Introduction
Clear cell renal cell carcinoma (ccRCC) is a
malignant tumor arising from the renal epithelium, accounting for
~85% of RCC cases (1). Compared
with other RCC subtypes, ccRCC is characterized by enhanced
invasion capacity and poorer prognosis (2). The latest grading system, developed
jointly by the World Health Organization (WHO) and International
Society of Urology, categorizes ccRCC into four grades (I–IV).
Grades I and II are considered low-grade tumors with a favorable
prognosis, while grades III and IV are characterized as high-grade
tumors associated with poor prognosis (3). Minimally invasive treatment
approaches, such as nephron sparing surgery, ablation, and even
active monitoring, are more suitable for the management of
low-grade ccRCC, while high-grade ccRCC can require radical surgery
(4). Currently, preoperative biopsy
remains the gold standard for the pathological grading of ccRCC.
However, biopsy is an invasive procedure that carries several
risks, such as dissemination and sampling errors (5). Therefore, non-invasive and accurate
grading of ccRCC using imaging techniques has emerged as a
prominent research direction in recent years.
It has been reported that magnetic resonance imaging
(MRI) can reveal several tumor characteristics, including tumor
size, necrosis, bleeding, enhancement pattern, and venous
thrombosis (4,6). However, MRI still lacks sufficient
information to aid radiologists in distinguishing the pathological
grade of ccRCC (7).
Radiomics is a science field that involves the
extraction, analysis, and interpretation of quantitative imaging
parameters. It utilizes computer-based post-processing technology
to convert medical images into high-dimensional and quantitative
imaging features. Utilizing model-building algorithms, these
imaging features can be associated with tumor tissue pathology and
heterogeneity, thus offering valuable insights for tumor
classification and grading, gene localization, as well as early
prediction of treatment response (4). Deep learning (DL) algorithms, a form
of radiomics, automatically learn and extract intricate patterns
and features from data via multi-level neural network models, thus
enabling efficient data analysis, prediction, and decision-making
(8). Currently, the association
between diffusion weighted imaging (DWI) and diffusion kurtosis
imaging (DKI) parameters and the pathological grade of ccRCC has
been extensively studied (2,9–14).
However, research on MRI-based DWI and DKI-based DL models for
predicting the pathological grade of ccRCC remains limited.
Therefore, the present study aimed to validate a DL
model based on MRI data for predicting the pathological grading of
ccRCC prior surgery.
Materials and methods
Patients
The present study was approved by the Medical Ethics
Committee of Zibo Central Hospital (approval no. 2017001), and all
patients provided signed written informed consent prior to
undergoing any examinations. The analysis included patients with
histologically confirmed ccRCC, who were treated at Zibo Central
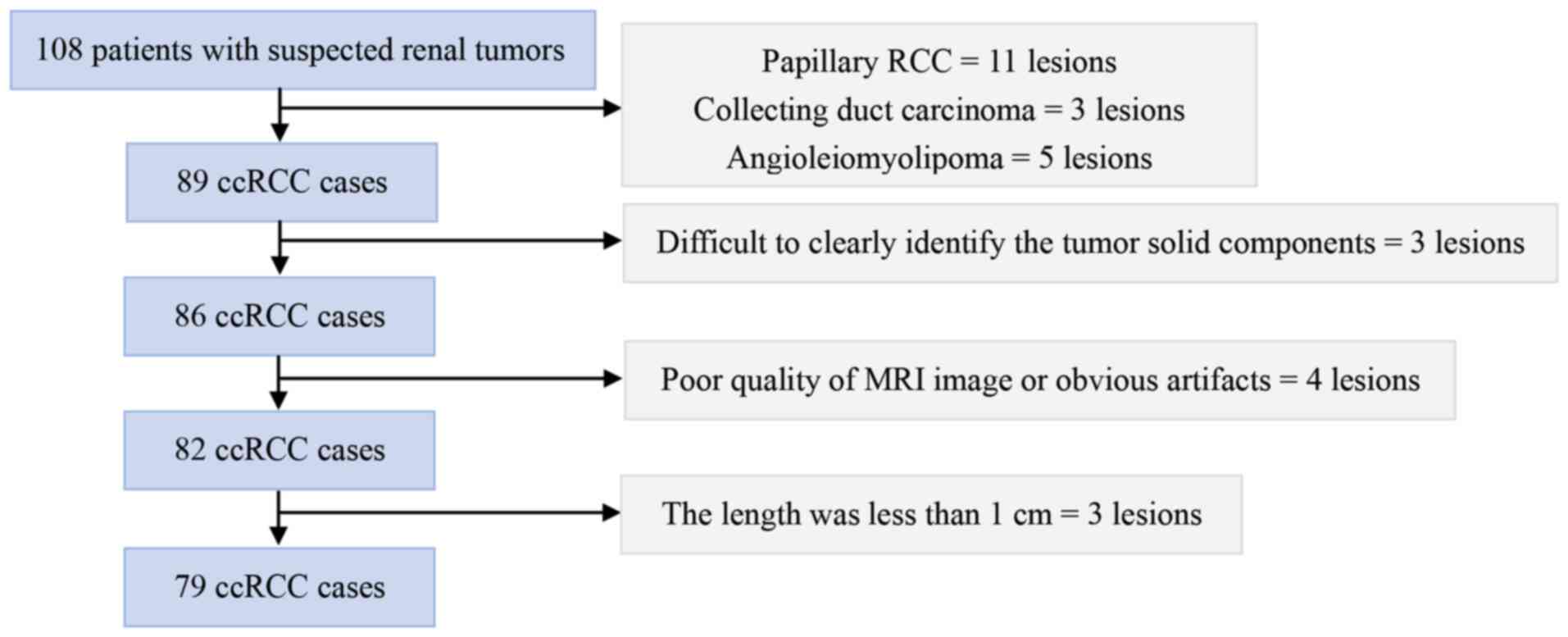
Hospital between March 2017 and November 2021. A total of 108
patients with suspected renal tumors underwent MRI scan 1–2 weeks
prior to surgery. The inclusion criteria were as follows: i)
Pathologically confirmed ccRCC via postoperative histologic
examination; ii) MRI protocol including both DWI and DKI; and iii)
no prior history of radiotherapy, chemotherapy, or percutaneous
biopsy prior to renal MRI scanning. The exclusion criteria were as
follows: i) Pathologically confirmed non-ccRCC (such as papillary
RCC, collecting duct carcinoma, angioleiomyolipoma, etc.); ii)
Renal MRI protocol lacking DWI or DKI, with incomplete imaging
data; iii) A history of radiotherapy, chemotherapy, or percutaneous
biopsy prior to renal MRI scanning; iv) Poor quality of MRI images
with obvious artifacts that interfere with the observation and
analysis of lesions; v) ccRCC lesions that are too small, in
special locations, or with other characteristics that make accurate
imaging classification or parameter measurement impossible.
