Introduction
Colorectal cancer (CRC) is the third most diagnosed
cancer and the third leading cause of cancer-related death
worldwide for both sexes combined (1). In the U.S, it is the second most
common cancer in men under 50 years of age, with an estimated total
in 2023 of 153,020 diagnosed cases and 52,550 deaths, including
19,550 cases and 3,750 deaths in individuals younger than 50 years
(2). In the E.U., CRC accounts for
520,000 new cases (12.9% of all cancer diagnoses) and 250,000
deaths (12.6% of cancer-related mortality) (3). The management of metastatic CRC (mCRC)
has advanced, particularly with monoclonal antibodies targeting the
epidermal growth factor receptor (anti-EGFR mAb) combined with
5FU-based chemotherapy. Anti-EGFR mAb therapy requires the absence
of somatic mutations in exons 2, 3, and 4 of KRAS and NRAS genes
(4). BRAF gene mutations are
routinely assessed for their prognostic significance and their
association with new therapeutic strategies combining anti-EGFR
mAbs and BRAF kinase inhibitors (5).
Tissue-based biopsy remains the gold standard method
for molecular analysis of cancer, but its invasiveness and
potential complications limit its frequent use (6).
In France, somatic mutations in KRAS, NRAS,
and BRAF are primarily analysed on INCa (Institut National
du Cancer-French Cancer Institute) certified molecular genetics
platforms (INCa platform) using formalin-fixed paraffin embedded
(FFPE) tissue from biopsies of primary or secondary lesions or
surgically excised primary tumours (7). Given the limitations of tissue-based
biopsies and the dynamic genetic evolution of tumours, there is a
growing demand for a less invasive and more precise alternative
like liquid biopsy. In real-world settings, challenges such as
insufficient sample quantity or quality for genotyping, or the
inability to retrieve specimens from external centres, further
underscore this need. Monocentric prospective biomarker studies
have demonstrated the clinical utility of circulating tumour DNA
(ctDNA) as a valuable marker in first-line mCRC treatment (8) and in primary CRC at surgery and during
post-surgery follow-up (9).
However, without standardized workflows, liquid biopsy must be
validated against standard-of-care tissue testing in real-world
settings for routine tumour molecular profiling and identification
of treatment response biomarkers, such as KRAS, NRAS, and
BRAF mutations in CRC.
Several ctDNA analysis techniques exist, with
studies confirming the feasibility of liquid biopsy for KRAS and
BRAF genotyping (10,11). Yet, few prospective real-world
studies have been conducted. The BEAMing (Beads, Emulsion,
Amplification and Magnetics) technique, a reference method used in
re-analyses of historical trials like CRYSTAL (12) and OPUS (13), and studies, such as FIRE3 (14).
The ColoBEAM protocol evaluated the real-world
feasibility of liquid biopsy as a standard for detecting KRAS,
NRAS, and BRAF mutations, comparing genotyping results
from blood samples with those from routine FFPE tissue
specimens.
Materials and methods
Patients
A total of 278 patients, aged 18 or older, with
pathologically confirmed mCRC were enrolled in this study from
March 2016 to May 2017 at 8 medical centres (Institut de
Cancérologie de Lorraine, Vandœuvre-lès-Nancy, France; Polyclinique
de Gentilly, Nancy, France; Hôpital Belle-Isle-Metz, Metz, France;
Centre Paul Strauss, Strasbourg, France; CHU Reims Hôpital Robert
Debré, Reims, France; CH Auxerre, Auxerre, France; CH Chalon Sur
Saône-William Morey, Chalon sur Saône, France; CH Besançon-Hôpital
Jean Minjoz, Besançon, France). Eligible patients were adults (≥18
years) diagnosed with metastatic colorectal cancer. Inclusion
required that a molecular analysis of KRAS, NRAS, and
BRAF mutations be clinically indicated as part of routine
disease management. All participants provided written informed
consent prior to enrolment, and were covered by the French national
health insurance system. Patients with metastatic colorectal cancer
(mCRC) were eligible for inclusion if they had not received prior
anti-EGFR monoclonal antibody therapy. This criterion was
established to ensure that the evaluation of RAS/BRAF mutational
status by BEAMing in plasma would reflect the untreated molecular
profile. As prior exposure to anti-EGFR agents may induce clonal
selection and alter the ctDNA landscape, excluding previously
treated patients was necessary to avoid potential confounding
effects on concordance analyses between tissue and liquid biopsy
results (15).
Exclusion criteria included non-metastatic CRC at
the time of initial tissue biopsy, local recurrence, exclusive
nodal metastases, contraindications to a 30 ml blood draw, receipt
of a blood transfusion within 15 days prior to blood collection,
other malignant tumours within the past 5 years, and pregnancy or
breastfeeding, and prior receipt of anti-EGFR therapy (n=4). Of the
included patients, 25 had no detectable metastases at the time of
blood sampling due to prior resection of metastatic lesions (e.g.
liver or lung metastases). Blood samples were collected without
strict timing constraints, including from patients who had received
chemotherapy or radiotherapy, to reflect real-world clinical
conditions.
The research protocol was approved by the Ethics
Committee (CPP Est III, Nancy, France; number 15.09.09), and
written informed consent was obtained from all patients. The
protocol was registered at ClinicalTrials.gov (NCT02751177).
