Introduction
Lung cancer, a type of cancer with the highest
incidence rate worldwide (accounting for 12.4% of all types of
cancer), is the leading cause of cancer-associated mortality
(accounting for 18.7% of all types of cancer) worldwide (1). With the increasing popularity of
low-dose computed tomography in lung cancer screening, the
detection rate of lung cancer has increased substantially (2). Non-small cell lung cancer (NSCLC) is a
major type of lung cancer that accounts for ~85% of lung cancer
cases, and >40% of patients diagnosed with NSCLC have
unresecTable disease that requires chemotherapy (3,4).
Platinum-based systemic chemotherapy is the cornerstone of adjuvant
or neoadjuvant therapy for patients with NSCLC (5). It is also an essential component of
comprehensive treatment for patients with locally advanced tumors.
Moreover, with advancements in immunotherapy, the combination of
platinum-based chemotherapy and immunotherapy can markedly increase
patient survival rates; thus, platinum-based chemotherapy will
continue to serve as the core therapeutic option in the future
(6–9).
A previous study has focused on enhancing the
efficacy of chemotherapy have suggested that factors such as
chemotherapeutic agents, regimens, cycles, drug species and
platinum drugs do not affect the long-term prognosis of patients
(10). However, multicenter
clinical-controlled studies focusing on the adverse effects (AEs)
of chemotherapy have not been performed. Although preventing
chemotherapy resistance in patients is clinically important,
mitigating AEs is equally important for safeguarding patient
efficacy and benefits; thus, more studies should explore AE
mitigation. Chemotherapy-associated AEs or side effects of
chemotherapy refer to the subjective discomfort and harmful and
undesired reactions observed in various body organ systems that
occur during the treatment or recovery period of patients with
cancer receiving normal doses of chemotherapeutic agents. Adverse
drug reactions occur in more than half of patients receiving
systemic anticancer treatments such as chemotherapy, and ~20% of
patients with cancer are readmitted to the hospital because of AEs
(11). Owing to the unpredicTable
occurrence timing and the delayed nature of chemotherapy-associated
AEs, clinicians can be passive in managing these severe side
effects, which affects systemic chemotherapy cycles, and patients
are likely to be affected by the interruption of chemotherapy,
ultimately affecting treatment efficacy. Therefore, predicting AEs
as early as possible will greatly contribute to overcoming the
aforementioned clinical problems.
Several existing studies have predicted adverse drug
reactions on the basis of genomics or drug databases; nevertheless,
these predictions have not been translated into clinical
applications (12–14). Real-world data can accurately
reflect the current state of cancer clinics and aid in addressing
clinical problems (15). With the
commissioning of electronic health record (EHR) systems and the
development of deep learning technology, predicting specific AEs is
possible (16,17). Moreover, deep learning models can be
used to accurately assess disease prognosis in numerous fields,
assisting clinicians in predicting AEs to intervene in advance
(18–20). To predict drug side effects,
numerous scholars have integrated various drug databases, drug
structural properties and protein-binding features, combined with
tumor- or drug-related human gene expression profiles and other
information, to train machine learning (ML) models, and their
performance has comprehensively surpassed that of traditional
methods (12–14,21).
In the present study, real-world data on
hematological indicators, chemotherapy-associated AEs and
interventions in patients with lung cancer before and after each
chemotherapy cycle was extracted from EHRs used and ML models were
used to develop predictive models for identifying common
chemotherapy-associated AEs. Finally, the performance of the
developed ML models was evaluated.
Materials and methods
Patients
The information of lung cancer patients admitted to
the First Affiliated Hospital, Zhejiang University School of
Medicine (Hangzhou, China) who received 2–4 cycles (3 weeks per
cycle) of adjuvant or neoadjuvant chemotherapy between January 2016
and February 2020 was extracted from a single-center EHR system in
December 2020. The inclusion criteria were as follows: i) A clear
pathological diagnosis of NSCLC; ii) a detailed medical history;
iii) regular chemotherapy cycles; iv) detailed hematological
indicators; and v) detailed records of interventions in patients
with NSCLC with AEs. A total of 403 patients were ultimately
included, totaling 2,062 admissions. The patient cohorts consisted
of 310 men and 93 women with a median age of 62 years (age range,
32–78 years). The final cohort comprised 1,659 single chemotherapy
cycles, of which 1,224 experienced grade ≥1 adverse events [Common
Terminology Criteria for AEs (CTCAE version 5.0)]. And 45
characteristics potentially associated with chemotherapy-associated
AEs were incorporated into the model. This yielded
events-per-variable (EPV)=1,224/45=27.2, markedly above the
traditional logistic-regression threshold of EPV and within the
recommended range of EPV for moderate-complexity algorithms such as
random forest (RF). The present study integrated multi-level data
(2,062 patient characteristic records +1,659 chemotherapy session
records). The use of longitudinal data analysis methods enhanced
statistical power, with this intensive measurement design partially
compensating for patient number limitations.
Response evaluation criteria in solid tumors
(RECIST; version 1.1) were used to evaluate the clinical efficacy
of chemotherapy. Follow-up was performed to evaluate AEs before
each cycle of chemotherapy. The survival of patients who completed
neoadjuvant chemotherapy and underwent surgery was followed up for
3 years by telephone and clinical re-examination, follow-up was
performed every 3 months. All procedures in the present
retrospective study involving human participants were performed in
accordance with the Declaration of Helsinki (as revised in 2013).
The patients were informed that the clinical information was stored
by the hospital and potentially used for scientific research, and
the need to obtain signed informed consent was waived by the Ethics
Committee of The First Affiliated Hospital, School of Medicine,
Zhejiang University, (Hangzhou, China). All patient cohorts
included in the present retrospective study underwent standardized
testing and treatment protocols. The relevant features and adverse
reaction data were uniformly recorded in the EHR system. For the
very few instances where data were incomplete, a complete-case
analysis was performed and these patients were excluded from the
study to maintain the integrity and robustness of the dataset.
Features
ML algorithms were used to predict AEs in real-world
patients with NSCLC who were receiving chemotherapy. The primary
task for the algorithms was to predict the probability of the next
severe AEs based on data from the present or previous chemotherapy
characterization. The targeted AEs predicted in the present study
were myelosuppression, low albumin (ALB) and hepatic impairment,
with the judgment criteria based on the CTCAE (v5.0), published by
the U.S. Department of Health and Human services.
The following chemotherapy-associated AE
characteristics were extracted as potential predictors for risk
prediction: i) Patient baseline characteristics [age, sex, history
of hypertension, diabetes and tumor history, family tumor history,
smoking status, drinking status, weight loss and body mass index
(BMI)]; ii) tumor-related features (tumor location, tumor size,
histology, grade and stage after surgery); iii)
chemotherapy-related features (first-line treatment,
chemotherapeutic agents and dose); iv) hematological indicators
[white blood cells (WBC), neutrophils, lymphocytes, monocytes,
hemoglobin (Hb), platelet (PLT), total protein (ToP), ALB, alanine
aminotransferase (ALT), aspartate aminotransferase, creatinine,
uric acid, triglyceride and cholesterol]; and v) clinical
intervention characteristics (recombinant-human granulocyte colony
stimulating factor, thymosin and reduced glutathione).
Identification of significant
features
The original dataset included 45 features generated
during hospitalization. A forward stepwise regression approach was
employed based on logistic regression (LR) for feature selection.
