Introduction
Post-translational modifications (PTMs) are
regulatory mechanisms that modulate protein function, serving roles
in cellular signaling and disease pathogenesis (1–4).
Chromatin is a dynamic nucleoprotein complex localized in cell
nuclei, consisting of DNA, histones, non-histone proteins and small
regulatory RNAs. It serves a role in essential nuclear processes,
including the regulation of gene expression, DNA replication and
repair and mitotic chromosome segregation (5–8). The
N-terminal tails of core histones represent the major sites for
post-translational chromatin modifications, which dynamically
regulate chromatin accessibility and function (9). Aberrant histone PTMs constitute a
fundamental epigenetic mechanism in oncogenesis that dysregulates
transcriptional networks, DNA repair fidelity and the fate of the
cell, which promotes tumor development and metastatic competence
(10–14).
Lactate is a key signaling molecule that mediates
intercellular communication, with protein lactylation, a novel PTM,
emerging as a central mechanism underlying lactate-dependent
regulation. This epigenetic modification promotes tumor progression
through lactate accumulation-dependent mechanisms and the
subsequent activation of specific lactylation-promoting
acyltransferases (15,16). First identified as a novel PTM in a
study by Zhang et al (17)
in 2019, histone lactylation is a ubiquitously present,
evolutionarily conserved and dynamically regulated lactate-mediated
epigenetic mark. Furthermore, it is preferentially enriched at gene
promoters and enhancers, serving as a potential indicator of
transcriptionally active chromatin (17,18).
As an epigenetic regulation, histone lactylation mediates
transcriptional control through lactyl group deposition on histone
lysines, which alters nucleosome dynamics and DNA-histone
interactions (19). Histone
lactylation serves a role in cancer progression and treatment
resistance, with aberrant histone lactylation levels serving as a
prognostic indicator for patients with cancer (20,21).
Targeting histone lactylation may potentially be used for precision
cancer therapy (22,23).
The present review discussed the current knowledge
on histone lactylation and emphasized its role as a pivotal
metabolic-epigenetic nexus in cancer. The present review described
its mechanistic contributions to tumor progression, immune evasion
and therapy resistance. Furthermore, the cascading interplay
between lactylation and other PTMs (histone acetylation and histone
methylation) were evaluated and emerging therapeutic strategies
targeting the lactylation axis were discussed.
Lactate metabolism and lactate
transport
During glycolysis, glucose undergoes sequential
enzymatic breakdown to yield pyruvate, which is subsequently
converted to lactate via NADH-dependent reduction. This process is
catalyzed by lactate dehydrogenase (LDH), resulting in lactate
accumulation (24). Lactate is
actively shuttled across cellular membranes via monocarboxylate
transporters (MCTs) (25). Under
conditions of adenosine triphosphate (ATP) depletion and hypoxia,
oxidative phosphorylation is compromised, leading to suppression of
the tricarboxylic acid cycle and the upregulation of glycolytic
flux to maintain cellular ATP levels (26). During physiological stress, such as
intense exercise, skeletal muscle cells exhibit marked lactate
production. This metabolic byproduct contributes to an increase in
the carbon dioxide concentrations, which stimulates the respiratory
center to enhance pulmonary ventilation, thereby meeting the
increased oxygen demands (27).
During inflammation, lactate functions as a pleiotropic signaling
molecule that orchestrates immune cell responses during
inflammation (28).
The core function of mitochondria is to generate
energy through oxidative phosphorylation. Under normal
physiological conditions, pyruvate enters the mitochondria and is
metabolized, resulting in minimal lactate production. However,
during mitochondrial dysfunction, the transport of pyruvate into
mitochondria is impaired, resulting in cells being reliant on
glycolysis, which leads to lactate accumulation. Therefore, the
metabolic state of mitochondria determines the intracellular levels
of lactate (29–31). Abnormal lactate accumulation
promotes tumorigenesis and progression through histone lactylation
(25,32,33).
In the Warburg effect, glucose is preferentially converted into
lactate to facilitate rapid energy production, which is concomitant
with metabolic reprogramming that is characterized by enhanced
aerobic glycolysis, lactate accumulation, suppressed mitochondrial
oxidative phosphorylation and compromised mitochondrial function
(34,35).
Histone lactylation in tumor
progression
Histones are highly conserved, nuclear-encoded basic
proteins characterized by their abundance of positively charged
amino acids (lysine and arginine residues) (16). As the core structural components of
nucleosomes, they organize and compact eukaryotic DNA into
chromatin (36). In addition to
their architectural role, histones dynamically regulate essential
nuclear processes including transcriptional control, DNA damage
response and mitotic chromosome segregation (36). Histones are classified into two
categories, namely core and linker histones. The core histones
(H2A, H2B, H3 and H4) form the octameric structural scaffold of
nucleosomes, while the H1 linker histone binds to the
inter-nucleosomal DNA regions (16,37,38).
Histone H3 lysine 18 lactylation
(H3K18la) in tumor progression
Multiple histone lactylation sites are identified on
histone H3 and H4 (39,40). Since its initial identification,
H3K18la is the most studied histone lactylation site (17). Previous studies demonstrate that
H3K18la influences tumor initiation, progression and metastasis
through transcriptional regulation (41,42).
In uveal melanoma, H3K18la transcriptionally upregulates YTH
N6-methyladenosine RNA-binding protein F2 (YTHDF2), which promotes
uveal melanoma pathogenesis. Mechanistically, YTHDF2 recognizes and
binds to N6-methyladenosine (m6A)-modified period circadian
regulator 1 and TP53 mRNAs, which facilitates their degradation and
accelerates uveal melanoma tumorigenesis (43). In ocular melanoma, increased levels
of H3K18la epigenetically activates the transcription of
α-ketoglutarate-dependent dioxygenase homolog 3 (ALKBH3). ALKBH3
mediates the N1-methyladenosine demethylation of SP100A mRNA, which
promotes malignant transformation (44). In hepatocellular carcinoma (HCC),
histone lactylation (H3K18la) and histone H3 lysine 27 acetylation
(H3K27ac) cooperatively regulate tumor progression through an
epigenetic cascade mechanism. H3K18la upregulates the expression of
the m6A reader protein YTH m6A RNA binding protein C1 (YTHDC1),
which stabilizes m6A-modified long non-coding RNA nuclear
paraspeckle assembly transcript 1 (NEAT1) and recruits histone
acetyltransferase p300 to the promoter region of the stearoyl-CoA
desaturase (SCD) gene. This leads to increased histone H3K27ac
levels, which activates the expression of SCD, induces fatty acid
metabolic reprogramming and promotes HCC progression (45). This exemplifies a complex cascade of
epigenetic modifications, in which histone lactylation (H3K18la)
acts as an upstream regulator that promotes downstream histone
acetylation (H3K27ac), and induces oncogenic metabolic
reprogramming. This exemplifies a complex cascade of epigenetic
modifications, in which histone lactylation acts as an upstream
regulator that directly promotes downstream histone acetylation,
thereby driving oncogenic metabolic reprogramming. It underscores
that histone PTMs do not function in isolation but can form a
hierarchical cascade, with upstream modifications dictating
downstream epigenetic events. In glioblastoma (GBM), the NF-κB
pathway promotes H3K18la via the Warburg effect, which enhances the
expression of the long non-coding RNA LINC01127. Subsequently,
LINC01127 activates the mitogen-activated protein kinase kinase
kinase kinase 4/JNK/NF-κB signaling axis, promoting self-renewal of
GBM stem cells (46).
Dysfunctional mitochondrial processes (including
impaired mitochondrial oxidative phosphorylation and dysregulated
mitophagy) and proteins [such as sirtuin (SIRT), Parkin and Numb]
serve as central therapeutic targets in various diseases (such as
neurodegenerative diseases, cancer and metabolic disorders)
(47–55). Glycolysis and oxidative
phosphorylation are the primary pathways for cellular energy
production. When mitochondrial oxidative phosphorylation is
impaired, glycolysis is enhanced, leading to an increased
accumulation of lactic acid in cells, which further influences
histone lactylation (34). SIRT 4,
a member of the mitochondrial SIRT family, serves a key role in
regulating mitochondrial function. As a mitochondrial protein,
SIRT4 deacetylates glycolytic enzyme α-enolase at lysine 358. This
promotes a metabolic shift from mitochondrial oxidative
phosphorylation to glycolysis and enhances lactate production. The
accumulated lactate subsequently induces histone lactylation
(H3K18la and H3K9la), which activates stemness-related pathways via
epigenetic reprogramming, and enhances the stem-like properties of
pancreatic cancer (PCA) cells (52).
Mitophagy is a highly conserved and crucial cellular
self-cleaning process. Its function is to maintain intracellular
homeostasis by removing damaged mitochondria. This ensures that
there is an energy supply and prevents the release of pro-apoptotic
factors (such as cytochrome c and second mitochondria-derived
activator of caspases) and excessive reactive oxygen species (ROS)
that could lead to cellular damage or death (53,54).
The regulation of mitophagy influences cancer development (47). In prostate and lung cancer, the cell
fate determinant Numb binds to and promotes Parkin-mediated
mitophagy, which maintains mitochondrial stability. When the
Numb/Parkin axis is impaired, damaged mitochondria accumulate,
which triggers a metabolic reprogramming toward aerobic glycolysis.
This shift increases lactate production, which subsequently induces
epigenetic reprogramming via histone lactylation (H3K18la),
activating the transcription of neuroendocrine-related genes such
as synaptophysin, neural cell adhesion molecule-1 and
neuron-specific enolase. Ultimately, this process promotes the
transformation of cancer cells toward a neuroendocrine phenotype
and confers therapy resistance (55).
H3K18la also functions as a key epigenetic promoter
of metastatic progression in multiple types of cancer, such as HCC,
CRC, endometrial carcinoma and BC (33,41,42,56,57). A
study by Wang et al (56)
demonstrates using in vitro and in vivo experiments
that pyrroline-5-carboxylate reductase 1 (PYCR1) increases insulin
receptor substrate 1 (IRS1) expression levels by promoting H3K18la
at the IRS1 promoter region. This PYCR1/H3K18la/IRS1 axis
subsequently activates the PI3K/Akt/mTOR signaling pathway, which
promotes the proliferation and migration of HCC cells. H3K18la
promotes colorectal cancer (CRC) liver metastasis (CRLM) by
upregulating the expression of cell division cycle 27.
Mechanistically, this is promoted by lactate accumulation, which
results from the ubiquitin-specific peptidase 3 (USP3)-antisense
RNA 1/USP3/MYC axis-mediated activation of the glycolytic pathway
(41). Bioinformatic analysis
reveals that histone lysine lactylation (Kla) serves as a poor
prognostic biomarker in breast cancer (BC), which under high
glycolytic flux promotes BC cell proliferation and migration by
increasing c-Myc expression levels, upregulating
serine/arginine-rich splicing factor 10 and consequently modulating
the alternative splicing of murine double minute 4 and Bcl-x
(33,57). In endometrial carcinoma, H3K18la
positively regulates the expression of USP39. Subsequently, USP39
activates the PI3K/AKT/hypoxia inducible factor 1α signaling
pathway, promoting lactate production, proliferation and metastasis
(42).
Histone lactylation at other
sites
In addition to the well-characterized H3K18la, other
known lactylation sites include H3K27la, H3K14la, H3K9la and
H3K56la on histone H3, as well as histone H4 lysine 5 lactylation
(H4K5la), H4K8la, H4K12la and H4K16la on histone H4 (39,40,58).
These modifications exhibit distinct regulatory roles in tumor
progression, with mechanisms that include: i) Inhibition of tumor
apoptosis (59,60); ii) regulation of tumor metastasis
and invasion (61,62); and iii) induction of cellular
senescence and telomerase inhibition (58).
Bioinformatics analysis and experimental validation
using clinical samples reveals H4K12la as a novel prognostic
biomarker for triple-negative BC (TNBC) (59). A study by Li et al (60) demonstrates that lactate mediates
H4K12la binding to the Schlafen 5 (SLFN5) promoter region, which
suppresses the transcription of SLFN5 and subsequently inhibits
apoptosis in TNBC. Hypoxia-induced H3K9la promotes migration and
invasion in esophageal squamous cell carcinoma (ESCC) by
upregulating the expression of laminin subunit g 2 (LAMC2).
Mechanistic study further revealed that LAMC2 enhances the
expression of vascular endothelial growth factor by activating the
PI3K/Akt signaling pathway (61).