Procedures
DNA extracted from FFPE tissue, collected at the
time of the initial biopsy, was analysed for KRAS, NRAS and
BRAF mutations using PCR or next-generation sequencing (NGS)
assays at INCa platform, as per standard-of-care guidelines. At
inclusion, three 10 ml blood samples were collected in DNA BCT
tubes (Streck, La Vista, NE, USA). Sample collection lacked strict
timing constraints and, in some instances, occurred long after the
initial biopsy to reflect real-world conditions. The samples were
shipped at room temperature to the Biopathology Department of the
Institut de Cancérologie de Lorraine for cell-free DNA (cfDNA)
extraction and centralized analysis, hereafter referred to as ‘ICL
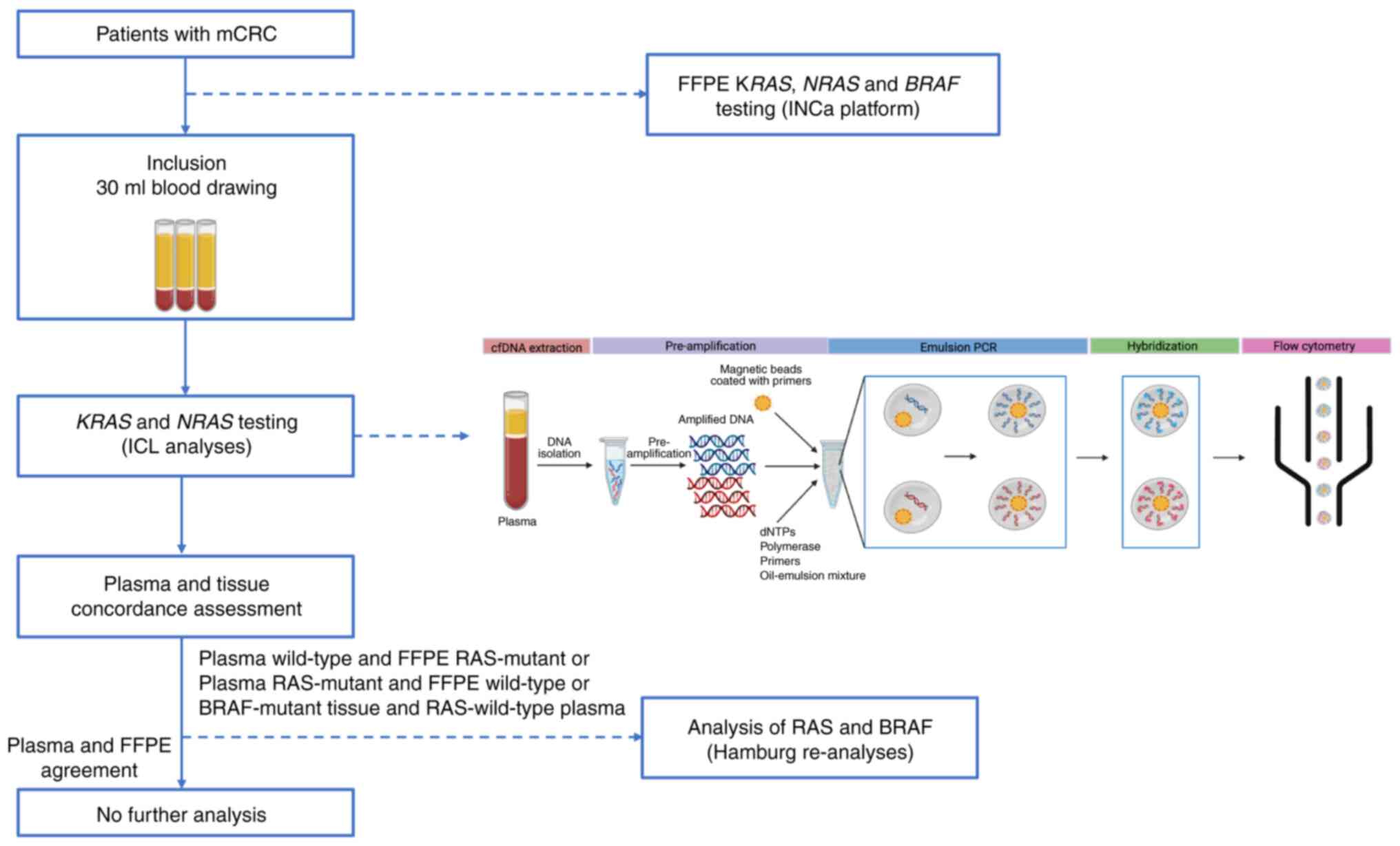
analyses’ (Fig. 1). Plasma was
obtained through double centrifugation (10 min at 1,600 × g,
followed by 10 min at 6,000 × g) and stored at −80°C until ctDNA
analysis. cfDNA was then extracted from 3–4 ml of plasma using the
QIAamp circulating nucleic acid kit (Qiagen, Hilden, Germany) and
analysed with the OncoBEAM™ assay (Sysmex, Norderstedt, Germany).
Plasma-derived RAS mutation results were then compared with
tissue-based results.
To ensure the robustness of mutational status
assignment and to resolve discordant cases, an independent blinded
re-analysis was conducted by the Sysmex service laboratory in
Hamburg-hereafter referred to as the ‘Hamburg re-analyses.’ This
laboratory served a dual role: it provided an external confirmation
of BRAF status (V600E mutation) for all relevant samples, and acted
as a fully blinded reference centre for the assessment of RAS
mutations, enabling an unbiased comparison with initial local
results. None of the named authors on the paper are affiliated with
Sysmex Inostics Inc., and the company did not have any input into
the planning or design of the experiments. The re-analysis focused
specifically on samples displaying discrepancies between tissue and
plasma results, including cases where i) the tumour tissue was
RAS-mutant but the corresponding plasma was wild-type (potential
false negatives), ii) the tumour was BRAF-mutant while plasma was
wild-type (potential false negatives), and iii) the tissue was RAS
wild-type but the plasma showed a RAS mutation (potential false
positives). For each of these cases, DNA was re-extracted from the
original FFPE tumour samples and ctDNA was isolated from a second
blood sample collected at the time of patient inclusion. Both DNA
sources were analysed in Hamburg using the BEAMing technology for
RAS and BRAF mutations. In the event of new discrepancies between
the initial analyses (‘ICL analyses’) and the Hamburg re-analyses,
the latter were considered the reference due to their blinded
nature and standardized quality procedures. The reconciled mutation
calls derived from this process were used as the final BEAMing
results in all statistical analyses, with FFPE tumour genotyping
from the INCa-certified platform consistently considered the gold
standard for tissue mutation status. This rigorous, multi-source
validation framework, based on blinded external testing with a
reference technology, ensured the reliability and clinical
relevance of the final dataset, particularly in evaluating the
performance of liquid biopsy in routine practice.
In accordance with routine clinical management of
metastatic colorectal cancer (mCRC), treatment response was
evaluated after three months using radiological imaging and
clinical assessment.
Data collection
All clinical and biological data at inclusion, along
with RAS and BRAF testing results, were recorded in
an electronic case report form (CleanWeb, Telemedicine
Technologies, Boulogne-Billancourt, France).
Samples analysis workflow
The sample analysis strategy is outlined in Fig. 1. All FFPE samples were analysed
using standard-of-care protocols established by INCa-certified
platforms, employing PCR-based or NGS-based assays (16–18).
RAS and BRAF results obtained from FFPE samples were
blinded to laboratory personnel during data analysis.
The BEAMing assay was used to analyse cfDNA samples,
targeting 34 mutations in codons 12, 13, 59, 61, 117, and 146 of
KRAS and NRAS gene. In brief, cfDNA underwent
pre-amplification, followed by emulsion PCR and hybridization, with
prepared samples analysed by flow cytometry per the manufacturer's
protocol.
