Introduction
Hepatocellular carcinoma (HCC) is a primary
malignancy of the liver and remains one of the most lethal cancers
worldwide (1). A previous study
indicated that >900,000 new cases are diagnosed annually, with
>830,000 mortalities attributed to HCC, underscoring its
critical burden on global health systems (2). Due to the fact that HCC is typically
discovered at an advanced stage, numerous patients are denied the
chance to have surgery (3).
Although a number of approaches are available for patients with HCC
in addition to surgery, including percutaneous ablation, liver
transplantation and immunotherapy, the clinical prognosis for
patients with HCC remains unfavorable, as <20% survive beyond 5
years (3). The main factors
contributing to the poor clinical outcomes in patients with HCC are
high heterogeneity, invasiveness and metastatic potential (4). Additionally, reliable biomarkers for
predicting prognosis and guiding personalized therapeutic decisions
in HCC remain scarce.
An important function of tumor cells is metabolic
reprogramming. The ‘Warburg effect’ refers to the concept that
tumor cells utilize ~80% of glucose for ATP synthesis by aerobic
glycolysis, which is also characterized by the production of lactic
acid, even in the presence of adequate oxygen (5). This aberrant glucose metabolism not
only sustains rapid cell proliferation but also serves pivotal
roles in cancer progression, metastasis and resistance to
chemotherapy (6). Enhanced
glycolytic activity has been identified as a hallmark of HCC, with
multiple glycolysis-related genes (GRGs) having been implicated in
its development and progression (7). By promoting glycolysis, a number of
genes are involved in the progression of HCC (8). Numerous intracellular and
extracellular proteins are susceptible to the effects of
lactylation (9). In addition,
histone lactylation is also involved in numerous processes,
including macrophage polarization under hypoxic conditions and
tumorigenesis (10).
Previous efforts have explored glycolysis-related
signatures (GRS) as prognostic tools across various malignancies,
including breast cancer (11),
bladder cancer (12) and HCC
(13). Due to the central role of
glucose metabolism in tumor biology, identifying
glycolysis-associated genes that predict patient outcomes and
therapeutic responses could offer valuable insights into HCC
management.
In the present study, transcriptomic data from The
Cancer Genome Atlas (TCGA) was used to construct a 7-GRS using 10
integrative machine learning algorithms. In addition, immune scores
and immune cell infiltration associated with the
glycolysis-associated gene signatures were investigated. The
present study aimed to fully explore the role of GRGs in the
prognosis of patients with HCC.
Materials and methods
Data acquisition
TCGA (n=329; http://portal.gdc.cancer.gov/), GSE72094 (n=90)
(14), ICGC (n=228; http://dcc.icgc.org/) and GSE14520 (n=218) (15) datasets provided the mRNA level data
of HCC. The present study excluded metastatic HCC cases. The
GSE91061 (melanoma; n=89) (16) and
IMvigor210 (bladder cancer; n=298) (17) datasets were used to investigate the
relationship between immunotherapy response and GRS. GRG lists
(Table SI) were acquired from
three studies (18–20) and hallmark gene sets of gene set
enrichment analysis (GSEA).
Integrative machine learning
algorithms constructed an optimal GRS
Differentially expressed genes (DEGs) were obtained
using the R package ‘limma’ (version 4.2.1; RStudio, Inc.)
(21), with a cut-off level of
LogFC ≥2. Univariate Cox analysis was carried out to determine the
potential prognostic biomarkers within GRGs. These potential
biomarkers were then utilized to develop a stable prognostic GRS
through integrative machine learning analysis. The process
encompassed 10 machine learning techniques: i) Random survival
forest; ii) survival support vector machine; iii) ridge; iv)
elastic network; v) supervised principal components; vi) stepwise
Cox; vii) generalized boosted regression modelling; viii) partial
least squares regression for Cox; ix) CoxBoost; and x) Least
Absolute Shrinkage and Selection Operator (LASSO). The
regularization parameter λ in the LASSO models was determined
through 10-fold cross-validation, while the tradeoff parameter α
was set between 0 and 1 (interval=0.1). When α is equal to 1, LASSO
is executed. The GRS was developed in the following four steps
using R scripts (https://github.com/Zaoqu-Liu/IRLS) obtained from a
previous study (22): i) In order
to investigate prognostic biomarkers in TCGA dataset, univariate
Cox regression was used; ii) after which, algorithm combinations
were fitted to the prediction model of TCGA dataset; iii) all
algorithm combinations were carried out in Gene Expression Omnibus
cohorts; and iv) the C-index was computed for each cohort. After
obtaining the GRS score of the patients with HCC, the R package
‘survminer’ (https://cran.r-project.org/web/packages/survminer/index.html)
containing the ‘surv_cutpoint’ function was used to identify the
optimal cut-off for separating patients with HCC into high and low
GRS score groups. Using the R package ‘rms’ (https://cran.r-project.org/web/packages/rms/index.html),
the C-index curves for prognostic signatures and clinical features
were also determined. The prognosis of HCC was examined using
univariate and multivariate Cox analyses to investigate potential
risk factors. Based on GRS score and additional clinical factors,
the R package ‘nomogramEx’ (https://cran.r-project.org/web/packages/nomogramEx/index.html)
was used to construct a predicting nomogram.
Immune infiltration analysis
Estimation of stromal and immune cells in malignant
tumors (ESTIMATE) score of patients with HCC was determined using
ESTIMATE analysis (23). The R
package ‘Immunedeconv’ (24), a
tool that integrates six algorithms, was utilized to explore the
association between immune cells and GRS scores. The single sample
GSEA method was applied to evaluate the levels of immune cells and
scores related to immune activities or functions. Additionally, the
R package ‘GSVA’ (25) was used to
calculate the scores for the ‘h.all.v7.4.symbols.gmt’ gene set
(https://data.broadinstitute.org/gsea-msigdb/msigdb/release/7.4/).
Drug sensitivity analysis
GRS may predict the potential benefits of
immunotherapy for patients with HCC and its ability to do this was
examined through various prediction scores, such as the Tumor
Immune Dysfunction and Exclusion (TIDE) score, immunophenoscore,
tumor escape score and tumor mutational burden (TMB) score.
Individuals with a lower TIDE score and a higher TMB score are more
likely to respond positively to immunotherapy and have reduced
chances of immune evasion. Subsequently, the IC50 values
for each drug in each HCC case were determined using the R package
‘oncoPredict’ (26).
Cell culture
The human normal liver cell line (THLE-2) was
maintained in bronchial epithelial cell growth medium (ScienCell
Research Laboratories, Inc.) and HCC cell lines (MHCC97H, HCCLM3
and SNU449) were maintained in DMEM (Thermo Fisher Scientific,
Inc.). The medium was supplemented with 10% fetal bovine serum
(Biological Industries; Sartorius AG) and 1%
penicillin/streptomycin. All cells were cultured in a humidity
incubator with 5% CO2 at 37°C.
Western blotting
Total protein was extracted by lysing cells in RIPA
buffer containing protease inhibitors (Soochow New Cell &
Molecular Biotech Co., Ltd.). Protein concentration was determined
using a BCA assay kit (Thermo Fisher Scientific, Inc.). Equal
amounts proteins (40 µg/lane) were then separated by SDS-PAGE on a
12% gel (Soochow New Cell & Molecular Biotech Co., Ltd.), and
further transferred onto PVDF membranes (Soochow New Cell &
Molecular Biotech Co., Ltd.). After blocking with 5% skimmed milk
at room temperature for 2 h, the membranes were incubated overnight
with specific primary antibodies at 4°C. The membranes were
detected using an ultra-sensitive ECL kit (Soochow New Cell &
Molecular Biotech Co., Ltd.), and the grayscale values of the
protein bands were analyzed using ImageJ 1.0 software (National
Institutes of Health). The following antibodies were used:
Anti-MCT1 (1:1,000; cat. no. 20139-1-AP; Proteintech Group, Inc.),
anti-epithelial cadherin (E-cadherin; 1:1,000; cat. no. 3195; Cell
Signaling Technology, Inc.), anti-neural cadherin (1:1,000; cat.
no. 13116; Cell Signaling Technology, Inc.), with anti-GAPDH
(1:2,000; cat. no. 5174; Cell Signaling Technology, Inc.) and
anti-heat shock protein 90 (1:1,000; cat. no. 4784; Cell Signaling
Technology, Inc.) used as the loading control.
