|---|
| 1. Metabolic
reprogramming | H3K18la | ccRCC | HIF, PDGFRβ | VHL loss → HIF
accumulation → lactate production → PDGFRβ activation → lactylation
↑ | Drives tumor
progression; poor prognosis | (50) |
|
| NUSAP1 K34 | PDAC | c-Myc, HIF-1α,
LDHA | TAM lactate →
NUSAP1 lactylation → LDHA transcription → glycolysis ↑ | Promotes
metastasis; therapy resistance | (51) |
| 2. Tumor growth and
metastasis | H3K56la | Liver cancer stem
cells | LDHA, p300 | Glycolysis ↑ →
lactate ↑ → H3K56la → stemness maintenance | Enhances
tumorigenicity; stemness | (53) |
|
| H3 lactylation | GBM | NF-κB | NF-κB → H3
lactylation → LINC01127 lncRNA → MAP4K4/JNK axis | Sustains cancer
stemness | (52) |
| 3. Angiogenesis
induction | HIF1α-K672la | Prostate
cancer | MCT1, HIF1α,
KIAA1199 | HIF1α lactylation →
KIAA1199 transcription → angiogenesis | Promotes vascular
mimicry; metastasis | (57) |
|
| NUSAP1 K34
(indirect) | PDAC | c-Myc, HIF-1α,
LDHA | NUSAP1 complex →
LDHA activation → lactate ↑ → angiogenesis | Drives
metastasis | (51) |
| 4. Polymorphic
microbiomes | H4K8la | CRC | YY1 | LPS → H3
lactylation at LINC00152 promoter → YY1 disruption → invasion | Promotes metastatic
dissemination | (60) |
| 5. Senescent
cells | H3K18la,
H3K27ac | HCC | GLIS1 | Glycolysis ↑ →
lactate ↑ → H3K18la/H3K27ac → chromatin relaxation | Facilitates
cellular reprogramming; immune evasion | (63,64) |
| 6. Tumor-Promoting
Inflammation | H3 lactylation |
Macrophages/TAMs | p300, SIRTs | Lactate → H3
lactylation → M2 polarization → VEGF/IL-10 secretion | Immunosuppression;
angiogenesis | (2,67) |
| 7. Evading immune
destruction | Moesin-K72la | Tregs | MCT1, LDHA,
LDHB | Lactylation →
moesin activation → TGF-β signaling ↑ → Treg suppression ↑ | Immune escape;
immunotherapy resistance | (44,100) |
|
| XRCC1-K247la | GBM | ALDH1A3, LDHA | Glycolysis ↑ →
XRCC1 lactylation → nuclear translocation → DNA repair ↑ |
Radio/chemoresistance | (71) |
|
| NBS1
lactylation | Pan-cancer | AARS1, LDHA
CBP | Lactylation →
enhanced DNA repair efficiency | Chemotherapy
resistance | (72) |
| 8. Genomic
instability | MRE11-K673la | Breast cancer |
| CBP-mediated
lactylation → MRE11-DNA binding ↑ → homologous recombination repair
↑ | Chemoresistance;
genomic instability | (73) |
| 9. Non-mutational
epigenetic reprogramming | Histone
methylation | Glioma, CRC | TET inhibition | Lactate ↑ → α-KG ↓
→ TET suppression → CpG hypermethylation (such as CDKN2A) | Silencing of
tumorsuppressors; therapy resistance | (76,77) |
| 10. Phenotypic
plasticity | LSD1-K503la | Melanoma | p300, GTPCS | Lactate → LSD1
lactylation → stabilization → TFRC suppression → ferroptosis
resistance | Targeted therapy
resistance | (80) |
|
| H3K18la | GBM | p300 | GTPCS → lactoyl-CoA
→ H3K18la → GDF15 ↑ → stemness/radioresistance | Radiation
resistance; recurrence | (81) |
Lactylation and cancer development
As research into lactylation progresses, evidence is
mounting that this modification, affecting both histones and
non-histone proteins, serves an oncogenic role across various
malignancies (85,86). Pan et al (87) demonstrated that LCSCs exhibited
higher lactylation levels compared with HCC cells. Lactate, as a
metabolic substrate, promotes histone lactylation, particularly at
H3K9la and H3K56la sites, driving LCSC proliferation and
tumorigenicity. Demethylzeylasteral, a triterpenoid from
Tripterygium wilfordii Hook F, curbs histone H3 lactylation
by inhibiting lactate production, thus restraining LCSC
proliferation, migration and inducing apoptosis.
In CRC, Li et al (88) revealed lactylation, especially
H3K18la, enhanced Rubicon like autophagy enhancer (RUBCNL) gene
transcription, associated with a worse CRC prognosis. Such
modifications boost RUBCNL/Pacer protein expression, crucial for
autophagosome maturation. This process aids cancer cell survival in
harsh conditions such as hypoxia. In renal cancer, Yang et
al (50) found a positive
association between lactylation and disease progression,
particularly in ccRCC. Inactive VHL triggers PDGFRβ transcription
via histone lactylation, driving ccRCC progression. PDGFRβ
signaling activation further stimulated histone lactylation,
creating a vicious cycle promoting ccRCC progression.
In bladder cancer, Xie et al (89) reported that the circular RNA,
circXRN2, suppressed cancer progression by activating the Hippo
signaling pathway, countering histone lactylation-driven cancer.
circXRN2 is downregulated in bladder cancer tissues and cell lines.