Specifically, at the beginning, univariate analysis was performed
for each feature, and feature A with the best predictive
performance for the outcome was selected and incorporated into the
model. Multivariate analysis involving two features was
subsequently performed on the basis of feature A and each of the
remaining features, and feature B, included in the combination with
the best predictive performance, was selected and added to the
model. This process was repeated step by step to incorporate
predictive factors until the model performance converged, at which
point the addition of factors was stopped, resulting in a set of
features beneficial for predicting AEs. The Shapley Additive
Explanation (SHAP) methodology was used to evaluate the
interpretability of the prediction models. Feature ranking was
achieved through the calculation of SHAP values, with features
being prioritized based on the mean absolute SHAP value for each.
The integration of machine learning techniques with SHAP offers a
clear and explicit interpretation of efficacy predictions.
Statistical algorithms
ML is a general term for a class of methods that
includes multiple algorithms with different technical principles,
each of which may have different performance on a particular task.
To select the optimal model, representative models of common ML
algorithms were used, namely, RF, multilayer perceptron (MLP) and
AdaBoost. For comparison, LR was also used, which is commonly used
in clinical medical research, as a benchmark for performance
comparison. The experiment was performed with five-fold
cross-validation, and the receiver operating characteristic (ROC)
curve, area under the ROC curve (AUC) value and calibration curve
were used to evaluate the performance of the model.
RF
An RF model is an integrated learning model that
comprises a number of independent decision trees (also referred to
as weak classifiers), each of which is trained on separate training
data; thus, each tree independently predicts the type of a new
sample. RF subsequently counts the results on the basis of the
predictions of each decision tree and ultimately determines the
specific type of a new sample and its corresponding probability on
the basis of the majority vote classification and mean. The
inclusion of numerous decision trees is the reason for the word
‘forest’ in the name. To avoid the lack of variability in the
trained decision trees due to the inclusion of the same training
data, RF uses a randomized strategy for selecting the training
data. Specifically, for an ‘N’ number of samples of the original
data, the model uses put-back sampling to randomly sample a set of
‘N’ from the original data to train a decision tree. The put-back
sampling strategy ensures that the generated sample will cover ~63%
of the original data; thus, the training data of each decision tree
differ, and there is no significant homogeneity among the resulting
decision trees.
MLP
An MLP is a type of basic neural network that can be
divided into input, hidden and output layers according to its
structure. The layers can be interconnected or not connected. After
data are input into an MLP through the input layer, each node in
the input layer inputs feature values in the data to the nodes in
the next layer with specific weights. The input strength of the
corresponding nodes in the next layer is the cumulative sum of the
output nodes in the previous layer, which is then processed by a
non-linear transformation function to continue to transfer
information to the next layer. These steps are repeated until the
prediction result is finally output at the output layer.
AdaBoost
AdaBoost is an ensemble learning model, similar to
an RF, that also uses numerous decision trees to complete
classification tasks. The decision trees used by AdaBoost are not
independent but are interrelated. Specifically, after the model has
trained the first decision tree, the second decision tree focuses
on the samples misclassified by the first decision tree, thus
ensuring classification accuracy. The third decision tree focuses
on samples misclassified by the former two decision trees. In the
testing phase, after new samples are accepted, AdaBoost runs all
the decision trees simultaneously and then calculates the average
of their outputs with specific weights to obtain the final
prediction results.
Building and environment
All the models used in the present study were
provided by sklearn (22), and
relevant statistical analysis was performed through SciPy (23). The specific experimental environment
was conducted on a Lenovo computer (Lenovo) including an Intel Xeon
E2520 (Mountain View; Intel Corporation), 32 GB of memory and two
Nvidia Titan V graphics cards (NVIDIA Corporation). All original
code has been deposited at GitHub (github.com/ZJU-BMI/cancer). Data
are available from the authors upon reasonable request and with
written permission; following the requirements of data supervision
regulations, these data were not uploaded to a public platform.
Statistical analysis
Comparisons of categorical data between
chemotherapy-related AEs in patients and clinical response to
chemotherapy were performed using χ2 or Fisher's exact
test. Survival curves were estimated using the Kaplan-Meier method
and analyzed using the log-rank test. Statistical analysis was
performed using Prism 10.3 (GraphPad; Dotmatics) and SPSS software
25.0 (IBM Corp.), and P<0.05 was considered to indicate a
statistically significant difference.
Results
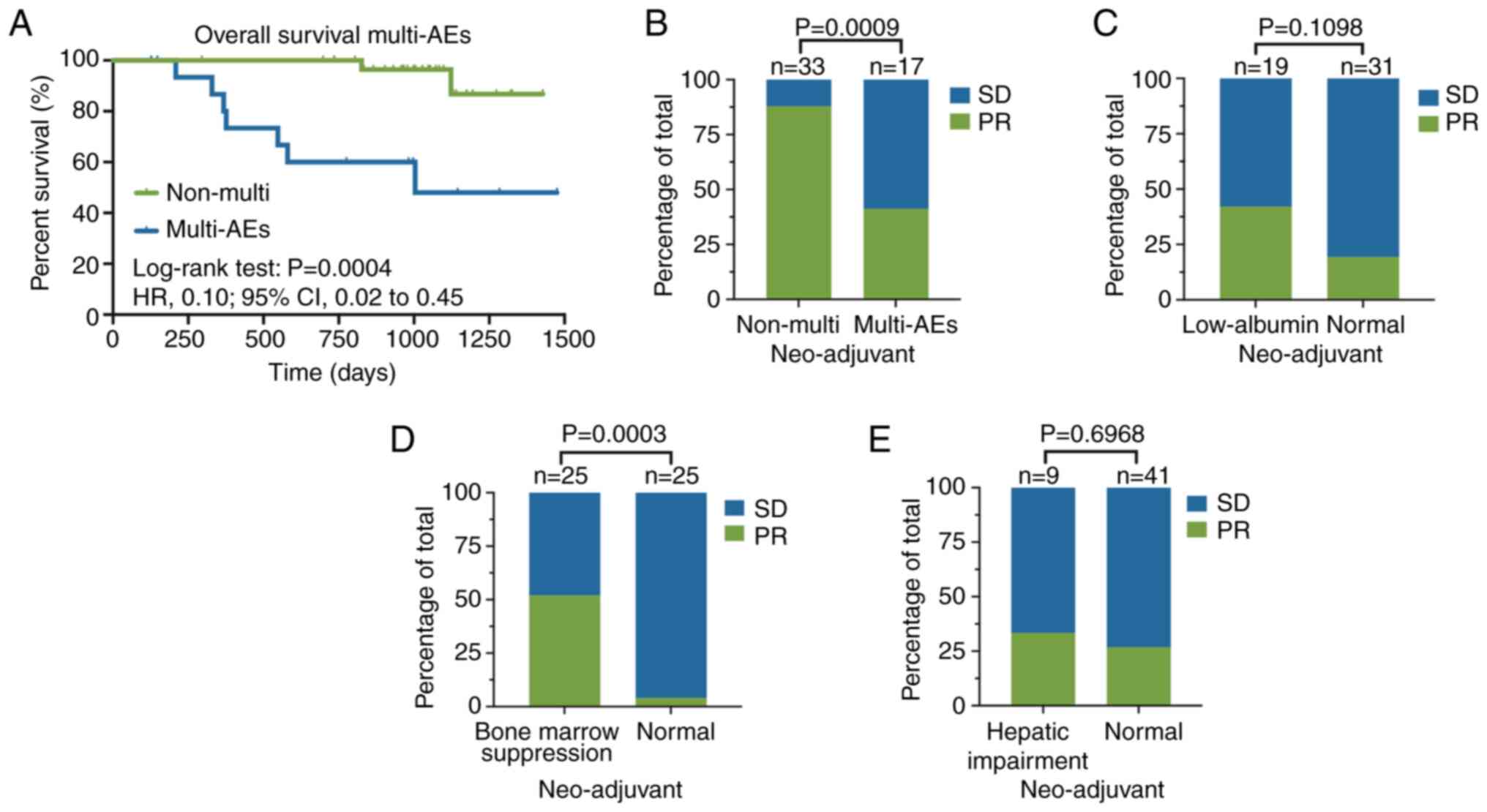
Impact of multi-AEs on patients with
NSCLC receiving chemotherapy
A total of 50 patients with NSCLC who completed
neoadjuvant chemotherapy and underwent surgery were analyzed. CTCAE
v5.0 was used to determine the extent of AEs in neoadjuvant
patients. All the AEs were found to be grade 1 or 2. Our previous
study revealed that clinical responses according to RECIST v1.1
could predict survival outcomes and an improved objective response
was notably associated with improved overall survival (OS)
(24). Patients who did not
experience multi-AEs (two or more adverse events) had improved OS
compared with those who experienced multi-AEs throughout the
chemotherapy period (hazard ratio, 0.10; 95% CI, 0.02 to 0.45;
Fig. 1A). Furthermore, multi-AEs
were significantly associated with the efficacy of neoadjuvant
chemotherapy (Fig. 1B). However,
there was no significant association between the single AEs,
including low ALB levels and hepatic impairment, and the objective
response (Fig. 1C and E; Table SI).