Lentiviral transduction for MCT1
knockdown
MHCC-97H cells were transduced with MCT1-targeting
lentivirus (Shanghai GeneChem Co., Ltd.) following the supplier's
protocol. Puromycin selection (2 µg/ml) was applied 48 h
post-transduction to establish stable knockdown clones. In the
maintenance phase, MHCC-97H cells were cultured in medium
containing puromycin (0.5 µg/ml). After cells were cultured for
7–10 days, subsequent experiments were continued. Knockdown
efficiency was demonstrated through western blotting. The short
hairpin RNA (shRNA) sequences were as follows: shRNA1 sense,
5′-GCTCCGTATTGTTTGAAACAT-3′; anti-sense,
5′-ATGTTTCAAACAATACGGAGC-3′; shRNA2 sense,
5′-GCAGGGAAAGATAAGTCTAAA-3′; anti-sense,
5′-TTTAGACTTATCTTTCCCTGC-3′; short hairpin control sense,
5′-TTCTCCGAACGTGTCACGT-3′; anti-sense,
5′-ACGTGACACGTTCGGAGAA-3′.
Colony formation assay
A total of 500 MHCC-97H cells were plated in each
well of a 6-well plate and incubated for 2 weeks in a humidity
incubator with 5% CO2 at 37°C. After removing culture
medium, the cells were fixed with 100% methanol at room temperature
for 20 min, and stained by 0.1% crystal violet (Beyotime
Biotechnology) at room temperature for 30 min. Finally, the
colonies defined as clusters containing >50 cells were counted
under a light microscope.
Flow cytometry
Flow cytometry was performed using a
CytoFLEX™ flow cytometer (Beckman Coulter, Inc.). For
cell cycle analysis, MHCC-97H cells were fixed with 70% ethanol at
−20°C overnight, and stained with 5 µg/ml PI (MultiSciences Biotech
Co., Ltd.) at room temperature in the dark for 30 min. The cycle
distribution of cells was analyzed using CytExpert 2.0 software
(Beckman Coulter, Inc.). The cell apoptosis was detected using the
Annexin V-FITC apoptosis assay kit (MultiSciences Biotech Co.,
Ltd.) according to the manufacturer's instructions. Cell apoptosis
rate was analyzed using CytExpert 2.0 software (Beckman Coulter,
Inc.).
Wound-healing assays
Cell migration was evaluated using a wound healing
assay. A total of 5×105 MHCC-97H cells were cultured in
each well of a 6-well plate until they reached ~90% confluency in a
humidified incubator with 5% CO2 at 37°C. Confluent
monolayers in 6-well plates were scratched with a 100 µl pipette
tip, and the medium was replaced with a complete medium containing
1% FBS (Biological Industries; Sartorius AG). At the time points of
0 h and 24 h, the gap distances after the wound were captured by a
light microscopy (magnification, ×10; Olympus Corporation). The
cell migration rate was calculated using the following formula:
Cell migration rate (%)=[(width at 0 h-width at 24 h)/width at 0 h]
×100%.
Transwell assay
Invasion was assessed using Matrigel-coated
Transwell chambers (Corning, Inc.). Matrigel (BD Biosciences) was
diluted with PBS buffer (1:8) at 4°C, with 100 µl evenly precoated
on the surface of polycarbonate membrane in upper chamber at 37°C
for 2 h. A total of 5×104 MHCC-97H cells were seeded in
the upper chamber with serum-free medium and 500 µl complete medium
containing 10% FBS (Biological Industries; Sartorius AG) was added
to the lower chamber. The cells were cultured in a humidity
incubator with 5% CO2 at 37°C for 24 h. Then the cells
invaded to bottom membrane of chamber were fixed with 4%
paraformaldehyde for 30 min and stained with 0.1% crystal violet
(Beyotime Biotechnology) at room temperature. A total of five
fields of vision were randomly selected and counted under an
inverted microscope (magnification, ×50; Olympus Corporation).
Statistical analysis All statistical analyses
were conducted using R software and GraphPad Prism (version 9.2.0;
Dotmatics). All results represent the mean ± standard deviation of
assays performed at least three independent experiments. A
two-sided unpaired Student's t-test was used for intergroup
comparison, while a one-way ANOVA with a Tukey's post-hoc test was
used for comparisons between multiple groups. The relationships
between two continuous variables were assessed through Pearson
correlation analysis. To compare differences in Kaplan-Meier
survival curves, the two-sided log-rank test was applied. P<0.05
was considered to indicate a statistically significant
difference.
Results
Integrative machine learning
algorithms develop an optimal prognostic GRS
Using LogFC ≥2 as the cut-off, 2,898 DEGs in total
were obtained in HCC (Fig. S1A)
and 215 differentially expressed GRGs were identified among these
DEGs (Fig. S1B). A total of 80
differentially expressed GRGs were shown to have a significant
association with the prognosis of patients with HCC according to
Cox univariate analysis (Fig. S1C;
P<0.05). Using 10 machine learning-based methods, 30 different
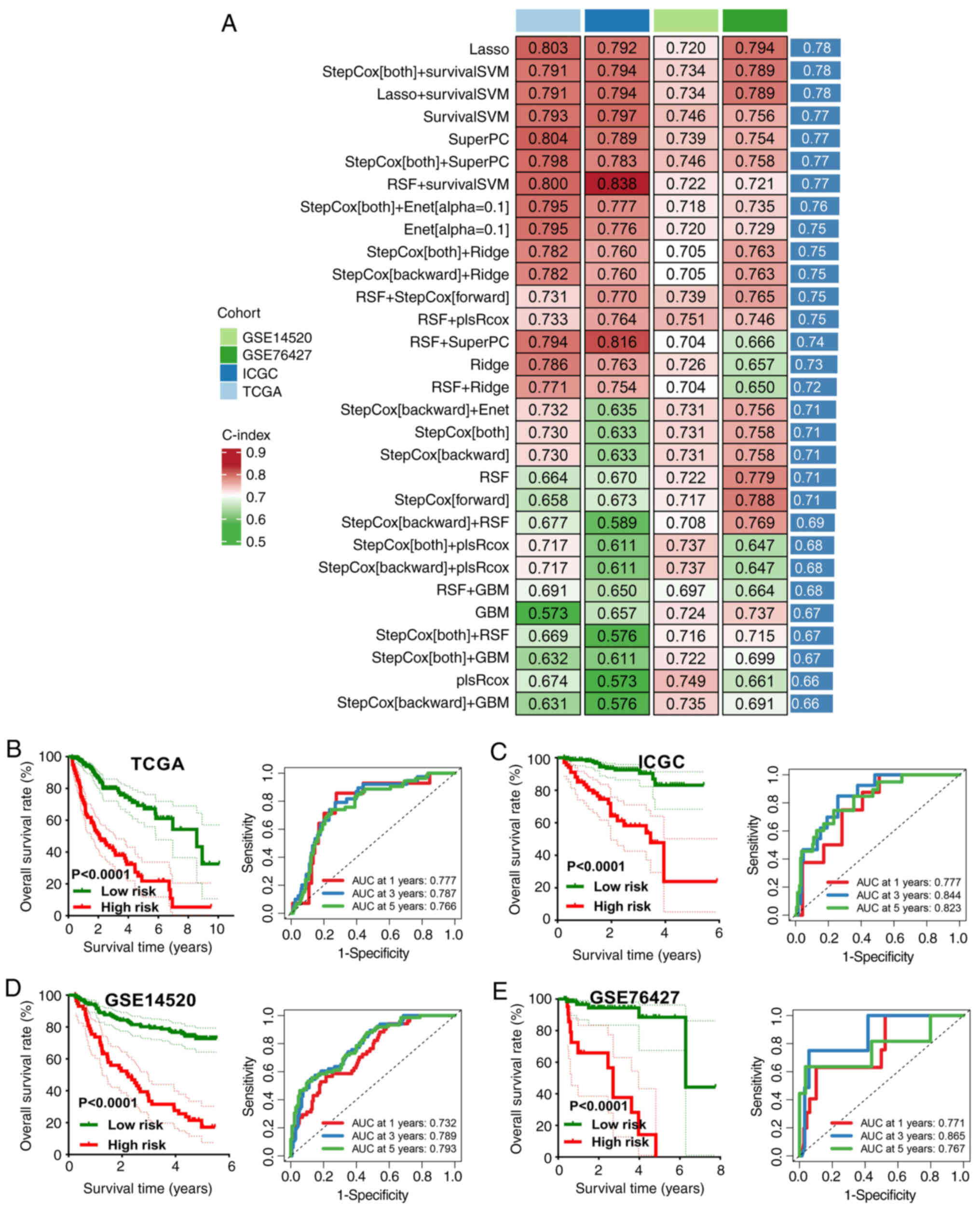
types of prognostic models were developed (Fig. 1A) and the C-index for each
prognostic model was determined. The model built using the LASSO
approach achieved the highest mean C-index of 0.78 and was selected
as the final signature. This final GRS comprised the following
seven genes: Treacle ribosome biogenesis factor 1 (TCOF1),
replication factor C subunit 4 (RFC4), ribonucleic acid export 1
(RAE1), kinesin family member 2C (KIF2C), DEP domain containing 1
(DEPDC1), activity dependent neuroprotector homeobox (ADNP) and
MCT1. The risk score was calculated using the following formula:
Risk score=(−0.0825 × TCOF1expression) + (0.0254 ×
RFC4expression) + (0.1454 × RAE1expression) +
(0.0158 × KIF2Cexpression) + (0.1254 × DEPDC1
expression) + (0.0354 × ADNP expression) +
(0.768 × MCT1 expression). HCC cases were divided into
high-risk and low-risk categories using the optimal cut-off. The
results showed 1, 3 and 5-year area under the curves (AUCs) of
0.777, 0.787 and 0.766 in TCGA cohort; 0.777, 0.844 and 0.823 in
the ICGC cohort; 0.732, 0.789 and 0.793 in the GSE14520 cohort and
0.771, 0.865 and 0.767 in the GSE76427 cohort. Furthermore, a poor
overall survival (OS) rate was found in patients with HCC with a
high-risk score in TCGA, ICGC, GSE14520 and GSE76427 cohorts (all
P<0.001; Fig. 1B-E).