Finally, a total of 79 patients with ccRCC were enrolled in the
current prospective study. In addition, a total of 19 patients were
excluded from the study due to non-ccRCC pathological types,
including papillary RCC (n=11), collecting duct carcinoma (n=3),
and angioleiomyolipoma (n=5). Among patients diagnosed with ccRCC,
three were excluded because the solid components of the lesions
were difficult to identify, four were excluded due to poor MRI
image quality or significant artifacts, and three were excluded
since the lesion size was <1 cm in length. The inclusion and
exclusion criteria are listed in Fig.
1.
MRI acquisition
All MRI examinations were carried using a 3-Tesla
(T) scanner (Signa™ HDx; GE Healthcare) and a commercial 16-channel
torso phased-array coil positioned at the renal level. All patients
underwent respiratory training prior to MRI examinations and were
scanned to the supine, feet-first position. Each patient was
subjected to two MRI sequence types, namely DWI and DKI. DWI was
carried out using a single-shot echo planar imaging sequence in the
axial plane, also incorporating imaging technology, and
respiratory-triggered acquisition, with b value of 0,800
s/mm2, a field of view (FOV) of 410 mm, and repetition
time (TR)/echo time (TE) of 5,400/64 ms. The layer thickness was
5.0 mm with 1.0-mm overlap, while a total of 19 slices were
acquired. Finally, 128×128 matrix images were produced, with a
total collection time of 1 min. For DKI, a single-shot echo planar
imaging sequence in the axial plane with respiratory-triggered
acquisition was performed. DKI images were captured with 30
diffusion gradient directions with three different b values for
each direction (0, 1,000, and 2,000 s/mm2). The
remaining MRI parameters were as follows: a FOV of 410 mm, TR/TE of
5,500/85 ms, and slice thickness of 5.0 mm with a 1.0-mm overlap.
The image matrix was 128×128 with a collection time of
approximately 3 min and 7 sec. The center level of the DKI was the
same as that of DWI. Finally, the raw data from the DWI and DKI
were transferred and processed using Functool software on the
Advantage Workstation (release 4.5). For DWI, apparent diffusion
coefficient (ADC) maps were generated, while for DKI, axial
kurtosis (Ka), fractional anisotropy (FA), radial kurtosis (Kr),
mean kurtosis (MK), and mean diffusivity (MD) maps were
calculated.
Image processing
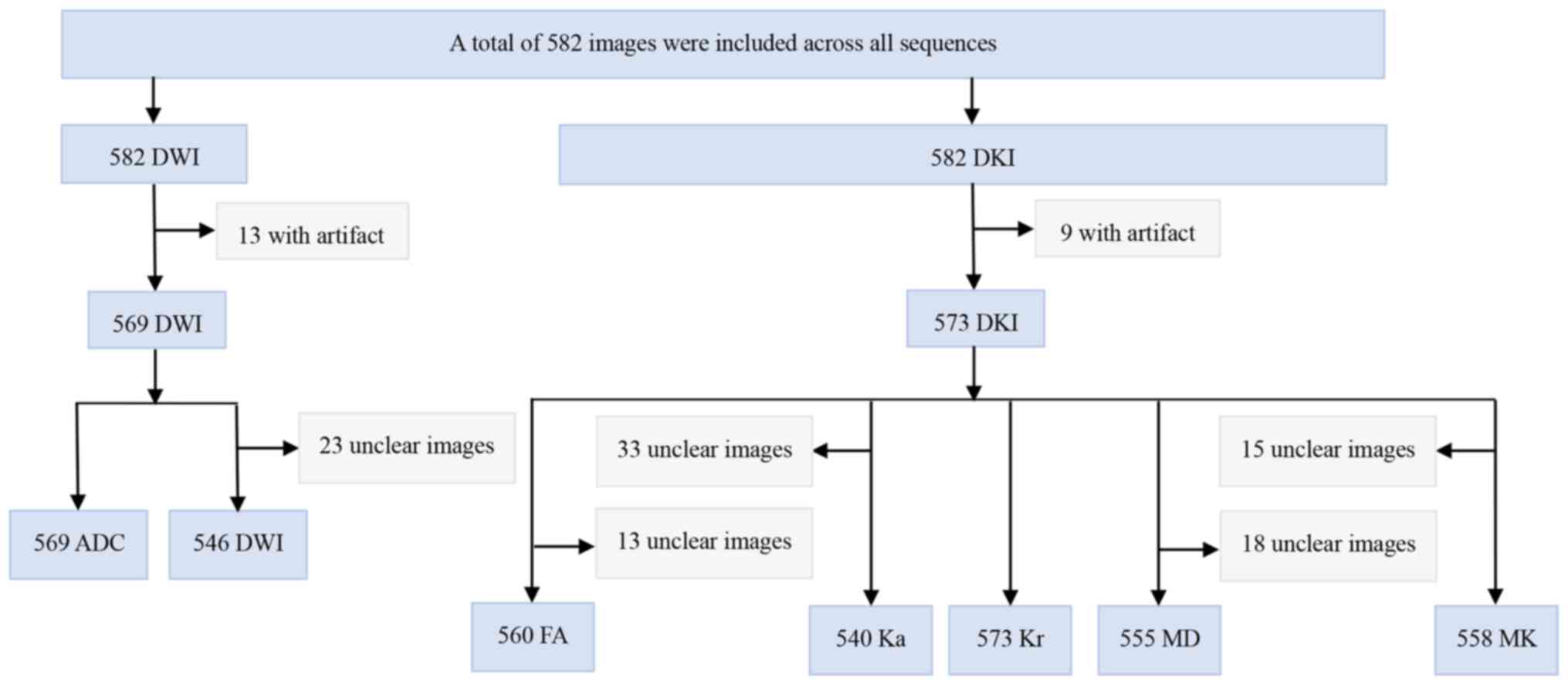
A total of 569 ADC images, including 385 in the
training set and 184 in the test set, were acquired. The program