Evaluation of the predictive
performance of the models
The characteristics of the single chemotherapy
treatments in the dataset are shown in Table I, and the baseline characteristics
of the patients in the experimental dataset are shown in Table II. A total of four independent
prediction models were developed and the performance of all models
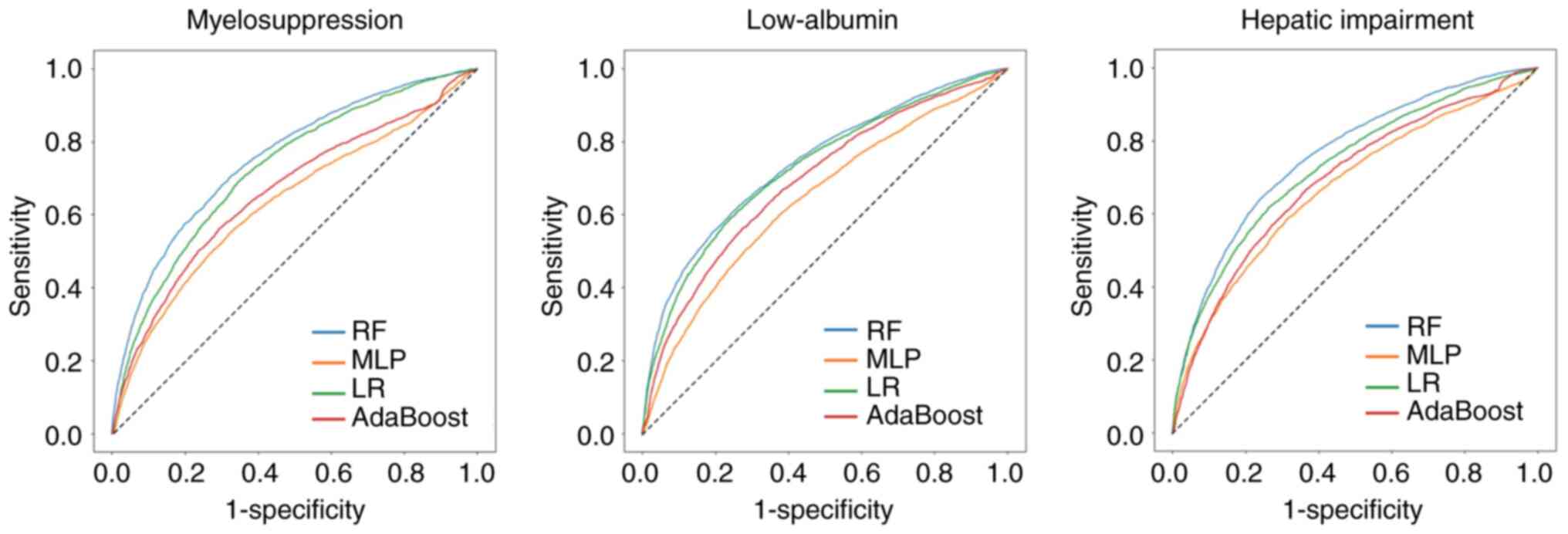
were evaluated and compared using ROC curves and AUCs. Among the
proposed models, the RF model exhibited the best performance in
both AE prediction tasks, followed by the LR model, and AdaBoost
outperformed the MLP (Table III).
When comparing the AUC, accuracy and precision between RF and the
other three models, RF outperformed other models. The ROC curves
exhibited a similar trend (Fig. 2),
and the ROC curves of the RF model were consistently higher
compared with those of the other three models for all three
prediction tasks, implying that at any cutoff point, the RF model
demonstrated improved results in terms of the true-positive rate
and false-positive rate simultaneously. With respect to specific
classification performance metrics, the accuracy, precision and
recall rates of the RF model were consistently higher compared with
those of the LR model.
 | Table I.Characteristics of single
chemotherapy treatment in data set. |
Table I.
Characteristics of single
chemotherapy treatment in data set.
|
Characteristics | No. of single
treatments (n=1,659) |
|---|
| First-line
treatment, n (mean dose, mg) |
|
| doc/cis
(DP) | 420 (114/41) |
| doc/lob
(DL) | 89 (116/16) |
| doc/oxa
(DOCOX) | 83 (99/64) |
| eto/cis
(EP) | 189 (171/65) |
| eto/lob
(EL) | 23 (155/14) |
| pem/cis
(PP) | 322 (834/46) |
|
pem/carbo (PC) | 52 (820/539) |
| pem/lob
(PL) | 194 (789/13) |
| pem/oxa
(POX) | 95 (758/123) |
| tax/cis
(TP) | 155 (315/33) |
| tax/lob
(TL) | 37 (313/15) |
| Blood test, mean
(unit) |
|
|
WBC | 6.12
(×109/l) |
|
NEU | 4.26
(×109/l) |
|
NEU% | 67.01 (%) |
|
LYM | 1.51
(×109/l) |
|
LYM% | 26.00 (%) |
| MO | 0.44
(×109/l) |
|
MO% | 5.89 (%) |
| Hb | 119.87 (g/l) |
|
PLT | 207.40
(×109/l) |
|
ToP | 65.91 (g/l) |
|
ALB | 42.99 (g/l) |
|
ALT | 28.48 (U/l) |
|
AST | 28.30 (U/l) |
| Cr | 70.11 (µmol/l) |
| UA | 289.29
(µmol/l) |
| TG | 2.27 (mmol/l) |
|
CHOL | 5.14 (mmol/l) |
| Adverse reactions,
n (%) |
|
| Bone
marrow suppression | 304 (18.3%) |
|
Cachexy | 543 (32.7%) |
| Liver
injury | 377 (22.7%) |
| Interventions, n
(%) |
|
|
rhG-CSF | 485 (29.2%) |
|
Thymosin | 168 (10.1%) |
| Reduced
glutathione | 1356 (81.7%) |
 | Table II.Baseline characteristics of patients
in experimental dataset. |
Table II.