In patients with neuroblastoma (NB), low expression
levels of DnaJ heat shock protein family member C12 (DNAJC12) is
associated with poor clinical outcomes. Functionally, DNAJC12
deficiency upregulates the expression of collagen type Iα1 through
H4K5la-mediated epigenetic regulation, which promotes NB cell
invasion and metastasis. Mechanistically, this process involves
DNAJC12 downregulation enhances the glycolytic pathway by
upregulating key enzymes such as hexokinase 2, pyruvate kinase
M1/M2 (PKM), lactate dehydrogenase A (LDHA) and LDHB. This
activation leads to lactate accumulation, which in turn promotes
H4K5la (62).
In lung adenocarcinoma (LUAD), liver kinase B1
regulates lactate metabolism to suppress H4K8la and H4K16la, which
inhibits telomerase reverse transcriptase gene expression and
represses telomerase activity, leading to senescence in LUAD cells
(58).
Histone lactylation, particularly H3K18la, promotes
oncogenesis across various types of cancer (such as HCC, CRC, BC,
GBM and LUAD) through epigenetic reprogramming, metabolic rewiring
and the activation of oncogenic signaling pathways (such as the
PI3K/Akt/mTOR signaling pathway and PI3K/AKT/HIF-1α signaling
pathway) by modulating chromatin architecture and transcriptional
regulation. Additional lactylation sites (H4K8la, H4K16la, H4K12la,
H3K9la, H4K5la) are implicated in tumor progression through diverse
mechanisms including apoptosis inhibition, metastasis promotion and
cellular senescence inhibition (Table
I). However, notable gaps in the knowledge remain regarding the
functional roles and specific downstream targets of H3K27la and
H3K56la in carcinogenesis. Furthermore, studies on histone
lactylation sites predominantly focus on a single type of cancer,
highlighting the need for systematic pan-cancer analyses (58–61).
 | Table I.Effects of histone lactylation on
tumor cells. |
Table I.
Effects of histone lactylation on
tumor cells.
| First author,
year | Type of cancer | Lactylation
site | Downstream
targets | Associated
molecules or pathways | Effects | (Refs.) |
|---|
| Zhou et al,
2025 | CRC | H3K18la | CDC27 | Not mentioned | Liver
metastases | (41) |
| Wei et al,
2024 | EC | H3K18la | USP39 | PI3K/AKT/HIF-1α
signaling pathway | Proliferation and
metastasis | (42) |
| Yu et al,
2021 | Uveal melanoma | H3K18la | YTHDF2 | PER1 and TP53 | Oncogenesis | (43) |
| Gu et al,
2024 | Ocular
melanoma | H3K18la | ALKBH3 | SP100A | Progression | (44) |
| Du et al,
2024 | HCC | H3K18la | YTHDC1 | NEAT1, p300 and
H3K27ac | Progression | (45) |
| Li et al,
2023 | GBM | H3K18la | LINC01127 | MAP4K4/JNK/NF-κB
axis | Stem cells
self-renew | (46) |
| Wang et al,
2024 | HCC | H3K18la | IRS1 | PI3K/Akt/mTOR
signaling pathway | Proliferation and
migration | (56) |
| Pandkar et
al, 2023 | BC | H3K18la | c-Myc | SRSF10, MDM4 and
Bcl-x | Proliferation and
migration | (33) |
| Liu et al,
2024 | LUAD | H4K8la and
H4K16la | TERT | Not mentioned | Cellular
senescence | (58) |
| Li et al,
2024 | TNBC | H4K12la | SLFN5 | Not mentioned | Progression | (60) |
| Zang et al,
2024 | ESCC | H3K9la | Not mentioned | PI3K/Akt signaling
pathway and VEGFA | Migration and
invasion | (61) |
| Yang et al,
2025 | NB | H4K5la | COL1A1 | Not mentioned | Migration and
invasion | (62) |
Histone lactylation modulates tumor
progression by remodeling the tumor microenvironment (TME)
Tumor-associated macrophages
(TAMs)
First revealed in macrophage research, histone
lactylation promotes the expression of M2-like
macrophage-associated genes (17).
Further studies reveal that histone lactylation promotes tumor
progression by inducing M2 polarization of TAMs (21,63–66).
A study by Li et al (21) demonstrates that lactate derived from
CRC cells promotes tumorigenesis by inducing H3K18la in
macrophages, which suppresses the expression of retinoic acid
receptor γ and activates the TNF receptor associated factor
6-IL-6-STAT3 signaling axis.
In bladder cancer (BLCA), combining single-cell RNA
sequencing with computational biology and functional assays, a
study by Deng et al (63)
reveals that H3K18la epigenetically enhances parkin RBR E3
ubiquitin protein ligase transcription. This promotes both
mitochondrial autophagy and M2-like polarization in TAMs, which
forms an immunosuppressive microenvironment that increases BLCA
progression. Additionally, under hypoxic conditions, lactate
produced by glioma cells promotes M2 polarization of TAMs through
the MCT1/H3K18la/TNF superfamily member 9 axis (64). In addition, H3K18la in macrophages
upregulates the expression of nuclear protein 1 (NUPR1), which
promotes M2-like polarization through the suppression of ERK and
JNK signaling pathways. This NUPR1-mediated reprogramming increases
the levels of immune checkpoint molecules programmed death ligand 1
(PD-L1) and signal regulatory protein α, which induces
CD8+ T-cell dysfunction and compromises the efficacy of
immunotherapy (65). In PCA,
lactate-induced H3K18la activates acetyl-CoA acetyltransferase 2
(ACAT2) at the transcriptional level, establishing a positive
feedback loop of lactate/H3K18la/ACAT2/mitochondrial carrier
homolog 2, which impairs mitochondrial oxidative phosphorylation
and promotes lactate accumulation. In addition, ACAT2 facilitates
cholesterol transport via small extracellular vesicles, which
promotes the M2 polarization of TAMs (66).
In addition to its role in macrophage polarization,
histone lactylation can also regulate the phagocytic capacity of
macrophages. In PTEN/p53-deficient aggressive PCA, tumor-derived
lactate suppresses the phagocytic function of TAMs via H3K18la,
which promotes tumor progression (67). Elevated H3K18la levels in
tumor-infiltrating myeloid cells upregulates the expression of
methyltransferase-like 3, which increases the RNA m6A modification.
This subsequently increases the mRNA translation efficiency of
Janus kinase 1 (JAK1) and activates the JAK1/STAT3 signaling
pathway, which amplifies the immunosuppressive function of
tumor-infiltrating myeloid cells in tumors (68).
Cancer-associated fibroblasts
(CAFs)
Functioning as critical microenvironmental
modulators, CAFs contribute to tumor pathogenesis (69). Lactate derived from lung cancer
cells promotes the expression of collagen triple helix
repeat-containing 1 (CTHRC1) in CAFs via H3K18la, which maintains
their CTHRC1+ CAF phenotype. These activated
CTHRC1+ CAFs subsequently upregulate the expression of
hexokinase 2 in tumor cells through the activation of TGF-β/Smad
signaling. This then induces a resistance to epidermal growth
factor receptor tyrosine kinase inhibitors and enhances glycolytic
metabolism in lung cancer cells (70).
Neutrophils
G protein-coupled receptor 37 promotes CRLM
progression through dual mechanisms: i) It activates the Hippo
signaling pathway to enhance tumor cell proliferation; and ii) it
upregulates the expression of chemokine (C-X-C motif) ligand (CXCL)
1 and 5 via H3K18la, which promotes neutrophil accumulation in the
TME. These coordinated actions establish a pro-metastatic niche
that facilitates CRLM development (71). Hypoxia-induced glycolytic activation
in neutrophils increases lactate-dependent histone lactylation,
which transcriptionally activates arginase-1 to enhance
immunosuppression. Inhibition of this lactylation pathway reverses
neutrophil-mediated immune evasion and improves the response of
brain tumors to immunotherapy (72).
CD8+ T cells
Histone lactylation exhibits dual roles in
modulating CD8+ T cell function in the TME. Tumor
cell-intrinsic histone lactylation induces CD8+ T cell
exhaustion; however, CD8+ T cell-autonomous histone
lactylation enhances their antitumor immunity. In head and neck
squamous cell carcinoma, H3K9la upregulates the expression of IL-11
in tumor cells. This tumor-derived IL-11 then induces
CD8+ T cell exhaustion via the JAK2/STAT3 signaling
pathway (73). A study using the
MB49 tumor model demonstrates that combination therapy with
anti-programmed cell death protein 1 (PD-1) and anti-cytotoxic
T-lymphocyte associated protein 4 (CTLA-4) antibodies markedly
elevates the H3K18la and H3K9la levels in tumor-infiltrating
CD8+ T cells, which is associated with marked tumor
growth inhibition (74).
In summary, histone lactylation, particularly
H3K18la, is a pivotal epigenetic mechanism that orchestrates the
immunosuppressive TME through multifaceted pathways including
immune cell reprogramming, stromal cell activation and therapy
resistance. Mechanistically, histone lactylation mediates
immunosuppression by polarizing myeloid cells toward an M2-like
phenotype, promoting CD8+ T cell exhaustion and
enhancing neutrophil-mediated immunosuppression. Additionally,
CD8+ T cell-intrinsic histone lactylation exhibits
context-dependent effects, potentiating antitumor activity when
induced by immune checkpoint blockades (Table II).
 | Table II.Effects of histone lactylation on the
tumor microenvironment. |
Table II.
Effects of histone lactylation on the
tumor microenvironment.
| First author,
year | Type of cancer | Cell type | Lactylation
site | Downstream
targets | Associated
molecules or pathways | Effects | (Refs.) |
|---|
| Li et al,
2024 | CRC | TAMs | H3K18la | RARγ |
TRAF6/IL-6/STAT3 | Oncogenesis | (21) |
| Deng et al,
2025 | BLCA | TAMs | H3K18la | PRKN | Not mentioned | Mitophagy and M2
polarization | (63) |
| Li et al,
2024 | GBM | TAMs | H3K18la | TNFSF9 | Not mentioned | M2
polarization | (64) |
| Cai et al,
2025 | HCC | TAMs | H3K18la | NUPR1 | ERK and JNK
signaling pathways | M2
polarization | (65) |
| Yang et al,
2025 | PCA | TAMs | H3K18la | ACAT2 | Not mentioned | M2
polarization | (66) |
| Chaudagar et
al, 2023 | Prostate
cancer | TAMs | H3K18la | Not mentioned | Not mentioned | Inhibition of
phagocytosis in TAMs | (67) |
| Xiong et al,
2022 | CRC | TIMs | H3K18la | METTL3 | JAK1/STAT3 | Tumor immune
evasion | (68) |
| Zhang et al,
2025 | Lung cancer | CAFs | H3K18la | Cthrc1 | TGF-β/Smad | Resistance to
EGFR-TKI | (70) |
| Zhou et al,
2023 | CRC | Neutrophils | H3K18la | CXCL1 and 5 | Not mentioned |
Immunosuppression | (71) |
| Ugolini et
al, 2025 | Brain tumor | TIMs | Not mentione2 | ARG1 | Not mentioned |
Immunosuppression | (72) |
| Wang et al,
2024 | HNSCC | CD8+ T
cells | H3K9la | IL-11 | JAK2/STAT3 | Dysfunction of
CD8+ T cells | (73) |
| Raychaudhuri et
al, 2024 | BLCA | CD8+ T
cells | H3K18la and
H3K9la | Not mentioned | Not mentioned | Tumor
suppression | (74) |
Histone lactylation and chemoresistance
Emerging evidence demonstrates that this epigenetic
modification regulates chemotherapeutic responses through multiple
molecular mechanisms, which contributes to chemoresistance in
malignant tumors.
Modulation of cellular autophagy
pathways
Compared with bevacizumab-sensitive patients, the
histone lactylation level is markedly higher in
bevacizumab-resistant patients with CRC. In patient-derived tumor
xenograft models and patient-derived organoids, the combination of
bevacizumab and histone lactylation inhibitor (oxamate) relieves
the bevacizumab resistance in CRC. Mechanistically, H3K18la
activates the transcription of the autophagy-enhancing gene
rubicon-like autophagy enhancer (RUBCNL). RUBCNL interacts with
Beclin 1 to mediate the recruitment and function of the class III
phosphatidylinositol 3-kinase complex, which promotes autophagosome
maturation (75).