Statistical analysis
Final BEAMing results for plasma samples were
determined after reconciling ICL analyses with Hamburg re-analyses.
FFPE tissue results from INCa-certified platforms served as the
gold standard. Mutation carriers were defined as patients with at
least one detected mutation in the KRAS, NRAS, or
BRAF genes. The sensitivity of the BEAMing test was
calculated as the proportion of mutation carriers identified by the
BEAMing test relative to those identified in FFPE samples, reported
with a 95% confidence interval.
Similarly, the specificity of the BEAMing assay was
calculated as the proportion of patients classified as non-mutation
carriers in plasma relative to those classified as non-mutation
carriers in FFPE samples, also reported with a 95% confidence
interval.
Results
Patient inclusion and sample
availability
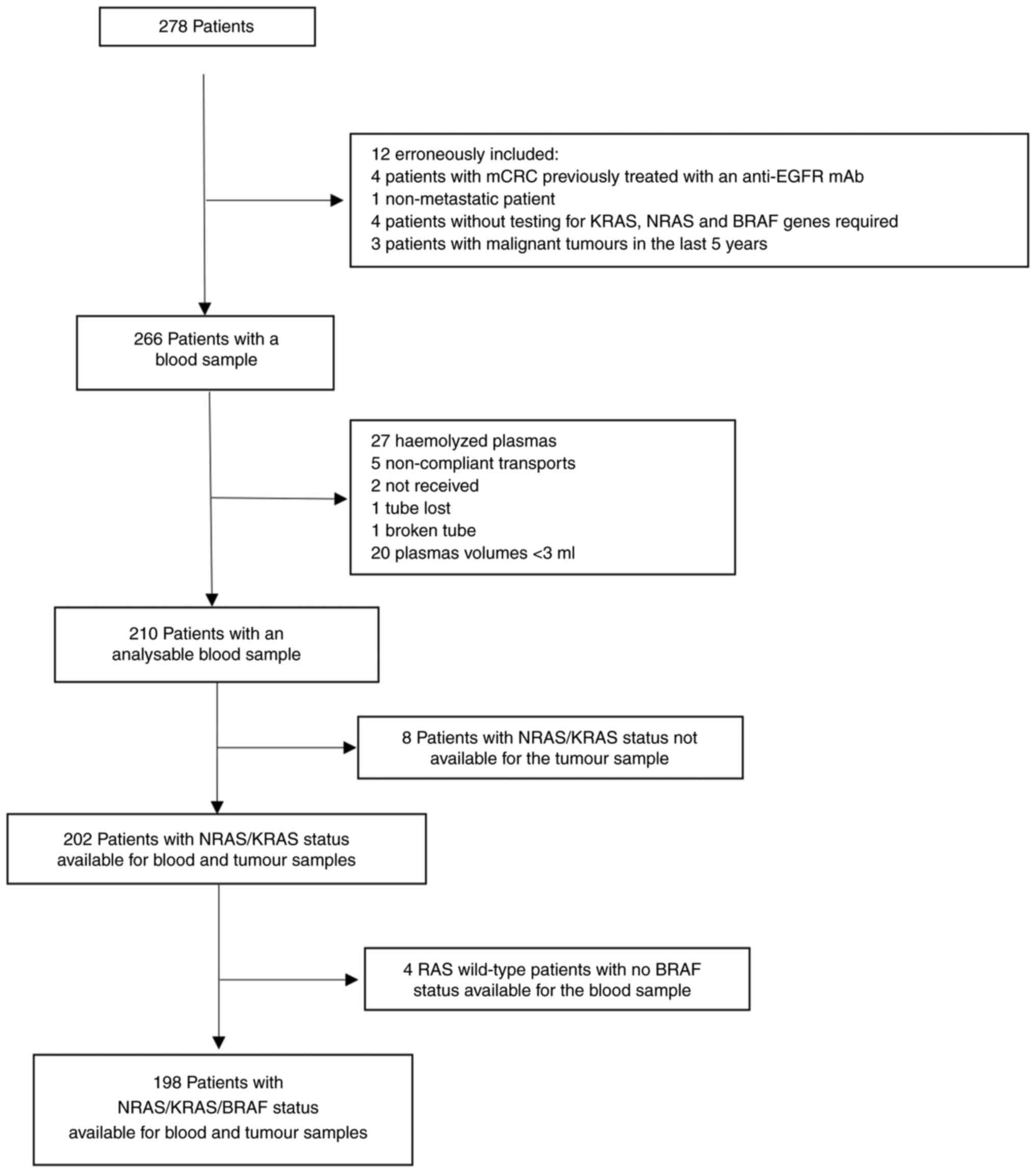
A total of 278 patients were initially enrolled in
the study. Twelve patients were excluded due to erroneous inclusion
(Fig. 2), resulting in a corrected
cohort of 266 patients. Among them, 56 blood samples were not
analysable due to failure to meet quality or processing criteria.
Of the remaining 210 patients with analysable blood samples,
KRAS/NRAS status from tumour tissue was unavailable in 8 cases.
Consequently, 202 patients had complete RAS genotyping data
available for both blood and tumour samples. Within this subgroup,
BRAF mutation status in plasma was unavailable for 4 patients,
resulting in 198 patients with complete RAS and BRAF data for both
sample types. In summary, 202 patients were included in the final
RAS concordance analysis and 198 patients in the combined RAS/BRAF
analysis (Fig. 2).
Discordant cases and external
re-analysis
After local testing (‘ICL analyses’), discordances
between plasma and tumour samples were identified in 50 patients
for NRAS status (refer to Data S1,
and Tables SI, SII and SIII). Among these, 46 patients had RAS
mutations detected in tumour tissue but classified as wild-type in
plasma (false negatives), and 4 patients had wild-type RAS in
tissue but were called mutated in plasma (false positives).
Additionally, 16 patients initially classified as RAS wild-type in
plasma by ICL were reclassified as RAS-mutated based on Hamburg
re-analyses.
Regarding the five BRAF-mutated tumour samples, BRAF
mutation status in plasma was confirmed in two cases, while data
were not available for two others (refer to Data S1).
Sample and tumour characteristics
Patient characteristics at the time of blood
sampling are detailed in Table I,
and the tumour sample availability is summarized in Table II. Among the 202 patients analysed,
the timing of blood collection relative to the initial tumour
biopsy was distributed as follows: 15.8% (n=32) within one month,
10.9% (n=22) between 1 and 3 months, 14.4% (n=29) between 3 months
and 1 year, 21.8% (n=44) between 1 and 2 years, 18.8% (n=38)
between 2 and 3 years, and 18.3% (n=37) three or more years after
biopsy.
 | Table I.Study population characteristics at
the time of blood sampling of the 202 analysed patients. |
Table I.