effectively processed all 569 ADC images. For the DKI/FA scans, a
total of 560 images, 363 in the training set and 197 in the test
set, were captured, which were all successfully read by the
program. In addition, for the DKI/Ka scans, there were a total of
540 effectively read images, with 351 in the training set and 189
in the test set. The DKI/Kr scans totaled 573 images, comprising
365 and 208 in the training and test sets, respectively. All images
were also successfully read by the program. Similarly, the DKI/MD
scans consisted of 555 effectively read images (363 in the training
set and 192 in the test set). For the DKI/MK scans, 558 images, 361
in the training set and 197 in the test set, were included, which
were all also successfully accessed. Finally, the DWI scans
consisted of 546 images, with 377 in the training set and 169 in
the test set, all of which were effectively read. The potential
reasons for inconsistencies among the different parameter maps are
illustrated in Fig. 2.
Data processing
During the collection of DWI mono-exponential mode
images, it was noticed that ccRCC lesions were primarily observed
around the central slice (Fig. 3A),
while the remaining slices provided limited diagnostic value.
Therefore, the appropriate slices, from where experts could
visually identify lesions, were manually selected. The filtering
process was carried out by two professional radiologists. More
particularly, the one radiologist filtered all images, and the
second reviewed the filtering results. In case of possible
disagreement, the final decision was made through consultation. In
addition, frequency distribution maps of the ADC and DWI sequences
indicated that although the intensity of mono-exponential mode
images varied, the high-frequency intensities were mainly
concentrated within the low-value range (Fig. 3B). To normalize image intensity
among different exponential imaging modes, the high-frequency range
of the ADC and DWI sequences was scaled to [0, 255]. More
particularly, the effective intensity ranges of the ADC and DWI
sequences were [-256,500] and [0,120], respectively. The
DKI-derived sequences remained unchanged.
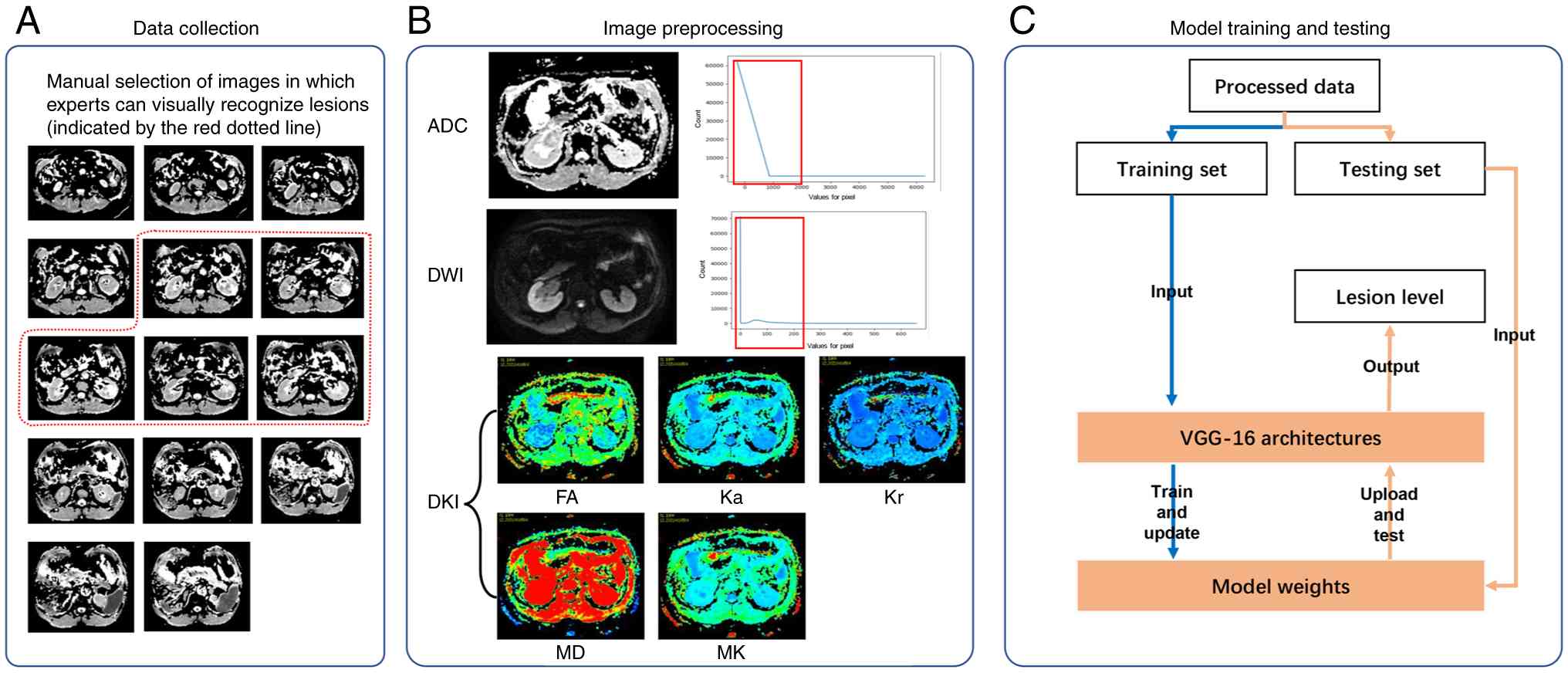 | Figure 3.Overview of data processing. (A) Data
collection, (B) image preprocessing, and (C) model training and
test. ADC, apparent diffusion coefficient; DKI, diffusion kurtosis
imaging; DWI, diffusion weighted imaging; FA, fractional
anisotropy; Ka, axial kurtosis; Kr, radial kurtosis; MD, mean
diffusivity; MK, mean kurtosis. |
Model training and test
A supervised DL model was used to predict the
