Introduction
Colorectal cancer (CRC) ranks third among the most
prevalent diseases and is the second deadliest malignant disease
globally (1,2). In 2020, there were ~1.9 million newly
reported cases of colorectal cancer (CRC) globally, with
>900,000 deaths attributed to CRC (3). Key factors contributing to its high
mortality include challenges in early diagnosis, tumor
heterogeneity and the ineffectiveness of late-stage treatments
(4–6). Tumor heterogeneity encompasses both
cellular heterogeneity within the tumor and heterogeneity of the
tumor microenvironment (7,8). A growing body of research indicates
that the heterogeneity of CRC is among the most notable in
malignant tumors and exerts a profound influence on the prognosis
and treatment of the disease (5,9,10).
Moreover, previous studies leveraging bioinformatics analysis for
molecular classification and prognostic prediction of CRC have
emerged, indicating its considerable clinical promise (11–13).
However, due to its heterogeneity, CRC progression is a
multifactorial, multistage and dynamic process, which imposes
certain limitations on existing models. Consequently, it is
imperative to explore novel molecular subtypes and models of CRC
for prognostic evaluation.
Tissue inhibitor of matrix metalloproteinase 1
(TIMP1), a member of the TIMP gene family, has been reported to
inhibit matrix metalloproteinase (MMP) activity (14). The TIMP1 gene is situated on human
chromosome Xp11.1-p11.4 and its mRNA codes for a protein composed
of 184 amino acids, exhibiting a molecular mass of 29–34 kDa
(15). Accumulating evidence
indicates that it interacts with several molecules to regulate
biological processes such as cell proliferation, apoptosis and
differentiation, and it is implicated in the pathological
mechanisms of several diseases, particularly cancer (16,17). A
total of >35 independent clinical studies have reported that
high TIMP1 expression is positively associated with poor prognosis
across several cancer types, including lung, brain, prostate,
breast and colon cancer (14).
Moreover, the levels of TIMP1 are elevated in advanced-stage tumors
(14). Tian et al (18) reported that pancreatic cancer
cell-derived TIMP1 activates CD63/PI3K/AKT signaling to promote
perineural invasion by stimulating Schwann cells. Moreover, TIMP1
loss has been reported to alter the senescence-associated secretory
phenotype of senescent tumor cells and enhance prostate cancer
metastasis via MMP activation (19). However, the role of TIMP1 in CRC
remains poorly understood. Therefore, the present study aimed to
explore the clinical and prognostic significance of TIMP1 in
CRC.
Materials and methods
The Cancer Genome Atlas (TCGA)
TCGA database (http://cancergenome.nih.gov/) was the primary resource
for acquiring CRC mRNA expression data and clinical information.
The present study analyzed multi-omics patient data, which were
systematically organized across multiple databases, including the
original TCGA datasets (http://cancergenome.nih.gov/). Subsequent databases,
such as the University of Alabama at Birmingham CANcer data
analysis Portal (UALCAN) (20,21)
and Gene Expression Profiling Interactive Analysis (GEPIA)
(22), provided comprehensive
insights into several aspects. The patient records from each
database were specifically utilized for diverse analyses.
Gene expression data processing
The UALCAN database (https://ualcan.path.uab.edu/) (20,21)
was used to analyze TIMP1 mRNA expression levels in pan-cancer and
in colon cancer samples (primary tumor, n=286; normal, n=41).
GEPIA2 (http://gepia2.cancer-pku.cn/#general) (21) was used to assess TIMP1 expression in
TCGA tumors by matching TCGA normal and Genotype-Tissue Expression
project data for CRC (tumor, n=275; normal, n=349). The GSE24550
dataset was used to analyze TIMP1 mRNA expression levels in CRC
tissues (G2, n=77) and normal tissues (G1, n=13) (23). In addition, the Clinical Proteomic
Tumor Analysis Consortium (CPTAC) database within UALCAN was used
to evaluate TIMP1 protein expression in CRC (normal, n=100; primary
tumor, n=97).