Study population characteristics at
the time of blood sampling of the 202 analysed patients.
| Variable | Value |
|---|
| Median age,
yearsa | 67.0
(58.0–76.0) |
| Weight, kg
(median)a | 72.0
(62.0–83.0) |
|
Missing | 5 |
| Location, n
(%) |
|
|
Colon | 110 (54.5) |
|
Left | 18 (16.4) |
|
Right | 50 (45.5) |
|
Hinge | 34 (30.9) |
|
Transverse | 8 (7.3) |
|
Rectosigmoid junction | 30 (14.9) |
|
Rectum | 59 (29.2) |
|
Other | 3 (1.5) |
| Presence of
metastasis at the time of blood sampling, n (%) |
|
| No | 25 (12.4) |
|
Yes | 177 (87.6) |
| Number of
metastatic sites, n (%) |
|
| 0 | 25 (12.4) |
| 1 | 78 (38.6) |
|
>1 | 99 (49.0) |
| Metastasis location
(non-exclusive), n (%) (n=177) |
|
|
Liver | 127 (62.9) |
|
Pulmonary | 94 (46.5) |
|
Peritoneal | 45 (22.3) |
| Ganglionary | 36 (17.8) |
|
Bone | 10 (5.0) |
|
Brain | 3 (1.5) |
|
Other | 15 (7.4) |
|
Chemotherapyb, n (%) |
|
|
Naive | 59 (29.2) |
|
Undergoing | 94 (46.5) |
|
Finished | 49 (24.3) |
| Chemotherapy
administration, n (%) (n=143) |
|
| Within
15 days of blood collection | 29 (20.9) |
| >15
days from blood collection | 110 (79.1) |
|
Missing | 4 |
| Primary tumour, n
(%) |
|
|
Present | 70 (34.7) |
|
Eradicated after
treatment | 132 (65.3) |
 | Table II.Tumour sample description
(n=202). |
Table II.
Tumour sample description
(n=202).
| Variable | Value |
|---|
| Nature of tissue, n
(%) |
|
|
Biopsy | 89 (44.1) |
| Surgery
specimen | 113 (55.9) |
| Tumour type, n
(%) |
|
| Primary
tumour | 164 (81.2) |
|
Metastasis | 38 (18.8) |
| Tumour cell
content, %a | 50 (30–70) |
| Routine results
from INCa platform, n (%) |
|
| Known
mutation status (FFPE tissue) | 202 (100) |
| KRAS
or NRAS gene mutation |
|
|
Mutated | 132 (65.4) |
|
Wild-type | 70 (34.6) |
|
BRAF gene mutation |
|
|
Mutated | 5 (2.5) |
|
Wild-type | 197 (97.5) |
|
KRAS/NRAS/BRAF gene mutation |
|
|
Mutated | 137 (67.8) |
|
Wild-type | 65 (32.2) |
Primary endpoint and overall
performance
The comparison of BEAMing plasma genotyping results
with tumour tissue genotyping from INCa-certified platforms is
presented in Table III. The
study's primary objective-evaluating the accuracy of BEAMing for
detecting KRAS, NRAS, and BRAF mutations using tumour genotyping as
the reference-was achieved. For RAS mutation detection, the
sensitivity was 77.3%, specificity was 94.3%, and overall
concordance was 83.2%. For combined RAS and BRAF mutations,
sensitivity was 77.0%, specificity 93.7%, and concordance 82.3%.
Despite high specificity (>93%) across analyses, overall
sensitivity remained moderate when evaluating tumour vs. plasma
mutation status globally.
 | Table III.KRAS, NRAS and BRAF
mutational status in plasma samples determined by beads emulsion
and magnetic digital PCR, compared with tumour genotyping performed
on formalin-fixed paraffin-embedded samples at the French National
Cancer Institute platform, used as the reference standard. |
Table III.
KRAS, NRAS and BRAF
mutational status in plasma samples determined by beads emulsion
and magnetic digital PCR, compared with tumour genotyping performed
on formalin-fixed paraffin-embedded samples at the French National
Cancer Institute platform, used as the reference standard.
| Mutations | No. of
patients | TP, n | TN, n | FN, n | FP, n | Sensitivity, % | Specificity, % | Concordance, % | Kappa (95% CI) |
|---|
| RAS | 202 | 102 | 66 | 30 | 4 | 77.3 | 94.3 | 83.2 | 0.658
(0.557–0.759) |
| RAS +
BRAF | 198 | 104 | 59 | 31 | 4 | 77.0 | 93.7 | 82.3 | 0.634
(0.529–0.740) |
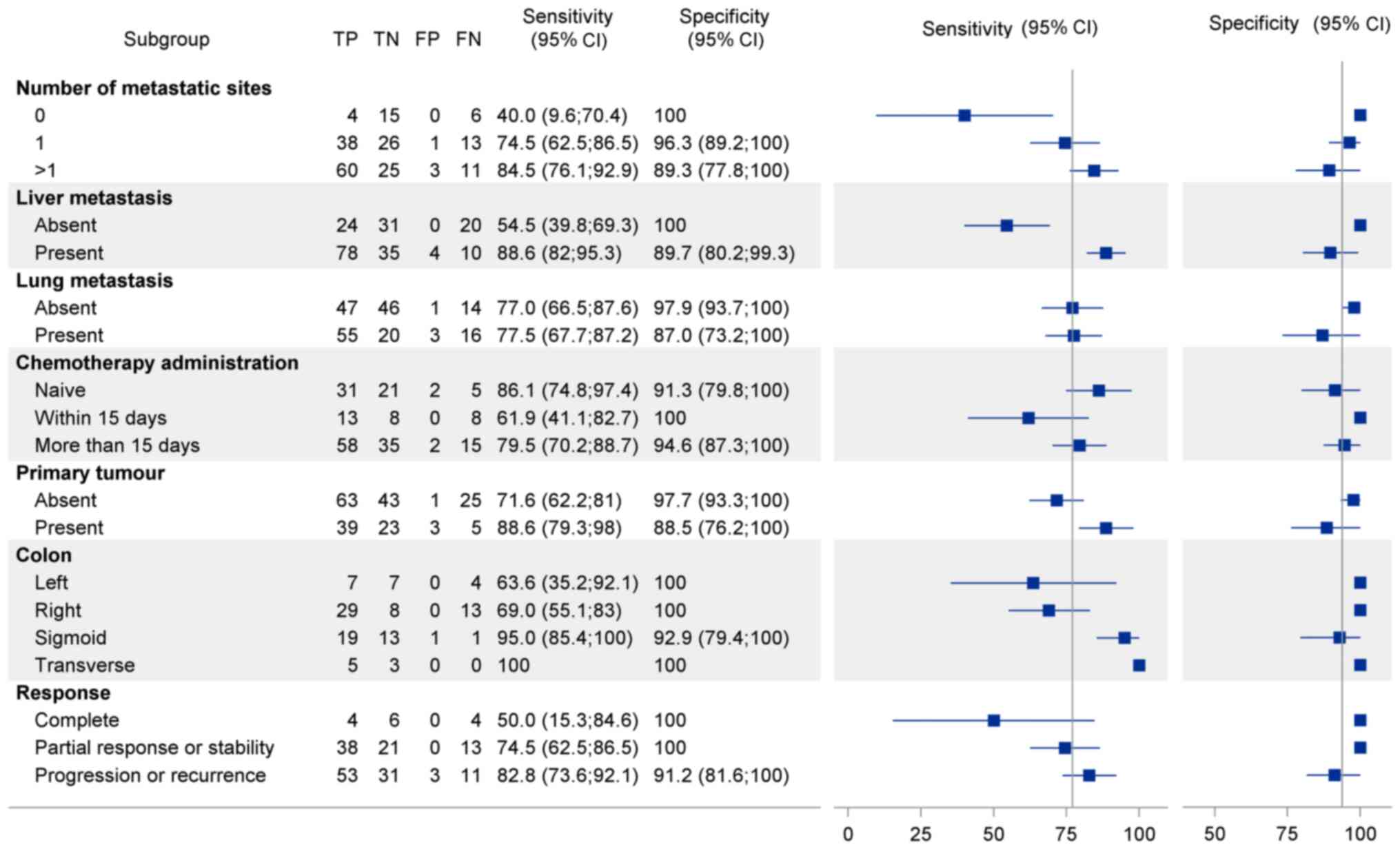
Subgroup analyses
Subgroup analyses are shown in Fig. 3 and account for key clinical
parameters at the time of blood sampling, including tumour
presence, metastatic status and location, chemotherapy exposure,
and treatment response.
Metastatic status and tumour
presence
Sensitivity improved significantly in patients with
liver metastases (sensitivity, 88.6%; specificity, 89.7%) and in
those with a visible primary tumour (sensitivity, 88.6%;
specificity, 88.5%). In contrast, patients without metastases