Regulation of transcriptional activity
and activation of proliferation-associated signaling pathways
In BLCA, H3K18la promotes cisplatin resistance by
facilitating the transcriptional induction of Y-Box binding protein
1 and Yin Yang 1 transcription factor (76). In HCC, H3K18la exacerbates
resistance to lenvatinib by promoting the transcription of the HECT
domain E3 ubiquitin protein ligase 2 (HECTD2) gene. HECTD2
acts as an E3 ubiquitin ligase for Kelch-like ECH associated
protein 1 (KEAP1), facilitating the degradation of KEAP1 protein
and activating the antioxidant response, which induces lenvatinib
resistance in HCC cells (77).
H3K14la promotes multi-drug (oxaliplatin and 5-fluorouracil)
resistance through neural precursor cell expressed, developmentally
downregulated protein 4 E3 ubiquitin protein ligase-mediated
phosphatase and PTEN ubiquitination and degradation, which leads to
the subsequent activation of the PI3K/Akt/mTOR signaling pathway in
HCC (78). However, the suppression
of histone lactylation enhances the sensitivity of cancer to
chemotherapy.
In GBM, peroxisome proliferator-activated receptor α
activates the p38 MAPK signaling pathway, which in turn upregulates
acyl-CoA oxidase 1 (ACOX1). ACOX1 suppresses lactate-mediated
H3K18la by promoting ROS-dependent downregulation of PKM2, which
enhances the sensitivity of GBM to temozolomide (TMZ) (79). Transfer RNA-derived small RNA
(tsRNA)-08614 enhances the sensitivity of CRC to oxaliplatin by
targeting aldehyde dehydrogenase 1 family member A3 (ALDH1A3) to
inhibit glycolysis and histone lactylation modifications.
Mechanistically, tsRNA-08614 downregulates the expression of
ALDH1A3, which reduces lactate production and decreases the level
of H3K18la. The decreased H3K18la level suppresses the
transcriptional activity of EF-hand domain family member D2, which
promotes the sensitivity of CRC to oxaliplatin (80).
Inhibition of ferroptosis
H4K12la in CRC stem cells suppresses ferroptosis
signaling by upregulating the expression of glutamate-cysteine
ligase, which enhances oxaliplatin resistance. This epigenetic
modification is dynamically regulated by p300-mediated lactylation
and histone deacetylase (HDAC) 1-mediated delactylation (81). Lactate derived from CAFs promotes
doxorubicin resistance in TNBC cells by enhancing the expression of
zinc finger protein 64 through H3K18la-mediated epigenetic
regulation, which suppresses ferroptosis (82).
Modulation of DNA damage repair
pathways
In lung cancer brain metastasis, aldo-keto reductase
family 1 member B10 enhances lactate accumulation. Lactate-induced
H4K12la transcriptionally activates the cell cycle gene cyclin B1,
which promotes DNA replication and cell cycle progression, inducing
an acquired resistance to pemetrexed (83). In ovarian cancer (OC), H4K12la
activates super-enhancers to recruit the oncogenic transcription
factor MYC, which upregulates the expression of the nucleotide
excision repair-related gene radiation sensitive protein (RAD)23
homolog A, nucleotide excision repair protein (84). Additionally, general control
non-depressible 5 (GCN5) promotes cisplatin resistance in OC by
upregulating H3K9la and RAD51 Kla to enhance homologous
recombination repair (85). H3K9la
promotes the transcription of LUC7-like 2, pre-mRNA splicing factor
(LUC7L2). However, in GBM, LUC7L2 downregulation reduces the
expression of MLH1, which suppresses mismatch repair and leads to
TMZ resistance (86).
In summary, histone lactylation acts as a crucial
mediator in chemoresistance across various types of cancer. It
promotes chemoresistance through multiple mechanisms, including
activation of autophagy-related pathways, modulation of
transcription factor activity, regulation of DNA repair,
interference with apoptosis, reprogramming of cellular metabolism
and stimulation of proliferation-related signaling pathways
(Table III).
 | Table III.An overview of the effects of histone
lactylation on chemotherapy. |
Table III.
An overview of the effects of histone
lactylation on chemotherapy.
| First author,
year | Type of cancer | Drug | Lactylation
site | Downstream
targets | Effects | (Refs.) |
|---|
| Li et al,
2024 | CRC | Bevacizumab | H3K18la | RUBCNL | Autophagosome
maturation | (75) |
| Li et al,
2024 | BLCA | Cisplatin | H3K18la | YBX1 and YY1 | Reduction in the
sensitivity of tumor cells to cisplatin | (76) |
| Dong et al,
2025 | HCC | Lenvatinib | H3K18la | HECTD2 | Reduction in the
sensitivity of tumor cells to lenvatinib | (77) |
| Zeng et al,
2025 | HCC | Oxaliplatin and
5-fluorouracil | H3K14la | The ubiquitin E3
ligase NEDD4 | Activation of
PI3K/Akt/mTOR signaling pathway | (78) |
| Wang et al,
2025 | GBM | Temozolomide | H3K18la | Not mentioned | Reduction in the
sensitivity of tumor cells to temozolomide | (79) |
| Chen et al,
2025 | CRC | Oxaliplatin | H3K18la | EFHD2 | Reduction in the
sensitivity of tumor cells to oxaliplatin | (80) |
| Deng et al,
2025 | CRC | Oxaliplatin | H4K12la | Glutamate-cysteine
ligase | Reduction in the
sensitivity of tumor cells to oxaliplatin | (81) |
| Zhang et al,
2025 | TNBC | Doxorubicin | H3K18la | ZFP64 | Reduction in
ferroptosis | (82) |
| Duan et al,
2023 | Lung cancer | Pemetrexed | H4K12la | CCNB1 | Increases in DNA
replication and the acceleration of the cell cycle | (83) |
| Lu et al,
2025 | Ovarian cancer | Niraparib | H4K12la | Super-enhancer | Increases in DNA
damage repair | (84) |
| Sun et al,
2025 | Ovarian cancer | Cisplatin | H3K9la | Not mentioned | Increases in
homologous recombination repair | (85) |
| Yue et al,
2024 | GBM | Temozolomide | H3K9la | LUC7L2 | Reductions in
mismatch repair | (86) |
Histone lactylation and immune checkpoint
inhibitors
Immune checkpoint inhibitors, such as CTLA-4 and
PD-1, function by blocking immune checkpoints to restore the
ability of the immune system to recognize and attack cancer cells
(87). Notable clinical success is
demonstrated with checkpoint proteins such as PD-1/PD-L1 and CTLA-4
in cancer treatment (88). Emerging
evidence reveals histone lactylation-mediated immune evasion
mechanisms across various types of cancer (Table IV). In non-small cell lung cancer
(NSCLC), H3K18la activates pore membrane protein 121 transcription,
which facilitates MYC nuclear translocation. This enhances MYC
binding to the CD274 promoter, which upregulates the expression of
PD-L1 and promotes immune evasion (89). In acute myeloid leukemia,
STAT5-induced lactate accumulation promotes E3 binding protein
nuclear translocation, which increases H4K5la levels. H4K15la
further elevates the expression of PD-L1, which establishes a
metabolic-epigenetic cascade linking lactate metabolism to immune
escape (90).
 | Table IV.An overview of the effects of histone
lactylation on immunotherapy. |
Table IV.
An overview of the effects of histone
lactylation on immunotherapy.
| First author,
year | Type of cancer | Lactylation
site | Downstream
targets | Immune
checkpoint | (Refs.) |
|---|
| Zhang et al,
2024 | Non-small cell lung
cancer | H3K18la | Pore membrane
protein 121 | PD-L1 | (89) |
| Huang et al,
2023 | Acute myeloid
leukemia | H4K5la | PD-L1 | PD-L1 | (90) |
| Li et al,
2024 | Gastric cancer | H3K18la | PD-L1 | PD-L1 | (91) |
| Chao et al,
2024 | Ovarian cancer | H3K18la | PD-L1 | PD-L1 | (92) |
| Ding et al,
2025 | HCC | H3K18la | PD-L1 | PD-L1 | (94) |
| Ma et al,
2025 | HCC | H3K18la | CD276 | CD276 | (95) |
| Wang et al,
2024 | Glioblastoma | Not mentioned | CD47 | CD47 | (98) |
CAF-derived lysyl oxidase activates the
TGF-β/insulin-like growth factor 1 signaling pathway to promote the
epithelial-mesenchymal transition and glycolysis in GC. The
resulting lactate accumulation enhances the H3K18la-mediated
expression of PD-L1 (91). Elevated
H3K18la levels are associated with poor prognosis in OC (92). LDHB, a subunit of lactate LDH,
facilitates immune evasion in OC by modulating H3K18la at the PD-L1
promoter to upregulate the expression of PD-L1 (93). Protein arginine methyltransferase 3
methylates pyruvate dehydrogenase kinase isoform 1 at residues R363
and R368, which enhances its kinase activity to promote glycolysis
and the accumulation of lactate. The resulting lactate induces
H3K18la, which directly binds to the PD-L1 promoter to upregulate
its expression, which promotes immune evasion in HCC (94). Histone lactylation modulates the
expression of PD-L1, which impairs the immune system recognition
and attack of cancer cells (91,93,94).
However, the role of histone lactylation in regulating CTLA-4
during cancer therapy is yet to be fully elucidated, which may be
an avenue for future investigation.
In addition to PD-L1 and CTLA-4, new immune
checkpoints, including B7 homologue 3 (B7-H3), are being
investigated (88). Lactate
upregulates the expression of B7-H3/CD276 in HCC cells via histone
lactylation modifications, particularly H3K18la, which suppresses
the proportion and function of tumor-infiltrating CD8+ T
cells and promotes tumor immune escape. Inhibiting lactate
metabolism reduces the expression of B7-H3 and synergizes with
anti-PD-1 therapy, which markedly suppresses tumor progression
(95).
CD47 (integrin-associated protein) is expressed on
the surface of various cell types, including tumor cells. Its
ligand, signal-regulatory protein α (SIRPα), is primarily expressed
on immune cells such as macrophages. When CD47 binds to SIRPα, a
signal is transmitted to macrophages, which inhibits their
phagocytic activity against the tumor cells (96,97).
In GBM stem cells, histone lactylation promotes the expression of
CD47, which inhibits the phagocytic activity of
microglia/macrophages against tumor cells and facilitates tumor
immune escape. Histone lactylation also enhances the substrate
specificity of histone acetyltransferase p300 for lactyl-CoA
through its interaction with heterochromatin protein chromobox
homolog 3 (CBX3), which further promotes the expression of
immunosuppressive cytokines (IL-4, IL-10 and IL-13) (98).
In summary, histone lactylation serves as a critical
epigenetic regulatory mechanism mediating tumor immune evasion.
This modification orchestrates the expression levels of immune
checkpoint molecules through multiple pathways, impairing immune
cell recognition and cytotoxic functions against tumor cells, which
leads to immune escape. Elucidating the precise mechanisms of
histone lactylation in tumor immune evasion may provide a
theoretical foundation for identifying novel immunotherapy targets
and may also facilitate the rational design of combination
therapies.
Current research status and future
directions of histone lactylation in cancer diagnosis or
treatment
Histone lactylation has the potential
to serve as a biomarker for cancer diagnosis and prognosis
The levels of histone lactylation modification show
marked differences in various types of cancer and may be a
potential diagnostic biomarker (57,99).
Combined with multi-omics analysis techniques, genes modified by
histone lactylation may also serve as novel biomarkers for
predicting the prognosis of cancer or evaluating therapeutic
efficacy (100).
The study by Hou et al (20) first reveals an association between
H3K18la and both severity and prognosis in PCA. The aforementioned
study demonstrates an elevated expression of H3K18la in PCA
tissues, which shows notable associations with serum lactate levels
as well as tumor markers such as carbohydrate antigen 19-9 and
carcinoembryonic antigen (CEA). These findings indicate that
H3K18la may be a promising novel biomarker for PCA diagnosis and
prognostic evaluation. A study by Zhu et al (99) performs a comprehensive evaluation of
the clinical significance of H4K5la in BC. The aforementioned study
demonstrates elevated H4K5la levels across multiple sample types,
including TNBC tissues, non-TNBC tissues and peripheral blood
mononuclear cells. Furthermore, the authors identify positive
associations between the expression of H4K5la and tumor progression
markers such as proliferation index Ki-67, CEA levels and lymph
node metastasis status. However, higher H4K5la expression levels
show a negative association with overall survival time, suggesting
its potential as both a prognostic indicator and therapeutic
monitoring marker in BC management. The study by He et al
(101) uses an integrated analysis
of transcriptomic and single-cell RNA sequencing data to identify
five histone lactylation-related prognostic genes (synuclein α
interacting protein, transmembrane protein 100, NLR family pyrin
domain containing 11, homeobox C11 and D10) in GBM. Based on these
molecular signatures, the aforementioned study presents a risk
prediction model that demonstrates robust prognostic performance in
patients with GBM.