Its overexpression inhibited cancer cell proliferation and
migration, acting as a negative regulator of glycolysis and lactate
production. In thyroid cancer, Wang et al (90) noted enhanced glycolysis in BRAFV600E
mutation-positive thyroid cancers raised intracellular lactate
levels, promoting histone lactylation, especially at H4K12la. This
lactylation activated cell cycle regulation-related genes,
fostering thyroid cancer proliferation. In ocular melanoma, Yu
et al (91) found higher
lactylation levels in ocular melanoma tissues compared with in
normal melanocyte tissues, associated with early recurrence and
increased cancer invasiveness. Lactylation promoted cancer
formation by increasing N6-methyladenosine RNA binding protein 2
(YTHDF2) protein expression. YTHDF2 was shown to recognize
m6A-modified RNAs and accelerate the degradation of PER1 and TP53
mRNA, two cancer suppressor genes, expediting ocular melanoma
formation. In non-small cell lung cancer (NSCLC), Jiang et
al (92) demonstrated that
lactate not only accumulates in lung cancer but was also associated
with cancer progression. Lactate adjusts the expression of specific
metabolic enzyme genes [such as hexokinase 1 and isocitrate
dehydrogenase [NAD(+)] (IDH) 3 non-catalytic subunit γ] by
increasing histone lactylation in their promoter regions, thereby
influencing NSCLC cell proliferation and migration.
Lactylation can both drive and suppress cancer. For
example, METTL16 lactylation in gastric cancer cells promoted m6A
modification on ferritin oxidoreductase protein 1 mRNA, inducing
copper death and curbing gastric cancer progression (93). In cervical cancer cells,
glucose-6-phosphate dehydrogenase (G6PD) lactylation at K45 may
lower its binding affinity with NADP+, dampening G6PD
enzyme activity and inhibiting cancer cell proliferation (94). Lactate, as a metabolic byproduct,
can markedly suppress uveal melanoma (UM) cell proliferation and
migration, and alter cellular metabolism by increasing oxidative
phosphorylation. Lactate-treated UM cells exhibited higher
heterochromatin content and a quiescent state compared to untreated
controls, which correlated with reduced proliferation and
metastatic capacity (95).
As lactylation research deepens, its clinical
importance becomes more evident. Lactylation levels can predict
patient survival, prognosis and aid in disease diagnosis. In
pancreatic cancer, Peng et al (96) identified a set of
lactylation-related genes associated with pancreatic cancer
prognosis by combining RNA-sequencing data and clinical
information. A prognostic model based on lactylation was developed
using intersection analysis of differentially expressed genes and
lactate-associated genes, along with univariate and multivariate
Cox regression models. In breast cancer, lactate levels were
notably associated with the Nottingham Prognostic Index (NPI).
Grade III breast cancer tissues showed markedly higher lactate
levels compared with grade II tissues, and were positively
associated with the NPI but negatively with LDHA levels (97). In liver cancer, specific lactylation
sites on ubiquitin specific peptidase 14 (USP14) and ATP binding
cassette subfamily F member 1 proteins can serve as diagnostic
markers for liver cancer and its metastasis (98).
The current understanding of lactylation-driven
tumorigenesis remains fragmented, primarily due to unresolved
tissue-specific differences and insufficient clinical research.
While lactylation promotes tumorigenesis in most malignancies (such
as H3K9la/H3K56la-mediated stemness maintenance in LCSCs), it
suppresses metastasis in UM by inducing metabolic quiescence and
heterochromatinization.
This bidirectional regulation implies the existence
of a ‘lactylation threshold’, which is likely determined by the
microenvironmental lactate concentration, substrate specificity
(such as differential effects of H3K56la vs. mitochondrial targets)
and spatiotemporal dynamics, factors inadequately integrated into
current mechanistic models. Reported causal relationships lack
molecular verification: The antitumor effect of circXRN2 in bladder
cancer is attributed to activation of the Hippo pathway, yet its
association with the lactylation machinery (for example,
YAP/TAZ-mediated suppression of p300 lactyltransferase activity)
remains undetermined. RUBCNL-driven autophagy in CRC is described
as lactylation-dependent, but quantitative evidence linking lactate
flux to H3K18la levels at the RUBCNL promoter is lacking.
Furthermore, findings, such as USP14 lactylation as a liver cancer
biomarker, lack actionable diagnostic frameworks. The present
review proposes mapping lactylation modification thresholds by
integrating lactate biosensors (such as Laconic) with single-cell
lactylomics. To functionally assess specific lactylation sites
identified through this approach, CRISPR-dCas9-mediated
site-specific delactylation will be implemented, exemplified by
targeting H4K12la in thyroid cancer models. The diagnostic
potential of key lactylation marks will subsequently be evaluated
using tissue microarrays and receiver operating characteristic
curve analysis; for instance, comparing the diagnostic efficacy of
USP14-Kla vs. α-fetoprotein for detecting liver cancer
metastasis.
Lactylation and cancer therapy
Due to the growing evidence that lactylation is
positively associated with carcinogenesis, lactate producers or
lactate transporters, such as LDHA and MCT1/4, have been proposed
as novel targets for tumor therapy, either alone or in combination
with other anticancer strategies. In the present review, the role
of lactate in targeted therapy, traditional therapy and
immunotherapy is discussed.
Lactylation and targeted
therapies
Targeting lactylation has emerged as a promising
anticancer strategy, offering novel therapeutic avenues for drug
development. Current approaches focus on key nodes in lactate
metabolism, transport and modification. Peng et al (96) demonstrated that inhibiting solute
carrier family 16 member 1, a lactate transporter that is
upregulated in pancreatic cancer and associated with lactylation
levels, reduced intracellular lactate levels, suppressed
lactylation and inhibited tumor proliferation and migration.