showed limited sensitivity (50.0%), while those with metastases had
improved sensitivity (75.0%), which increased with the number of
metastatic sites. Metastatic site analysis revealed higher
sensitivity for liver metastases (~88%) than for pulmonary
metastases (77%).
Tumour location
Sensitivity also varied with primary tumour
location: 100% for transverse colon, 95% for sigmoid, 69% for left
colon, and 63% for right colon tumours.
Chemotherapy exposure
BEAMing sensitivity was highest in
chemotherapy-naive patients (sensitivity, 86.1%; specificity,
91.3%) and declined with recent chemotherapy. Sensitivity was
acceptable (79.0%) when chemotherapy was administered more than 15
days before sampling but dropped to 61.0% when treatment occurred
within 15 days.
Treatment response
Sensitivity was highest in patients with progressive
or recurrent disease (82.8%), intermediate in those with stable
disease or partial response (74.5%), and lowest in patients
achieving complete response (50.0%). Regardless of response status,
specificity remained excellent (91.2 to 100%).
Discussion
The ColoBEAM study was designed as a real-world
study to evaluate the feasibility of using liquid biopsy to detect
KRAS, NRAS, and BRAF mutations in patients with mCRC,
potentially replacing tissue biopsy. Each patient underwent routine
tissue analysis, with plasma derived from whole blood analysed
using the OncoBEAMTM assay (19,20).
Compared to previous studies, such as the prospective multicentre
real-world comparison of OncoBEAM-based liquid biopsy and tissue
analysis for RAS mutations in mCRC (21), or a multi-institutional study
(22), the ColoBEAM study provides
additional insights by simultaneously assessing RAS and
BRAF mutations in ctDNA from mCRC patients.
Of the 278 enrolled patients, data from 202 patients
with RAS status and 198 patients with both RAS and
BRAF status were analysed (4 patients with wild-type
RAS lacked BRAF status in plasma samples, as shown in
Fig. 2) after exclusions. To assess
concordance, results were compared, with discordant cases sent to
Sysmex in Germany for blinded external re-analysis. Some samples
were excluded due to quality issues, such as haemolysis during
transport.
This study evaluated BEAMing for detecting KRAS,
NRAS, and BRAF mutations in plasma from 202 CRC patients, achieving
a sensitivity of 77.3%, specificity of 94.3%, and concordance of
83.2% for RAS mutations, with similar metrics for combined RAS/BRAF
analysis (sensitivity 77.0%, specificity 93.7%, concordance 82.3%).
Compared to studies using various liquid biopsy methods, our
sensitivity is slightly lower than Bettegowda et al
(23) at 87.2% with digital PCR in
CRC, likely due to our mixed cohort including non-metastatic cases,
and Thierry et al (24) at
85% in metastatic CRC (mCRC). Focusing on BEAMing-specific studies,
García-Foncillas et al (21)
reported 93.3% concordance for OncoBEAM RAS testing in mCRC, and
Vivancos et al (25) found
89% concordance (kappa 0.770) in 236 mCRC patients, with
re-analysis improving to 92%; our lower sensitivity in
non-metastatic cases (50.0%) and post-chemotherapy settings echoes
Grasselli et al (26) on
reduced BEAMing sensitivity with lower tumour burden. The ColoBEAM
study provides unique value by simultaneously evaluating KRAS,
NRAS, and BRAF mutations in a real-world setting with a
diverse patient cohort, including non-metastatic cases, which may
explain the slightly lower concordance (83.2% for RAS mutations)
compared to Vidal et al (27) (93%) and García-Foncillas et
al (21) (89%). These studies
primarily focused on metastatic patients, whereas our broader
inclusion criteria likely introduced variability in ctDNA shedding,
impacting sensitivity, as noted in our subgroup analyses (e.g.,
50.0% sensitivity in non-metastatic cases vs. 88.6% in patients
with liver metastases). The high specificity (>93%) supports
BEAMing's reliability for ruling out mutations, as seen in Bando
et al (28) with 90.4%
positive agreement, reinforcing its utility for guiding anti-EGFR
therapy in mCRC.