CRC clinical pathological
parameters
STAR-counts data and the corresponding clinical
information for CRC tumors were downloaded from TCGA database
(24). The data in transcripts per
million (TPM) format were extracted and normalized using
log2(TPM+1) transformation. Following filtering of the
samples with both RNA sequencing (RNAseq) and clinical data, CRC
samples with high TIMP1 expression (top 25%, n=156) and low TIMP1
expression (top 25%, n=157) were selected for analysis (Table I). Statistical analysis was
performed using R software (version v4.0.3; The R Foundation).
P<0.05 was considered to indicate a statistically significant
difference.
 | Table I.Tissue inhibitor of matrix
metalloproteinase 1 expression and clinical features in 313 samples
from patients with colorectal cancer. |
Table I.
Tissue inhibitor of matrix
metalloproteinase 1 expression and clinical features in 313 samples
from patients with colorectal cancer.
|
| TIMP1 expression
level |
|
|---|
|
|
|
|
|---|
| Characteristic | High (n=156) | Low (n=157) | P-value |
|---|
| Status, n |
|
| 0.0019 |
|
Alive | 109 (34.8) | 133 (42.5) |
|
|
Dead | 47 (15.0) | 24 (7.7) |
|
| Age |
|
|
|
| Mean ±
SD | 66.7±12.8 | 64.9±13.4 | 0.4630 |
| Median
(min, max) | 68 (31, 90) | 66 (31, 90) | 0.2150 |
| Sex, n |
|
| 0.4978 |
|
Female | 77 (24.6) | 71 (22.7) |
|
|
Male | 79 (25.2) | 86 (27.5) |
|
|
Ethnicitya |
|
| 0.0404 |
|
American Indian | 1 (0.3) | 0 (0.0) |
|
|
Asian | 4 (1.3) | 2 (0.6) |
|
|
Black | 12 (3.8) | 25 (8.0) |
|
|
White | 89 (28.4) | 69 (22.0) |
|
| pT
stagea |
|
| 0.1457 |
| T1 | 6 (1.9) | 6 (1.9) |
|
| T2 | 14 (4.5) | 26 (8.3) |
|
| T3 | 107 (34.2) | 109 (34.8) |
|
| T4 | 13 (4.2) | 9 (2.9) |
|
|
T4a | 12 (3.8) | 4 (1.3) |
|
|
T4b | 3 (1.0) | 2 (0.6) |
|
|
Tis | 0 (0.0) | 1 (0.3) |
|
| pN
stagea |
|
| 0.1345 |
| N0 | 85 (27.2) | 103 (32.9) |
|
| N1 | 23 (7.3) | 22 (7.0) |
|
|
N1a | 6 (1.9) | 5 (1.6) |
|
|
N1b | 7 (2.2) | 2 (0.6) |
|
|
N1c | 1 (0.3) | 2 (0.6) |
|
| N2 | 18 (5.8) | 19 (6.1) |
|
|
N2a | 3 (1.0) | 2 (0.6) |
|
|
N2b | 11 (3.5) | 2 (0.6) |
|
| NX | 1 (0.3) | 0 (0.0) |
|
| pM
stagea |
|
| 0.1457 |
| M0 | 116 (37.1) | 114 (36.4) |
|
| M1 | 16 (5.1) | 17 (5.4) |
|
|
M1a | 6 (1.9) | 1 (0.3) |
|
| MX | 14 (4.5) | 22 (7.0) |
|
| pTNM
stagea |
|
| 0.2748 |
| I | 15 (4.8) | 27 (8.6) |
|
| II | 10 (3.2) | 9 (2.9) |
|
|
IIA | 56 (17.9) | 51 (16.3) |
|
|
IIB | 2 (0.6) | 7 (2.2) |
|
|
III | 2 (0.6) | 6 (1.9) |
|
|
IIIA | 5 (1.6) | 2 (0.6) |
|
|
IIIB | 22 (7.0) | 19 (6.1) |
|
|
IIIC | 15 (4.8) | 12 (3.8) |
|
| IV | 15 (4.8) | 14 (4.5) |
|
|
IVA | 7 (2.2) | 4 (1.3) |
|
| IA | 0 (0.0) | 1 (0.3) |
|
Survival prognosis analysis
The GEPIA2 database was used for the analysis of the
association between TIMP1 expression, overall survival and
disease-free survival in patients with CRC. In addition, the UALCAN
database was used for the analysis of the association between TIMP1
expression and overall survival. CRC samples with high TIMP1
expression (median, n=309) and low TIMP1 expression (median, n=310)
from TCGA database were used for further analysis. For survival
analysis, the log-rank test was employed to compare survival
differences between the two groups in the Kaplan-Meier analysis,
obtaining the P-value, hazard ratio (HR) and 95% confidence
interval (CI) using the log-rank test and univariate Cox
regression. To perform Cox regression analysis, univariate and
multivariate Cox proportional hazards regression analyses were
performed. The ‘forestplot’ package was used to visually present
the P-value, HR and 95% CI for each variable using a forest plot.