pathological grades of ccRCC based on the aforementioned data. As
shown in Fig. 3C, the training set
was used to optimize the model parameters and to update the model
weights. However, the optimal model weights were retained. During
the testing phase, model weights were accordingly loaded into the
model architecture to predict the lesion grades of the test
samples. The detailed model architecture is depicted in Fig. 4. Due to the limited data, the VGG-16
model was selected as the backbone architecture. The model
comprised of five convolutions and three linear classification
layers. The first two convolutional blocks consisted of two
convolutional modules, each including a convolution layer, a batch
normalization (BatchNorm) layer and a ReLU function module, and a
max-pooling layer. The remaining three convolutional layers
consisted of three convolutional modules and a max-pooling layer.
The number of convolutional channels were 64, 128, 256, 512, and
512 respectively. The classification layers primarily included a
linear-mapping layer, a BatchNorm module and a ReLU activation
module. In the present study, patients diagnosed with grade I and
II tumors were regarded as low-grade patients, marked as 0, while
those with grade III and IV tumors were classified as high-grade,
marked as 1. Therefore, the grading of ccRCC lesions was considered
as a binary classification problem. The final linear classification
channel was set to 2, followed by a softmax classification.
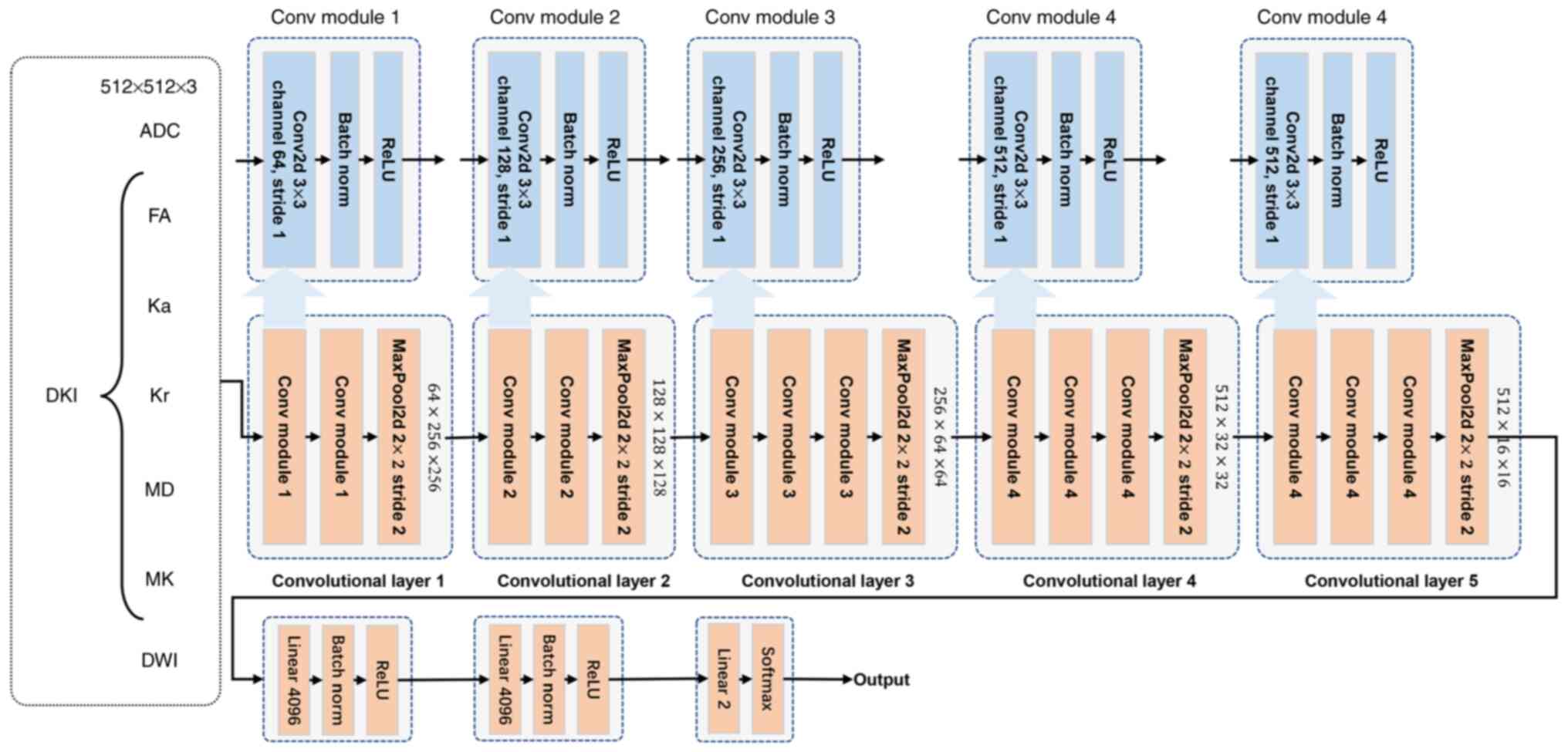 | Figure 4.Detailed model architecture. ADC,
apparent diffusion coefficient; DKI, diffusion kurtosis imaging;
DWI, diffusion weighted imaging; FA, fractional anisotropy; Ka,
axial kurtosis; Kr, radial kurtosis; MD, mean diffusivity; MK, mean
kurtosis; Conv, convolution; Conv2d, 2D convolution; MaxPool2d, 2D
max-pooling. |
Statistical analysis
Data analyses were performed using the IBM SPSS
Statistics for Windows, version 22.0 (IBM Corp.). The Shapiro-Wilk
test for normality and Levene's test for homogeneity of variances
were carried out on the age variable. Both tests verified that the
data met the assumptions of normality and homogeneity of variance.
The differences in age between patients with low- and high-grade
ccRCC were compared utilizing an independent samples (unpaired)
t-test. The χ2 test was employed to compare the
differences in sex between low- and high-grade tumors. For tumor
side, Fisher's exact test was used instead, as the assumption of
expected counts for the χ2 test was not met. P<0.05
was considered to indicate a statistically significant