Subgroup analysis revealed that the presence of
metastases aligns with existing literature, which indicates that
patients with disease progression exhibit higher ctDNA levels,
enhancing the sensitivity and effectiveness of ctDNA detection
(29–31). In our study, the concordance between
KRAS, NRAS and BRAF mutation status in ctDNA and FFPE
tissue was higher in patients with progressive disease or liver
metastases (sensitivity, 82.8%; specificity, 91.2%), consistent
with prior findings (21),
reporting greater concordance in patients with liver metastases
(94.5–94.8%) comparted to those without (83.8%; P=0.040). The ‘lung
metastasis’ subgroup, consisting of patients with unique lung
metastases only, showed the lowest concordance rate of 68.8%. The
inclusion of 25 patients with no detectable metastases at the time
of blood sampling, due to prior resection of metastatic lesions,
likely contributed to the lower overall concordance (83.2% for RAS
mutations) compared to studies focused exclusively on patients with
active metastases (27). These
patients exhibited a reduced sensitivity of 50.0% for detecting
KRAS, NRAS, and BRAF mutations in plasma (Fig. 3), likely due to decreased
circulating tumour DNA (ctDNA) shedding in the absence of
metastatic burden, as supported by prior studies (32). This finding highlights the influence
of metastatic status on the performance of liquid biopsy and
underscores the need to consider disease stage when implementing
the BEAMing assay in real-world clinical settings.
These findings have significant implications,
supporting the strategic timing of blood-based mutation testing,
ideally before treatment initiation as per current guidelines, to
maximize sensitivity and clinical utility. The differential
sensitivity in chemotherapy-naive patients vs. those receiving
chemotherapy suggests a dynamic interplay between treatment and
ctDNA release or clearance.
Although ctDNA offers high specificity, cfDNA
profiling has broader applications, reflecting diverse cellular
process in the body. This precision medicine strategy supports
liquid biopsy not only as a potential alternative to tissue biopsy
but also as a complementary tool that captures the temporal and
spatial heterogeneity of tumours. As demonstrated in lung and
breast cancer, ctDNA mutation tracking has proven effective for
guiding therapeutic adjustments throughout disease progression
(33,34).
Our findings are consistent highlighting the utility
of circulating tumour DNA (ctDNA) as a highly specific biomarker
for cancer detection, disease progression monitoring, adjuvant
therapy guidance, and potentially reducing unnecessary toxicity
(35). Notably, studies such as
CIRCULATE-Japan have established a foundation for understanding
liquid biopsy in cancer treatment and recurrence monitoring
(36).
Despite the promising results, this study
acknowledges limitations, including the influence of recent
chemotherapy and metastatic burden on test sensitivity. Another
notable limitation of our study was the exclusion of 56 out of 266
blood samples (21%) due to preanalytical issues, such as haemolysis
or insufficient blood volume, despite the use of DNA BCT tubes
designed to stabilize cfDNA. These issues were likely exacerbated
by initial sample transport at room temperature and, in some
instances, accidental freezing during shipment, which compromised
sample integrity. These challenges, observed in our multicentre
real-world setting, highlight the critical need for standardized
preanalytical protocols, including controlled transport conditions
to prevent temperature fluctuations, to ensure reliable ctDNA
analysis. Future studies should prioritize optimized sample
collection, handling, and transport workflows to minimize such
issues and enhance the clinical applicability of liquid biopsy
techniques like the BEAMing assay.
A notable finding in our study was the
reclassification of 16 patients from RAS wild-type to RAS-mutated
in plasma following centralized re-analysis in Hamburg,
highlighting differences in sensitivity between local (ICL) and
centralized analyses. Several factors may explain this discrepancy.
First, the Hamburg laboratory, as a centralized facility with
extensive experience in OncoBEAM™ assay implementation, likely
benefited from optimized workflows and greater technical expertise,
enhancing mutation detection. Second, the Hamburg re-analysis may
not have strictly followed the same procedure as the local ICL
analyses, with potential differences in assay optimization or
quality control measures. These findings underscore the importance
of centralized laboratory expertise and standardized procedures to
maximize the sensitivity of ctDNA analysis in real-world settings.
Another limitation of our study is that only discrepant cases
underwent re-analysis using BEAMing. It is therefore possible that
additional discrepancies would have been identified if all samples
had been retested, particularly in cases classified as RAS
or BRAF wild-type in tissue. The higher sensitivity of
BEAMing may have revealed ultra-low frequency subclonal mutations,
potentially leading to a reclassification and further highlighting
the complexity of tumour heterogeneity.
The timing of blood collection relative to initial
tissue biopsy may also influence the concordance between plasma and
tissue-based mutation detection, as tumour evolution or changes in
circulating tumour DNA (ctDNA) shedding could occur over time. In
our study, blood sampling occurred at varying intervals post-biopsy
(15.8% within one month, 21.8% between 1 and 2 years, and 18.3%
three or more years, as shown in Table
I), reflecting the real-world setting of the ColoBEAM study.
While we did not perform a specific statistical analysis of this
variable's impact, longer intervals may contribute to discordance
due to tumour heterogeneity or disease progression, particularly in
patients with mCRC. This observation aligns with prior studies
suggesting that temporal discrepancies between tissue and plasma
sampling can affect ctDNA detection (15). Future studies should quantify the
impact of sampling timing on concordance to optimize liquid biopsy
protocols, particularly in real-world clinical settings.
Integrating ctDNA analysis with other biomarkers,
such as microsatellite instability (MSI) status (6), could facilitate the development of
innovative checkpoint inhibitor therapies (37,38).
This synergy may advance the identification of a comprehensive set
of routine biomarkers to support personalized medicine.
In conclusion, our study demonstrates that
blood-based testing for detecting mutations in patients with mCRC
is feasible. Although the test exhibited high accuracy in
identifying mutations compared to tissue biopsies, factors such as
recent chemotherapy and tumour location may reduce its sensitivity.
However, adherence to guideline recommendations (4), may mitigate these limitations, as
testing is ideally performed before treatment initiation to
minimize the impact of factors like recent chemotherapy. ctDNA
analysis is now included in guidelines for CRC progression after
therapy, at diagnosis in patients with limited access to tissue
biopsies, or when tissue specimens are inadequate due to
insufficient quantity or quality or are unavailable for molecular
analysis (39). Despite these
limitations, our results indicate that blood-based mutation testing
provides a less invasive and valuable option in real-world
settings.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Frederick S.
Jones and Dr Dan Edelstein from Sysmex Inostics Inc (Baltimore, MD,
USA) for their support in facilitating the contractual arrangements
and organizing the Hamburg re-analyses.
Funding
The present study was supported by a research collaboration
agreement between Sysmex Inostics Inc. and the Institut de
Cancérologie de Lorraine, under which kits were provided.