Evaluation of the performance of
GRS
Multivariate and univariate Cox regression analyses
demonstrated that GRS was an independent prognostic factor across
all four datasets (all P<0.05; Fig.
2A and B). When compared with conventional clinical features
such as age, sex and tumor stage, the GRS-based risk score achieved
superior prognostic discrimination as reflected by its higher
C-index value (Fig. 2C-F). Notably,
among 31 existing HCC prognostic signatures, GRS outperformed all
others with regards to the C-index value (Table SII; Fig. 2G).
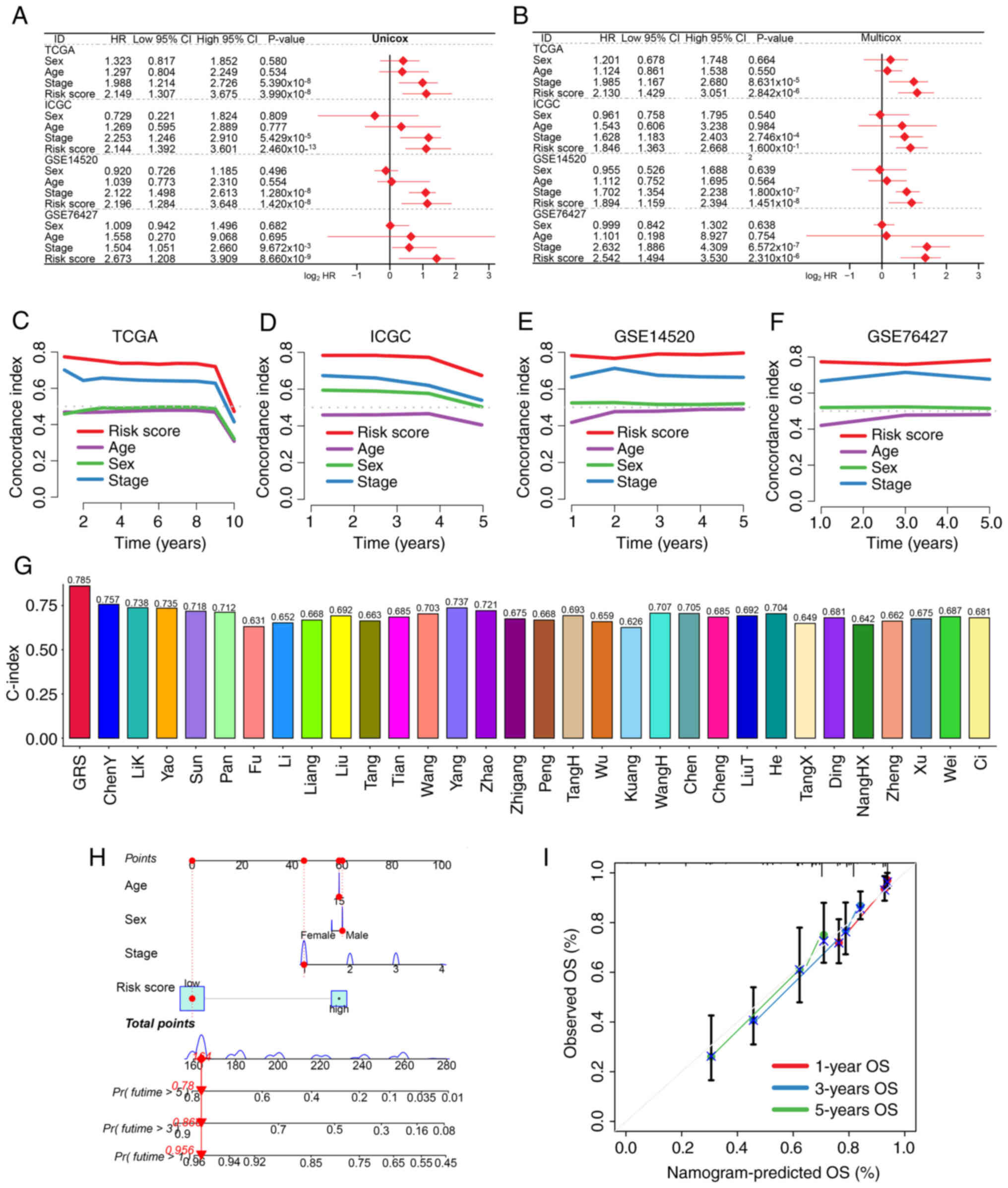 | Figure 2.Assessment of the predictive
performance of the GRS. (A) Univariate and (B) multivariate Cox
regression analyses identified risk factors associated with HCC
prognosis. C-index curves comparing GRS with clinical parameters
across (C) TCGA, (D) ICGC, (E) GSE14520 and (F) GSE76427 training
and testing datasets. (G) Comparison of C-index values between GRS
and previously published HCC prognostic models. (H) Prognostic
nomogram integrating age, sex, clinical stage and GRS score. (I)
Calibration plots showing agreement between predicted and actual 1,
3 and 5-year survival probabilities. GRS, glycolysis-related
signature; HCC, hepatocellular carcinoma; TCGA, The Cancer Genome
Atlas; ICGC, International Cancer Genome Consortium; HR, hazard
ratio; OS, overall survival. |
A predictive nomogram was also constructed
incorporating the GRS and clinical variables to estimate 1, 3 and
5-year survival probabilities (Fig.
2H). Calibration plots consistency between predicted and
observed outcomes (Fig. 2I),
further validating the clinical utility of the model.