Baseline characteristics of patients
in experimental dataset.
|
Characteristics | No. of patients
(n=403) |
|---|
| Sex, n (%) |
|
|
Male | 310 (77.3) |
|
Female | 93 (22.7) |
| Age, mean (SD) | 60.95 (8.26) |
| BMI, mean (SD) | 22.58 (2.88) |
| History of present
illness, n (%) |
|
|
Hypertension | 121 (30.0) |
|
Diabetes mellitus | 33 (8.2) |
| History
of other cancer | 20 (5.0) |
| Weight
loss | 48 (11.9) |
| Personal history, n
(%) |
|
|
Smoking | 252 (62.5) |
|
Drinking | 139 (34.5) |
| Family tumor
history, n (%) | 83 (20.6) |
| Lung cancer Stage,
n (%) |
|
| Early
stage (stage I to II) | 191 (47.4) |
|
Advanced stage (stage III to
IV) | 212 (52.6) |
| Histology, n
(%) |
|
|
NSCLC | 367 (91.1) |
|
SCLC | 36 (8.9) |
| Grade, n (%) |
|
| High
(G1 and G2) | 91 (22.6) |
| Low (G3
and missing) | 312 (77.4) |
| Tumor location, n
(%) |
|
|
Left | 179 (44.4) |
|
Right | 224 (55.6) |
 | Table III.Performance of machine learning
models for AEs prediction. |
Table III.
Performance of machine learning
models for AEs prediction.
| Task | Model | AUC | ACC | Precision | Recall |
|---|
|
Myelosuppressive | RF | 0.754±0.037 | 0.709±0.065 | 0.365±0.076 | 0.699±0.102 |
|
| MLP | 0.663±0.047 | 0.683±0.097 | 0.316±0.079 | 0.558±0.195 |
|
| LR | 0.733±0.039 | 0.690±0.074 | 0.344±0.063 | 0.691±0.118 |
|
| AdaBoost | 0.671±0.051 | 0.689±0.074 | 0.326±0.081 | 0.569±0.136 |
| Low-ALB | RF | 0.742±0.026 | 0.721±0.036 | 0.583±0.080 | 0.608±0.093 |
|
| MLP | 0.645±0.043 | 0.659±0.044 | 0.495±0.068 | 0.553±0.128 |
|
| LR | 0.725±0.035 | 0.712±0.034 | 0.563±0.069 | 0.614±0.097 |
|
| AdaBoost | 0.691±0.037 | 0.649±0.052 | 0.495±0.085 | 0.617±0.144 |
| Hepatic
impairment | RF | 0.762±0.034 | 0.724±0.051 | 0.443±0.071 | 0.692±0.089 |
|
| MLP | 0.680±0.044 | 0.653±0.088 | 0.371±0.067 | 0.656±0.099 |
|
| LR | 0.732±0.043 | 0.712±0.069 | 0.431±0.074 | 0.650±0.102 |
|
| AdaBoost | 0.694±0.046 | 0.674±0.069 | 0.386±0.077 | 0.611±0.138 |
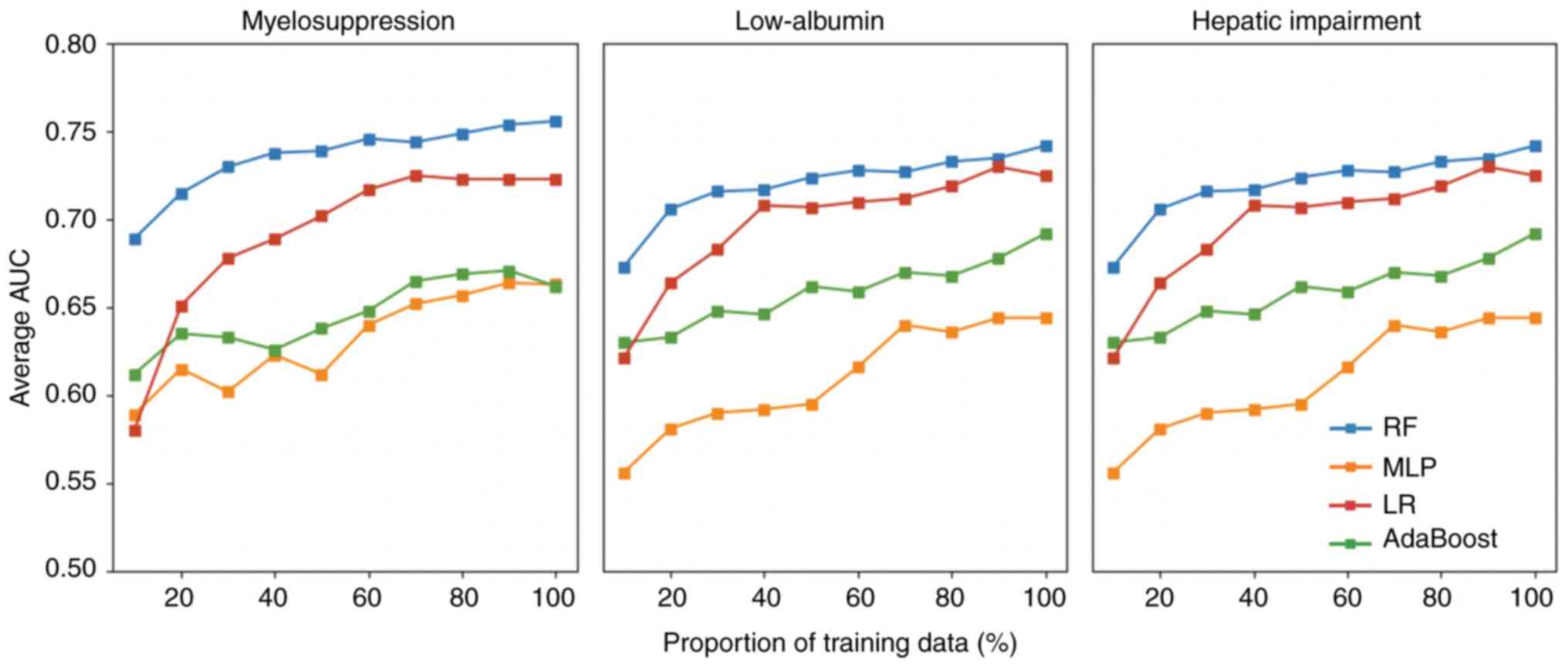
Effect of the number of training sets
on prediction model performance
As shown in Fig. 3,
each model included in the present study exhibited several
instances of performance degradation when the volume of training
data increased in each prediction task; however, overall, a more
pronounced performance of the four models was observed for the
three prediction tasks after increasing the training data volume.
With the exception of the LR model, no significant performance
convergence was observed, indicating performance saturation for
myelosuppression and liver impairment. Thus, we hypothesized that
if more patient data were used, the models could achieve higher
prediction accuracies. Again, the RF model exhibited the best
performance among all four models in any training environment.
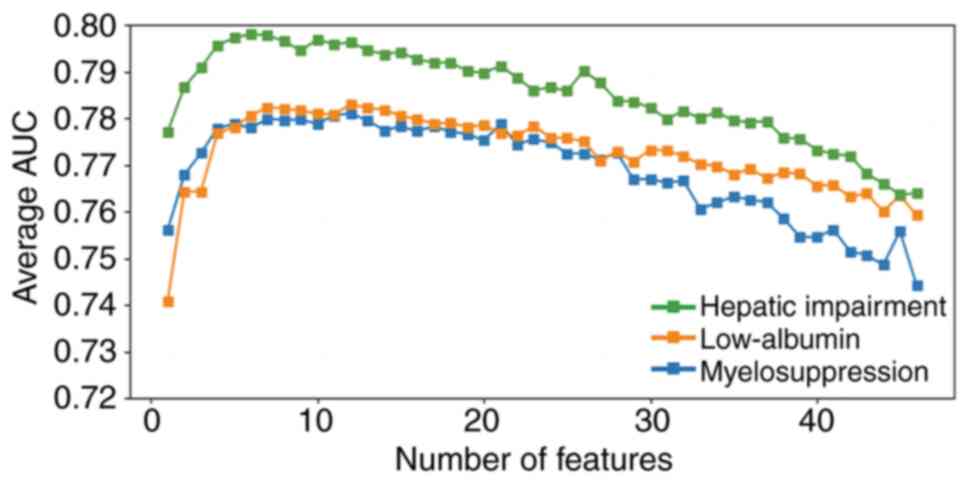
Effect of the number of incorporated
features on the predictive performance of the models
The effect of the number of incorporated features on
the predictive performance of the models was assessed by
individually selecting and overlaying the number of features. Each
feature was numbered from 0 to 45, these and their corresponding
meanings are shown in Table IV.