Additional support came from the private research fund of the
Institut de Cancérologie de Lorraine.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
AH, AL and JLM wrote the original protocol, analysed
the data and wrote the manuscript. CG was the principal
investigator. CG, OB, MBA, JP, DS, AB, FG, ALV and CB were
investigators. CG, OB, MBA, JP, DS, AB, FG, ALV, CB and PG
contributed to data acquisition and critically revised the
manuscript for important intellectual content. MR and MH analysed
the samples. JS analysed the data and all statistics. AH and JS
confirm the authenticity of all the raw data. All authors have read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
All patients gave their informed and written consent
to participate. The study is registered under number NCT02751177
and has been approved by the ethical committee Comité de Protection
des Personnes Est (CHRU de Nancy, Hôpital de Brabois,
Vandoeuvre-Lès-Nancy, France) under the number 15.09.09, ID RCB
2015-A01272-47.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, artificial
intelligence tools (OpenAI Chat GPT 4.0) were used to improve the
readability and language of the manuscript, and subsequently, the
authors revised and edited the content produced by the artificial
intelligence tools as necessary, taking full responsibility for the
ultimate content of the present manuscript.
Glossary
Abbreviations
Abbreviations:
|
BEAMing
|
beads emulsion and magnetics digital
PCR
|
|
cfDNA
|
cell-free DNA
|
|
ctDNA
|
circulating tumour DNA
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
INCa
|
Institut National du Cancer (French
National Cancer Institute)
|
|
mCRC
|
metastatic colorectal cancer
|
|
NCT
|
National Clinical Trial
|
|
NGS
|
next-generation sequencing
|
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Siegel RL, Wagle NS, Cercek A, Smith RA
and Jemal A: Colorectal cancer statistics, 2023. CA Cancer J Clin.
73:233–254. 2023.PubMed/NCBI
|
|
3
|
Dyba T, Randi G, Bray F, Martos C, Giusti
F, Nicholson N, Gavin A, Flego M, Neamtiu L, Dimitrova N, et al:
The European cancer burden in 2020: Incidence and mortality
estimates for 40 countries and 25 major cancers. Eur J Cancer.
157:308–347. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cervantes A, Adam R, Roselló S, Arnold D,
Normanno N, Taïeb J, Seligmann J, De Baere T, Osterlund P, Yoshino
T, et al: Metastatic colorectal cancer: ESMO Clinical Practice
Guideline for diagnosis, treatment and follow-up. Ann Oncol.
34:10–32. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tabernero J, Grothey A, Van Cutsem E,
Yaeger R, Wasan H, Yoshino T, Desai J, Ciardiello F, Loupakis F,
Hong YS, et al: Encorafenib Plus Cetuximab as a new standard of
care for previously treated BRAF V600E-mutant metastatic colorectal
cancer: Updated survival results and subgroup analyses from the
BEACON study. J Clin Oncol. 39:273–284. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Santini D, Botticelli A, Galvano A,
Iuliani M, Incorvaia L, Gristina V, Taffon C, Foderaro S,
Paccagnella E, Simonetti S, et al: Network approach in liquidomics
landscape. J Exp Clin Cancer Res. 42:1932023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lièvre A, Merlin JL, Sabourin JC, Artru P,
Tong S, Libert L, Audhuy F, Gicquel C, Moureau-Zabotto L, Ossendza
RA, et al: RAS mutation testing in patients with metastatic
colorectal cancer in French clinical practice: A status report in
2014. Dig Liver Dis. 50:507–512. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thomsen CB, Hansen TF, Andersen RF,
Lindebjerg J, Jensen LH and Jakobsen A: Monitoring the effect of
first line treatment in RAS/RAF mutated metastatic colorectal
cancer by serial analysis of tumor specific DNA in plasma. J Exp
Clin Cancer Res. 37:552018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Allegretti M, Cottone G, Carboni F,
Cotroneo E, Casini B, Giordani E, Amoreo CA, Buglioni S, Diodoro M,
Pescarmona E, et al: Cross-sectional analysis of circulating tumor
DNA in primary colorectal cancer at surgery and during post-surgery
follow-up by liquid biopsy. J Exp Clin Cancer Res. 39:692020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taly V, Pekin D, Benhaim L, Kotsopoulos
SK, Le Corre D, Li X, Atochin I, Link DR, Griffiths AD, Pallier K,
et al: Multiplex picodroplet digital PCR to detect KRAS mutations
in circulating DNA from the plasma of colorectal cancer patients.
Clin Chem. 59:1722–1731. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thierry AR, Mouliere F, El Messaoudi S,
Mollevi C, Lopez-Crapez E, Rolet F, Gillet B, Gongora C, Dechelotte
P, Robert B, et al: Clinical validation of the detection of KRAS
and BRAF mutations from circulating tumor DNA. Nat Med. 20:430–435.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Van Cutsem E, Lenz HJ, Köhne CH, Heinemann
V, Tejpar S, Melezínek I, Beier F, Stroh C, Rougier P, van Krieken
JH and Ciardiello F: Fluorouracil, leucovorin, and irinotecan plus
cetuximab treatment and RAS mutations in colorectal cancer. J Clin
Oncol. 33:692–700. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bokemeyer C, Bondarenko I, Hartmann JT, de
Braud F, Schuch G, Zubel A, Celik I, Schlichting M and Koralewski
P: Efficacy according to biomarker status of cetuximab plus
FOLFOX-4 as first-line treatment for metastatic colorectal cancer:
The OPUS study. Ann Oncol. 22:1535–1546. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Heinemann V, von Weikersthal LF, Decker T,
Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmüller
C, Kahl C, Seipelt G, et al: FOLFIRI plus cetuximab versus FOLFIRI
plus bevacizumab as first-line treatment for patients with
metastatic colorectal cancer (FIRE-3): A randomised, open-label,
phase 3 trial. Lancet Oncol. 15:1065–1075. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Siravegna G, Marsoni S, Siena S and
Bardelli A: Integrating liquid biopsies into the management of
cancer. Nat Rev Clin Oncol. 14:531–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Franczak C, Witz A, Geoffroy K, Demange J,
Rouyer M, Husson M, Massard V, Gavoille C, Lambert A, Gilson P, et
al: Evaluation of KRAS, NRAS and BRAF mutations detection in plasma
using an automated system for patients with metastatic colorectal
cancer. PLoS One. 15:e02272942020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Franczak C, Dubouis L, Gilson P, Husson M,
Rouyer M, Demange J, Leroux A, Merlin JL and Harlé A: Integrated
routine workflow using next-generation sequencing and a
fully-automated platform for the detection of KRAS, NRAS and BRAF
mutations in formalin-fixed paraffin embedded samples with poor DNA
quality in patients with colorectal carcinoma. PLoS One.