Similarly, BRAFV600E inhibitors decreased glycolysis and
lactylation, inducing cell cycle arrest in thyroid cancer,
suggesting potential synergy with existing targeted therapies
through modulation of a glycolysis-lactylation axis (94).
LDHA, the key enzyme converting pyruvate to lactate,
represents another critical target. Its upregulation is associated
with a poor prognosis across several types of cancer. The
competitive inhibitor oxamate reduced cancer proliferation by
blocking glycolysis (99), while
gossypin directly inhibited LDHA activity, decreasing lactate
accumulation and promoting apoptosis (100). However, LDHA inhibitors face
notable limitations: Off-target effects and unpredictable side
effects from non-selective inhibition necessitate strategies to
improve drug specificity and a deeper mechanistic understanding.
Emerging agents aim to address these challenges through more
precise modulation. Simeprevir and oxamate inhibit LDHA to reduce
NBS1-K388 lactylation, whereas L-alanine competes with lactate for
AARS1 binding. These targeted approaches show notable potential for
advancing lactylation-based cancer therapies.
Lactylation and conventional
therapy
Tumor resistance remains a major barrier to
effective cancer therapy, with emerging evidence implicating
lactylation as a key mediator. Chen et al (73) demonstrated that MRE11 lactylation
enhances DNA-binding capacity, promoting homologous recombination
repair and chemoresistance in a lactate-abundant TME. Similarly,
NBS1 lactylation markedly increases DNA repair efficiency, while
XRCC1-K247 lactylation facilitated nuclear translocation to confer
chemo/radioresistance in GBM (71,72).
These findings establish lactylation as a promising therapeutic
target to overcome treatment resistance.
Proof-of-concept studies validate this approach; a
cell-penetrating peptide blocking MRE11-K673 lactylation sensitized
tumors to cisplatin and PARP inhibitors by inhibiting DNA repair
(73). Additionally, the small
molecule D34-919 disrupted an aldehyde dehydrogenase 1 family,
member A3-pyruvate kinase M1/2 interaction, reducing lactate
production and XRCC1 lactylation, thus restoring radiosensitivity
in GBM (71). These targeted
strategies demonstrate that inhibiting specific lactylation events
can reverse treatment resistance mechanisms, providing a rational
foundation for combining lactylation-directed therapies with
conventional cancer treatments. Further investigations should focus
on optimizing lactylation-targeted agents for clinical
translation.
Lactylation and immunotherapy
Emerging evidence has established lactylation as a
pivotal epigenetic regulator of resistance to cancer immunotherapy.
Tumor-derived lactate drives immunosuppression within the TME
through lactylation-dependent mechanisms, including moesin-K72
lactylation, which enhances Treg immunosuppressive activity
(101). Critically, reducing TME
lactate levels improves anti-PD-1 efficacy, positioning lactate
modulation as a promising immunotherapeutic adjuvant (101). In HCC, lactate activates the
MCT1/NF-κB/cyclooxygenase 2 (COX-2) axis to generate
PD-L1+ neutrophils that mediate immune evasion, a
phenotype reversed by COX-2 inhibition with celecoxib, synergizing
with Lenvatinib (45). Similarly,
STAT5-driven glycolytic flux in acute myeloid leukemia (AML)
elevates histone lactylation, upregulating PD-L1 and impairing
CD8+ T cell function. PD-1/PD-L1 blockade restores T
cell activity in STAT5-high AML models, supporting biomarker-guided
immunotherapy for glycolytic subtypes (102).
Pharmacological inhibition of lactate transport via
AZD3965 (an MCT1 inhibitor) disrupts immunosuppression and
attenuates tumor growth (103),
while genetic lactylation inhibition overcomes VEGF resistance in
CRC (88). These findings highlight
the therapeutic promise of dual-targeting lactylation pathways and
immune checkpoints. Current strategies targeting lactate synthesis
(LDHA inhibitors), transport (MCT blockers) and lactylation
modifiers require improved precision. Future efforts should
prioritize developing site-specific lactylation inhibitors and
combinatorial regimens leveraging metabolic-immune crosstalk.
Despite promising preclinical validation of the chemosensitizing
role of lactylation, translational challenges, including
tissue-specific dynamics and off-target effects, necessitate deeper
mechanistic exploration to improve clinical translation.
Current therapeutic strategies targeting lactylation
focused on three pillars: Disrupting lactate metabolism (LDHA/MCT
inhibition), reversing treatment resistance (blocking lactylation
of DNA repair proteins) and overcoming immunosuppression
(reprogramming Tregs/myeloid cells). However, notable challenges
remain in utilizing these approaches clinically. Firstly, the
mechanistic understanding in vitro and in animal models
often fails to translate effectively, as exemplified by LDHA
inhibitors paradoxically inducing protective autophagy, which
enhances drug resistance. Secondly, targeting specificity conflicts
with physiological complexity, as demonstrated by MCT1 inhibitor
AZD3965 simultaneously impairing T cell antitumor functions.
Thirdly, static interventions ignore the spatiotemporal
heterogeneity of lactylation dynamics, particularly regarding
optimal dosing windows. Notably, lactate metabolism targeting (such
as oxamate inhibiting LDHA) frequently overlooks compensatory
metabolic pathways. Resistance-reversal strategies (such as
targeting MRE11-K673la) lack tissue selectivity, risking collateral
damage to normal DNA repair mechanisms. Immunotherapy combinations
often disregard the dual regulatory roles of lactylation in the
TME, where suppressing Treg lactylation may destabilize immune
homeostasis.