Subsequently, based on the multivariate Cox proportional hazard
model results, a Nomogram was constructed using the ‘rms’ package
to predict the total recurrence rate over X years. All statistical
analyses were performed using R software (v4.0.3).
Immune correlation analysis
To reliably evaluate immune scores in CRC,
‘immunedeconv’ was used, which is an R package (v4.1.3) that
combines two advanced algorithms, including Tumor IMune Estimation
Resource (TIMER) and Estimate the Proportion of Immune and Cancer
(EPIC), applied to TCGA CRC data (n=620) (25,26).
Each algorithm has been benchmarked for its unique strengths. For
result analysis and visualization, the R package ‘ggClusterNet’ was
employed. In addition, Spearman's correlation analysis was
performed to assess the relationship between TIMP1 and immune cells
(CD4+ and CD8+ T cells) in CRC using TCGA
data (n=620).
Association analysis of TIMP1
expression and tumor mutation burden (TMB)/microsatellite
instability (MSI)
RNAseq data and clinical information of CRC (n=620)
were obtained from TCGA database. Spearman's correlation analysis
was used to assess the correlation between TIMP1 and TMB/MSI
(25,26).
Association analysis of TIMP1
expression and pathway scores
CRC RNAseq data and the clinical information (n=620)
were retrieved from TCGA database. Following compilation of the
genes from the relevant pathways, the ‘GSVA’ package was used in R
software with the parameter method ‘ssgsea’ used to perform
single-sample gene set enrichment analysis (ssGSEA). Finally, the
correlation between gene expression and pathway scores was assessed
using Spearman's correlation analysis (27–29).
CRC specimens and immunohistochemical
(IHC) staining
A colon cancer tissue microarray (TMA) was purchased
from Hunan Aifang Biotechnology Co., Ltd., and the relevant
follow-up data were included. The TMA comprised 78 pairs of colon
cancer and normal tissues. The patient characteristics are
summarized in Table SI. The
present study was approved by the Medical Ethics Committee of
Soochow University's First Affiliated Hospital (approval no.
2021-327).
IHC staining was performed as follows (30): Tissue samples were subjected to
fixation utilizing a 4% paraformaldehyde solution (Beyotime
Institute of Biotechnology) maintained at an ambient temperature of
25°C for a duration of 24 h. Subsequently, the specimens were
processed for paraffin embedding. Sections, each with a thickness
of 5 µm, excised from the paraffin-embedded tissue blocks,
underwent deparaffinization and rehydration procedures. Following
antigen retrieval conducted with a 10 mM sodium citrate buffer (pH
6.0; Beyotime Institute of Biotechnology), the sections were
treated with a 3% hydrogen peroxide solution at room temperature
for 10 min to inhibit endogenous peroxidase activity and to
mitigate non-specific protein interactions. The sections were then
incubated with anti-TIMP1 antibodies (1:300 dilution; cat. no.