14:e02128012019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Witz A, Dardare J, Betz M, Gilson P,
Merlin JL and Harlé A: Tumor-derived cell-free DNA and circulating
tumor cells: Partners or rivals in metastasis formation? Clin Exp
Med. 24:22024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Van Cutsem E, Köhne CH, Hitre E, Zaluski
J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G,
et al: Cetuximab and chemotherapy as initial treatment for
metastatic colorectal cancer. N Engl J Med. 360:1408–1417. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bokemeyer C, Bondarenko I, Makhson A,
Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G,
Stroh C, et al: Fluorouracil, leucovorin, and oxaliplatin with and
without cetuximab in the first-line treatment of metastatic
colorectal cancer. J Clin Oncol. 27:663–671. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
García-Foncillas J, Tabernero J, Élez E,
Aranda E, Benavides M, Camps C, Jantus-Lewintre E, López R,
Muinelo-Romay L, Montagut C, et al: Prospective multicenter
real-world RAS mutation comparison between OncoBEAM-based liquid
biopsy and tissue analysis in metastatic colorectal cancer. Br J
Cancer. 119:1464–1470. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Osumi H, Takashima A, Ooki A, Yoshinari Y,
Wakatsuki T, Hirano H, Nakayama I, Okita N, Sawada R, Ouchi K, et
al: A multi-institutional observational study evaluating the
incidence and the clinicopathological characteristics of NeoRAS
wild-type metastatic colorectal cancer. Transl Oncol.
35:1017182023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thierry AR, El Messaoudi S, Mollevi C,
Raoul JL, Guimbaud R, Pezet D, Artru P, Assenat E, Borg C,
Mathonnet M, et al: Clinical utility of circulating DNA analysis
for rapid detection of actionable mutations to select metastatic
colorectal patients for anti-EGFR treatment. Ann Oncol.
28:2149–2159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vivancos A, Aranda E, Benavides M, Élez E,
Gómez-España MA, Toledano M, Alvarez M, Parrado MRC,
García-Barberán V and Diaz-Rubio E: Comparison of the clinical
sensitivity of the Idylla platform and the OncoBEAM RAS CRC assay
for KRAS Mutation detection in liquid biopsy samples. Sci Rep.
9:89762019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grasselli J, Elez E, Caratù G, Matito J,
Santos C, Macarulla T, Vidal J, Garcia M, Viéitez JM, Paéz D, et
al: Concordance of blood- and tumor-based detection of RAS
mutations to guide anti-EGFR therapy in metastatic colorectal
cancer. Ann Oncol. 28:1294–1301. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vidal J, Muinelo L, Dalmases A, Jones F,
Edelstein D, Iglesias M, Orrillo M, Abalo A, Rodríguez C, Brozos E,
et al: Plasma ctDNA RAS mutation analysis for the diagnosis and
treatment monitoring of metastatic colorectal cancer patients. Ann
Oncol. 28:1325–1332. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bando H, Kagawa Y, Kato T, Akagi K, Denda
T, Nishina T, Komatsu Y, Oki E, Kudo T, Kumamoto H, et al: A
multicentre, prospective study of plasma circulating tumour DNA
test for detecting RAS mutation in patients with metastatic
colorectal cancer. Br J Cancer. 120:982–986. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Velimirovic M, Juric D, Niemierko A,
Spring L, Vidula N, Wander SA, Medford A, Parikh A, Malvarosa G,
Yuen M, et al: Rising circulating tumor DNA as a molecular
biomarker of early disease progression in metastatic breast cancer.
JCO Precis Oncol. 4:1246–1262. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mauri G, Vitiello PP, Sogari A, Crisafulli
G, Sartore-Bianchi A, Marsoni S, Siena S and Bardelli A: Liquid
biopsies to monitor and direct cancer treatment in colorectal
cancer. Br J Cancer. 127:394–407. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Loupakis F, Sharma S, Derouazi M, Murgioni
S, Biason P, Rizzato MD, Rasola C, Renner D, Shchegrova S, Koyen
Malashevich A, et al: Detection of molecular residual disease using
personalized circulating tumor DNA assay in patients with
colorectal cancer undergoing resection of metastases. JCO Precis
Oncol. 5:PO.21.00101. 2021.PubMed/NCBI
|
|
32
|
Diehl F, Schmidt K, Choti MA, Romans K,
Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al:
Circulating mutant DNA to assess tumor dynamics. Nat Med.
14:985–990. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Turner NC, Swift C, Jenkins B, Kilburn L,
Coakley M, Beaney M, Fox L, Goddard K, Garcia-Murillas I, Proszek
P, et al: Results of the c-TRAK TN trial: A clinical trial
utilising ctDNA mutation tracking to detect molecular residual
disease and trigger intervention in patients with moderate- and
high-risk early-stage triple-negative breast cancer. Ann Oncol.
34:200–211. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Powles T, Assaf ZJ, Davarpanah N,
Banchereau R, Szabados BE, Yuen KC, Grivas P, Hussain M, Oudard S,
Gschwend JE, et al: ctDNA guiding adjuvant immunotherapy in
urothelial carcinoma. Nature. 595:432–743. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tie J, Cohen JD, Lahouel K, Lo SN, Wang Y,
Kosmider S, Wong R, Shapiro J, Lee M, Harris S, et al: Circulating
tumor DNA analysis guiding adjuvant therapy in stage II colon
cancer. N Engl J Med. 386:2261–2272. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Taniguchi H, Nakamura Y, Kotani D, Yukami
H, Mishima S, Sawada K, Shirasu H, Ebi H, Yamanaka T, Aleshin A, et
al: CIRCULATE-Japan: Circulating tumor DNA-guided adaptive platform
trials to refine adjuvant therapy for colorectal cancer. Cancer
Sci. 112:2915–2920. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
André T, Shiu KK, Kim TW, Jensen BV,
Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs
P, et al: Pembrolizumab in microsatellite-instability-high advanced
colorectal cancer. N Engl J Med. 383:2207–2218. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD-1 blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pascual J, Attard G, Bidard FC, Curigliano
G, Mattos-Arruda LD, Diehn M, Italiano A, Lindberg J, Merker JD,
Montagut C, et al: ESMO recommendations on the use of circulating
tumour DNA assays for patients with cancer: A report from the ESMO
Precision Medicine Working Group. Ann Oncol. 33:750–768. 2022.
View Article : Google Scholar : PubMed/NCBI
|