GRS-based distinct immune
microenvironment in HCC
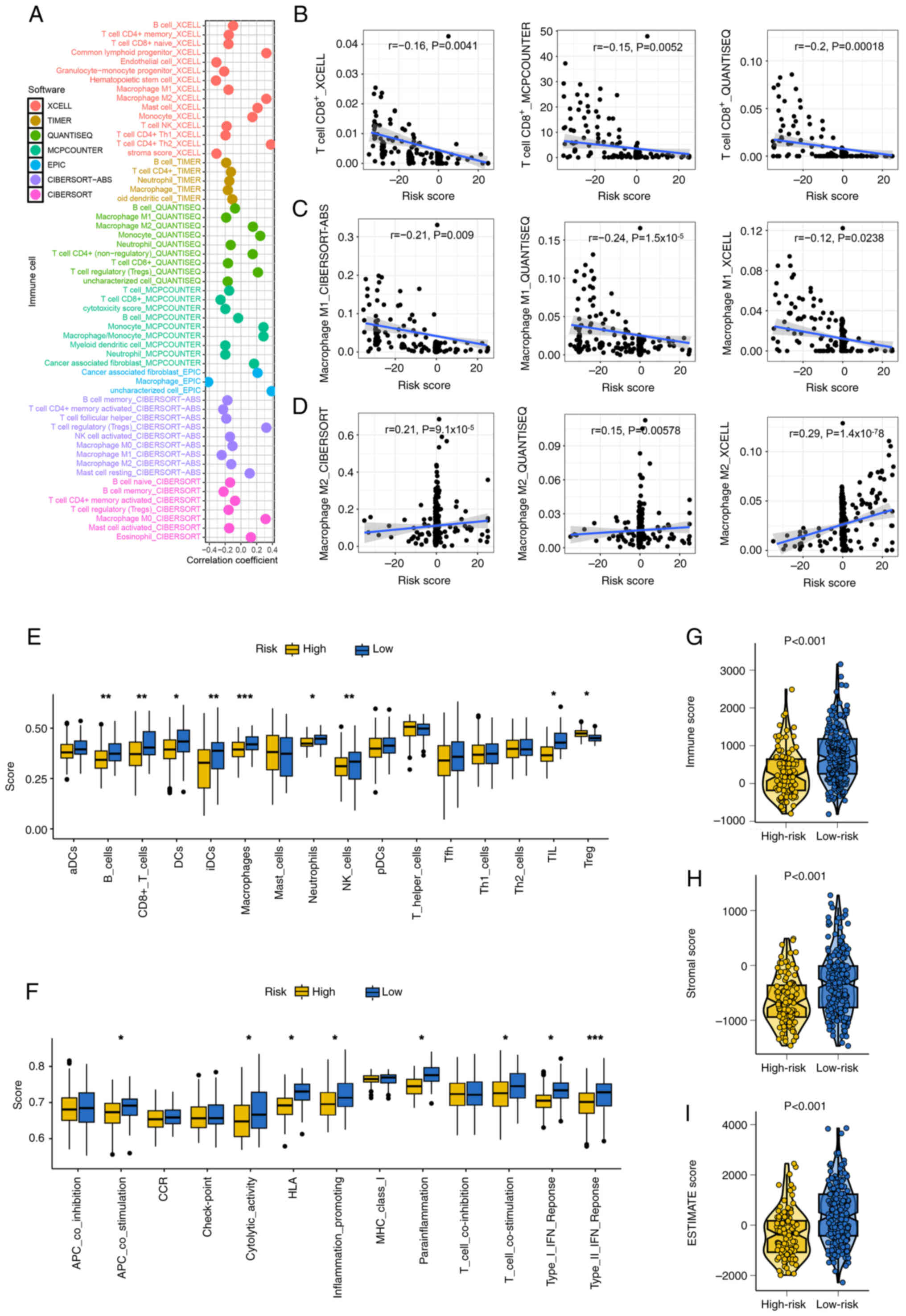
Immune microenvironment analysis revealed notable
correlations between GRS and immune cell infiltration. Risk scores
were positively or negatively associated with specific immune
populations, including CD8+ T cells, M1 and M2
macrophages (Fig. 3A-D). Patients
in the low-risk group showed elevated levels of immune-activated
cell types, including B cells, natural killer cells, neutrophils,
tumor-infiltrating lymphocytes and CD8+ T cells,
compared with the high-risk group (Fig.
3E). Moreover, patients in the low-risk group also demonstrated
higher enrichment scores in immune-related functions such as
antigen-presenting cell co-stimulation, cytolytic activity, human
leukocyte antigen (HLA) molecule expression, inflammation promotion
and T cell co-stimulation (Fig.
3F). Additionally, stromal score, immune score and ESTIMATE
scores were significantly higher in the low-risk group compared
with the high-risk group, indicating a more active and
immunologically enriched tumor microenvironment (TME; Fig. 3G-I; all P<0.05).
GRS acts as an indicator for drug
sensitivity in HCC
Predictive efficacy of the GRS-based risk score in
immunotherapy responses was assessed using a number of methods.
Firstly, TMB acted as a biomarker of immunotherapy response
(27). In patients with HCC, the
low-risk score group had a higher TMB score compared with the
high-risk score group (Fig. 4A;
P<0.001). In addition, the immunophenoscore acted as a predictor
of the response to checkpoint blockade. Patients with HCC that had
a low-risk score demonstrated a higher programmed cell death
protein 1 (PD-1) immunophenoscore and cytotoxic
T-lymphocyte-associated protein 4 (CTLA4) immunophenoscore
(Fig. 4B; all P<0.05). Moreover,
low-risk score patients with HCC had a lower immune escape score
(Fig. 4C; P=0.02). Furthermore,
TIDE scores can predict responses to cancer immunotherapy (28). As shown in Fig. 4D, patients with HCC that had a
low-risk score demonstrated a lower T cell dysfunction score, T
cell exclusion score and TIDE score (all P<0.05). Additionally,
HLA serves a vital role in antigen processing and antitumor
immunity (29), with a high risk
score indicating a higher level of HLA-related genes (Fig. 4E). Immunological checkpoints are
essential in promoting self-tolerance and stifling immunological
responses in malignancy.
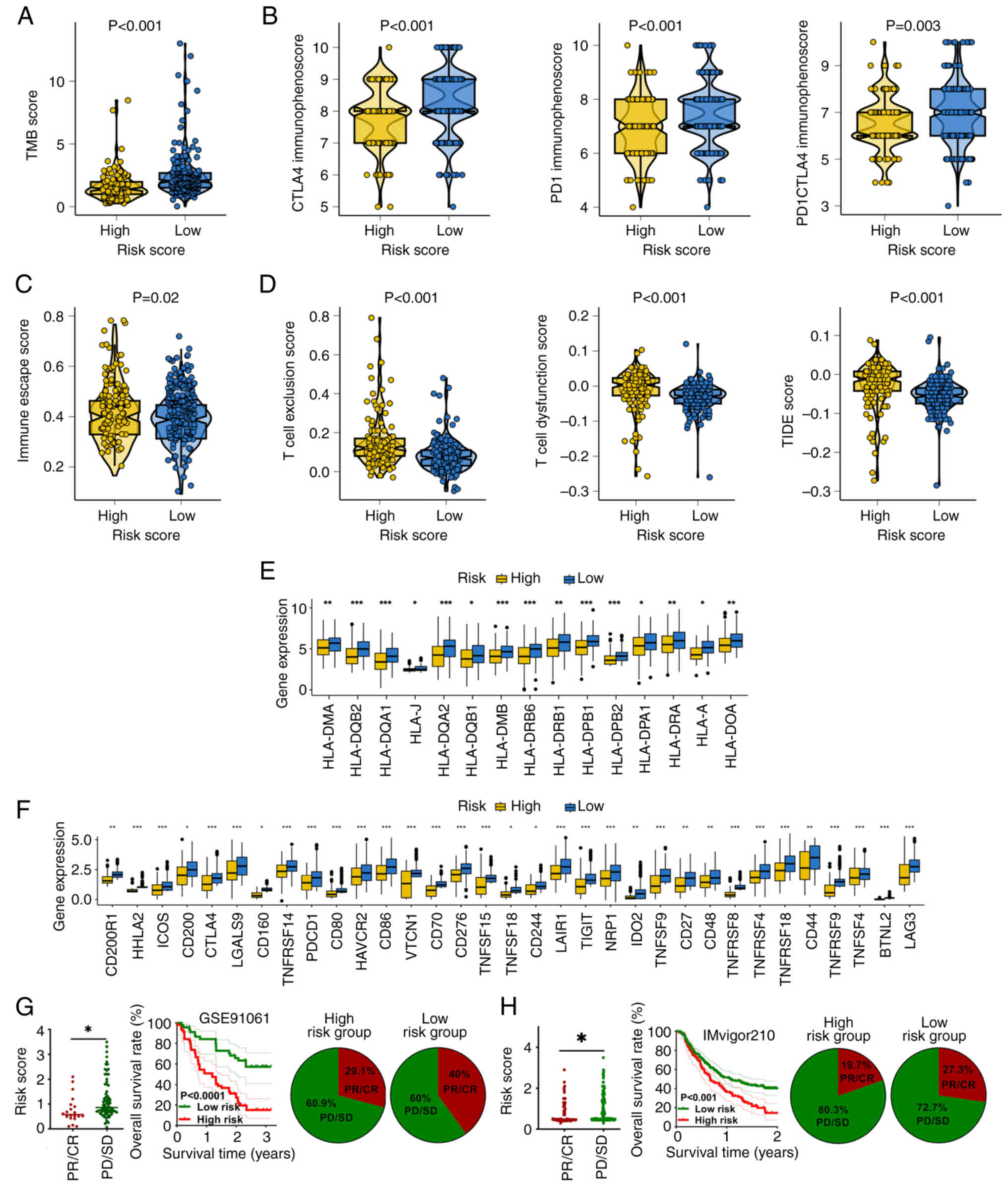 | Figure 4.Predictive role of the GRS in
immunotherapy responses among patients with hepatocellular
carcinoma. (A) TMB and (B) immunophenoscore distributions between
high and low-GRS groups. (C and D) Differences in immune escape
metrics including TIDE, T cell dysfunction and exclusion scores.