16644-1-AP; Proteintech Group, Inc.) overnight at 4°C, succeeded by
incubation with a biotinylated goat anti-rabbit secondary antibody
working solution (1:500 dilution; cat. no. SA1020; Boster
Biological Technology Co. Ltd.) at 37°C for 30 min. Immunodetection
was subsequently executed employing the Dako EnVision detection
system (Agilent Technologies, Inc.). The prepared slides were
examined and images were captured under a fluorescence microscope
(Leica Microsystems). The IHC score was determined by multiplying
the staining intensity (negative, 0; mild, 1; moderate, 2; and
strong, 3) by the percentage of the stained area (0%, 0; ≤25%, 1;
25–50%, 2; 50–75%, 3; and >75%, 4).
Statistical analysis
R software (version 4.0.3) and GraphPad Prism 8.0
(Dotmatics) were used for both data analysis and visualization. The
data are presented as the mean ± SD from a minimum of three
replicates. Differences between two groups were assessed using an
unpaired t-test, Wilcoxon rank-sum test or paired t-test, as
appropriate. Fisher's exact test or χ2 test were used to
analyze the associations between the expression of TIMP1 and
clinicopathological features. Spearman's correlation analysis was
performed to assess the correlation between TIMP1 and immune cells
(CD4+ and CD8+ T cells) in CRC using TCGA
data (n=620). P<0.05 was considered to indicate a statistically
significant difference.