Future research must advance toward dynamic
precision modulation: Developing spatiotemporally responsive
delivery systems (such as pH-sensitive nanocarriers with SIRT2
activators) to inhibit oncogenic lactylation (for example
MRE11-K673la) while sparing normal tissues; integrating multi-omic
biomarkers (such as plasma exosomal H3K18la or PET-based p300
activity tracing) for patient stratification and triple-targeting
(production-transportation-‘reader’) combination therapies; and
pioneering innovative tools such as CRISPR-dCas9-mediated
site-specific delactylation and microbiome-engineered bacteria
modulating gut lactylation. Collectively, these advances will
transition lactylation-targeted therapy from single-node inhibition
toward artificial intelligence-guided reprogramming of
epigenetic-metabolic networks, ultimately establishing curative
paradigms for cancer treatment.
Conclusions
Despite revealing the critical roles of lactylation
in tumor metabolic reprogramming, immune microenvironment
remodeling and resistance to therapy, three fundamental challenges
persist: i) An incomplete mechanistic understanding, exemplified by
tissue-specific functional contradictions (pro-tumorigenic vs.
suppressive effects); ii) technical limitations in capturing
dynamic modifications and discriminating D/L-lactylation isomers;
and iii) inadequate spatiotemporally precise intervention
strategies. Future breakthroughs require reconstructing research
paradigms through the following integrated approaches: i) Deepening
mechanistic insights by decoding lactylation context-dependent
regulatory logic (such as concentration-dependent modification
thresholds and substrate specificity) and quantifying
microenvironment-modification dynamics using metabolic gradient
organoid models; ii) advancing technical innovation through
single-cell spatiotemporal lactylomics and development of
isomer-specific probes for D/L-lactylation; and iii) developing
precision targeting via microenvironment-responsive delivery
systems (such as pH-responsive nanocarriers loaded with SIRT2
activators) to modulate specific effectors such as DNA repair
proteins (MRE11-K673la) or epigenetic regulators (H4K12la),
combined with metabolic-immune dual-targeting strategies (such as
MCT4 inhibitors + anti-PD-1) to overcome resistance. Only by
contextualizing lactylation within dynamic modification networks
can this biological concept transform clinical oncology
practice.
Acknowledgements
Not applicable.
Funding
The funding was received from Lishui City Technology Application
Research Project (grant no. 2023GYX61) and Zhejiang Public Welfare
Technology Research Program (grant no. LGF21H160001).
Availability of data and materials
Not applicable.
Authors' contributions
YF drafted the manuscript and ZC revised the
manuscript. LD collected the literature and revised the manuscript.
JL designed the whole review and provided funding support. Data
authentication is not applicable. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wan N, Wang N, Yu S, Zhang H, Tang S, Wang
D, Lu W, Li H, Delafield DG, Kong Y, et al: Cyclic immonium ion of
lactyllysine reveals widespread lactylation in the human proteome.
Nat Methods. 19:854–864. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang D, Tang Z, Huang H, Zhou G, Cui C,
Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al: Metabolic
regulation of gene expression by histone lactylation. Nature.
574:575–580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qu J, Li P and Sun Z: Histone lactylation
regulates cancer progression by reshaping the tumor
microenvironment. Front Immunol. 14:12843442023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Izzo LT and Wellen KE: Histone lactylation
links metabolism and gene regulation. Nature. 574:492–493. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Niu Z, Chen C, Wang S, Lu C, Wu Z, Wang A,
Mo J, Zhang J, Han Y, Yuan Y, et al: HBO1 catalyzes lysine
lactylation and mediates histone H3K9la to regulate gene
transcription. Nat Commun. 15:35612024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rabinowitz JD and Enerbäck S: Lactate: The
ugly duckling of energy metabolism. Nat Metab. 2:566–571. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brooks GA: The science and translation of
lactate shuttle theory. Cell Metab. 27:757–785. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang W, Wang G, Xu ZG, Tu H, Hu F, Dai J,
Chang Y, Chen Y, Lu Y, Zeng H, et al: Lactate is a natural
suppressor of RLR signaling by targeting MAVS. Cell. 178:176–189.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jain M, Aggarwal S, Nagar P, Tiwari R and
Mustafiz A: A D-lactate dehydrogenase from rice is involved in
conferring tolerance to multiple abiotic stresses by maintaining
cellular homeostasis. Sci Rep. 10:128352020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang H, Yang L, Liu M and Luo J: Protein
post-translational modifications in the regulation of cancer
hallmarks. Cancer Gene Ther. 30:529–547. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Millán-Zambrano G, Burton A, Bannister AJ
and Schneider R: Histone post-translational modifications-cause and
consequence of genome function. Nat Rev Genet. 23:563–580. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Q, Cao L and Xu K: Role and
mechanism of lactylation in cancer. Zhongguo Fei Ai Za Zhi.
27:471–479. 2024.(In Chinese). PubMed/NCBI
|
|
14
|
Li F, Si W, Xia L, Yin D, Wei T, Tao M,
Cui X, Yang J, Hong T and Wei R: Positive feedback regulation
between glycolysis and histone lactylation drives oncogenesis in
pancreatic ductal adenocarcinoma. Mol Cancer. 23:902024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang Y, Wen J, Lou S, Han Y, Pan Y, Zhong
Y, He Q, Zhang Y, Mo X, Ma J and Shen N: DNAJC12 downregulation
induces neuroblastoma progression via increased histone H4K5
lactylation. J Mol Cell Biol. 16:mjae0562024. View Article : Google Scholar
|
|
16
|
Zhang N, Jiang N, Yu L, Guan T, Sang X,
Feng Y, Chen R and Chen Q: Protein lactylation critically regulates
energy metabolism in the Protozoan Parasite Trypanosoma brucei.