(E) Expression levels of HLA-associated genes and (F) immune
checkpoint molecules across risk groups. Immunotherapy response
rate and survival outcomes in (G) GSE91061 and (H) IMvigor210
cohorts stratified by GRS. *P<0.05, **P<0.01 and
***P<0.001. GRS, glycolysis-related signature; TMB, tumor
mutational burden; HLA, human leukocyte antigen; TIDE, Tumor Immune
Dysfunction and Exclusion; CTLA, cytotoxic T-lymphocyte associated
protein 4; CR, complete response; PR; partial response; SD, stable
disease; PD, progressive disease. |
Patients with HCC that had a low-risk score in the
present study demonstrated higher levels of major immunological
checkpoints (Fig. 4F; all
P<0.05). These data suggested that patients with HCC that had a
low-risk score would be more responsive to immunotherapy.
Additionally, two immunotherapy cohorts were used to demonstrate
the aforementioned findings. In patients that received
immunotherapy, non-responders had a higher risk score (Fig. 4G), with this high-risk score
indicating a poor OS rate (Fig.
4G). Additionally, there was a notably decreased response rate
in patients with a high-risk score (Fig. 4G). The IMvigor210 cohort also showed
similar outcomes (Fig. 4H). The
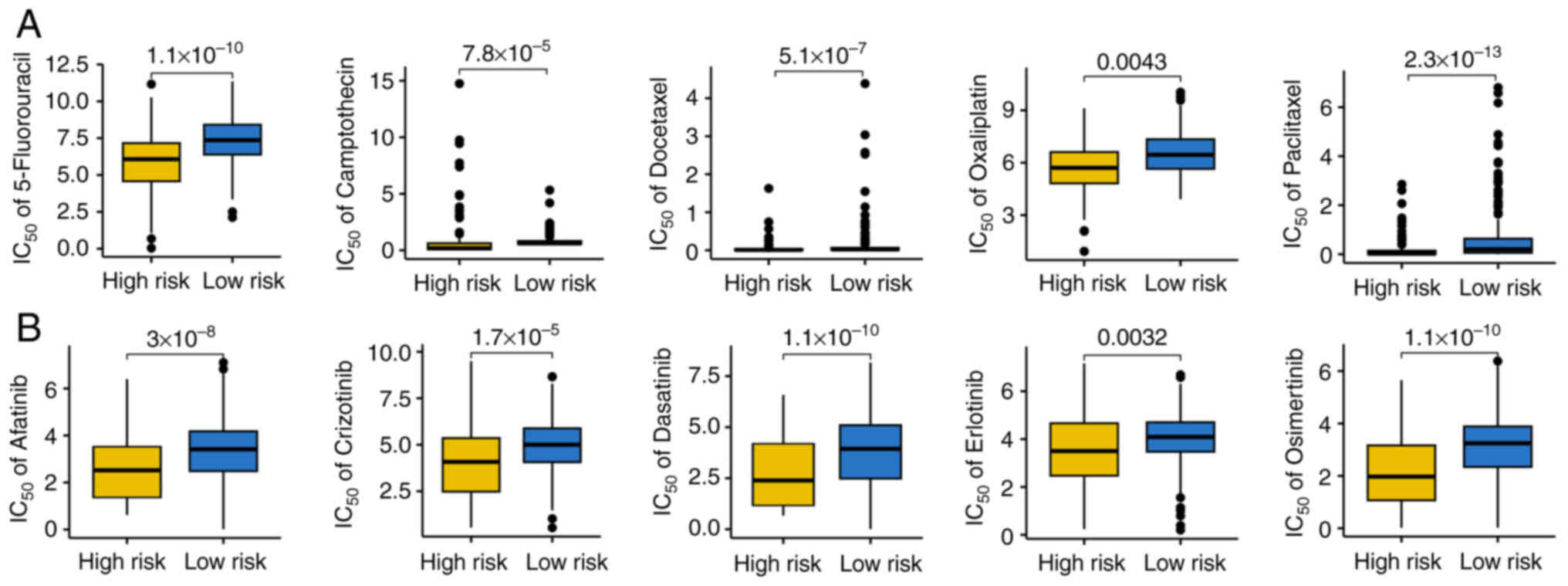
IC50 value of common medications used in targeted
therapy and chemotherapy for HCC was also explored. Findings
indicated that patients with high-risk scores for HCC had lower
IC50 values for chemotherapy drugs, such as
5-fluorouracil, camptothecin, docetaxel, oxaliplatin and paclitaxel
and targeted therapy drugs, such as afatinib, crizotinib,
dasatinib, erlotinib and osimertinib (Fig. 5A and B). Consequently, patients with
HCC and a high-risk score may respond more favorably to targeted
therapy and chemotherapy.
Distinct differences in cancer-related
hallmarks in different GRS-based risk score groups
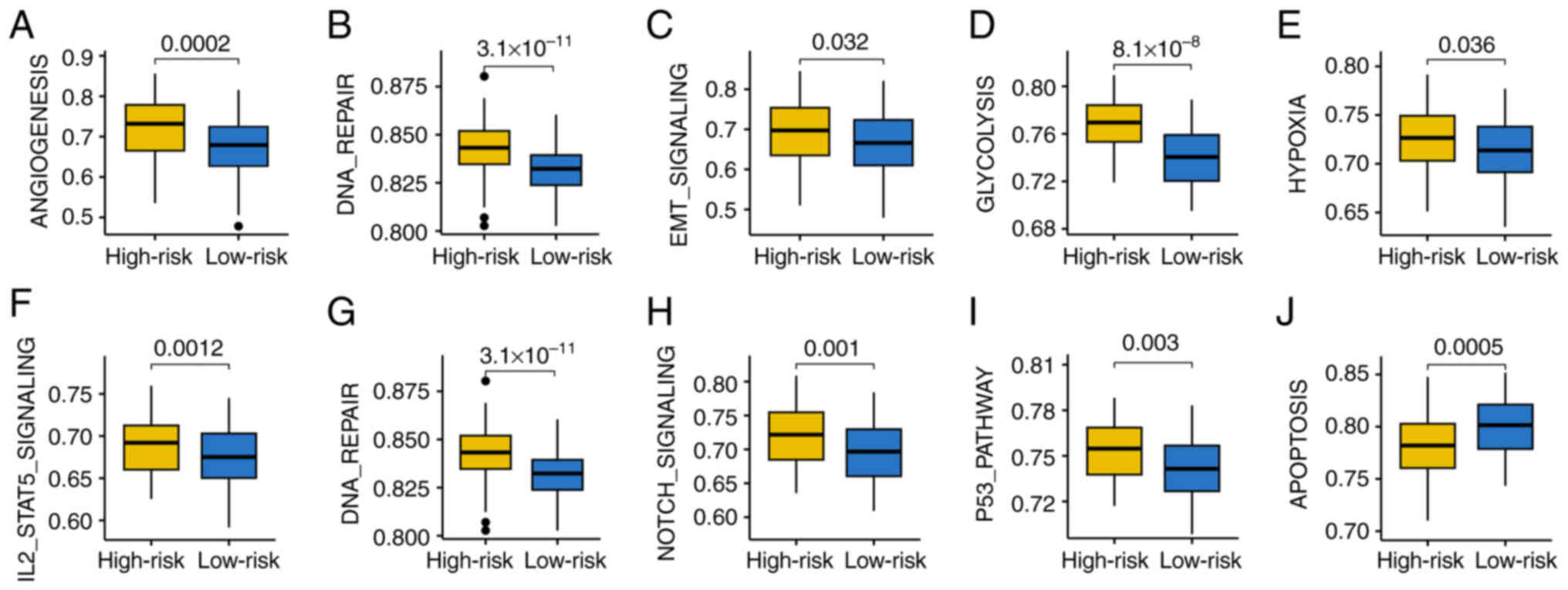
In order to investigate the molecular mechanisms in
patients with HCC with varying GRS scores, GSEA was performed. Gene
set score was associated with angiogenesis, DNA repair, epithelial
mesenchymal transition (EMT) signaling, glycolysis, hypoxia,
IL2-STAT5 signaling, mTORC1 signaling and the p53 pathway, and were
all higher in patients with high-risk HCC (Fig. 6A-I). Patients with HCC that had a
high-risk score indicated a higher apoptosis gene set score
(Fig. 6J). This suggests that these
biological processes may be crucial in the initiation and
progression of HCC tumors.