The results revealed that ≤10 features (Table SII) significantly improved the
performance (Fig. 4). With an
increasing number of incorporated features, the models exhibited
noTable overfitting (25),
resulting in a decreased average AUC. The features in each
predictive model of AEs were ranked based on importance as follows:
i) Myelosuppression: 27, 35, 18, 44, 36, 4, 14, 34, 23, 17, 2, 45,
41, 40, 25, 39, 33, 11, 19, 30, 31, 21, 10, 38, 29, 6, 12, 0, 15,
7, 3, 5, 32, 8, 26, 20, 43, 13, 42, 9, 16, 37, 28, 22, 24, 1, 2;
ii) low-ALB: 36, 23, 1, 26, 10, 30, 31, 35, 45, 34, 37, 9, 28, 29,
39, 6, 42, 19, 18, 20, 16, 4, 5, 21, 41, 33, 12, 40, 0, 15, 3, 7,
38, 43, 27, 44, 32, 17, 25, 13, 24, 2, 11, 22, 8, 14, 3; and iii)
hepatic impairment: 38, 21, 16, 10, 25, 20, 12, 24, 17, 1, 22, 2,
43, 31, 6, 29, 4, 18, 9, 34, 14, 0, 32, 26, 37, 39, 36, 8, 23, 13,
45, 15, 28, 27, 44, 33, 5, 7, 30, 3, 42, 19, 35, 41, 11, 40.
 | Table IV.Meaning of feature number. |
Table IV.
Meaning of feature number.
| Number | Feature |
|---|
| 0 | Sex |
| 1 | Age |
| 2 | History of
hypertension |
| 3 | History of
diabetes |
| 4 | Tumor history |
| 5 | Family tumor
history |
| 6 | Smoking |
| 7 | Drinking |
| 8 | Weight loss |
| 9 | Tumor location |
| 10 | Tumor
histology |
| 11 | Tumor grade |
| 12 | Tumor stage |
| 13 | Treatment
interval |
| 14 | doc/lob (DL) |
| 15 | doc/oxa
(DOCOX) |
| 16 | doc/cis (DP) |
| 17 | eto/lob (EL) |
| 18 | eto/cis (EP) |
| 19 | pem/carbo (PC) |
| 20 | pem/lob (PL) |
| 21 | pem/oxa (POX) |
| 22 | pem/cis (PP) |
| 23 | Non-complete
cycle |
| 24 | tax/lob (TL) |
| 25 | tax/cis (TP) |
| 26 | Body mass
index |
| 27 | WBC |
| 28 | NEU |
| 29 | NEU% |
| 30 | LYM |
| 31 | LYM% |
| 32 | MO |
| 33 | MO% |
| 34 | Hb |
| 35 | PLT |
| 36 | ToP |
| 37 | ALB |
| 38 | ALT |
| 39 | AST |
| 40 | Cr |
| 41 | UA |
| 42 | TG |
| 43 | CHOL |
| 44 | Weight (kg) |
| 45 | Clinical
interventions (associated with particular tasks) |
Calibration curve for the predictive
performance of the models
Metric calibration is crucial for evaluating the
accuracy of a model in predicting the probability of an AE
occurring in an individual in the future. This reflects the extent
to which the theoretical risk predicted by a model agrees with the
observed risk. Issues were observed in the MLP and AdaBoost model
calibrations because their calibration curves for predicting
myelosuppression, low ALB levels and hepatic impairment differed
markedly from the optimal values (Fig.
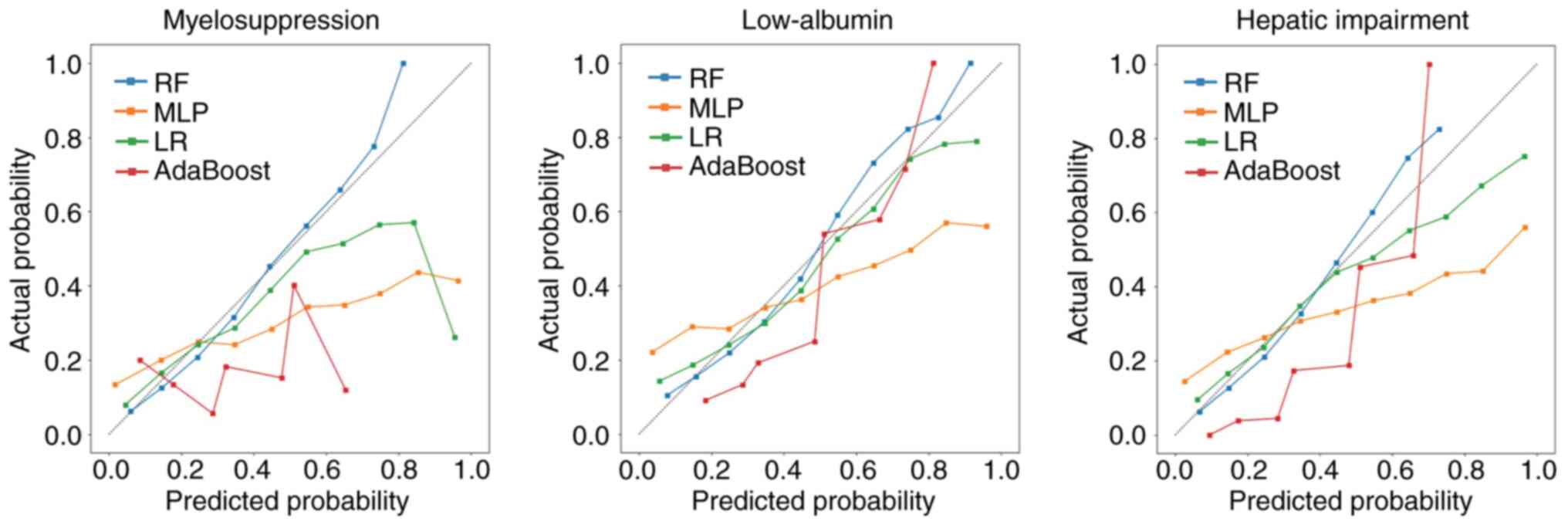
5). Comparatively, the calibration and optimization curves of
the RF model had improved fit for the three prediction tasks,
indicating that the probability of predicting the risk of side
effects in patients suggested by the RF model represented the true
value to a considerable extent. Conversely, the LR model was
significantly under-calibrated for the myelosuppression side
effect, whereas the other two models exhibited calibration degrees
that were similar to that of the RF model.
Explanation of predictive models with
SHAP values
To improve the transparency and interpretability of
the model, the SHAP algorithm was used to elucidate the model's
output. The SHAP value of each of the most important features on RF
model output was calculated (Fig.
6). Based on the importance ranking derived from the average
absolute SHAP values, the top five features (WBC, PLT, Weight, ToP,
Hb) were identified as the most significant variables for
predicting myelosuppression. It was demonstrated that ‘ToP’, ‘ALB’,
‘BMI’, ‘Age’ and ‘Hb’ were the five most influential features in
predicting low-ALB. Additionally, ‘ALT’ was identified as the most
significant variable for predicting hepatic impairment. Fig. 6B presents a violin plot for each
feature, illustrating the association between the feature values
and their corresponding SHAP values. The horizontal position
indicates whether a particular feature value contributes to a
higher or lower model prediction. The color gradient reflects
whether the variable value is high (red) or low (blue) for a given
observation. A larger absolute SHAP value indicates a stronger
influence of that feature on the predictions of the RF-based model.