Front Cell Dev Biol. 9:7197202021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mao Y, Zhang J, Zhou Q, He X, Zheng Z, Wei
Y, Zhou K, Lin Y, Yu H, Zhang H, et al: Hypoxia induces
mitochondrial protein lactylation to limit oxidative
phosphorylation. Cell Res. 34:13–30. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao M, Zhang N and Liang W: Systematic
analysis of lysine lactylation in the plant fungal pathogen
botrytis cinerea. Front Microbiol. 11:5947432020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Z, Yan C, Ma J, Peng P, Ren X, Cai S,
Shen X, Wu Y, Zhang S, Wang X, et al: Lactylome analysis suggests
lactylation-dependent mechanisms of metabolic adaptation in
hepatocellular carcinoma. Nat Metab. 5:61–79. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hao B, Dong H, Xiong R, Song C, Xu C, Li N
and Geng Q: Identification of SLC2A1 as a predictive biomarker for
survival and response to immunotherapy in lung squamous cell
carcinoma. Comput Biol Med. 171:1081832024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Q, Yuan H, Bronze MS and Li M:
Targeting lactylation and the STAT3/CCL2 axis to overcome
immunotherapy resistance in pancreatic ductal adenocarcinoma. J
Clin Invest. 135:e1914222025. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Peng M, Oyang L, Shen M, Li S,
Jiang X, Ren Z, Peng Q, Xu X, Tan S, et al: Mechanism and
application of lactylation in cancers. Cell Biosci. 15:762025.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Peng X and Du J: Histone and non-histone
lactylation: Molecular mechanisms, biological functions, diseases,
and therapeutic targets. Mol Biomed. 6:382025. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mi L, Cai Y, Qi J, Chen L, Li Y, Zhang S,
Ran H, Qi Q, Zhang C, Wu H, et al: Elevated nonhomologous
end-joining by AATF enables efficient DNA damage repair and
therapeutic resistance in glioblastoma. Nat Commun. 16:49412025.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cui H, Xie N, Banerjee S, Ge J, Jiang D,
Dey T, Matthews QL, Liu RM and Liu G: Lung myofibroblasts promote
macrophage profibrotic activity through lactate-induced histone
lactylation. Am J Respir Cell Mol Biol. 64:115–125. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen Q, Yang B, Liu X, Zhang XD, Zhang L
and Liu T: Histone acetyltransferases CBP/p300 in tumorigenesis and
CBP/p300 inhibitors as promising novel anticancer agents.
Theranostics. 12:4935–4948. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Polich J: Updating P300: An integrative
theory of P3a and P3b. Clin Neurophysiol. 118:2128–2148. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xiao Y, Li W, Yang H, Pan L, Zhang L, Lu
L, Chen J, Wei W, Ye J, Li J, et al: HBO1 is a versatile histone
acyltransferase critical for promoter histone acylations. Nucleic
Acids Res. 49:8037–8059. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moreno-Yruela C, Zhang D, Wei W, Bæk M,
Liu W, Gao J, Danková D, Nielsen AL, Bolding JE, Yang L, et al:
Class I histone deacetylases (HDAC1-3) are histone lysine
delactylases. Sci Adv. 8:eabi66962022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zu H, Li C, Dai C, Pan Y, Ding C, Sun H,
Zhang X, Yao X, Zang J and Mo X: SIRT2 functions as a histone
delactylase and inhibits the proliferation and migration of
neuroblastoma cells. Cell Discov. 8:542022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jin J, Bai L, Wang D, Ding W, Cao Z, Yan
P, Li Y, Xi L, Wang Y, Zheng X, et al: SIRT3-dependent
delactylation of cyclin E2 prevents hepatocellular carcinoma
growth. EMBO Rep. 24:e560522023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu X, Huang X, Yang Y, Sun Y, Zhao Y,
Zhang Z, Qiu D, Wu Y, Wu G and Lei L: Dux activates
metabolism-lactylation-MET network during early iPSC reprogramming
with Brg1 as the histone lactylation reader. Nucleic Acids Res.
52:5529–5548. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang P, Lin K, Huang D, Jiang Z, Liao L
and Wang X: The regulatory role of protein lactylation in various
diseases: Special focus on the regulatory role of non-histone
lactylation. Gene. 963:1495952025. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu X, Wu X, Jin D, Ji J, Wu T, Huang M,
Zhao J, Shi Z, Zhou L, He X, et al: Lactylation: The regulatory
code of cellular life activity and a barometer of diseases. Cell
Oncol (Dordr). 16:10.1007/s13402–025-01083-4. 2025.
|
|
35
|
Sun Y, Wang H, Cui Z, Yu T, Song Y, Gao H,
Tang R, Wang X, Li B, Li W and Wang Z: Lactylation in cancer
progression and drug resistance. Drug Resist Updat. 81:1012482025.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhu W, Fan C, Hou Y and Zhang Y:
Lactylation in tumor microenvironment and immunotherapy resistance:
New mechanisms and challenges. Cancer Lett. 627:2178352025.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wan J, Liu H and Ming L: Lysine
crotonylation is involved in hepatocellular carcinoma progression.
Biomed Pharmacother. 111:976–982. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yuan H, Wu X, Wu Q, Chatoff A, Megill E,
Gao J, Huang T, Duan T, Yang K, Jin C, et al: Lysine catabolism
reprograms tumour immunity through histone crotonylation. Nature.