Potential therapeutic strategies
targeting histone lactylation
Inhibition of histone lactylation through
targeting lactate transportation and glycolysis
MCTs, particularly MCT1 and MCT4, are key lactate
transporters that maintain cellular metabolic balance. MCT1
mediates bidirectional lactate transport and MCT4 primarily
facilitates lactate efflux, especially in highly glycolytic cells
such as cancer cells and immune cells (102). The MCT1 inhibitor AZD3965 can
increase the accumulation of lactate (103). At present, AZD3965, as the
first-in-class drug targeting tumor lactate metabolism, has entered
phase I/II clinical trials for the treatment of diffuse large
B-cell lymphoma and neuroendocrine tumors (NCT01791595) (104). The effects of MCT1 inhibition on
histone lactylation exhibit notable cell-type specificity and
microenvironmental dependency (105).
Glycolysis, a central metabolic pathway that
converts glucose into pyruvate and lactate to generate ATP and key
intermediates, promotes lactate accumulation in cells with a
Warburg effect. This links cellular metabolic status to histone
lactylation modifications (106).
Inhibition of glycolysis-related genes reduces lactate production,
which decreases histone lactylation levels. LDHA inhibitors
suppress tumor growth and synergize with immunotherapy (107). A study by Guo et al
(23) demonstrates that Fargesin
inhibits aerobic glycolysis in NSCLC cell lines by targeting
glycolysis-related genes such as LDHA, solute carrier family 2
member 1, and PKM2. The aforementioned study also reveals that
Fargesin suppresses the lactylation of histone H3, which inhibits
the growth of NSCLC. Tanshinone I, a bioactive diterpenoid derived
from Salvia miltiorrhiza (Danshen), suppresses OC
progression by downregulating glycolysis-related genes and
subsequently reducing H3K18la levels (22). Additionally, administration of
sodium dichloroacetate, the pyruvate dehydrogenase kinase 1
inhibitor, attenuates histone lactylation modifications (17).
Writers, erasers and readers of
histone lactylation
Histone lactylation is a dynamically regulated
process that is installed and removed by regulatory enzymes,
instead of being a spontaneous chemical reaction. It involves
‘writers’, ‘erasers’ and ‘readers’. These regulatory enzymes
regulate the dynamic balance of histone lactylation, which
influences gene expression levels and cellular functions (108–110).
The ‘writers’ refer to enzymes that add lactate
groups to lysine residues on histones. At present, known ‘writers’
for histone lactylation include acetyltransferase p300 (17,111),
GCN5 (also known as lysine acetyltransferase 2A), lysine
acetyltransferase 2A (KAT2A) (81,112,113), histone acetyltransferase binding
to ORC1 (HBO1; also known as KAT7) (114), KAT5 and KAT8 (115). After histone lactylation was first
demonstrated in 2019, p300 emerged as a candidate ‘writer’
(17). Since identifying p300 as a
histone acetyltransferase, a key question is how it specifically
catalyzes histone lactylation without affecting histone
acetylation. In GBM, CBX3 enhances the specificity of p300 for
lactyl-CoA through their interaction, which specifically catalyzes
histone lactylation without affecting histone acetylation. This
promotes tumor progression (99).
In atherosclerosis, the histone chaperone protein anti-silencing
function 1A histone chaperone acts as a cofactor for p300, and
regulates the enrichment of H3K18la at the snail family
transcriptional repressor 1 (SNAI1) promoter region to activate the
transcription of SNAI. This in turn induces the
endothelial-to-mesenchymal transition, and the use of the p300
agonist cholera toxin B markedly enhances H3K18la levels (116).
A study by Niu et al (114) demonstrates that HBO1 functions as
a lactyltransferase that preferentially catalyzes H3K9la to enhance
malignant behaviors, including proliferation, migration and
invasion in various cancer cell lines [including HeLa, HepG2 (HCC),
U87MG (gliomas), KYSE-30 (ESCC), MDA-MB-231 (BC), HCT116 (CRC) and
H460 (NSCLC)]. In GBM, it is KAT2A, instead of p300, that is the
‘writer’ and it promotes endothelial growth factor-induced H3K14la
and H3K18la. In addition, acyl-CoA synthetase short chain family
member 2 (ACSS2) binds to KAT2A, which then acts as a
lactyltransferase to catalyze the lactylation of histone H3 at K14
and K18 residues. This regulates gene expression levels and
promotes the proliferation and immune evasion of GBM cells. KAT2A
carried out distinct histone acylation in different protein
complexes and the interaction between ACSS2 and KAT2A promotes the
catalytic activity of KAT2A toward histone lactylation instead of
acetylation (113). A study by Zou
et al (115) demonstrates
that KAT5 and KAT8 function synergistically as ‘writers’ to mediate
lactate-induced H4K12la in macrophages. The aforementioned study
demonstrates that H4K12la is not influenced by other ‘writers’,
such as p300 and KAT2A.
‘Erasers’ are enzymes that remove lactylation
modifications from histones. Previous studies indicate that a
number of deacetylases, such as HDAC1-3 and SIRT1-3, act as
‘erasers’ to remove lactyl modifications (109,110,117–120). A study by Moreno-Yruela et
al (110) reports that HDAC1-3
function as ‘erasers’ to remove histone lactylation modifications.
At the H3K9 and H4K12 sites, HDAC3 has greater delactylation
activity compared with its deacetylation activity. Additionally,
HDAC1 and HDAC3, but not HDAC2, appear to preferentially regulate
H4K5la. A study by Xu et al (121) demonstrates that HDAC2 and HDAC3
function as ‘erasers’ for H3 Kla, participating in the regulation
of H3K18la-mediated BC cell proliferation. However, the
aforementioned study, reveals that HDAC1 levels show no association
with H3K18la. SIRT1 acts as a histone delactylase, inhibiting the
oncogenic positive feedback loop of long non-coding RNA
H19-glycolysis-H3K18la. Moreover, the combined use of the
SIRT1-specific activator SRT2104 and the LDHA inhibitor oxamate
markedly suppresses the growth of GC cells, with limited effects on
normal GC cells; thus, potentially providing a novel strategy for
GC treatment (122). A study by
Yang et al (62) also
demonstrates that SIRT2 and p300/CBP act as the ‘eraser’ and
‘writer’ of H4K5la, respectively, to regulate the metastatic
phenotype of NB cells. In a study on esophageal cancer, SIRT3 is
found to act as a specific ‘eraser’ of H3K9la, which inhibits the
progression of ESCC cells (123).
In addition, a study by Fan et al (109) suggests that SIRT3 also acts as an
‘eraser’ for H4K16la. The development of small-molecule modulators
targeting erasers may potentially be useful in cancer therapy.
‘Readers’ are proteins that recognize and bind to
lactylated histones, regulating gene expression levels and other
cellular processes, such as chromatin remodeling and accessibility
regulation, cell survival and tumorigenesis, and gene transcription
program regulation. Known histone lactylation ‘readers’ include
brahma-related gene 1 (Brg1) (108), double plant homeodomain finger 2
(DPF2) (124) and tripartite motif
containing 33 (TRIM33) (125). In
2024, the study by Hu et al (108) first reports that Brg1 interacts
with H3K18la and functions as a ‘reader’ of H3K18la and is
potentially involved in facilitating the accessibility of chromatin
regions-thereby promoting an open chromatin state and enhancing
gene expression. DPF2 recognizes H3K14la and binds to it through
its DPF domain, which modulates gene transcription and cell
survival. In cervical cancer, the accumulation of lactate leads to
increased levels of H3K14la, and DPF2 promotes the expression of
oncogenes by recognizing H3K14la, which promotes tumorigenesis
(124). The bromodomain of TRIM33
specifically recognizes Kla on histones. It is demonstrated that
the bromodomain of TRIM33 selectively binds to Kla via a unique
glutamic acid residue (E981), establishing it as a Kla-specific
‘reader’ protein (125). However,
a previous study, focusing on H3K18la as the primary subject of
investigation, did not demonstrate direct evidence regarding
whether TRIM33 functions as a ‘reader’ at other histone sites
(125).
In summary, the primary strategies for targeting
histone lactylation include: i) Modulation of lactate transport;
ii) regulation of lactate production; and iii) inhibition or
activation of enzymes mediating histone lactylation modifications.
The present evidence regarding the ‘readers’, ‘erasers’ and
‘writers’ of histone lactylation is yet to be fully elucidated.
Further identification and validation of specific targets are
required to enhance therapeutic precision. Moreover, there may be a
potential for the development of small-molecule inhibitors and
activators targeting these regulatory proteins (Fig. 1).
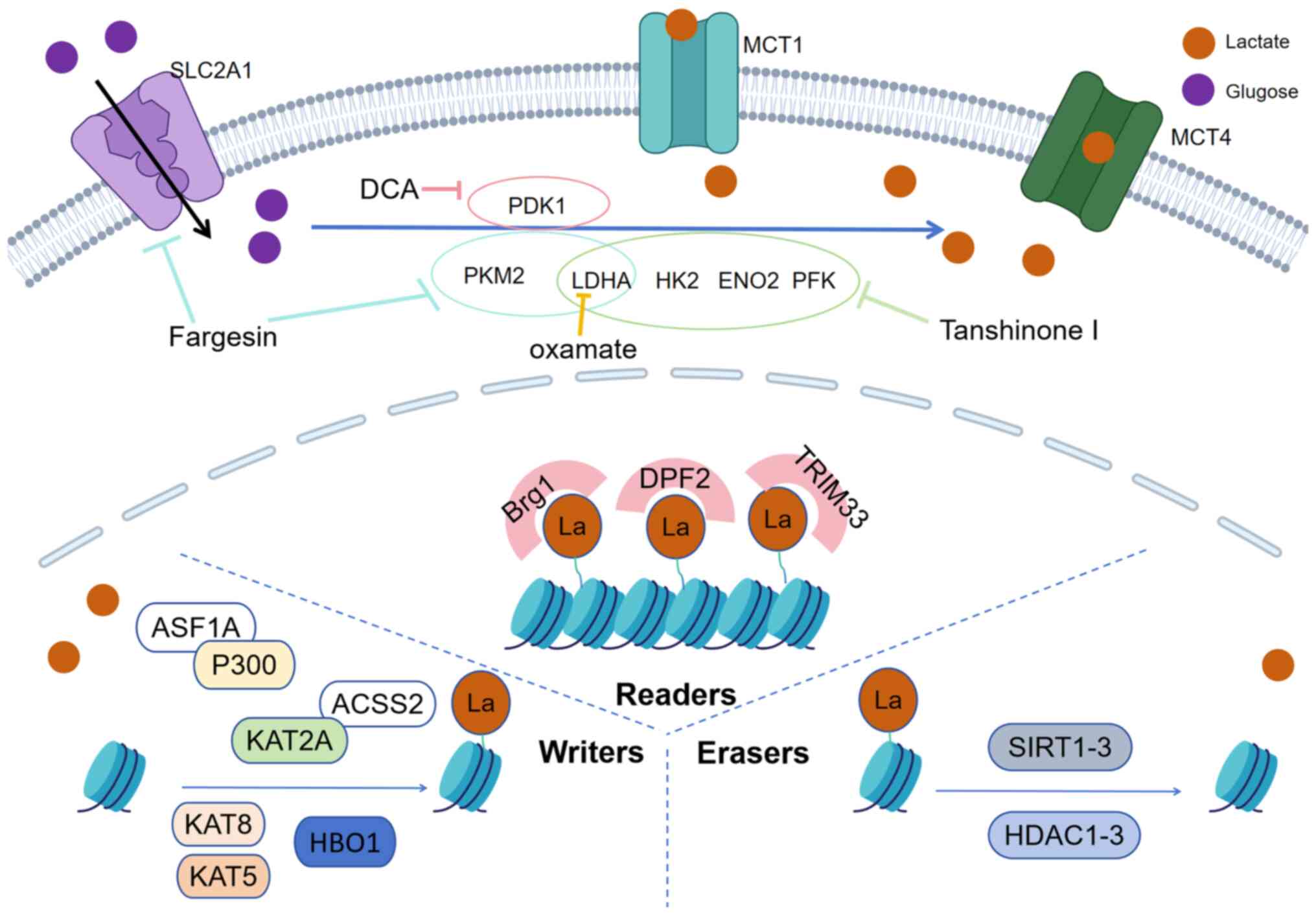 | Figure 1.Potential therapeutic strategies
targeting histone lactylation. Key strategies to modulate histone
lactylation include: i) Downregulating lactate flux (via MCT1/4
inhibition); ii) inhibition of the expression of glycolytic genes
(such as HK2, LDHA, ENO2, PFK, PKM2, PDK1 and
SLC2A1); iii) blocking lactylation-recognizing ‘readers’
(e.g., Brg1, DPF2, TRIM33) and modifying ‘writers’ (e.g., P300,
KAT2A, KAT5, KAT8, HBO1); and iv) enhancing the enzymatic activity
of ‘erasers’ (e.g., SIRT1-3, HDAC1-3). ‘T’ shaped arrows denote the
inhibitions of the pathways. DCA, sodium dichloroacetate; SLC2A1,
solute carrier family 2 member 1; MCT1, monocarboxylate transporter
1; MCT4, monocarboxylate transporter 4; HK2, hexokinase 2; LDHA,
lactate dehydrogenase; ENO2, enolase 2; PFK, phosphofructokinase;
PKM2, pyruvate kinase isozyme type M2; PDK1, pyruvate dehydrogenase
kinase 1; HBO1, home box office 1; La, lactate; Brg1,
brahma-related gene 1; DPF2, double PHD fingers 2; TRIM33,
tripartite motif containing 33; ASF1A, anti-silencing function 1A
histone chaperone; P300, E1A-binding protein; ACSS2, acyl-CoA
synthetase short chain family member 2; KAT, lysine
acetyltransferase; HDAC, histone deacetylase; SIRT, sirtuin. |
Conclusions and research perspectives
Histone lactylation is an epigenetic modification
that links cellular metabolism with gene regulation, serving a role
in cancer initiation, progression, metastasis, drug resistance and
immune evasion. The present review highlighted several key
findings. Histone lactylation, particularly at H3K18la, regulates
oncogenic pathways and metabolic reprogramming. Other lactylation
sites (such as H4K12la, H3K9la and H4K5la) contribute to
tumorigenesis in a context-dependent manner, influencing apoptosis
suppression, immune evasion and therapy resistance.