Lower WBC and lower PLT were associated with a higher predicted
probability of myelosuppression. And lower ToP were associated with
a higher predicted probability of low-ALB. It was also observed
that increases in the ALT had a positive influence, directing the
prediction toward hepatic impairment. The SHAP algorithm was also
used to elucidate the other model's output. (Fig. S1, Fig.
S2, Fig. S3). For predictive
tasks assessed using the LR model, ‘WBC’ and ‘PLT’ were found to be
the two most important features in predicting myelosuppression,
‘ToP’ was the most important feature in predicting low-ALB and
‘ALT’ was identified as the most significant variable for
predicting hepatic impairment (Fig.
S1). For predictive tasks using AdaBoost, ‘WBC’, ‘ToP’ or ‘ALT’
were the most influential features in predicting myelosuppression,
low-ALB and hepatic impairment, respectively (Fig. S2). For predictive tasks using MLP,
‘PLT’, ‘WBC’, ‘Hb’, ‘Top’, ‘Weight’ and ‘HTN’ were the six most
influential features in predicting myelosuppression. It was also
demonstrated that ‘ToP’, ‘ALB’, ‘BMI’, ‘Age’, ‘Hb’, ‘PLT’, ‘LYM’
and ‘LYM%’ were the eight most influential features in predicting
low-ALB. Additionally, ‘ALT’, ‘DP’ and ‘Tumor histology’ were
identified as the most significant variable for predicting hepatic
impairment (Fig. S3).
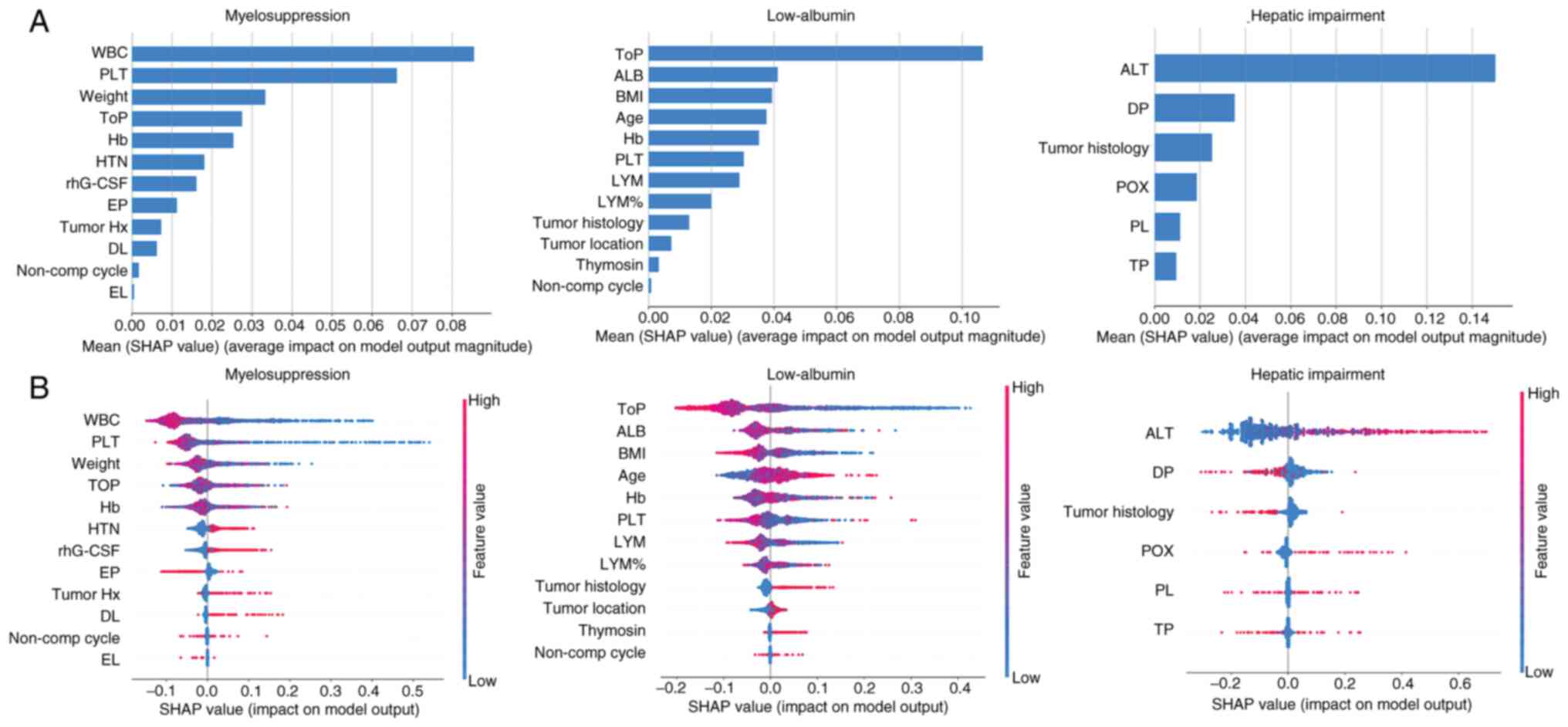 | Figure 6.SHAP values and feature interaction
scores in machine learning-based prediction. (A) The most important
features for the prediction of chemotherapy-associated adverse
effects (ranked from most to least important). (B) The distribution
of the impacts of each of the most important features on model
output. The horizontal location shows whether the effect of that
value is associated with a higher or lower prediction. The colors
represent the feature values: Red for larger values and blue for
smaller values. SHAP, Shapley Additive Explanation; WBC, white
blood cell; PLT, platelet; ToP, total protein; Hb, hemoglobin; HTN,
hypertension; rhG-CSF, recombinant-human granulocyte colony
stimulating factor; EP, etoposide/cisplatin; Tumor Hx, tumor
history; DL, docetaxel/lobaplatin; non-comp, non-complete; EL,
etoposide/lobaplatin; ALB, albumin; BMI, body mass index; LYM,
lymphocytes; ALT, alanine aminotransferase; DP,
docetaxel/cisplatin; POX, pemetrexed/oxaliplatin; PL,
pemetrexed/lobaplatin; TP, paclitaxel/ cisplatin. |
Discussion
Chemotherapy-associated side effects are among the
major concerns for clinicians, in addition to the efficacy of
treatment. The AEs of chemotherapy agents for lung cancer involve
numerous organ systems (11). The
AEs of chemotherapeutic drugs are complex, their side effects vary
from person to person, and their side effects do not occur
immediately after taking the drugs. Untimely and incomplete
interventions worsen common AEs, thereby affecting the routine
chemotherapy cycle of patients and aggravating socioeconomic
burdens on patients. Therefore, developing an effective method for
predicting chemotherapy-associated AEs to guide clinicians to
intervene in patients promptly is imperative, and the importance
and necessity of an effective and accurate tool for predicting the
side effects of chemotherapeutic drugs are clear.
It is difficult to predict AEs promptly with
traditional statistical techniques, and it is feasible to use
genomics and biomarkers to identify individuals who are susceptible
to AEs (26). However, late-stage
prognosis prediction may be less accurate due to the tumor
heterogeneity induced by chemotherapy. At present, several scholars
are employing data mining, ML or artificial intelligence (AI)
methods to predict potential adverse drug reactions. Numerous
scholars integrate the indications, known adverse drug reactions,
chemical structures and biological properties of drugs in various
drug databases, combined with tumor- or drug-related human gene
expression features, and use ML algorithms to predict the potential
side effects of drugs, which is helpful for guiding drug clinical
trials and monitoring the AEs of existing commercial drugs
(12–14). Among the emerging novel methods, ML
methods have comprehensively outperformed traditional methods in
predicting the side effects of chemotherapeutic drugs. Predictive
models developed for drugs, targets and AEs using deep learning
techniques, knowledge graphs and biomedicine outperform traditional
methods. However, databases developed for accumulating information
on drug side effects contain complex, limited and unauthorized
information.