difference.
Results
Patient and tumor characteristics
Based on the pathological results, a total of 79
patients with ccRCC were enrolled, including 40 cases of low-grade
disease (six cases of grade I and 34 cases of grade II) and 39
cases of high-grade disease (25 cases of grade III and 14 cases of
grade IV). The baseline clinical characteristics of the included
patients are listed in Table I.
Therefore, no statistical differences were obtained between
patients with low- and high-grade ccRCC.
 | Table I.Baseline clinical characteristics of
the study cohort. |
Table I.
Baseline clinical characteristics of
the study cohort.
| Characteristics | Low-grade (grade I
and II; N=40) | High-grade (grade III
and IV; N=39) | T/χ2 | P-value |
|---|
| Mean age ± SD, years
(range) | 57.25±13.04
(36–80) | 60.33±10.45
(27–79) | 1.158 | 0.250 |
| Sex, n (%) |
|
| 1.764 | 0.241 |
| Male | 23 (57.5) | 28 (71.8) |
|
|
|
Female | 17 (42.5) | 11 (28.2) |
|
|
| Side, n (%) |
|
| 1.143 | 0.736 |
| Left | 19 (47.5) | 17 (43.6) |
|
|
|
Right | 20 (50.0) | 22 (56.4) |
|
|
| Both
sides | 1 (2.5) | 0 (0.0) |
|
|
| Grade, n (%) |
|
|
|
|
| I | 6 (15.0) | - |
|
|
| II | 34 (85.0) | - |
|
|
|
III | - | 25 (64.1) |
|
|
| IV | - | 14 (35.9) |
|
|
Model performance
The VGG-16 model was used for binary classification,
and the particular experimental results are presented in Table II. Among the investigated imaging
parameters, MK achieved the highest accuracy, followed by Kr and
Ka. The precision, recall, F1-score, and accuracy values for MK
were 81.48, 76.04, 74.08, and 76.04%, respectively. In addition,
those for Kr were 75.51, 75.36, 75.42, and 75.36%, respectively.
Finally, for Ka the corresponding rates for precision, recall,
F1-score and accuracy were 81.39, 71.81, 68.31, and 71.81%,
respectively.
 | Table II.Results of the binary classification
experiment for clear cell renal cell carcinoma. |
Table II.
Results of the binary classification
experiment for clear cell renal cell carcinoma.
| Imaging
parameter | Precision, % | Recall, % | F1-score, % | Accuracy, % |
|---|
| ADC | 69.96 | 69.32 | 65.99 | 69.32 |
| DKI/FA | 71.21 | 71.35 | 70.94 | 71.35 |
| DKI/Ka | 81.39 | 71.81 | 68.31 | 71.81 |
| DKI/Kr | 75.51 | 75.36 | 75.42 | 75.36 |
| DKI/MD | 78.13 | 71.74 | 65.36 | 71.74 |
| DKI/MK | 81.48 | 76.04 | 74.08 | 76.04 |
| DWI | 70.79 | 70.24 | 70.44 | 70.24 |
Discussion
The current study aimed to evaluate the
effectiveness of different MRI sequences for a binary DL model that
was established for the preoperative pathological grading of ccRCC.