Hypoxia and mitochondrial dysfunction often lead to
the accumulation of lactic acid within cells. By acting as a
metabolic ‘gatekeeper’ controlling lactate metabolism,
mitochondrial targets serve as upstream regulators of histone
lactylation. At present, it is indicated that SIRT4 enhances
histone lactylation by promoting a metabolic shift from
mitochondrial oxidative phosphorylation to glycolysis, which
increases lactate accumulation (52). When the Numb/Parkin axis is
impaired, the accumulation of damaged mitochondria triggers
metabolic reprogramming toward aerobic glycolysis, which leads to
lactate accumulation and the promotion of histone lactylation
(55). In addition, histone
lactylation can also influence mitochondrial function, which
affects intracellular lactate accumulation and forms a positive
feedback loop. H3K18la disrupts mitochondrial oxidative
phosphorylation by promoting the transcriptional activation of
ACAT2, exacerbates lactate accumulation and forms a protumorigenic
positive feedback loop (66). In
Alzheimer's disease, the shift from oxidative phosphorylation to
glycolysis in microglia promotes proinflammatory activation. During
this process, the glycolysis/H4K12la/PKM2 positive feedback loop
exacerbates microglial dysfunction (126).
In the context of cancer, targeting mitochondria
with regards to histone lactylation is yet to be fully elucidated.
However, understanding its implications in non-cancer contexts
(such as inflammation and hypoxic pulmonary hypertension) may be
significant (127,128). In the context of inflammation,
mitochondrial fragmentation promotes a metabolic shift that
increases lactate, which in turn promotes histone lactylation,
enhancing macrophage phagocytosis and inflammation resolution
(127). Hypoxia-induced
mitochondrial ROS stabilizes hypoxia-inducible factor-1α (HIF-1α)
via inhibition of its hydroxylation, which upregulates pyruvate
dehydrogenase kinase 1/2 to promote glycolytic switching and
lactate accumulation in pulmonary arterial smooth muscle cells
(PASMCs). H3K18la, induced by accumulated lactate, activates HIF-1α
target genes, which facilitates PASMC proliferation and vascular
remodeling (128). Selenium
deficiency induces mitochondrial dysfunction via the ROS/HIF-1α
pathway, leading to enhanced glycolysis and lactate production. The
accumulated lactate promotes H3K18la, which activates the NLRP3
inflammasome promoter and induces pyroptosis and the release of
inflammatory factors (including TNF-α, IL-6, IL-8, IFN-γ) (129). Methyltransferase-like 15
deficiency impairs mitochondrial ribosome function, leading to
reduced oxidative phosphorylation, increased ROS and diminished
membrane potential. This promotes enhanced glycolysis and lactate
secretion, and leads to a notable increase in histone lactylation
at the H4K12 and H3K9 sites (130). In summary, mitochondrial
dysfunction is a key regulator of histone lactylation, primarily
through its impact on glycolysis and intracellular lactate levels.
Enhanced glycolysis fuels lactate production, which in turn
promotes histone lactylation by substrate provision. This
establishes a key mechanistic link through which mitochondrial
dysfunction influences histone lactylation (34,118,126,131).
The present review highlighted that histone PTMs do
not function in isolation. A key emerging concept is the cascading
interplay between lactylation and other PTMs, which regulate
oncogenic programs. For example, in HCC, H3K18la upregulates the
expression of YTHDC1, stabilizes NEAT1 RNA and recruits p300 to the
SCD gene promoter. This subsequently promotes H3K27ac, forming a
cascade of epigenetic modifications that promotes tumor progression
by activating fatty acid metabolic reprogramming (45). This ‘lactylation-acetylation axis’
links lactate metabolism to lipid metabolic reprogramming and tumor
progression. In addition to acetylation, the potential crosstalk
between lactylation and lysine methylation warrants further
investigation. A study by Yang et al (132) reveals that H3K18la upregulates the
expression of suppressor of zeste 12 (SUZ12), which enhances the
level of trimethylated histone H3 at K27 (H3K27me3). Subsequently,
SUZ12/H3K27me3 inhibits the transcription of Krüppel-like factor 4
(KLF4). The downregulation of KLF4 relieves its inhibitory effect
on LDHA, a key enzyme in glycolysis, leading to lactate production.
In turn, the accumulated lactate further promotes histone
lactylation, thus forming a self-amplifying positive feedback loop
that promotes the Warburg effect and malignant progression of
retinoblastoma. Elucidating these cascading interactions may be
important to understand the full scope of histone lactylation in
cancer and may contribute to clinical translation strategies from
single target to ‘PTM cascade’ targeting therapies.
Lactate-induced H3K18la in TAMs and myeloid cells
promotes an immunosuppressive TME via PD-L1 upregulation, M2
polarization and CD8+ T cell exhaustion. However,
CD8+ T cell-intrinsic lactylation may enhance antitumor
immunity after induction by immune checkpoint blockade, which
suggests a dual role in immunotherapy. Histone lactylation promotes
resistance to chemotherapy such as oxaliplatin, cisplatin,
lenvatinib and targeted therapies through autophagy activation, DNA
repair enhancement and ferroptosis suppression.
Lactylation-mediated immune checkpoint upregulation (such as via
PD-L1, CD47 and B7-H3) contributes to immunotherapy resistance,
which highlights the potential of combination strategies.
However, despite notable progress in associated
research, numerous questions are yet to be investigated. At
present, a hypothesis regarding the process of histone lactylation
is that lactate condenses with CoA within the cell to form lac-CoA.
Lac-CoA is then acted upon by a ‘writer’, which transfers the
lactyl group to lysine residues. Subsequently, the lactylated
histones are recognized by a ‘reader’, leading to changes in the
transcription levels of downstream genes. Finally, after the
modification is complete, an ‘eraser’ recognizes and removes the
lactyl group from the lysine residues, restoring the normal
chromatin structure (19). Previous
studies demonstrate direct experimental evidence for non-enzymatic
histone lactylation. Non-enzymatic Kla is experimentally
demonstrated on glycolytic enzymes in vitro (45,133).
The study by Zhang et al (17) provides evidence for its occurrence
on histones and demonstrate the modification after incubating
histones with sodium lactate in the absence of enzymes.
Subsequently, the study by Tan et al (134) demonstrates that lactoyl-CoA and
lactoylglutathione are the direct inducers of this spontaneous
reaction. It is hypothesized that in cellular environments with
high lactate and low pH (such as the TME), lactate accumulation may
promote histone lactylation through similar non-enzymatic
mechanisms (133,135,136). However, at present, it is
challenging for the majority of cellular studies to delineate
whether lactylation at a specific site arises from enzymatic
catalysis or non-enzymatic mechanisms, which is a notable
limitation in the field. Therefore, non-enzymatic modification
should be considered as a potential confounding factor when
interpreting associated physiological or pathological data. Future
studies should aim to resolve this by developing site-specific
probes or constructing enzyme-deficient models.
At present, multiple ‘writers’ of histone
lactylation (such as P300, GCN5, HBO1, KAT5 and KAT8) and ‘erasers’
(such as HDAC1-3 and SIRT1-3) are identified. Previous studies
suggest that HBO1 preferentially catalyzes H3K9la. Furthermore,
compared with HDAC2, HDAC1 and 3 more preferentially regulate
H4K5la. These findings indicate that future studies on histone
lactylation should select appropriate regulatory enzymes based on
specific sites. However, at present, studies on regulatory enzymes
for histone lactylation are limited. Therefore, further studies are
needed to provide evidence regarding whether these enzymes
specifically or preferentially regulate lactylation at particular
histone sites. In addition, the specific functions and mechanisms
of these enzymes in different types of cancer are yet to be fully
elucidated.
Additionally, studies on the ‘readers’ of histone
lactylation is limited, and their functions and regulatory
mechanisms in cancer still requires thorough investigation. Due to
the high degree of conservation and acetyl-lysine specificity of
bromodomain modules, and the structural, functional and
evolutionary parallels between histone lactylation and acetylation,
it is hypothesized that bromodomain-containing proteins may
function as ‘readers’ of histone lactylation (108,125). It is suggested that future studies
further identify and validate the specific targets of these enzymes
and possibly develop small-molecule inhibitors or modulators
targeting them to potentially achieve more precise cancer
therapy.
At present, the majority of studies are still in
the basic research stage, and further translational research is
required to verify the safety and efficacy of histone
lactylation-targeting therapeutic strategies in cancer. Histone
lactylation represents a dynamic link between metabolism and
epigenetics and may potentially offer novel insights into cancer
biology and therapy. Future studies may potentially investigate the
enzymatic regulation and functional outcomes of lactylation,
develop lactylation-targeted drugs and rational combinations, and
validate lactylation as a biomarker and therapeutic target in
clinical trials.
Acknowledgements
Not applicable.
Funding
The present work was supported by the Scientific Research
Foundation of Jilin province (grant no. 20240402001GH) and the
National Nature and Science Foundation of China (grant no.
82372690).
Availability of data and materials
Not applicable.
Authors' contributions
ZJ and PG conceived and designed the present study.