617:818–826. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dai SK, Liu PP, Li X, Jiao LF, Teng ZQ and
Liu CM: Dynamic profiling and functional interpretation of histone
lysine crotonylation and lactylation during neural development.
Development. 149:dev2000492022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Feng J, Chen X, Li R, Xie Y, Zhang X, Guo
X, Zhao L, Xu Z, Song Y, Song J and Bi H: Lactylome analysis
reveals potential target modified proteins in the retina of
form-deprivation myopia. iScience. 27:1106062024. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang Y, Luo G, Peng K, Song Y, Wang Y,
Zhang H, Li J, Qiu X, Pu M, Liu X, et al: Lactylation stabilizes
TFEB to elevate autophagy and lysosomal activity. J Cell Biol.
223:e2023080992024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vaupel P and Multhoff G: The warburg
effect: Historical dogma versus current rationale. Adv Exp Med
Biol. 1269:169–177. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kumagai S, Koyama S, Itahashi K,
Tanegashima T, Lin YT, Togashi Y, Kamada T, Irie T, Okumura G, Kono
H, et al: Lactic acid promotes PD-1 expression in regulatory T
cells in highly glycolytic tumor microenvironments. Cancer Cell.
40:201–218. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Deng H, Kan A, Lyu N, He M, Huang X, Qiao
S, Li S, Lu W, Xie Q, Chen H, et al: Tumor-derived lactate inhibit
the efficacy of lenvatinib through regulating PD-L1 expression on
neutrophil in hepatocellular carcinoma. J Immunother Cancer.
9:e0023052021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Martinez-Outschoorn UE, Lin Z, Trimmer C,
Flomenberg N, Wang C, Pavlides S, Pestell RG, Howell A, Sotgia F
and Lisanti MP: Cancer cells metabolically ‘fertilize’ the tumor
microenvironment with hydrogen peroxide, driving the Warburg
effect: Implications for PET imaging of human tumors. Cell Cycle.
10:2504–2520. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liang L, Li W, Li X, Jin X, Liao Q, Li Y
and Zhou Y: ‘Reverse Warburg effect’ of cancer-associated
fibroblasts (Review). Int J Oncol. 60:672022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang J, Luo L, Zhao C, Li X, Wang Z, Zeng
Z, Yang X, Zheng X, Jie H, Kang L, et al: A positive feedback loop
between inactive VHL-triggered histone lactylation and PDGFRβ
signaling drives clear cell renal cell carcinoma progression. Int J
Biol Sci. 18:3470–3483. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen M, Cen K, Song Y, Zhang X, Liou YC,
Liu P, Huang J, Ruan J, He J, Ye W, et al:
NUSAP1-LDHA-Glycolysis-Lactate feedforward loop promotes Warburg
effect and metastasis in pancreatic ductal adenocarcinoma. Cancer
Lett. 567:2162852023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li L, Li Z, Meng X, Wang X, Song D, Liu Y,
Xu T, Qin J, Sun N, Tian K, et al: Histone lactylation-derived
LINC01127 promotes the self-renewal of glioblastoma stem cells via
the cis-regulating the MAP4K4 to activate JNK pathway. Cancer Lett.
579:2164672023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Feng F, Wu J, Chi Q, Wang S, Liu W, Yang
L, Song G, Pan L, Xu K and Wang C: lactylome analysis unveils
lactylation-dependent mechanisms of stemness remodeling in the
liver cancer stem cells. Adv Sci (Weinh). 11:e24059752024.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sharma D, Singh M and Rani R: Role of LDH
in tumor glycolysis: Regulation of LDHA by small molecules for
cancer therapeutics. Semin Cancer Biol. 87:184–195. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Brown TP and Ganapathy V: Lactate/GPR81
signaling and proton motive force in cancer: Role in angiogenesis,
immune escape, nutrition, and Warburg phenomenon. Pharmacol Ther.
206:1074512020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Haaga JR and Haaga R: Acidic lactate
sequentially induced lymphogenesis, phlebogenesis, and
arteriogenesis (ALPHA) hypothesis: Lactate-triggered glycolytic
vasculogenesis that occurs in normoxia or hypoxia and complements
the traditional concept of hypoxia-based vasculogenesis. Surgery.
154:632–637. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Luo Y, Yang Z, Yu Y and Zhang P: HIF1α
lactylation enhances KIAA1199 transcription to promote angiogenesis
and vasculogenic mimicry in prostate cancer. Int J Biol Macromol.
222:2225–2243. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dzutsev A, Badger JH, Perez-Chanona E, Roy
S, Salcedo R, Smith CK and Trinchieri G: Microbes and cancer. Annu
Rev Immunol. 35:199–228. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Helmink BA, Khan MAW, Hermann A,
Gopalakrishnan V and Wargo JA: The microbiome, cancer, and cancer
therapy. Nat Med. 25:377–388. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang J, Liu Z, Xu Y, Wang Y, Wang F, Zhang
Q, Ni C, Zhen Y, Xu R, Liu Q, et al: Enterobacterial LPS-inducible
LINC00152 is regulated by histone lactylation and promotes cancer
cells invasion and migration. Front Cell Infect Microbiol.