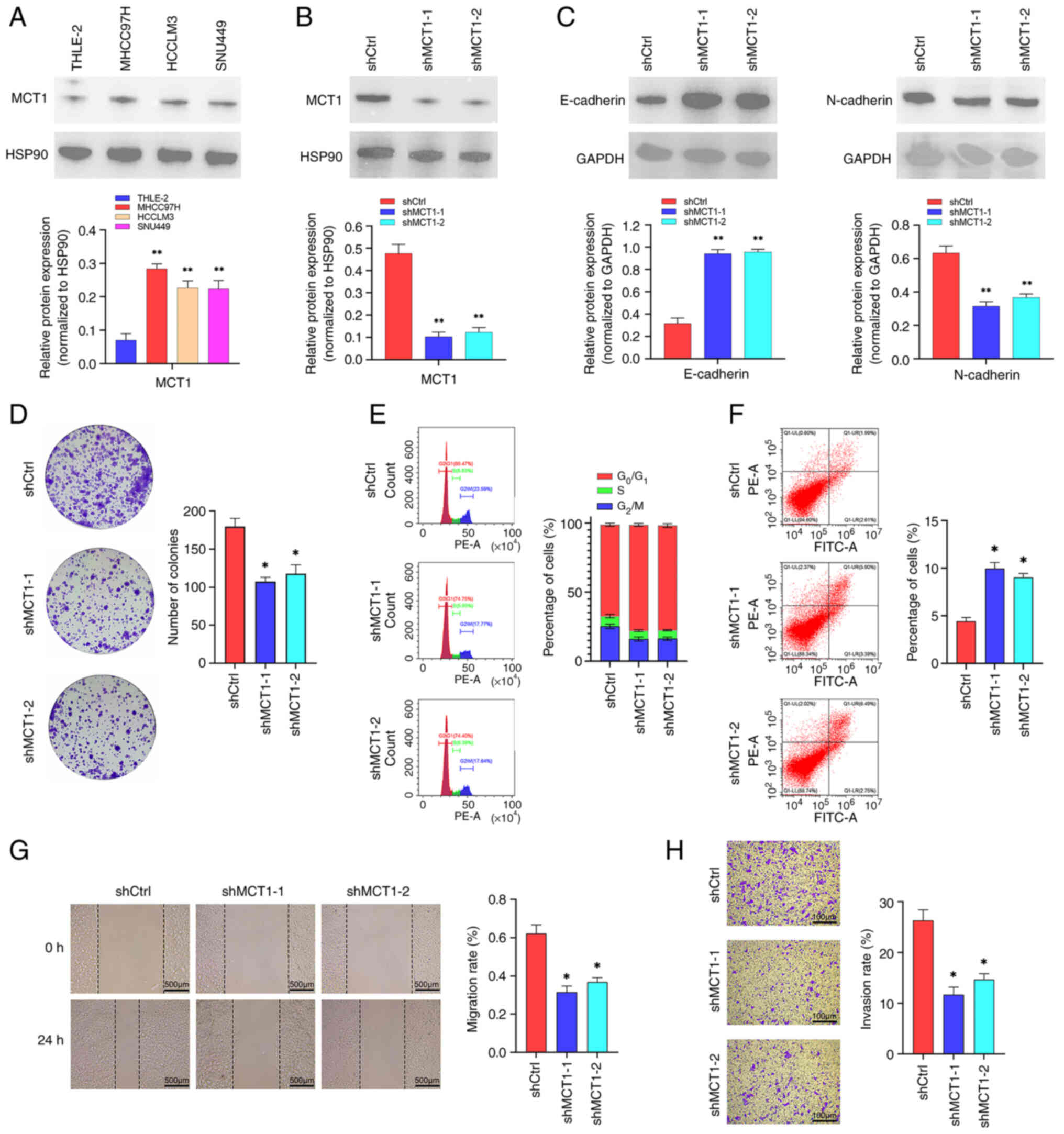
Biological functions of MCT1 in
HCC
MCT1, the gene with the maximum coefficient in the
risk score calculation formula for further analysis, was selected.
As shown in Fig. 7A, MCT1 was
upregulated in HCC cell lines, including MHCC97, HCCLM3 and SNU449,
compared with the normal liver cell line, THLE-2. Subsequently,
MCT1 was knocked down in MHCC97H cell lines (Fig. 7B). EMT-related proteins were also
detected, and the results indicated that downregulation of MCT1
inhibited EMT in MHCC97H cell line (Fig. 7C). The results showed that the
downregulation of MCT1 inhibited colony formation in HCC and
resulted in cell cycle arrest and apoptosis (Fig. 7D-F). Moreover, downregulation of
MCT1 inhibited the migration and invasion of MHCC97H cell line
(Fig. 7G and H).
Discussion
In the present study, a GRS was established by
integrating 10 machine learning algorithms, and its prognostic
value was evaluated across three independent cohorts. Among these,
the model constructed using the LASSO method demonstrated a
superior performance and was selected as the optimal prognostic
tool. Patients with elevated GRS scores exhibited notably worse
clinical outcomes, as reflected by 1, 3 and 5-year AUCs of 0.777,
0.787 and 0.766, respectively. In addition to the prognostic value,
the GRS also proved effective in predicting therapeutic
responsiveness in HCC.
The final GRS was comprised of seven GRGs: TCOF1,
RFC4, RAE1, KIF2C, DEPDC1, ADNP and MCT1. TCOF1 has been reported
to drive tumorigenesis by regulating ribosomal RNA synthesis and
promoting oncogenic pathways (30).
RFC4 acts as a powerful prognostic indicator and serves a role in
cell proliferation within HCC (31). Moreover, RFC4 serves as a novel
biomarker in pan-cancer due to its association with immune
infiltration and drug response (32). A previous study also demonstrated
that RAE1 was a prognostic marker for HCC and showed an association
with clinicopathological characteristics (33). In addition, the silencing of KIF2C
has been shown to enhance chemosensitivity through the
PI3K/AKT/MAPK axis (34), and
DEPDC1 promotes proliferation, invasion and angiogenesis in HCC
through the C-C motif chemokine ligand 20/C-C motif chemokine
receptor 6 pathway (35,36). Furthermore, ADNP is linked to both
immune modulation and radiosensitivity in liver cancer (37). Finally, MCT1, a key glycolytic
transporter, has been implicated in autophagy-mediated metastasis
and metabolic reprogramming through activation of the Wnt/β-catenin
pathway (38).
Immunotherapy is a promising treatment alternative
for patients who are unable to undergo surgery (39). However, reliable biomarkers that
predict immunotherapy responses remain limited. The efficacy of GRS
in predicting the benefits a patient may receive from immunotherapy
was examined in the present study, with findings indicating that
the GRS may offer predictive value in this context. Patients with
HCC and higher TIDE scores, indicative of immune evasion, benefited
less from immunotherapy (40), with
an improved immunotherapy benefit being indicated by a higher TMB
score (41). In patients with HCC
that had a low-risk score, a higher TMB score as well as lower TIDE
and tumor escape scores were observed. The immunophenoscore was
established to forecast patient reactions to immune checkpoint
inhibitor therapies, mainly using TCGA RNA-sequencing data
(42). A higher immunophenoscore
supports the concept that immunotherapy is more effective in
patients with HCC and a low-risk score, which was in line with the
findings of the present study. A worse response to immunotherapy
may therefore be indicated by a higher GRS-based risk score in
HCC.
The potential mechanisms underlying the association
between patients with HCC with a high GRS score and poor
immunotherapy benefits must be further explored. Notably, the
findings of the present study align with prior studies by Zhang
et al (43) and Peng et
al (44), which systematically
outlined the role of altered glycolysis in mediating drug
resistance across diverse cancer types and other disease. Zhang
et al (43) found that the
N6-methyladenosine demethylase fat mass and obesity-associated
protein (FTO) attenuates cardiac dysfunction by regulating glucose
uptake and glycolysis in mice with pressure overload-induced heart
failure, suggesting FTO is a potential target for heart failure
prevention and treatment. Peng et al (44) suggested that glycolysis-driven
suppression of IFN-γ signaling may contribute to immunotherapy
resistance. By contrast, excessive glycolytic activity driven by
STAT5-induced lactate production may also sensitize certain tumors
to a PD-1/programmed death-ligand 1 (PD-L1) blockade, indicating a
complex relationship between metabolic status and immune response
(45). These studies emphasized
that excessive lactate production and acidification of the TME
impairs drug delivery and fosters immune evasion, both of which
contribute to reduced therapeutic efficacy.