Among the MRI-based approaches applied, DKI/MK displayed the
highest grading accuracy. The best-performing DL model showed
strong diagnostic performance, with accuracy and precision rates of
76.0 and 81.5% respectively. These results indicated that the DL
method was effective for ccRCC grading evaluation, thus offering an
non-invasive and valuable approach for ccRCC grading, and providing
a significant tool that could support clinicians in guiding
individualized treatment strategies. In recent decades, several
studies have investigated the association between DWI, DKI, and
radiomics, and the pathological grading of ccRCC, thus supporting
the increasing attention in this field.
DWI is a non-invasive method used to detect the
diffusion movement of water molecules within living tissues. The
net movement of water molecules can be quantified using the
apparent ADC value (15). It has
been reported that the ADC values of ccRCC tends to decrease as the
pathological grading increases (2,11). DKI
is an advanced functional MRI technology developed based on DWI,
incorporating a non-Gaussian model. This method is based on the
fact that water molecule diffusion in living tissues follows a
non-Gaussian distribution due to the effect of different factors,
such as cell membranes and intercellular organelles. DKI can
provide a more accurate assessment of microstructure changes via
calculating the deviation from the Gaussian distribution between
real and ideal diffusion states (2,16). The
kurtosis parameters of DKI include MK, Ka and Kr, while the
diffusion parameters include MD, FA, axial diffusivity, and radial
diffusivity (17).
MK represents the mean value of diffusion kurtosis
values measured in each direction. It serves as an average index of
the limitations experienced by water molecule diffusion within
tissues, thereby indirectly reflecting the complexity of the tissue
structure (12). Additionally, Kr
and Ka are used to elucidate tissue diffusion patterns in distinct
orientations, with Kr reflecting the mean kurtosis along the
principal axis of the diffusion tensor, and Ka perpendicular to the
principal axis. Both metrics are considered as indicative markers
of tissue complexity (18).
However, there are only a few reports on the
application of DKI in ccRCC classification. Wu et al
(14) explored the potential of DKI
in grading ccRCC among 91 patients. The results showed that MK
could more accurately grade ccRCC. These findings were consistent
with the results of the present study. In addition, Cao et
al (2) evaluated 89 patients
with confirmed ccRCC using DKI on a 3-T MRI scanner. Consistent
with the results of the current study, the above study reported
that MK exhibited the highest area under the curve (AUC).
In addition, Ye et al (13) utilized a 3.0-T MRI scanner to
examine 148 patients with ccRCC, using the DKI approach with three
b values (0, 1,000, and 2,000 s/mm2) and 30 diffusion
directions. The results demonstrated that the AUC value for MD was
the highest, thus suggesting that MD could be the most valuable
parameter for grading ccRCC using DKI. Similarly, Zou et al
(12) evaluated 108 patients with
confirmed ccRCC with DKI (b values, 0, 1,000, and 2,000
s/mm2) utilizing a 3.0-T MRI scanner. The results of
this study consistently showed that the MD values displayed the
highest diagnostic performance for ccRCC grading. Additionally, in
the study by Cheng et al (9), a total of 65 patients with
pathologically confirmed ccRCC were assessed using DKI sequences
with three b values (0, 500, 1,000 s/mm2), also using a
3.0-T MRI scanner. Therefore, this study also showed that MD values
exhibit a good diagnostic efficiency for ccRCC grading. However,
the results of the aforementioned studies were not consistent with
those observed in the current study. This could be due to the
following reasons: i) MK indirectly reflects the complexity of the
tissue structure, while MD characterizes the overall diffusion of
water molecules. In high-grade ccRCC, the presence of necrotic
areas can lead to increased restriction on water molecule
diffusion, potentially resulting in falsely enhanced MD values.
This could obscure the actual microstructural differences resulting
from cellular density and architectural disorders, thus
contributing to the superior performance of MK compared with MD;
ii) In the study by Cheng et al (9) a maximum b value of only 1,000
s/mm2 was employed, which could be suboptimal.
Rosenkrantz et al (10)
suggested that in order to ensure reliable results, the maximum b
value for kidney DKI should not be less than 1,500
s/mm2; and iii) In the aforementioned studies, including
the current one, the majority of patients with ccRCC undergone
radical or partial nephrectomy. Therefore, the number of cases with
high-grade and advanced ccRCC was relatively small, while the
sample size between the two groups was quite different, thus
introducing selection bias and affecting the comparisons between
groups.
Radiomics, a significant scientific field, is
associated with the extraction, analysis, and interpretation of
quantitative imaging parameters. It utilizes computer
post-processing technology to transform conventional imaging data
into high-dimensional and quantitative imaging data. Core
radiomic-related methods include texture analysis (TA), machine
learning (ML), and DL (4). TA is a
quantitative imaging technique that is used to extract additional
information from the grayscale distribution of pixels or voxels
within medical images, thus providing quantitative statistical
parameters (19). ML and DL,
subfields of artificial intelligence, primarily focus on developing
algorithms that can learn from and improve via data analysis
without requiring explicit programming in advance (20). ML models commonly use TA-derived
parameters as input features, thus enhancing the sensitivity of
medical imaging-mediated diagnosis (21). By contrast, DL models do not require
manual feature selection, as the algorithm itself can autonomously
determine which features are the most suitable for model
development (22). As a form of
radiomics, DL algorithm can automatically learn and extract complex
patterns and features from data via using multi-level neural
network models, thus enabling efficient data analysis, prediction
and decision making (8). Although
several studies have been conducted on the pathological grading of
ccRCC using MRI-based approaches (7,23,24),
to the best of our knowledge, no studies have yet employed the
DKI-based approach.