12:9138152022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Dong H, Zhang J, Zhang H, Han Y, Lu C,
Chen C, Tan X, Wang S, Bai X, Zhai G, et al: YiaC and CobB regulate
lysine lactylation in Escherichia coli. Nat Commun. 13:66282022.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Maekawa M and Yamanaka S: Glis1, a unique
pro-reprogramming factor, may facilitate clinical applications of
iPSC technology. Cell Cycle. 10:3613–3614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li L, Chen K, Wang T, Wu Y, Xing G, Chen
M, Hao Z, Zhang C, Zhang J, Ma B, et al: Glis1 facilitates
induction of pluripotency via an epigenome-metabolome-epigenome
signalling cascade. Nat Metab. 2:882–892. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Rong D, Wang Y, Liu L, Cao H, Huang T, Liu
H, Hao X, Sun G, Sun G, Zheng Z, et al: GLIS1 intervention enhances
anti-PD1 therapy for hepatocellular carcinoma by targeting
SGK1-STAT3-PD1 pathway. J Immunother Cancer. 11:e0051262023.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang L, Li S, Luo H, Lu Q and Yu S: PCSK9
promotes the progression and metastasis of colon cancer cells
through regulation of EMT and PI3K/AKT signaling in tumor cells and
phenotypic polarization of macrophages. J Exp Clin Cancer Res.
41:3032022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Han S, Bao X, Zou Y, Wang L, Li Y, Yang L,
Liao A, Zhang X, Jiang X, Liang D, et al: d-lactate modulates M2
tumor-associated macrophages and remodels immunosuppressive tumor
microenvironment for hepatocellular carcinoma. Sci Adv.
9:eadg26972023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu N, Luo J, Kuang D, Xu S, Duan Y, Xia
Y, Wei Z, Xie X, Yin B, Chen F, et al: Lactate inhibits ATP6V0d2
expression in tumor-associated macrophages to promote
HIF-2α-mediated tumor progression. J Clin Invest. 129:631–646.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ivashkiv LB: The hypoxia-lactate axis
tempers inflammation. Nat Rev Immunol. 20:85–86. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Siska PJ, Singer K, Evert K, Renner K and
Kreutz M: The immunological Warburg effect: Can a
metabolic-tumor-stroma score (MeTS) guide cancer immunotherapy?
Immunol Rev. 295:187–202. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Husain Z, Huang Y, Seth P and Sukhatme VP:
Tumor-derived lactate modifies antitumor immune response: Effect on
myeloid-derived suppressor cells and NK cells. J Immunol.
191:1486–1495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li G, Wang D, Zhai Y, Pan C, Zhang J, Wang
C, Huang R, Yu M, Li Y, Li X, et al: Glycometabolic
reprogramming-induced XRCC1 lactylation confers therapeutic
resistance in ALDH1A3-overexpressing glioblastoma. Cell Metab.
36:1696–1710. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chen H, Li Y, Li H, Chen X, Fu H, Mao D,
Chen W, Lan L, Wang C, Hu K, et al: NBS1 lactylation is required
for efficient DNA repair and chemotherapy resistance. Nature.
631:663–669. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Chen Y, Wu J, Zhai L, Zhang T, Yin H, Gao
H, Zhao F, Wang Z, Yang X, Jin M, et al: Metabolic regulation of
homologous recombination repair by MRE11 lactylation. Cell.
187:294–311. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zong Z, Xie F, Wang S, Wu X, Zhang Z, Yang
B and Zhou F: Alanyl-tRNA synthetase, AARS1, is a lactate sensor
and lactyltransferase that lactylates p53 and contributes to
tumorigenesis. Cell. 187:2375–2392. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Liu C, Wu J, Zhu J, Kuei C, Yu J, Shelton
J, Sutton SW, Li X, Yun SJ, Mirzadegan T, et al: Lactate inhibits
lipolysis in fat cells through activation of an orphan
G-protein-coupled receptor, GPR81. J Biol Chem. 284:2811–2822.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Thienpont B, Van Dyck L and Lambrechts D:
Tumors smother their epigenome. Mol Cell Oncol. 3:e12405492016.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yang JH, Hayano M, Griffin PT, Amorim JA,
Bonkowski MS, Apostolides JK, Salfati EL, Blanchette M, Munding EM,
Bhakta M, et al: Loss of epigenetic information as a cause of
mammalian aging. Cell. 186:305–326. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Webb BA, Chimenti M, Jacobson MP and
Barber DL: Dysregulated pH: A perfect storm for cancer progression.
Nat Rev Cancer. 11:671–677. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ippolito L, Morandi A, Taddei ML, Parri M,
Comito G, Iscaro A, Raspollini MR, Magherini F, Rapizzi E,
Masquelier J, et al: Cancer-associated fibroblasts promote prostate
cancer malignancy via metabolic rewiring and mitochondrial
transfer. Oncogene. 38:5339–5355. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li A, Gong Z, Long Y, Li Y, Liu C, Lu X,
Li Q, He X, Lu H, Wu K, et al: Lactylation of LSD1 is an acquired
epigenetic vulnerability of BRAFi/MEKi-resistant melanoma. Dev
Cell. 60:1974–1990. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Liu R, Ren X, Park YE, Feng H, Sheng X,
Song X, AminiTabrizi R, Shah H, Li L, Zhang Y, et al: Nuclear
GTPSCS functions as a lactyl-CoA synthetase to promote histone
lactylation and gliomagenesis. Cell Metab. 37:377–394. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang Y, Zhang YC, Ding ZQ, Xu SY, Zhang
YR, Zhu HJ, Huang C, Wang JN, Zhu M, Ji JD, et al: Feedback
regulation between histone lactylation and ALKBH3-mediated
glycolysis regulates age-related macular degeneration pathology.