ZJ, SL and ZW drafted the initial manuscript. ZJ prepared the
figures and tables. PG revised figures, tables and manuscript. All
authors read and approved the final version of the manuscript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
ACOX1
|
acyl-CoA oxidase 1
|
|
ACSS2
|
acyl-CoA synthetase short chain
family member 2
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family
member A3
|
|
BC
|
breast cancer BLCA, bladder
cancer
|
|
B7-H3
|
B7 homologue 3
|
|
Brg1
|
brahma-related gene 1
|
|
CEA
|
carcinoembryonic antigen
|
|
CRLM
|
colorectal cancer liver
metastasis
|
|
CRC
|
colorectal cancer
|
|
CAFs
|
cancer-associated fibroblasts
|
|
CXCL
|
chemokine (C-X-C motif) ligand
|
|
CBX3
|
chromobox homolog 3
|
|
CTHRC1
|
collagen triple helix
repeat-containing 1
|
|
DNAJC12
|
DnaJ heat shock protein family
(Hsp40) member C12
|
|
DPF2
|
double plant homeodomain finger 2
|
|
ESCC
|
esophageal squamous cell
carcinoma
|
|
GBM
|
glioblastoma
|
|
GCN5
|
general control non-depressible 5
|
|
HCC
|
hepatocellular carcinoma
|
|
H3K27ac
|
histone H3 lysine 27 acetylation
|
|
H3K18la
|
histone H3 lysine 18 lactylation
|
|
HECTD2
|
HECT domain E3 ubiquitin protein
ligase 2
|
|
HDAC
|
histone deacetylase
|
|
IRS1
|
insulin receptor substrate 1
|
|
Kla
|
lysine lactylation
|
|
KEAP1
|
Kelch-like ECH associated protein
1
|
|
KAT2A
|
lysine acetyltransferase 2A
|
|
LDH
|
lactate dehydrogenase
|
|
LUAD
|
lung adenocarcinoma
|
|
LAMC2
|
laminin subunit γ 2
|
|
MCTs
|
monocarboxylate transporters
|
|
NEAT1
|
nuclear paraspeckle assembly
transcript 1
|
|
NB
|
neuroblastoma
|
|
NSCLC
|
non-small cell lung cancer
|
|
NUPR1
|
nuclear protein 1
|
|
OC
|
ovarian cancer
|
|
PTMs
|
post-translational modifications
|
|
PYCR1
|
pyrroline-5-carboxylate reductase
1
|
|
PCA
|
pancreatic cancer
|
|
PKM2
|
pyruvate kinase M2
|
|
RUBCNL
|
rubicon-like autophagy enhancer
|
|
SCD
|
stearoyl-CoA desaturase
|
|
SLFN5
|
Schlafen 5
|
|
SIRT
|
sirtuin
|
|
TME
|
tumor microenvironment
|
|
TNBC
|
triple-negative breast cancer
|
|
TAMs
|
tumor-associated macrophages
|
|
TMZ
|
temozolomide
|
|
TRIM33
|
tripartite motif containing 33
|
|
USP
|
ubiquitin-specific peptidase
|
|
YTHDF2
|
YTH N6-methyladenosine
RNA-binding protein F2
|
References
|
1
|
Zhong Q, Xiao X, Qiu Y, Xu Z, Chen C,
Chong B, Zhao X, Hai S, Li S, An Z and Dai L: Protein
posttranslational modifications in health and diseases: Functions,
regulatory mechanisms, and therapeutic implications. MedComm
(2020). 4:e2612023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee JM, Hammaren HM, Savitski MM and Baek
SH: Control of protein stability by Post-translational
modifications. Nat Commun. 14:2012023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shu F, Xiao H, Li QN, Ren XS, Liu ZG, Hu
BW, Wang HS, Wang H and Jiang GM: Epigenetic and Post-translational
modifications in autophagy: Biological functions and therapeutic
targets. Signal Transduct Target Ther. 8:322023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li Z, Chen J, Huang H, Zhan Q, Wang F,
Chen Z, Lu X and Sun G: Post-translational modifications in
diabetic cardiomyopathy. J Cell Mol Med. 28:e181582024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Winston DJ, Pakrasi A and Busuttil RW:
Prophylactic fluconazole in liver transplant recipients. A
randomized, double-blind, placebo-controlled trial. Ann Intern Med.
131:729–737. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Durrant JD, de Oliveira CA and McCammon
JA: POVME: An algorithm for measuring Binding-pocket volumes. J Mol
Graph Model. 29:773–776. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Silman I, Roth E, Paz A, Triquigneaux MM,
Ehrenshaft M, Xu Y, Shnyrov VL, Sussman JL, Deterding LJ, Ashani Y,
et al: The specific interaction of the photosensitizer methylene
blue with acetylcholinesterase provides a model system for studying
the molecular consequences of photodynamic therapy. Chem Biol
Interact. 203:63–66. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hanson G and Coller J: Codon optimality,
bias and usage in translation and mRNA decay. Nat Rev Mol Cell
Biol. 19:20–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jenuwein T and Allis CD: Translating the
histone code. Science. 293:1074–1080. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mansour MR, Abraham BJ, Anders L,
Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan
SE, Silverman LB, et al: Oncogene regulation. An oncogenic
super-enhancer formed through somatic mutation of a noncoding
intergenic element. Science. 346:1373–1377. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu XY, Guo CH, Xi ZY, Xu XQ, Zhao QY, Li
LS and Wang Y: Histone methylation in pancreatic cancer and its
clinical implications. World J Gastroenterol. 27:6004–6024. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Komar D and Juszczynski P: Rebelled
epigenome: Histone H3S10 phosphorylation and H3S10 kinases in
cancer biology and therapy. Clin Epigenetics. 12:1472020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tamburri S, Conway E and Pasini D:
Polycomb-dependent histone H2A ubiquitination links developmental
disorders with cancer. Trends Genet. 38:333–352. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Duan X, Xing Z, Qiao L, Qin S, Zhao X,
Gong Y and Li X: The role of histone post-translational
modifications in cancer and cancer immunity: Functions, mechanisms
and therapeutic implications. Front Immunol. 15:14952212024.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Yang Y, Zhang B, Lin X, Fu X, An Y,
Zou Y, Wang JX, Wang Z and Yu T: Lactate metabolism in human health
and disease. Signal Transduct Target Ther. 7:3052022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao Y and Hong J: Purification of
recombinant histones and mononucleosome assembly. Curr Protoc.
5:e701552025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang D, Tang Z, Huang H, Zhou G, Cui C,
Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al: Metabolic
regulation of gene expression by histone lactylation. Nature.
574:575–580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu Y, He Z, Li Z, Wang Y, Wu N, Sun H,
Zhou Z, Hu Q and Cong X: Lactylation: The novel histone
modification influence on gene expression, protein function, and
disease. Clin Epigenetics. 16:722024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu X, Yang J, Xu J, Pan H, Wang W, Yu X
and Shi S: Histone lactylation: From tumor lactate metabolism to
epigenetic regulation. Int J Biol Sci. 20:1833–1854. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hou J, Guo M, Li Y and Liao Y: Lactylated
histone H3K18 as a potential biomarker for the diagnosis and
prediction of the severity of pancreatic cancer. Clinics (Sao
Paulo). 80:1005442025. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li XM, Yang Y, Jiang FQ, Hu G, Wan S, Yan
WY, He XS, Xiao F, Yang XM, Guo X, et al: Histone lactylation
inhibits RARgamma expression in macrophages to promote colorectal
tumorigenesis through activation of TRAF6-IL-6-STAT3 signaling.
Cell Rep. 43:1136882024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jin Z, Yun L and Cheng P: Tanshinone I
reprograms glycolysis metabolism to regulate histone H3 lysine 18
lactylation (H3K18la) and inhibits cancer cell growth in ovarian
cancer. Int J Biol Macromol. 291:1390722025. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo Z, Tang Y, Wang S, Huang Y, Chi Q, Xu
K and Xue L: Natural product fargesin interferes with H3 histone
lactylation via targeting PKM2 to inhibit non-small cell lung
cancer tumorigenesis. Biofactors. 50:592–607. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rabinowitz JD and Enerback S: Lactate: The
ugly duckling of energy metabolism. Nat Metabo. 2:566–571. 2020.
View Article : Google Scholar
|
|
25
|
Chen S, Xu Y, Zhuo W and Zhang L: The
emerging role of lactate in tumor microenvironment and its clinical
relevance. Cancer Lett. 590:2168372024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gatenby RA and Gillies RJ: Why do cancers
have high aerobic glycolysis? Nat Rev Cancer. 4:891–899. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brooks GA, Osmond AD, Arevalo JA, Duong
JJ, Curl CC, Moreno-Santillan DD and Leija RG: Lactate as a myokine
and exerkine: Drivers and signals of physiology and metabolism. J
Appl Physiol (1985). 134:529–548. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fang Y, Li Z, Yang L, Li W, Wang Y, Kong
Z, Miao J, Chen Y, Bian Y and Zeng L: Emerging roles of lactate in
acute and chronic inflammation. Cell Commun Signal. 22:2762024.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chang X, Zhou S, Liu J, Wang Y, Guan X, Wu
Q, Liu Z and Liu R: Zishenhuoxue decoction-induced myocardial
protection against ischemic injury through TMBIM6-VDAC1-mediated
regulation of calcium homeostasis and mitochondrial quality
surveillance. Phytomedicine. 132:1553312024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang X, Zhang Q, Huang Y, Liu J, Wang Y,
Guan X, Wu Q, Liu Z and Liu R: Quercetin inhibits necroptosis in
cardiomyocytes after ischemia-reperfusion via
DNA-PKcs-SIRT5-orchestrated mitochondrial quality control.
Phytother Res. 38:2496–2517. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nedel WL and Portela LV: Lactate levels in
sepsis: Don't forget the mitochondria. Intensive Care Med.
50:1202–1203. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Paul S, Ghosh S and Kumar S: Tumor
glycolysis, an essential sweet tooth of tumor cells. Semin Cancer
Biol. 86:1216–1230. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pandkar MR, Sinha S, Samaiya A and Shukla
S: Oncometabolite lactate enhances breast cancer progression by
orchestrating histone lactylation-dependent c-Myc expression.
Transl Oncol. 37:1017582023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma F and Yu W: The roles of lactate and
lactylation in diseases related to mitochondrial dysfunction. Int J
Mol Sci. 26:71492025. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y and Patti GJ: The Warburg effect: A
signature of mitochondrial overload. Trends Cell Biol.
33:1014–1020. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shechter D, Dormann HL, Allis CD and Hake
SB: Extraction, purification and analysis of histones. Nat Protoc.
2:1445–1457. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Doenecke D, Albig W, Bode C, Drabent B,
Franke K, Gavenis K and Witt O: Histones: Genetic diversity and
Tissue-specific gene expression. Histochem Cell Biol. 107:1–10.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Crane-Robinson C: Linker histones: History
and current perspectives. Biochim Biophys Acta. 1859:431–435. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu K, Zhang K, Wang Y and Gu Y:
Comprehensive review of histone lactylation: Structure, function,
and therapeutic targets. Biochem Pharmacol. 225:1163312024.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li H, Sun L, Gao P and Hu H: Lactylation
in cancer: Current understanding and challenges. Cancer Cell.
42:1803–1807. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou JM, Dai WX, Wang RJ, Xu WQ, Xiang Z,
Wang YX, Zhang T, Zhao YM, Wang L and Mao AR: Organoid modeling
identifies USP3-AS1 as a novel promoter in colorectal cancer liver
metastasis through increasing glucose-driven histone lactylation.
Acta Pharmacol Sinica. 46:1404–1418. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wei S, Zhang J, Zhao R, Shi R, An L, Yu Z,
Zhang Q, Zhang J, Yao Y, Li H and Wang H: Histone lactylation
promotes malignant progression by facilitating USP39 expression to
target PI3K/AKT/HIF-1α signal pathway in endometrial carcinoma.
Cell Death Discov. 10:1212024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu J, Chai P, Xie M, Ge S, Ruan J, Fan X
and Jia R: Histone lactylation drives oncogenesis by facilitating
m6A reader protein YTHDF2 expression in ocular melanoma. Genome
Biol. 22:852021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gu X, Zhuang A, Yu J, Yang L, Ge S, Ruan
J, Jia R, Fan X and Chai P: Histone lactylation-boosted ALKBH3
potentiates tumor progression and diminished promyelocytic leukemia
protein nuclear condensates by m1A demethylation of SP100A. Nucleic
Acids Res. 52:2273–2289. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Du W, Tan S, Peng Y, Lin S, Wu Y, Ding K,
Chen C, Liu R, Cao Y, Li Z, et al: Histone Lactylation-driven
YTHDC1 promotes hepatocellular carcinoma progression via lipid
metabolism remodeling. Cancer Lett. 611:2174262024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li L, Li Z, Meng X, Wang X, Song D, Liu Y,
Xu T, Qin J, Sun N, Tian K, et al: Histone lactylation-derived
LINC01127 promotes the self-renewal of glioblastoma stem cells via
the cis-regulating the MAP4K4 to activate JNK pathway. Cancer Lett.