In a previous study, Pan et al (24) extracted MRI texture features from 89
patients with ccRCC. Therefore, conventional sequences, such as
T1-weighted imaging (T1WI), T2WI and contrast-enhanced T1WI
(CE-T1WI) were classified as conventional MRI (cMRI), while
Dixon-MRI, blood oxygen level-dependent MRI and SWI were classified
as functional MRI (fMRI). The accuracy rates of cMRI and fMRI were
49.56 and 70.80%, respectively. However, their approach primarily
relied on manually defined features, and therefore not all imaging
data could be captured. By contrast, DL can artificially outline
regions of interest (ROI), and automatically extract image
features, thus compensating for the lack of TA (23). Chen et al (7) combined T1WI, T2WI, CE-T1WI, and DWI
sequences to establish a ML model for the pathological grading of
ccRCC, including 99 patients. The training and validation
accuracies were 95.9 and 86.2%, respectively. However, ML depends
on manual mapping of ROIs, which can result in manual errors in the
boundary of the lesion. By contrast, DL can directly identify the
characteristics of the lesion from the model. In addition, Zhao
et al (23) used T1-enhanced
and T2WI sequences to establish DL models using data from 430
patients with ccRCC. The reported accuracy rates for T1 enhanced,
T2WI, and their combination were 71, 71, and 90%, respectively. In
the present study, the accuracy of the established DL model, using
the DKI/MK, DKI/Kr, DKI/Ka, DKI/FA, and DKI/MD parameters, was
higher compared with that reported for the individual sequence
model by Zhao et al (23).
Different pathological grades of ccRCC can be
associated with different surgical approaches and prognoses
(25,26). Currently, preoperative biopsy
remains the gold standard for determining the pathological grade of
ccRCC. However, biopsy is an invasive examination, which is
associated with particular risk factors, such as tumor seeding and
sampling error (5). By contrast, DL
models can predict the pathological grade of ccRCC under
non-invasive conditions, thus assisting clinicians in developing
personalized treatment strategies for patients with ccRCC.
However, this study has several limitations.
Firstly, the number of patients with ccRCC included in the study
was relatively small. Therefore, a multicenter study with a larger
patient cohort should be performed in the future. In addition, due
to the limited number of patients with grade I and IV ccRCC, binary
classification was employed. Future studies with more patients of
this grade and a four-classification approach should be considered
to improve the specificity of pathological grading.
Overall, the present study validated a DL-based
model using DWI and DKI imaging to accurately and non-invasively
distinguish between low- and high-grade ccRCC. Therefore, this
model could assist clinicians in guiding individualized clinical
treatment of patients with ccRCC under non-invasive conditions.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JC and WZ conceived and designed the experiments.
XL, YaL, CX, ZD and WZ performed the experiments. KL, YuL, XS, DK
and PS analyzed the data. JC, CX and ZD contributed to the
provision of materials and analysis tools. YuL, XS and WZ confirm
the authenticity of all the raw data. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Medical
Ethics Committee of Zibo Central Hospital (approval no. 2017001;
Zibo, China). Written informed consent was obtained from all
patients prior to enrollment.
Patient consent for publication
All patients or their next of kin consented to
publication of the article.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li Q, Zeng K, Chen Q, Han C, Wang X, Li B,
Miao J, Zheng B, Liu J, Yuan X and Liu B: Atractylenolide I
inhibits angiogenesis and reverses sunitinib resistance in clear
cell renal cell carcinoma through ATP6V0D2-mediated autophagic
degradation of EPAS1/HIF2α. Autophagy. 21:619–638. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cao J, Luo X, Zhou Z, Duan Y, Xiao L, Sun
X, Shang Q, Gong X, Hou Z, Kong D and He B: Comparison of
diffusion-weighted imaging mono-exponential mode with diffusion
kurtosis imaging for predicting pathological grades of clear cell
renal cell carcinoma. Eur J Radiol. 130:1091952020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fan X, Fu F, Liang R, Xue E, Zhang H, Zhu
Y and Ye Q: Associations between contrast-enhanced ultrasound
features and WHO/ISUP grade of clear cell renal cell carcinoma: A
retrospective study. Int Urol Nephrol. 56:1157–1164. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li Q, Liu YJ, Dong D, Bai X, Huang QB, Guo
AT, Ye HY, Tian J and Wang HY: Multiparametric MRI radiomic model
for preoperative predicting WHO/ISUP nuclear grade of clear cell
renal cell carcinoma. J Magn Reson Imaging. 52:1557–1566. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choi JW, Hu R, Zhao Y, Purkayastha S, Wu
J, McGirr AJ, Stavropoulos SW, Silva AC, Soulen MC, Palmer MB, et
al: Preoperative prediction of the stage, size, grade, and necrosis
score in clear cell renal cell carcinoma using MRI-based radiomics.