Results
TIMP1 is highly expressed in CRC
tissues
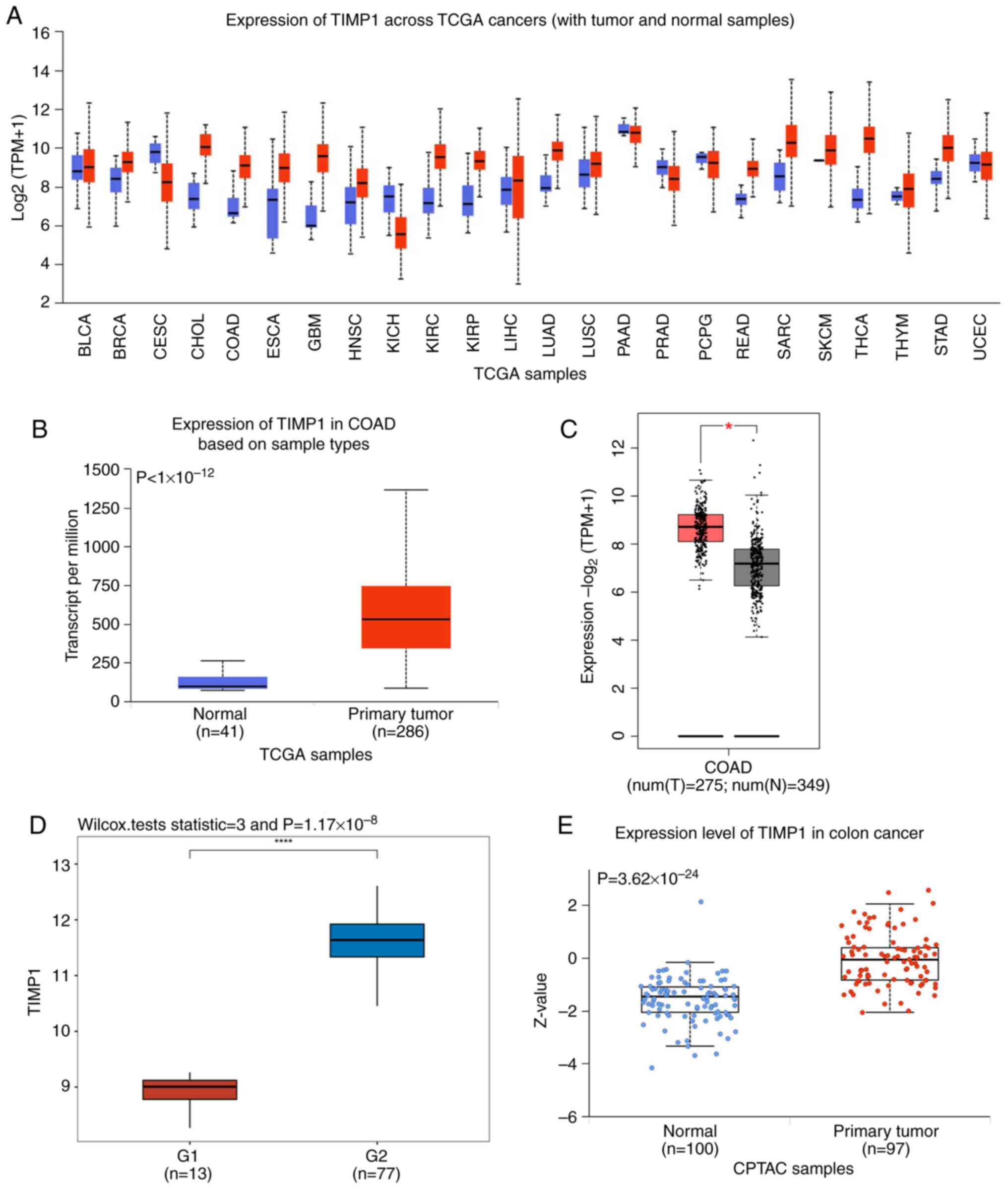
Using the UALCAN database, TIMP1 mRNA expression was
assessed across TCGA cancer tissues. The results indicated that,
compared with that in normal tissues, its expression was markedly
upregulated in several tumor types, including CRC. In 15/24 tumors,
TIMP1 was overexpressed (Fig. 1A).
Based on a database visualization website of UALCAN with TCGA, the
mRNA expression levels of TIMP1 were also significantly higher in
colon cancer samples compared with those in normal samples
(Fig. 1B). The GSE24550 dataset was
also evaluated, where the specimens of patients with colon cancer
exhibited high TIMP1 expression compared with those of normal
controls (Fig. 1C). Furthermore,
TCGA data analysis of GEPIA confirmed this result (Fig. 1D). Moreover, CPTAC data revealed
that the protein expression of TIMP1 exhibited significant
upregulation specifically in the tumor tissue, in comparison with
that in normal tissues (Fig.
1E).
Associations between TIMP1 and
clinicopathological features
To assess the association between TIMP1 expression
levels in CRC tissues and the clinicopathological characteristics
of patients with CRC, STAR-counts data and relevant clinical
information were obtained from TCGA database. The results revealed
that TIMP1 expression was significantly associated with survival