579:2164672023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang J, Zhuang H, Yang X, Guo Z, Zhou K,
Liu N, An Y, Chen Y, Zhang Z, Wang M, et al: Exploring the
mechanism of ferroptosis induction by sappanone A in cancer:
Insights into the mitochondrial dysfunction mediated by
NRF2/xCT/GPX4 axis. Int J Biol Sci. 20:5145–5161. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pu X, Zhang Q, Liu J, Wang Y, Guan X, Wu
Q, Liu Z, Liu R and Chang X: Ginsenoside Rb1 ameliorates heart
failure through DUSP-1-TMBIM-6-mediated mitochondrial quality
control and gut flora interactions. Phytomedicine. 132:1558802024.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pang B, Dong G, Pang T, Sun X, Liu X, Nie
Y and Chang X: Emerging insights into the pathogenesis and
therapeutic strategies for vascular endothelial injury-associated
diseases: Focus on mitochondrial dysfunction. Angiogenesis.
27:623–639. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kenny TC and Birsoy K: Mitochondria and
cancer. Cold Spring Harb Perspect Med. 14:a0415342024. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu Y, Sun Y, Guo Y, Shi X, Chen X, Feng
W, Wu LL, Zhang J, Yu S, Wang Y and Shi Y: An overview: The
diversified role of mitochondria in cancer metabolism. Int J Biol
Sci. 19:897–915. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lv M, Yang X, Xu C, Song Q, Zhao H, Sun T,
Liu J, Zhang Y, Sun G, Xue Y and Zhang Z: SIRT4 promotes pancreatic
cancer stemness by enhancing histone lactylation and epigenetic
reprogramming stimulated by calcium signaling. Adv Sci (Weinh).
12:e24125532025. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chang X, Li Y, Liu J, Wang Y, Guan X, Wu
Q, Zhou Y, Zhang X, Chen Y, Huang Y and Liu R: ß-tubulin
contributes to Tongyang Huoxue decoction-induced protection against
hypoxia/reoxygenation-induced injury of sinoatrial node cells
through SIRT1-mediated regulation of mitochondrial quality
surveillance. Phytomedicine. 108:1545022023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chang X, Liu R, Li R, Peng Y, Zhu P and
Zhou H: Molecular mechanisms of mitochondrial quality control in
ischemic cardiomyopathy. Int J Biol Sci. 19:426–448. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
He Y, Ji Z, Gong Y, Fan L, Xu P, Chen X,
Miao J, Zhang K, Zhang W, Ma P, et al: Numb/Parkin-directed
mitochondrial fitness governs cancer cell fate via metabolic
regulation of histone lactylation. Cell Rep. 42:1120332023.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang H, Xu M, Zhang T, Pan J, Li C, Pan B,
Zhou L, Huang Y, Gao C, He M, et al: PYCR1 promotes liver cancer
cell growth and metastasis by regulating IRS1 expression through
lactylation modification. Clin Transl Med. 14:e700452024.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Deng J and Liao X: Lysine lactylation
(Kla) might be a novel therapeutic target for breast cancer. BMC
Med Genomics. 16:2832023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liu M, Gu L, Zhang Y, Li Y, Zhang L, Xin
Y, Wang Y and Xu ZX: LKB1 inhibits telomerase activity resulting in
cellular senescence through histone lactylation in lung
adenocarcinoma. Cancer Lett. 595:2170252024. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cui Z, Li Y, Lin Y, Zheng C, Luo L, Hu D,
Chen Y, Xiao Z and Sun Y: Lactylproteome analysis indicates histone
H4K12 lactylation as a novel biomarker in triple-negative breast
cancer. Front Endocrinol (Lausanne). 15:13286792024. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li J, Chen Z, Jin M, Gu X, Wang Y, Huang
G, Zhao W and Lu C: Histone H4K12 lactylation promotes malignancy
progression in triple-negative breast cancer through SLFN5
downregulation. Cell Signal. 124:1114682024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zang Y, Wang A, Zhang J, Xia M, Jiang Z,
Jia B, Lu C, Chen C, Wang S, Zhang Y, et al: Hypoxia promotes
histone H3K9 lactylation to enhance LAMC2 transcription in
esophageal squamous cell carcinoma. iScience. 27:1101882024.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yang Y, Wen J, Lou S, Han Y, Pan Y, Zhong
Y, He Q, Zhang Y, Mo X, Ma J and Shen N: DNAJC12 downregulation
induces neuroblastoma progression via increased histone H4K5
lactylation. J Mol Cell Biol. 16:mjae0562025. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Deng X, Huang Y, Zhang J, Chen Y, Jiang F,
Zhang Z, Li T, Hou L, Tan W and Li F: Histone lactylation regulates
PRKN-Mediated mitophagy to promote M2 Macrophage polarization in
bladder cancer. Int Immunopharmacol. 148:1141192025. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li M, Sun P, Tu B, Deng G, Li D and He W:
Hypoxia conduces the glioma progression by inducing M2 macrophage
polarization via elevating TNFSF9 level in a
Histone-lactylation-dependent manner. Am J Physiol Cell Physiol.
327:C487–C504. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Cai J, Zhang P, Cai Y, Zhu G, Chen S, Song
L, Du J, Wang B, Dai W, Zhou J, et al: Lactylation-Driven NUPR1
promotes immunosuppression of Tumor-infiltrating macrophages in
hepatocellular carcinoma. Adv Sci (Weinh). 12:e24130952025.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yang J and Yu X, Xiao M, Xu H, Tan Z, Lei
Y, Guo Y, Wang W, Xu J, Shi S and Yu X: Histone lactylation-driven
feedback loop modulates cholesterol-linked immunosuppression in
pancreatic cancer. Gut. 74:1859–1872. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Chaudagar K, Hieromnimon HM, Kelley A,
Labadie B, Shafran J, Rameshbabu S, Drovetsky C, Bynoe K, Solanki
A, Markiewicz E, et al: Suppression of tumor cell
lactate-generating signaling pathways eradicates murine
PTEN/p53-deficient Aggressive-variant prostate cancer via
macrophage phagocytosis. Clin Cancer Res. 29:4930–4940. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xiong J, He J, Zhu J, Pan J, Liao W, Ye H,
Wang H, Song Y, Du Y, Cui B, et al: Lactylation-driven
METTL3-mediated RNA m6A modification promotes immunosuppression of
tumor-infiltrating myeloid cells. Mol Cell. 82:1660–1677.e10. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yamamoto Y, Kasashima H, Fukui Y, Tsujio
G, Yashiro M and Maeda K: The heterogeneity of cancer-associated
fibroblast subpopulations: Their origins, biomarkers, and roles in
the tumor microenvironment. Cancer Sci. 114:16–24. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang C, Zhou W, Xu H, Xu J, Li J, Liu X,
Lu X, Dai J, Jiang Y, Wang W, et al: Cancer-associated fibroblasts
promote EGFR-TKI resistance via the CTHRC1/glycolysis/H3K18la
positive feedback loop. Oncogene. 44:1400–1414. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhou J, Xu W, Wu Y, Wang M, Zhang N, Wang
L, Feng Y, Zhang T, Wang L and Mao A: GPR37 promotes colorectal
cancer liver metastases by enhancing the glycolysis and histone
lactylation via hippo pathway. Oncogene. 42:3319–3330. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ugolini A, De Leo A, Yu X, Scirocchi F,
Liu X, Peixoto B, Scocozza D, Pace A, Perego M, Gardini A, et al:
Functional reprogramming of neutrophils within the brain tumor
microenvironment by Hypoxia-driven histone lactylation. Cancer
Discov. 15:1270–1296. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang R, Li C, Cheng Z, Li M, Shi J, Zhang
Z, Jin S and Ma H: H3K9 lactylation in malignant cells facilitates
CD8(+) T cell dysfunction and poor immunotherapy response. Cell
Rep. 43:1146862024. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Raychaudhuri D, Singh P, Chakraborty B,
Hennessey M, Tannir AJ, Byregowda S, Natarajan SM, Trujillo-Ocampo
A, Im JS and Goswami S: Histone lactylation drives CD8+ T cell
metabolism and function. Nat Immunol. 25:2140–2151. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li W, Zhou C, Yu L, Hou Z, Liu H, Kong L,
Xu Y, He J, Lan J, Ou Q, et al: Tumor-derived lactate promotes
resistance to bevacizumab treatment by facilitating autophagy
enhancer protein RUBCNL expression through histone H3 lysine 18
lactylation (H3K18la) in colorectal cancer. Autophagy. 20:114–130.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Li F, Zhang H, Huang Y, Li D, Zheng Z, Xie
K, Cao C, Wang Q, Zhao X, Huang Z, et al: Single-cell transcriptome
analysis reveals the association between histone lactylation and
cisplatin resistance in bladder cancer. Drug Resist Updat.
73:1010592024. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Dong R, Fei Y, He Y, Gao P, Zhang B, Zhu
M, Wang Z, Wu L, Wu S, Wang X, et al: Lactylation-driven HECTD2
limits the response of hepatocellular carcinoma to lenvatinib. Adv
Sci (Weinh). 12:e24125592025. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zeng Y, Jiang H, Chen Z, Xu J, Zhang X,
Cai W, Zeng X, Ma P, Lin R, Yu H, et al: Histone lactylation
promotes multidrug resistance in hepatocellular carcinoma by
forming a positive feedback loop with PTEN. Cell Death Dis.
16:592025. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang ZC, Li C, Zhang Z, Lu S, Liu YM, Qi
P, Chen X, Wang YB, Feng WJ, Pan CL, et al: Targeting PPARα
activation sensitizes glioblastoma cells to temozolomide and
reverses acquired resistance by inhibiting H3K18 lactylation. Acta
Pharmacol Sin. 46:3071–3086. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Chen Z, Zhang Y, Yan F, Zhao G and Wang Y:
tsRNA-08614 inhibits glycolysis and histone lactylation by ALDH1A3
to confer oxaliplatin sensitivity in colorectal cancer. Transl
Oncol. 58:1024272025. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Deng J, Li Y, Yin L, Liu S, Li Y, Liao W,
Mu L, Luo X and Qin J: Histone lactylation enhances GCLC expression
and thus promotes chemoresistance of colorectal cancer stem cells
through inhibiting ferroptosis. Cell Death Dis. 16:1932025.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang K, Guo L, Li X, Hu Y and Luo N:
Cancer-associated fibroblasts promote doxorubicin resistance in
triple-negative breast cancer through enhancing ZFP64 histone
lactylation to regulate ferroptosis. J Transl Med. 23:2472025.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Duan W, Liu W, Xia S, Zhou Y, Tang M, Xu
M, Lin M, Li X and Wang Q: Warburg effect enhanced by AKR1B10
promotes acquired resistance to pemetrexed in lung cancer-derived
brain metastasis. J Transl Med. 21:5472023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Lu B, Chen S, Guan X, Chen X, Du Y, Yuan
J, Wang J, Wu Q, Zhou L, Huang X and Zhao Y: Lactate accumulation
induces H4K12la to activate super-enhancer-driven RAD23A expression
and promote niraparib resistance in ovarian cancer. Mol Cancer.
24:832025. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sun C, Li X, Teng Q, Liu X, Song L,
Schiöth HB, Wu H, Ma X, Zhang Z, Qi C, et al: Targeting
platinum-resistant ovarian cancer by disrupting histone and RAD51
lactylation. Theranostics. 15:3055–3075. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Yue Q, Wang Z, Shen Y, Lan Y, Zhong X, Luo
X, Yang T, Zhang M, Zuo B, Zeng T, et al: Histone H3K9 lactylation
confers temozolomide resistance in glioblastoma via luc7l2-mediated
MLH1 intron retention. Adv Sci (Weinh). 11:e23092902024. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ling SP, Ming LC, Dhaliwal JS, Gupta M,
Ardianto C, Goh KW, Hussain Z and Shafqat N: Role of immunotherapy
in the treatment of cancer: A systematic review. Cancers (Basel).