In general, few studies have investigated the
prediction of AEs of chemotherapy (27,28).
Dranitsaris et al (29)
performed a study specifically focused on chemotherapy-induced
nausea and vomiting. Boudali and Messaoud (30) developed ML models to predict
chemotherapy-related toxicity. Most studies use limited variables
that are not closely related to clinical work. Chemotherapeutic
drugs have been used for a long time, and the types of adverse
reactions associated with them are almost universally known.
However, accurately predicting AEs that may occur in patients
during chemotherapy is impossible in the clinical setting.
Recently, Shandong University researchers developed four ML models
using 11 clinical variables that predicted chemotherapy-associated
AEs with an overall AUC of 0.88 and greater accuracy for specific
toxicities in patients with colorectal cancer (31). Additionally, some common AEs,
including nausea, vomiting, diarrhea, anaphylaxis, kidney injury
and liver injury, despite timely intervention, cannot be
effectively avoided, and in some cases, they can be aggravated
during chemotherapy. On the other hand, the potential of ML for
accurate prediction is often compromised by inherent discrepancies
between training data and real-world clinical environments
(32).
While AI models may demonstrate strong statistical
performance, they frequently fall short in practical clinical
applications (33). Thus, the use
of real-world clinical data can accurately reflect the current
clinical situation and help address clinical problems (15). Thus, in the present study, the
characteristic information of patients with lung cancer were fully
incorporated, including baseline features, lung cancer features,
chemotherapeutic agent features, blood marker features and adverse
reaction interventions. An ML-based prediction model for
chemotherapy-associated AEs was constructed and the AEs of patients
with lung cancer during several cycles of chemotherapy were
monitored. Clinical data and ML methods were used to solve the
aforementioned clinical issues, providing novel insights into
chemotherapy-associated research on lung cancer. ML was
demonstrated to be an important method to solve the clinical
problems in the future. Compared with classical statistical
regression models, ML techniques are capable of capturing complex
nonlinear relationships among predictors, handling high-dimensional
data with intricate interactions and providing accurate
personalized predictions.
In addition to the LR model, the present study
employed several ML algorithms, including RF, MLP and AdaBoost, to
predict chemotherapy-associated AEs in patients with lung cancer.
The results demonstrated that the RF model outperformed the other
models, exhibiting the highest stability and alignment with
clinical intuition. Current research has evolved from exploring
single data sources and simple models to integrating multi-modal
data and complex model architectures, continuously improving
prediction performance and gradually enhancing interpretability.
However, challenges such as model generalization ability,
interpretability and ethical compliance still exist and require
multidisciplinary cooperation to resolve. We hypothesize that the
future trend will be the integration of multi-modal data, such as
unified database combining EHR, imaging, pathology, genomics and
real-time monitoring data processed by Transformer or graph-based
architectures to capture cross-modal interactions (34,35).
At the same time, successful clinical integration needs to be
clinical needs-oriented and seamlessly embedded into workflows.
At present, numerous hospitals have implemented
IT-based system, such as computerized physician order entry systems
or unreasonable medical orders monitoring system, to reduce
prescribing errors. ML is gradually being implemented in clinical
practice within the field of cardiovascular diseases (36). The integration of these innovative
models holds noTable promise for predicting the severity of atrial
fibrillation substrates and in-hospital mortality (37). The chemotherapy-associated AEs
prediction model could be automatically triggered when a physician
enters chemotherapy orders into the system, which is called
pre-chemotherapy planning monitoring. For multi-cycle chemotherapy,
the condition of a patient may evolve over time. Prior to each new
cycle, the system could re-run the model using the most up-to-date
clinical data to update risk predictions and assist oncologists in
making informed treatment adjustments. ML models could stratify
patients into high- and medium-risk categories, proactively
initiating appropriate follow-up actions and additional laboratory
tests. These interventions align with authoritative guidelines such
as those from the National Comprehensive Cancer Network, enabling
timely and appropriate responses to potential side effects.
Furthermore, close collaboration with hospital IT departments and
EHR vendors is essential to overcome technical challenges,
including data standardization and system integration. We
hypothesize that the ML-based prediction model will help clinicians
in everyday practice manage patients treated with chemotherapy.
Nevertheless, ML is neither an omnipotent tool nor
the perfect solution. Ideally, an appropriate method should be
selected according to the problems to be addressed. Moreover,
inputting high raw medical data volumes into ML algorithms without
analysis cannot yield the expected results (38). Instead, more attention should be
paid to cross-disciplinary research, fully integrating the
knowledge of multiple disciplines and selecting appropriate
algorithms to improve algorithms to advance future AI-assisted
clinical decision-making.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present work was supported by the Zhejiang Province
Traditional Chinese Medicine Science and Technology Plan Project
(grant no. 2026ZL0474), ‘Leading Goose’ Research and Development
Program of Zhejiang (grant no. 2025C02057) and the National Natural
Science Foundation of China (grant no. 82372773).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The original code and data
have been deposited at GitHub (https://github.com/ZJU-BMI/cancer).
Authors' contributions
SH, ZFH, ZXH and JH conceptualized and deigned the
present study. SH and ZJS wrote the original draft of the
manuscript and were involved in graph drawing. SH and ZWH designed
and performed the critical additional experiments and revised the
manuscript. The manuscript was reviewed and edited by TAX, ZXH and
ZFH. JH was responsible for raw data collection, critical revision
of the manuscript, supervision and project administration.
Provision of study materials or patients was performed by SH, ZJS
and TAX. Data collection and assembly were performed by XYZ and SL.
ZJS, TAX and ZXH were involved in data analysis and interpretation.
JH and ZFH confirm the authenticity of all raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Clinical
Research Ethics Committee of The First Affiliated Hospital, School
of Medicine, Zhejiang University (approval no. IIT20200016A).
Patients were informed that the clinical information were stored by
the hospital and potentially used for scientific research, and
signed informed consent to participants was waived by the Ethics
Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AEs
|
adverse effects
|
|
ALB
|
albumin
|
|
ALT
|
alanine aminotransferase
|
|
AUC
|
area under the curve
|
|
AI
|
artificial intelligence
|
|
BMI
|
body mass index
|
|
EHR
|
electronic health record
|
|
EPV
|
events-per-variable
|
|
Hb
|
hemoglobin
|
|
LR
|
logistic regression
|
|
ML
|
machine learning
|
|
MLP
|
multi-layer perceptron
|
|
NSCLC
|
non-small cell lung cancer
|
|
PLT
|
platelet
|
|
ROC
|
receiver operating characteristic
curve
|
|
RECIST
|
Response Evaluation Criteria in Solid
Tumors
|
|
RF
|
random forest
|
|
ToP
|
total protein
|
|
WBC
|
white blood cells
|
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI
|
|
3
|
Cao M and Chen W: Epidemiology of lung
cancer in China. Thorac Cancer. 10:3–7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Asamura H, Nishimura KK, Giroux DJ,
Chansky K, Hoering A, Rusch V and Rami-Porta R; Members of the
IASLC Staging, Prognostic Factors Committee of the Advisory Boards,
Participating Institutions, : IASLC Lung cancer staging project:
The new database to inform revisions in the ninth edition of the
TNM classification of lung cancer. J Thorac Oncol. 18:564–575.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pirker R: Chemotherapy remains a
cornerstone in the treatment of nonsmall cell lung cancer. Curr
Opin Oncol. 32:63–67. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Powell SF, Rodríguez-Abreu D, Langer CJ,
Tafreshi A, Paz-Ares L, Kopp HG, Rodríguez-Cid J, Kowalski DM,
Cheng Y, Kurata T, et al: Outcomes with pembrolizumab plus
platinum-based chemotherapy for patients with NSCLC, sTable brain
metastases: Pooled analysis of KEYNOTE-021, −189, and −407. J
Thorac Oncol. 16:1883–1892. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Forde PM, Spicer J, Lu S, Provencio M,
Mitsudomi T, Awad MM, Felip E, Broderick SR, Brahmer JR, Swanson
SJ, et al: Neoadjuvant nivolumab plus chemotherapy in resecTable
lung cancer. N Engl J Med. 386:1973–1985. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang C, Qiao W, Jiang Y, Zhu M, Shao J,
Wang T, Liu D and Li W: The landscape of immune checkpoint
inhibitor plus chemotherapy versus immunotherapy for advanced
non-small-cell lung cancer: A systematic review and meta-analysis.