(P=0019) and ethnicity (P=0.0404); however, no significant
associations were noted between TIMP1 levels and age, sex,
pathological (p) tumor (T)-node (N)-metastasis (M) stage, pT stage,
pN stage or pM stage (Table I).
Association between TIMP1 expression
and prognosis
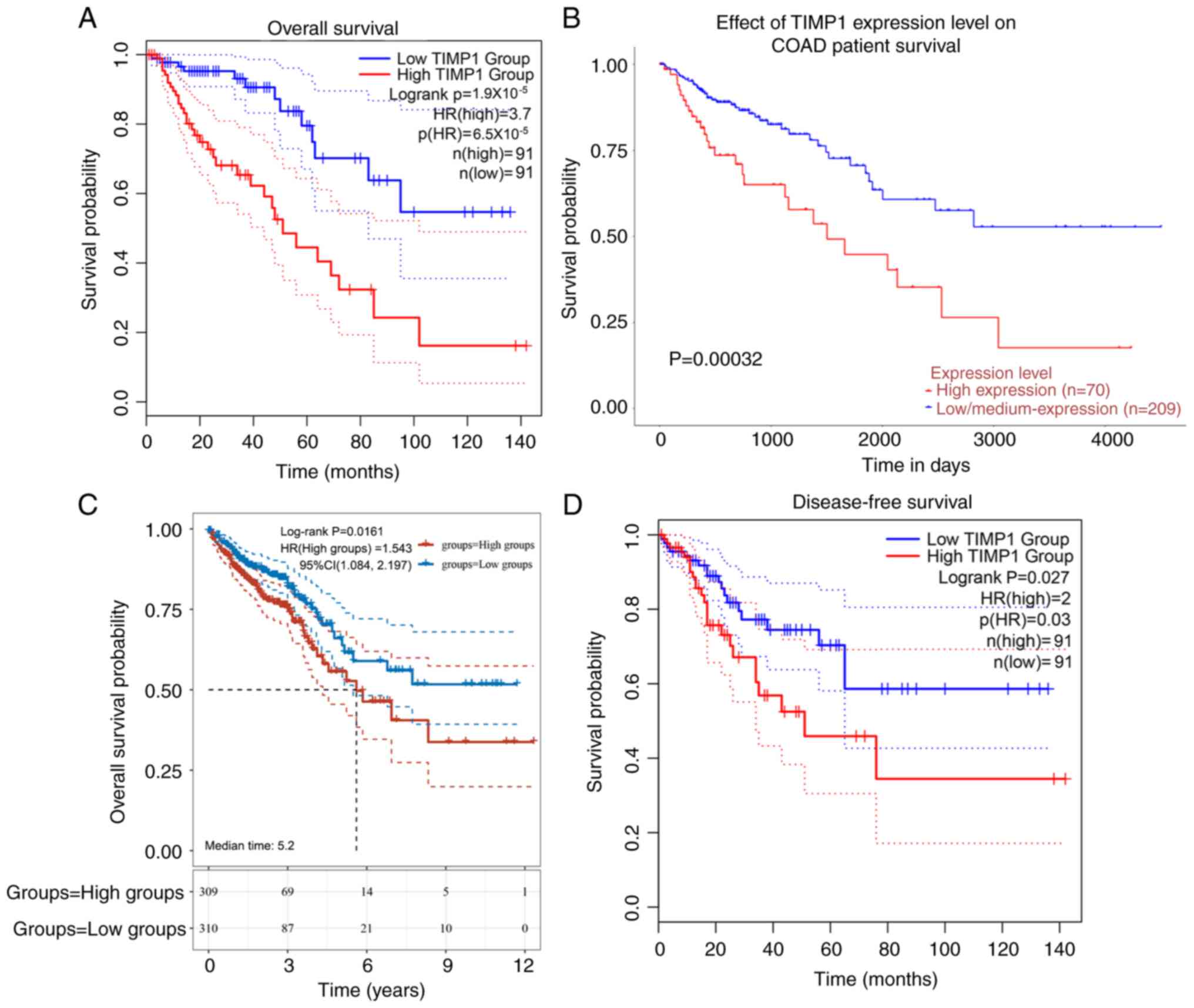
Patients with CRC and high TIMP1 expression
exhibited shorter overall survival compared with those with low
expression (Fig. 2A-C). Notably,
TIMP1 expression was significantly associated with disease-free
survival (Fig. 2D). Univariate Cox
regression analysis identified TIMP1 expression, age, pT stage (T4
vs. T1), pN stage (N1/2 vs. N0), pM stage (M1/X vs. M0) and pTNM
stage (III vs. I and IV vs. I) as significant predictors of overall
survival in patients with CRC (Fig.
3A). The results of multivariate Cox regression analysis
further confirmed that TIMP1 expression, age, pN stage (N1/2 vs.
N0) and pTNM stage (III vs. I, IV vs. I) were independent
prognostic factors for overall survival (Fig. 3B). Based on these factors, nomograms
were constructed using TCGA datasets to calculate prognostic scores
for patients with CRC, yielding a survival probability of 0.715
(Fig. 3C). The calibration curves
for TCGA cohorts were closely aligned with the 45-degree diagonal
line (Fig. 3D), indicating optimal
concordance between predicted and actual survival
probabilities.