14:52052022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Arafat Hossain M: A comprehensive review
of immune checkpoint inhibitors for cancer treatment. Int
Immunopharmacol. 143:1133652024. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zhang C, Zhou L, Zhang M, Du Y, Li C, Ren
H and Zheng L: H3K18 lactylation potentiates immune escape of
Non-small cell lung cancer. Cancer Res. 84:3589–3601. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Huang ZW, Zhang XN, Zhang L, Liu LL, Zhang
JW, Sun YX, Xu JQ, Liu Q and Long ZJ: STAT5 promotes PD-L1
expression by facilitating histone lactylation to drive
immunosuppression in acute myeloid leukemia. Signal Transduct
Target Ther. 8:3912023. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Li Z, Liang P, Chen Z, Chen Z, Jin T, He
F, Chen X and Yang K: CAF-secreted LOX promotes PD-L1 expression
via histone Lactylation and regulates tumor EMT through TGFβ/IGF1
signaling in gastric cancer. Cell Signal. 124:1114622024.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chao J, Chen GD, Huang ST, Gu H, Liu YY,
Luo Y, Lin Z, Chen ZZ, Li X, Zhang B, et al: High histone H3K18
lactylation level is correlated with poor prognosis in epithelial
ovarian cancer. Neoplasma. 71:319–332. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Hu X, Huang Z and Li L: LDHB mediates
histone lactylation to activate PD-L1 and promote ovarian cancer
immune escape. Cancer Invest. 43:70–79. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Ding CH, Yan FZ, Xu BN, Qian H, Hong XL,
Liu SQ, Luo YY, Wu SH, Cai LY, Zhang X and Xie WF: PRMT3 drives
PD-L1-mediated immune escape through activating PDHK1-regulated
glycolysis in hepatocellular carcinoma. Cell Death Dis. 16:1582025.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Ma Z, Yang J, Jia W, Li L, Li Y, Hu J, Luo
W, Li R, Ye D and Lan P: Histone Lactylation-driven B7-H3
expression promotes tumor immune evasion. Theranostics.
15:2338–2359. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Jia X, Yan B, Tian X, Liu Q, Jin J, Shi J
and Hou Y: CD47/SIRPα pathway mediates cancer immune escape and
immunotherapy. Int J Biol Sci. 17:3281–3287. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Liu Y, Weng L, Wang Y, Zhang J, Wu Q, Zhao
P, Shi Y, Wang P and Fang L: Deciphering the role of CD47 in cancer
immunotherapy. J Adv Res. 63:129–158. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Wang S, Huang T, Wu Q, Yuan H, Wu X, Yuan
F, Duan T, Taori S, Zhao Y, Snyder NW, et al: Lactate reprograms
glioblastoma immunity through CBX3-regulated histone lactylation. J
Clin Invest. 134:e1768512024. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhu Y, Fu Y, Liu F, Yan S and Yu R:
Appraising histone H4 lysine 5 lactylation as a novel biomarker in
breast cancer. Sci Rep. 15:82052025. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wu Q, Li X, Long M, Xie X and Liu Q:
Integrated analysis of histone lysine lactylation (Kla)-specific
genes suggests that NR6A1, OSBP2 and UNC119B are novel therapeutic
targets for hepatocellular carcinoma. Sci Rep. 13:186422023.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
He W, Chen R, Chen G, Zhang L, Qian Y,
Zhou J, Peng J, Wong VKW and Jiang Y: Identification and validation
of prognostic genes related to histone lactylation modification in
Glioblastoma: An integrated analysis of transcriptome and
Single-cell RNA sequencing. J Cancer. 16:2145–2166. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhang H, Yang X, Xue Y, Huang Y, Mo Y,
Huang Y, Zhang H, Zhang X, Zhao W, Jia B, et al: A basigin antibody
modulates MCTs to impact tumor metabolism and immunity. Cell
Discov. 11:442025. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Silva A, Antunes B, Batista A,
Pinto-Ribeiro F, Baltazar F and Afonso J: In vivo anticancer
activity of AZD3965: A systematic review. Molecules. 27:1812021.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Chen J, Huang Z, Chen Y, Tian H, Chai P,
Shen Y, Yao Y, Xu S, Ge S and Jia R: Lactate and lactylation in
cancer. Signal Transduct Target Ther. 10:382025. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhang Q, Cao L and Xu K: Role and
mechanism of lactylation in cancer. Zhongguo Fei Ai Za Zhi.
27:471–479. 2024.(In Chinese). PubMed/NCBI
|
|
106
|
Yao W, Hu X and Wang X: Crossing
epigenetic frontiers: The intersection of novel histone
modifications and diseases. Signal Transduct Target Ther.
9:2322024. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Sharma D, Singh M and Rani R: Role of LDH
in tumor glycolysis: Regulation of LDHA by small molecules for
cancer therapeutics. Semin Cancer Biol. 87:184–195. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Hu X, Huang X, Yang Y, Sun Y, Zhao Y,
Zhang Z, Qiu D, Wu Y, Wu G and Lei L: Dux activates
metabolism-lactylation-MET network during early iPSC reprogramming
with Brg1 as the histone lactylation reader. Nucleic Acids Res.
52:5529–5548. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Fan Z, Liu Z, Zhang N, Wei W, Cheng K, Sun
H and Hao Q: Identification of SIRT3 as an eraser of H4K16la.
iScience. 26:1077572023. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Moreno-Yruela C, Zhang D, Wei W, Bæk M,
Liu W, Gao J, Danková D, Nielsen AL, Bolding JE, Yang L, et al:
Class I histone deacetylases (HDAC1-3) are histone lysine
delactylases. Sci Adv. 8:eabi66962022. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Qin Q, Wang D, Qu Y, Li J, An K, Mao Z, Li
J, Xiong Y, Min Z and Xue Z: Enhanced glycolysis-derived lactate
promotes microglial activation in Parkinson's disease via histone
lactylation. NPJ Parkinsons Dis. 11:32025. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Wang N, Wang W, Wang X, Mang G, Chen J,
Yan X, Tong Z, Yang Q, Wang M, Chen L, et al: Histone lactylation
boosts reparative gene activation Post-myocardial infarction. Circ
Res. 131:893–908. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhu R, Ye X, Lu X, Xiao L, Yuan M, Zhao H,
Guo D, Meng Y, Han H, Luo S, et al: ACSS2 acts as a lactyl-CoA
synthetase and couples KAT2A to function as a lactyltransferase for
histone lactylation and tumor immune evasion. Cell Metab.
37:361–376.e7. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Niu Z, Chen C, Wang S, Lu C, Wu Z, Wang A,
Mo J, Zhang J, Han Y, Yuan Y, et al: HBO1 catalyzes lysine
lactylation and mediates histone H3K9la to regulate gene
transcription. Nat Commun. 15:35612024. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zou Y, Cao M, Tai M, Zhou H, Tao L, Wu S,
Yang K, Zhang Y, Ge Y, Wang H, et al: A Feedback loop driven by
H4K12 lactylation and HDAC3 in macrophages regulates
Lactate-induced collagen synthesis in fibroblasts via the TGF-β
signaling. Adv Sci (Weinh). 12:e24114082025. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Dong M, Zhang Y, Chen M, Tan Y, Min J, He
X, Liu F, Gu J, Jiang H, Zheng L, et al: ASF1A-dependent
P300-mediated histone H3 lysine 18 lactylation promotes
atherosclerosis by regulating EndMT. Acta Pharm Sin B.
14:3027–3048. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Sun Y, Sun Y, Yue S, Wang Y and Lu F:
Histone deacetylase inhibitors in cancer therapy. Curr Top Med
Chem. 18:2420–2428. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Li F, Si W, Xia L, Yin D, Wei T, Tao M,
Cui X, Yang J, Hong T and Wei R: Positive feedback regulation
between glycolysis and histone lactylation drives oncogenesis in
pancreatic ductal adenocarcinoma. Mol Cancer. 23:902024. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Du R, Gao Y, Yan C, Ren X, Qi S, Liu G,
Guo X, Song X, Wang H, Rao J, et al: Sirtuin 1/sirtuin 3 are robust
lysine delactylases and sirtuin 1-mediated delactylation regulates
glycolysis. iScience. 27:1109112024. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zu H, Li C, Dai C, Pan Y, Ding C, Sun H,
Zhang X, Yao X, Zang J and Mo X: SIRT2 functions as a histone
delactylase and inhibits the proliferation and migration of
neuroblastoma cells. Cell Discov. 8:542022. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Xu Y, Meng W, Dai Y, Xu L, Ding N, Zhang J
and Zhuang X: Anaerobic metabolism promotes breast cancer survival
via Histone-3 Lysine-18 lactylation mediating PPARD axis. Cell
Death Discov. 11:542025. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Tsukihara S, Akiyama Y, Shimada S, Hatano
M, Igarashi Y, Taniai T, Tanji Y, Kodera K, Yasukawa K, Umeura K,
et al: Delactylase effects of SIRT1 on a positive feedback loop
involving the H19-glycolysis-histone lactylation in gastric cancer.
Oncogene. 44:724–738. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Chen C, Zhang Y, Zang Y, Fan Z, Han Y, Bai
X, Wang A, Zhang J, Wang J, Zhang K, et al: SIRT3 functions as an
eraser of histone H3K9 lactylation to modulate transcription for
inhibiting the progression of esophageal cancer. Mol Cell
Proteomics. 24:1009732025. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zhai G, Niu Z, Jiang Z, Zhao F, Wang S,
Chen C, Zheng W, Wang A, Zang Y, Han Y and Zhang K: DPF2 reads
histone lactylation to drive transcription and tumorigenesis. Proc
Natl Acad Sci USA. 121:e24214961212024. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Nunez R, Sidlowski PFW, Steen EA,
Wynia-Smith SL, Sprague DJ, Keyes RF and Smith BC: The TRIM33
bromodomain recognizes histone lysine lactylation. ACS Chem Biol.
19:2418–2428. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Pan RY, He L, Zhang J, Liu X, Liao Y, Gao
J, Liao Y, Yan Y, Li Q, Zhou X, et al: Positive feedback regulation
of microglial glucose metabolism by histone H4 lysine 12
lactylation in Alzheimer's disease. Cell Metab. 34:634–648.e6.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Susser LI, Nguyen MA, Geoffrion M, Emerton
C, Ouimet M, Khacho M and Rayner KJ: Mitochondrial fragmentation
promotes inflammation resolution responses in macrophages via
histone lactylation. Mol Cell Biol. 43:531–546. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Chen J, Zhang M, Liu Y, Zhao S, Wang Y,
Wang M, Niu W, Jin F and Li Z: Histone lactylation driven by
mROS-mediated glycolytic shift promotes hypoxic pulmonary
hypertension. J Mol Cell Biol. 14:mjac0732023. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
An Z, Hu H, Wang Q, Qiu Y, Chu J, Xia Y
and Li S: Dietary Selenium deficiency activates the NLRP3
inflammasome to induce gallbladder pyroptosis by regulating
glycolysis and histone lactylation through ROS/HIF-1α pathway. J
Nutr Biochem. 147:1101182025. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Lv M, Zhou W, Hao Y, Li F, Zhang H, Yao X,
Shi Y and Zhang L: Structural insights into the specific
recognition of mitochondrial ribosome-binding factor hsRBFA and 12
S rRNA by methyltransferase METTL15. Cell Discov. 10:112024.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Li X, Chen M, Chen X, He X, Li X, Wei H,
Tan Y, Min J, Azam T, Xue M, et al: TRAP1 drives smooth muscle cell
senescence and promotes atherosclerosis via HDAC3-primed histone H4
lysine 12 lactylation. Eur Heart J. 45:4219–4235. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Yang L, Zuo S, Jia R, Gu X, Liao Q, Hua Y,
Ge S, He M, Fan J, Tong X, et al: Lactylation-boosted polycomb
repression of KLF4 elicits glycolysis in retinoblastoma: A positive
feedback circuit between histone modifications. Cancer Lett.
625:2178042025. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Gaffney DO, Jennings EQ, Anderson CC,
Marentette JO, Shi T, Schou Oxvig AM, Streeter MD, Johannsen M,
Spiegel DA, Chapman E, et al: Non-enzymatic lysine lactoylation of
glycolytic enzymes. Cell Chem Biol. 27:206–213.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Tan Q, Liu M and Tao X: Targeting
lactylation: From metabolic reprogramming to precision therapeutics
in liver diseases. Biomolecules. 15:11782025. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Varner EL, Trefely S, Bartee D, von
Krusenstiern E, Izzo L, Bekeova C, O'Connor RS, Seifert EL, Wellen
KE, Meier JL and Snyder NW: Quantification of lactoyl-CoA
(lactyl-CoA) by liquid chromatography mass spectrometry in
mammalian cells and tissues. Open Biol. 10:2001872020. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhang D, Gao J, Zhu Z, Mao Q, Xu Z, Singh
PK, Rimayi CC, Moreno-Yruela C, Xu S, Li G, et al: Lysine
L-lactylation is the dominant lactylation isomer induced by
glycolysis. Nat Chem Biol. 21:91–99. 2025. View Article : Google Scholar : PubMed/NCBI
|