J Cell Physiol. 235:4913–4927. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang J, Wang Y, Gao Y, Sugimura H,
Minervini F, Uchino J, Halmos B, Yendamuri S, Velotta JB and Li M:
Neoadjuvant immunotherapy or chemoimmunotherapy in non-small cell
lung cancer: A systematic review and meta-analysis. Transl Lung
Cancer Res. 11:277–294. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
NSCLC Meta-analysis Collaborative Group, .
Preoperative chemotherapy for non-small-cell lung cancer: A
systematic review and meta-analysis of individual participant data.
Lancet. 383:1561–1571. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lavan AH, O'Mahony D, Buckley M, O'Mahony
D and Gallagher P: Adverse drug reactions in an oncological
population: Prevalence, predictability, and preventability.
Oncologist. 24:e968–e977. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu M, Wu Y, Chen Y, Sun J, Zhao Z, Chen
XW, Matheny ME and Xu H: Large-scale prediction of adverse drug
reactions using chemical, biological, and phenotypic properties of
drugs. J Am Med Inform Assoc. 19:e28–e35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Z, Clark NR and Ma'ayan A:
Drug-induced adverse events prediction with the LINCS L1000 data.
Bioinformatics. 32:2338–2345. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo H, Fokoue-Nkoutche A, Singh N, Yang L,
Hu J and Zhang P: Molecular docking for prediction and
interpretation of adverse drug reactions. Comb Chem High Throughput
Screen. 21:314–322. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Blonde L, Khunti K, Harris SB, Meizinger C
and Skolnik NS: Interpretation and impact of real-world clinical
data for the practicing clinician. Adv Ther. 35:1763–1774. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shickel B, Tighe PJ, Bihorac A and Rashidi
P: Deep EHR: A survey of recent advances in deep learning
techniques for Electronic Health Record (EHR) analysis. IEEE J
Biomed Health Inform. 22:1589–1604. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rajkomar A, Oren E, Chen K, Dai AM, Hajaj
N, Hardt M, Liu PJ, Liu X, Marcus J, Sun M, et al: Scalable and
accurate deep learning with electronic health records. NPJ Digit
Med. 1:182018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Choi E, Schuetz A, Stewart WF and Sun J:
Using recurrent neural network models for early detection of heart
failure onset. J Am Med Inform Assoc. 24:361–370. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bernardini M, Romeo L, Misericordia P and
Frontoni E: Discovering the type 2 diabetes in electronic health
records using the sparse balanced support vector machine. IEEE J
Biomed Health Inform. 24:235–246. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tseng PY, Chen YT, Wang CH, Chiu KM, Peng
YS, Hsu SP, Chen KL, Yang CY and Lee OK: Prediction of the
development of acute kidney injury following cardiac surgery by
machine learning. Crit Care. 24:4782020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu X, Zheng D, Zhong Y, Xia Z, Luo H and
Weng Z: Machine-learning prediction of oral drug-induced liver
injury (DILI) via multiple features and endpoints. Biomed Res Int.
2020:47951402020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pedregosa F, Varoquaux G, Gramfort A,
Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R,
Dubourg V, et al: Scikit-learn: Machine learning in python. J Mach
Learn Res. 12:2825–2830. 2011.
|
|
23
|
Virtanen P, Gommers R, Oliphant TE,
Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P,
Weckesser W, Bright J, et al: Author correction: SciPy 1.0:
Fundamental algorithms for scientific computing in python. Nat
Methods. 17:3522020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang S, He T, Yang S, Sheng H, Tang X,
Bao F, Wang Y, Lin X, Yu W, Cheng F, et al: Metformin reverses
chemoresistance in non-small cell lung cancer via accelerating
ubiquitination-mediated degradation of Nrf2. Transl Lung Cancer
Res. 9:2337–2355. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Murphy KP: Machine learning: A
probabilistic perspective. The MIT Press; Cambridge, Massachusetts:
2012
|
|
26
|
Carr DF and Pirmohamed M: Biomarkers of
adverse drug reactions. Exp Biol Med (Maywood). 243:291–299. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kheifetz Y and Scholz M: Individual
prediction of thrombocytopenia at next chemotherapy cycle:
Evaluation of dynamic model performances. Br J Clin Pharmacol.
87:3127–3138. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Zhang R, Shen Y, Su L, Dong B and
Hao Q: Prediction of chemotherapy adverse reactions and mortality
in older patients with primary lung cancer through frailty index
based on routine laboratory data. Clin Interv Aging. 14:1187–1197.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dranitsaris G, Molassiotis A, Clemons M,
Roeland E, Schwartzberg L, Dielenseger P, Jordan K, Young A and
Aapro M: The development of a prediction tool to identify cancer
patients at high risk for chemotherapy-induced nausea and vomiting.
Ann Oncol. 28:1260–1267. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Boudali I and Messaoud IB: Machine
learning models for toxicity prediction in chemotherapy.
Intelligent Systems Design and Applications. Springer Nature
Switzerland Cham; pp. 350–364. 2023, View Article : Google Scholar
|
|
31
|
Wu Y, Zhao W, Zhang L, Wang Y, Wen Y and
Liu L: Machine learning models for predicting chemotherapy-induced
adverse drug reactions in colorectal cancer patients. Dig Liver
Dis. 57:1845–1852. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Polevikov S: Advancing AI in healthcare: A
comprehensive review of best practices. Clin Chim Acta.
548:1175192023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Adlung L, Cohen Y, Mor U and Elinav E:
Machine learning in clinical decision making. Med. 2:642–665. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kline A, Wang H, Li Y, Dennis S, Hutch M,
Xu Z, Wang F, Cheng F and Luo Y: Multimodal machine learning in
precision health: A scoping review. NPJ Digit Med. 5:1712022.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zheng S, Zhu Z, Liu Z, Guo Z, Liu Y, Yang
Y and Zhao Y: Multi-modal graph learning for disease prediction.
IEEE Trans Med Imaging. 41:2207–2216. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Luscher TF, Wenzl FA, D'Ascenzo F,
Friedman PA and Antoniades C: Artificial intelligence in
cardiovascular medicine: Clinical applications. Eur Heart J.
45:4291–4304. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Petzl AM, Jabbour G, Cadrin-Tourigny J,
Pürerfellner H, Macle L, Khairy P, Avram R and Tadros R: Innovative
approaches to atrial fibrillation prediction: Should polygenic
scores and machine learning be implemented in clinical practice?
Europace. 26:euae2012024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Couckuyt A, Seurinck R, Emmaneel A,
Quintelier K, Novak D, Van Gassen S and Saeys Y: Challenges in
translational machine learning. Human Genetics. 141:1451–1466.
2022. View Article : Google Scholar : PubMed/NCBI
|