In the present study, high-risk patients
demonstrated elevated glycolytic activity and upregulation of MCT1,
a monocarboxylate transporter responsible for lactate export,
supporting the hypothesis that glycolysis-driven acidosis may
underlie poor immunotherapy and chemotherapy responses. The
upregulation of glycolytic genes and enrichment of pathways such as
EMT, mTORC1 and hypoxia signaling in high-risk HCC, further
reinforces the mechanistic links between metabolic reprogramming
and treatment resistance. These observations underscore the
translational potential of combining glycolysis inhibitors, such as
MCT1 antagonists, with existing therapeutic regimens to overcome
resistance and improve outcomes in HCC.
Functional enrichment analysis further demonstrated
that several oncogenic pathways, including angiogenesis,
glycolysis, hypoxia, mTORC1, EMT and p53 signaling, were
upregulated in patients with a high-risk score. These biological
programs promote tumor development, immune evasion and treatment
resistance in HCC. Angiogenesis is important in the development of
HCC (46) and hypoxia is associated
with innate immunity and tumor progression in HCC (47). In addition, glycolysis is notably
associated with the prognosis of HCC (48) and phenylalanyl-tRNA synthetase
subunit β has been shown to promote tumor progression by activating
the mTORC1 signaling pathway (49).
Notably, HCC progression may also be associated with
therapeutic resistance. EMT is a mechanism in which E-cadherin
expression is lost during tumor progression (50). E-cadherins are crucial cell-cell
adhesion proteins with tumor suppression properties (51). Loss of E-cadherin during EMT is
associated with tumor progression and broad-spectrum treatment
resistance. In multiple cancers, EMT programs, occasionally
precipitated or reinforced during exposure to cytotoxic agents such
as cisplatin or paclitaxel, are associated with reduced drug
sensitivity and increased survival of motile, detached cells
(52). Recent structural and
single-molecule work by Xie et al (53) identified 66E8, a
conformation-specific monoclonal antibody against E-cadherin that
stabilizes the adhesive strand-swap dimer of E-cadherin by
strengthening electrostatic contacts around the N-terminal swapped
β-strand and its hydrophobic pocket, thereby impeding
conformational changes that favor dimer dissociation under force
(53). The ‘swapped β-strand’
refers to the N-terminal β-strand in the extracellular cadherin
domain 1 that exchanges between two E-cadherin protomers to form
the strand-swap dimer, the high-affinity adhesive state that
underpins epithelial cell-cell junctions. Functionally, stabilizing
cadherin-mediated adhesion is expected to reduce detachment and
anoikis resistance, blunt EMT-like migratory programs and
re-sensitize tumors to therapy by restoring mechanotransduction and
immune engagement at tumor-immune interfaces. Complementing this,
pro-inflammatory IL-17A can downregulate E-cadherin and upregulate
PD-L1, fostering immune evasion (54). Thus, therapeutic angles for future
studies in HCC include: i) Adhesion-stabilizing biologics (such as
66E8 or E-cadherin-stabilizing peptides) to curb dissemination and
enhance chemotherapy efficacy; ii) combination strategies pairing
adhesion stabilization with immune checkpoint blockade to counter
IL-17A-driven PD-L1 induction; and iii) rational combinations with
metabolic modulators (such as inhibitors targeting glycolysis/MCT1
in GRS-high, EMT-enriched tumors) to simultaneously relieve
acidosis-driven EMT and reinforce junctional adhesion.
Collectively, these approaches support the concept that enhancing
E-cadherin adhesion may inhibit HCC progression and improve
therapeutic benefits.
There are several limitations of the present study
which must be acknowledged. Firstly, although the expression and
function of MCT1 was evaluated in vitro, in vivo experiments
are required to demonstrate its role in tumor progression.
Secondly, the analyses were primarily based on transcriptomic data,
which may not fully capture protein-level or functional dynamics.
Lastly, external validation within an in-house or prospective
cohort would further strengthen the generalizability of the
findings of the present study.
In conclusion, the present study developed a novel
GRS for HCC, serving as an indicator for predicting clinical
outcomes and immunotherapeutic responses.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the Hunan Provincial
Natural Science Foundation (grant nos. 2023JJ40387 and 2024JJ6276),
Hunan Provincial Key Field Research and Development Program (grant
no. 2022SK2162) and the Natural Science Foundation of Changsha
(grant no. kq2208120).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding authors.
Authors' contributions
YZ contributed to writing, original draft
preparation, bioinformatics analysis and experimental research. WL
contributed to writing and conceptualization and methodology. ZF
contributed to the writing, software used and data collection. KZ
contributed to methodology and visualization. XH contributed to
conception, writing, reviewing and editing. SL contributed to the
software used and acquisition of data. WZ and QW were involved in
validation and analysis of data. XC and ZX contributed to study
design and supervision. YZ and WL confirm the authenticity of all
the raw data. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vogel A, Meyer T, Sapisochin G, Salem R
and Saborowski A: Hepatocellular carcinoma. Lancet. 400:1345–1362.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
3
|
Chidambaranathan-Reghupaty S, Fisher PB
and Sarkar D: Hepatocellular carcinoma (HCC): Epidemiology,
etiology and molecular classification. Adv Cancer Res. 149:1–61.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Y, Yang Y, Zhao Z, Sun H, Luo D,
Huttad L, Zhang B and Han B: A new nomogram model for prognosis of
hepatocellular carcinoma based on novel gene signature that
regulates cross-talk between immune and tumor cells. BMC Cancer.
22:3792022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Icard P, Shulman S, Farhat D, Steyaert JM,
Alifano M and Lincet H: How the Warburg effect supports
aggressiveness and drug resistance of cancer cells? Drug Resist
Updat. 38:1–11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han X, Ren C, Yang T, Qiao P, Wang L,
Jiang A, Meng Y, Liu Z, Du Y and Yu Z: Negative regulation of
AMPKα1 by PIM2 promotes aerobic glycolysis and tumorigenesis in
endometrial cancer. Oncogene. 38:6537–6549. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang D, Tang Z, Huang H, Zhou G, Cui C,
Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al: Metabolic
regulation of gene expression by histone lactylation. Nature.
574:575–580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang H, Chen K, Zhu Y, Hu Z, Wang Y, Chen
J, Li Y, Li D and Wei P: A multi-dimensional approach to unravel
the intricacies of lactylation related signature for prognostic and
therapeutic insight in colorectal cancer. J Transl Med. 22:2112024.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang R, Li Y, Lin K, Zheng L, Zhu X,
Huang L and Ma Y: A novel glycolysis-related gene signature for
predicting prognosis and immunotherapy efficacy in breast cancer.
Front Immunol. 16:15128592025. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen C, Suo Y, Guo J, Su W, Zhang Z, Yang
S, Wu Z, Fan Z, Zhou X and Hu H: Development and validation of a
glycolysis-associated gene signature for predicting the prognosis,
immune landscape, and drug sensitivity in bladder cancer. Front
Immunol. 15:14305832024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu Q, Miao D, Song X, Chen Z, Zeng L, Zhao
L, Xu J, Lin Z and Yu F: Glycolysis-related gene signature can
predict survival and immune status of hepatocellular carcinoma. Ann
Surg Oncol. 29:3963–3976. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grinchuk OV, Yenamandra SP, Iyer R, Singh
M, Lee HK, Lim KH, Chow PK and Kuznetsov VA: Tumor-adjacent tissue
co-expression profile analysis reveals pro-oncogenic ribosomal gene
signature for prognosis of resectable hepatocellular carcinoma. Mol
Oncol. 12:89–113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX and Wang XW: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Riaz N, Havel JJ, Makarov V, Desrichard A,
Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH,
et al: Tumor and microenvironment evolution during immunotherapy
with nivolumab. Cell. 171:934–949.e16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rosenberg JE, Galsky MD, Powles T,
Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J,
Perez-Gracia JL, van der Heijden MS, et al: Atezolizumab
monotherapy for metastatic urothelial carcinoma: Final analysis
from the phase II IMvigor210 trial. ESMO Open. 9:1039722024.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng Z, Huang H, Li M, Liang X, Tan Y and
Chen Y: Lactylation-related gene signature effectively predicts
prognosis and treatment responsiveness in hepatocellular carcinoma.