Abdom Radiol (NY). 46:2656–2664. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rizzo A, Racca M, Dall'Armellina S,
Rescigno P, Banna GL, Albano D, Dondi F, Bertagna F, Annunziata S
and Treglia G: The emerging role of PET/CT with PSMA-Targeting
radiopharmaceuticals in clear cell renal cancer: An updated
systematic review. Cancers (Basel). 15:3552023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen XY, Zhang Y, Chen YX, Huang ZQ, Xia
XY, Yan YX, Xu MP, Chen W, Wang XL and Chen QL: MRI-based grading
of clear cell renal cell carcinoma using a machine learning
classifier. Front Oncol. 11:7086552021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yasaka K, Kamagata K, Ogawa T, Hatano T,
Takeshige-Amano H, Ogaki K, Andica C, Akai H, Kunimatsu A, Uchida
W, et al: Parkinson's disease: Deep learning with a
parameter-weighted structural connectome matrix for diagnosis and
neural circuit disorder investigation. Neuroradiology.
63:1451–1462. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cheng Q, Ren A, Xu X, Meng Z, Feng X,
Pylypenko D, Dou W and Yu D: Application of DKI and IVIM imaging in
evaluating histologic grades and clinical stages of clear cell
renal cell carcinoma. Front Oncol. 13:12039222023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rosenkrantz AB, Padhani AR, Chenevert TL,
Koh DM, De Keyzer F, Taouli B and Le Bihan D: Body diffusion
kurtosis imaging: Basic principles, applications, and
considerations for clinical practice. J Magn Reson Imaging.
42:1190–1202. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu Q, Zhu W, Wu J, Chen W, Ye J and Ling
J: Comparative study of conventional diffusion-weighted imaging and
introvoxel incoherent motion in assessment of pathological grade of
clear cell renal cell carcinoma. Br J Radiol. 95:202104852022.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zou J, Ye J, Zhu W, Wu J, Chen W, Chen R
and Zhu Q: Diffusion-weighted and diffusion kurtosis imaging
analysis of microstructural differences in clear cell renal cell
carcinoma: A comparative study. Br J Radiol. 96:202301462023.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ye J, Xu Q, Wang SA, Zheng J and Dou WQ:
Quantitative evaluation of intravoxel incoherent motion and
diffusion kurtosis imaging in assessment of pathological grade of
clear cell renal cell carcinoma. Acad Radiol. 27:e176–e182. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu G, Zhao Z, Yao Q, Kong W, Xu J, Zhang
J, Liu G and Dai Y: The study of clear cell renal cell carcinoma
with MR diffusion kurtosis tensor imaging and its histopathologic
correlation. Acad Radiol. 25:430–438. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meyer HJ, Martin M and Denecke T: DWI of
the Breast-possibilities and limitations. Rofo. 194:966–974. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li S, He K, Yuan G, Yong X, Meng X, Feng
C, Zhang Y, Kamel IR and Li Z: WHO/ISUP grade and pathological T
stage of clear cell renal cell carcinoma: Value of ZOOMit diffusion
kurtosis imaging and chemical exchange saturation transfer imaging.
Eur Radiol. 33:4429–4439. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Duan Z, Tao J, Liu W, Liu Y, Fang S, Yang
Y, Liu X, Deng X, Song Y and Wang S: Correlation of IVIM/DKI
parameters with hypoxia biomarkers in fibrosarcoma murine models:
Direct control of MRI and pathological sections. Acad Radiol.
31:1014–1023. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song Q, Dong W, Tian S, Xie L, Chen L, Wei
Q and Liu A: Diffusion kurtosis imaging with multiple quantitative
parameters for predicting microsatellite instability status of
endometrial carcinoma. Abdom Radiol (NY). 48:3746–3756. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Litvin AA, Burkin DA, Kropinov AA and
Paramzin FN: Radiomics and digital image texture analysis in
oncology (review). Sovrem Tehnologii Med. 13:972021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li D, Hu J, Zhang L, Li L, Yin Q, Shi J,
Guo H, Zhang Y and Zhuang P: Deep learning and machine
intelligence: New computational modeling techniques for discovery
of the combination rules and pharmacodynamic characteristics of
Traditional Chinese Medicine. Eur J Pharmacol. 933:1752602022.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hügle T, Boedecker J, van Laar JM,
Boedecker J and Hügle T: Applied machine learning and artificial
intelligence in rheumatology. Rheumatol Adv Pract. 4:rkaa0052020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yousef R, Gupta G, Yousef N and Khari M: A
holistic overview of deep learning approach in medical imaging.
Multimed Syst. 28:881–914. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao Y, Chang M, Wang R, Xi IL, Chang K,
Huang RY, Vallières M, Habibollahi P, Dagli MS, Palmer M, et al:
Deep learning based on MRI for differentiation of Low- and
High-Grade in Low-stage renal cell carcinoma. J Magn Reson Imaging.
52:1542–1549. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pan L, Chen M, Sun J, Jin P, Ding J, Cai
P, Chen J and Xing W: Prediction of Fuhrman grade of renal clear
cell carcinoma by multimodal MRI radiomics: A retrospective study.
Clin Radiol. 79:e273–e281. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Lih TM, Dhanasekaran SM, Mannan R,
Chen L, Cieslik M, Wu Y, Lu RJ, Clark DJ, Kołodziejczak I, et al:
Histopathologic and proteogenomic heterogeneity reveals features of
clear cell renal cell carcinoma aggressiveness. Cancer Cell.
41:139–163.e17. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nezami BG and MacLennan GT: Clear cell
renal cell carcinoma: A comprehensive review of its histopathology,
genetics, and differential diagnosis. Int J Surg Pathol.
33:265–280. 2025. View Article : Google Scholar : PubMed/NCBI
|