Proc Natl Acad Sci USA. 122:e24160461222025. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ren H, Tang Y and Zhang D: The emerging
role of protein L-lactylation in metabolic regulation and cell
signalling. Nat Metab. 7:647–664. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Mi K, Chen Z, He J, Jiang C, Xia Y and
Peng J: P300-Mediated ARRB1 lactylation promotes mitochondrial
dysfunction and neuronal apoptosis in subarachnoid hemorrhage via
upregulating S100A9. Neurochem Res. 50:1742025. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Su J, Zheng Z, Bian C, Chang S, Bao J, Yu
H, Xin Y and Jiang X: Functions and mechanisms of lactylation in
carcinogenesis and immunosuppression. Front Immunol.
14:12530642023. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang Y, Peng Q, Zheng J, Yang Y, Zhang X,
Ma A, Qin Y, Qin Z and Zheng X: The function and mechanism of
lactate and lactylation in tumor metabolism and microenvironment.
Genes Dis. 10:2029–2037. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Pan L, Feng F, Wu J, Fan S, Han J, Wang S,
Yang L, Liu W, Wang C and Xu K: Demethylzeylasteral targets lactate
by inhibiting histone lactylation to suppress the tumorigenicity of
liver cancer stem cells. Pharmacol Res. 181:1062702022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Li W, Zhou C, Yu L, Hou Z, Liu H, Kong L,
Xu Y, He J, Lan J, Ou Q, et al: Tumor-derived lactate promotes
resistance to bevacizumab treatment by facilitating autophagy
enhancer protein RUBCNL expression through histone H3 lysine 18
lactylation (H3K18la) in colorectal cancer. Autophagy. 20:114–130.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Xie B, Lin J, Chen X, Zhou X, Zhang Y, Fan
M, Xiang J, He N, Hu Z and Wang F: CircXRN2 suppresses tumor
progression driven by histone lactylation through activating the
Hippo pathway in human bladder cancer. Mol Cancer. 22:1512023.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wang X, Ying T, Yuan J, Wang Y, Su X, Chen
S, Zhao Y, Zhao Y, Sheng J, Teng L, et al: BRAFV600E restructures
cellular lactylation to promote anaplastic thyroid cancer
proliferation. Endocr Relat Cancer. 30:e2203442023. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Yu J, Chai P, Xie M, Ge S, Ruan J, Fan X
and Jia R: Histone lactylation drives oncogenesis by facilitating
m6A reader protein YTHDF2 expression in ocular melanoma.
Genome Biol. 22:852021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Jiang J, Huang D, Jiang Y, Hou J, Tian M,
Li J, Sun L, Zhang Y, Zhang T, Li Z, et al: Lactate modulates
cellular metabolism through histone lactylation-mediated gene
expression in non-small cell lung cancer. Front Oncol.
11:6475592021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Sun L, Zhang Y, Yang B, Sun S, Zhang P,
Luo Z, Feng T, Cui Z, Zhu T, Li Y, et al: Lactylation of METTL16
promotes cuproptosis via m6A-modification on FDX1 mRNA
in gastric cancer. Nat Commun. 14:65232023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Meng Q, Zhang Y, Sun H, Yang X, Hao S, Liu
B, Zhou H, Wang Y and Xu ZX: Human papillomavirus-16 E6 activates
the pentose phosphate pathway to promote cervical cancer cell
proliferation by inhibiting G6PD lactylation. Redox Biol.
71:1031082024. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Longhitano L, Giallongo S, Orlando L,
Broggi G, Longo A, Russo A, Caltabiano R, Giallongo C, Barbagallo
I, Di Rosa M, et al: Lactate rewrites the metabolic reprogramming
of uveal melanoma cells and induces quiescence phenotype. Int J Mol
Sci. 24:242022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Peng T, Sun F, Yang JC, Cai MH, Huai MX,
Pan JX, Zhang FY and Xu LM: Novel lactylation-related signature to
predict prognosis for pancreatic adenocarcinoma. World J
Gastroenterol. 30:2575–2602. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Cheung SM, Husain E, Masannat Y, Miller
ID, Wahle K, Heys SD and He J: Lactate concentration in breast
cancer using advanced magnetic resonance spectroscopy. Br J Cancer.
123:261–267. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wu X: In-depth discovery of protein
lactylation in hepatocellular carcinoma. Proteomics.
23:e23000032023. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhao Z, Han F, Yang S, Wu J and Zhan W:
Oxamate-mediated inhibition of lactate dehydrogenase induces
protective autophagy in gastric cancer cells: Involvement of the
Akt-mTOR signaling pathway. Cancer Lett. 358:17–26. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Manerba M, Vettraino M, Fiume L, Di
Stefano G, Sartini A, Giacomini E, Buonfiglio R, Roberti M and
Recanatini M: Galloflavin (CAS 568-80-9): A novel inhibitor of
lactate dehydrogenase. ChemMedChem. 7:311–317. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Gu J, Zhou J, Chen Q, Xu X, Gao J, Li X,
Shao Q, Zhou B, Zhou H, Wei S, et al: Tumor metabolite lactate
promotes tumorigenesis by modulating MOESIN lactylation and
enhancing TGF-β signaling in regulatory T cells. Cell Rep.
39:1109862022. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Huang ZW, Zhang XN, Zhang L, Liu LL, Zhang
JW, Sun YX, Xu JQ, Liu Q and Long ZJ: STAT5 promotes PD-L1
expression by facilitating histone lactylation to drive
immunosuppression in acute myeloid leukemia. Signal Transduct
Target Ther. 8:3912023. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Huang T, Feng Q, Wang Z, Li W, Sun Z,
Wilhelm J, Huang G, Vo T, Sumer BD and Gao J: Tumor-Targeted
inhibition of monocarboxylate transporter 1 improves T-cell
immunotherapy of solid tumors. Adv Healthc Mater. 10:e20005492021.
View Article : Google Scholar : PubMed/NCBI
|