Pharmaceuticals (Basel). 16:6442023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang X, Li X, Cheng Y, Zhou J, Shen B,
Zhao L and Wang J: Comprehensive analysis of the glycolysis-related
gene prognostic signature and immune infiltration in endometrial
cancer. Front Cell Dev Biol. 9:7978262022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Li Y and Chen Y: Development of a
comprehensive gene signature linking hypoxia, glycolysis,
lactylation, and metabolomic insights in gastric cancer through the
integration of bulk and single-cell RNA-Seq data. Biomedicines.
11:29482023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Z, Liu L, Weng S, Guo C, Dang Q, Xu H,
Wang L, Lu T, Zhang Y, Sun Z and Han X: Machine learning-based
integration develops an immune-derived lncRNA signature for
improving outcomes in colorectal cancer. Nat Commun. 13:8162022.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sturm G, Finotello F and List M:
Immunedeconv: An R package for unified access to computational
methods for estimating immune cell fractions from bulk
RNA-sequencing data. Methods Mol Biol. 2120:223–232. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Maeser D, Gruener RF and Huang RS:
oncoPredict: an R package for predicting in vivo or cancer patient
drug response and biomarkers from cell line screening data. Brief
Bioinform. 22:bbab2602021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ansari A, Ray SK, Sharma M, Rawal R and
Singh P: Tumor mutational burden as a biomarker of immunotherapy
response: An immunogram approach in onco-immunology. Curr Mol Med.
24:1461–1469. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maggs L, Sadagopan A, Moghaddam AS and
Ferrone S: HLA class I antigen processing machinery defects in
antitumor immunity and immunotherapy. Trends Cancer. 7:1089–1101.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu C, Xia D, Wang D, Wang S, Sun Z, Xu B
and Zhang D: TCOF1 coordinates oncogenic activation and rRNA
production and promotes tumorigenesis in HCC. Cancer Sci.
113:553–564. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen P, Liu Y, Ma X, Li Q, Zhang Y, Xiong
Q and Song T: Replication factor C4 in human hepatocellular
carcinoma: A potent prognostic factor associated with cell
proliferation. Biosci Trends. 15:249–256. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu L, Li J, Zhang M, Li Y, Bai J, Liu P,
Yan J and Wang C: Identification of RFC4 as a potential biomarker
for pan-cancer involving prognosis, tumour immune microenvironment
and drugs. J Cell Mol Med. 28:e184782024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chi G, Pei JH and Li XQ: RAE1 is a
prognostic biomarker and is correlated with clinicopathological
characteristics of patients with hepatocellular carcinoma. BMC
Bioinformatics. 23:2522022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wei S, Lu C, Mo S, Huang H, Chen M, Li S,
Kong L, Zhang H, Hoa PTT, Han C and Luo X: Silencing of KIF2C
enhances the sensitivity of hepatocellular carcinoma cells to
cisplatin through regulating the PI3K/AKT/MAPK signaling pathway.
Anticancer Drugs. 35:237–250. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Amisaki M, Yagyu T, Uchinaka EI, Morimoto
M, Hanaki T, Watanabe J, Tokuyasu N, Sakamoto T, Honjo S and
Fujiwara Y: Prognostic value of DEPDC1 expression in tumor and
non-tumor tissue of patients with hepatocellular carcinoma.
Anticancer Res. 39:4423–4430. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo W, Li H, Liu H, Ma X, Yang S and Wang
Z: DEPDC1 drives hepatocellular carcinoma cell proliferation,
invasion and angiogenesis by regulating the CCL20/CCR6 signaling
pathway. Oncol Rep. 42:1075–1089. 2019.PubMed/NCBI
|
|
37
|
Wang X, Peng H, Zhang G, Li Z, Du Z, Peng
B and Cao P: ADNP is associated with immune infiltration and
radiosensitivity in hepatocellular carcinoma for predicting the
prognosis. BMC Med Genomics. 16:1782023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fan Q, Yang L, Zhang X, Ma Y, Li Y, Dong
L, Zong Z, Hua X, Su D, Li H and Liu J: Autophagy promotes
metastasis and glycolysis by upregulating MCT1 expression and
Wnt/β-catenin signaling pathway activation in hepatocellular
carcinoma cells. J Exp Clin Cancer Res. 37:92018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Riley RS, June CH, Langer R and Mitchell
MJ: Delivery technologies for cancer immunotherapy. Nat Rev Drug
Discov. 18:175–196. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang
P and Liu XS: Large-scale public data reuse to model immunotherapy
response and resistance. Genome Med. 12:212020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu L, Bai X, Wang J, Tang XR, Wu DH, Du
SS, Du XJ, Zhang YW, Zhu HB, Fang Y, et al: Combination of TMB and
CNA stratifies prognostic and predictive responses to immunotherapy
across metastatic cancer. Clin Cancer Res. 25:7413–7423. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zanfardino M, Pane K, Mirabelli P,
Salvatore M and Franzese M: TCGA-TCIA impact on radiogenomics
cancer research: A systematic review. Int J Mol Sci. 20:60332019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang B, Jiang H, Wu J, Cai Y, Dong Z,
Zhao Y, Hu Q, Hu K, Sun A and Ge J: m6A demethylase FTO attenuates
cardiac dysfunction by regulating glucose uptake and glycolysis in
mice with pressure overload-induced heart failure. Signal Transduct
Target Ther. 6:3772021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Peng J, Cui Y, Xu S, Wu X, Huang Y, Zhou
W, Wang S, Fu Z and Xie H: Altered glycolysis results in
drug-resistant in clinical tumor therapy. Oncol Lett. 21:3692021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang ZW, Zhang XN, Zhang L, Liu LL, Zhang
JW, Sun YX, Xu JQ, Liu Q and Long ZJ: STAT5 promotes PD-L1
expression by facilitating histone lactylation to drive
immunosuppression in acute myeloid leukemia. Signal Transduct
Target Ther. 8:3912023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang F, Wang B, Zhang W, Xu Y, Zhang C
and Xue X: Transcription factor MAZ potentiates the upregulated
NEIL3-mediated aerobic glycolysis, thereby promoting angiogenesis
in hepatocellular carcinoma. Curr Cancer Drug Targets.
24:1235–1249. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yuen VW and Wong CC: Hypoxia-inducible
factors and innate immunity in liver cancer. J Clin Invest.
130:5052–5062. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang B and Pu R: Association between
glycolysis markers and prognosis of liver cancer: A systematic
review and meta-analysis. World J Surg Oncol. 21:3902023.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang Y, Wang G, Hu S, Yin C, Zhao P, Zhou
X, Shao S, Liu R, Hu W, Liu GL, et al: FARSB facilitates
hepatocellular carcinoma progression by activating the mTORC1
signaling pathway. Int J Mol Sci. 24:167092023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kaszak I, Witkowska-Piłaszewicz O,
Niewiadomska Z, Dworecka-Kaszak B, Ngosa Toka F and Jurka P: Role
of cadherins in cancer-a review. Int J Mol Sci. 21:76242020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lee G, Wong C, Cho A, West JJ, Crawford
AJ, Russo GC, Si BR, Kim J, Hoffner L, Jang C, et al: E-cadherin
induces serine synthesis to support progression and metastasis of
breast cancer. Cancer Res. 84:2820–2835. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hashemi M, Arani HZ, Orouei S, Fallah S,
Ghorbani A, Khaledabadi M, Kakavand A, Tavakolpournegari A, Saebfar
H, Heidari H, et al: EMT mechanism in breast cancer metastasis and
drug resistance: Revisiting molecular interactions and biological
functions. Biomed Pharmacother. 155:1137742022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xie B, Xu S, Schecterson L, Gumbiner BM
and Sivasankar S: Strengthening E-cadherin adhesion via
antibody-mediated binding. Structure. 32:217–227.e3. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liao H, Chang X, Gao L, Ye C, Qiao Y, Xie
L, Lin J, Cai S and Dong H: IL-17A promotes tumorigenesis and
upregulates PD-L1 expression in non-small cell lung cancer. J
Transl Med. 21:8282023. View Article : Google Scholar : PubMed/NCBI
|