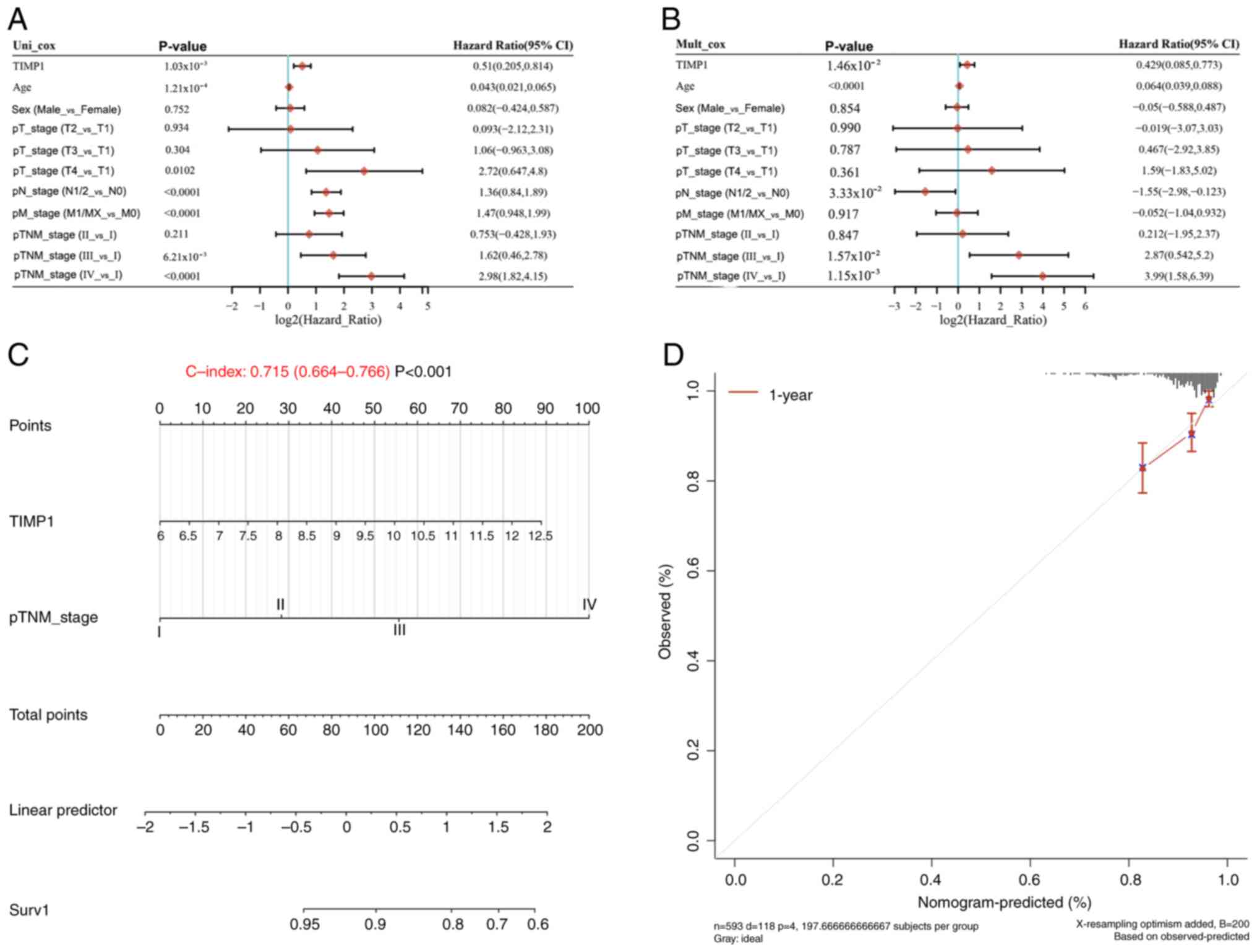 | Figure 3.TIMP1 serves as a prognostic factor
for overall survival in CRC. (A) Univariate and (B) multivariate
Cox analyses of TCGA data of patients with CRC for TIMP1 expression
and clinical characteristics. (C) Nomogram formulated to predict
1-year overall survival for patients with CRC from TCGA data. (D)
Calibration curve for the overall survival nomogram model in the
discovery cohort, where the diagonal dashed line denotes the ideal
nomogram, and the red line represent the observed 1-year nomograms.
TIMP1, tissue inhibitor of matrix metalloproteinase 1; CRC,
colorectal cancer; TCGA, The Cancer Genome Atlas; HR, hazard ratio;
CI, confidence interval; p, pathological; T, tumor; N, node; M,
metastasis. |
Association between TIMP1 and
immunity
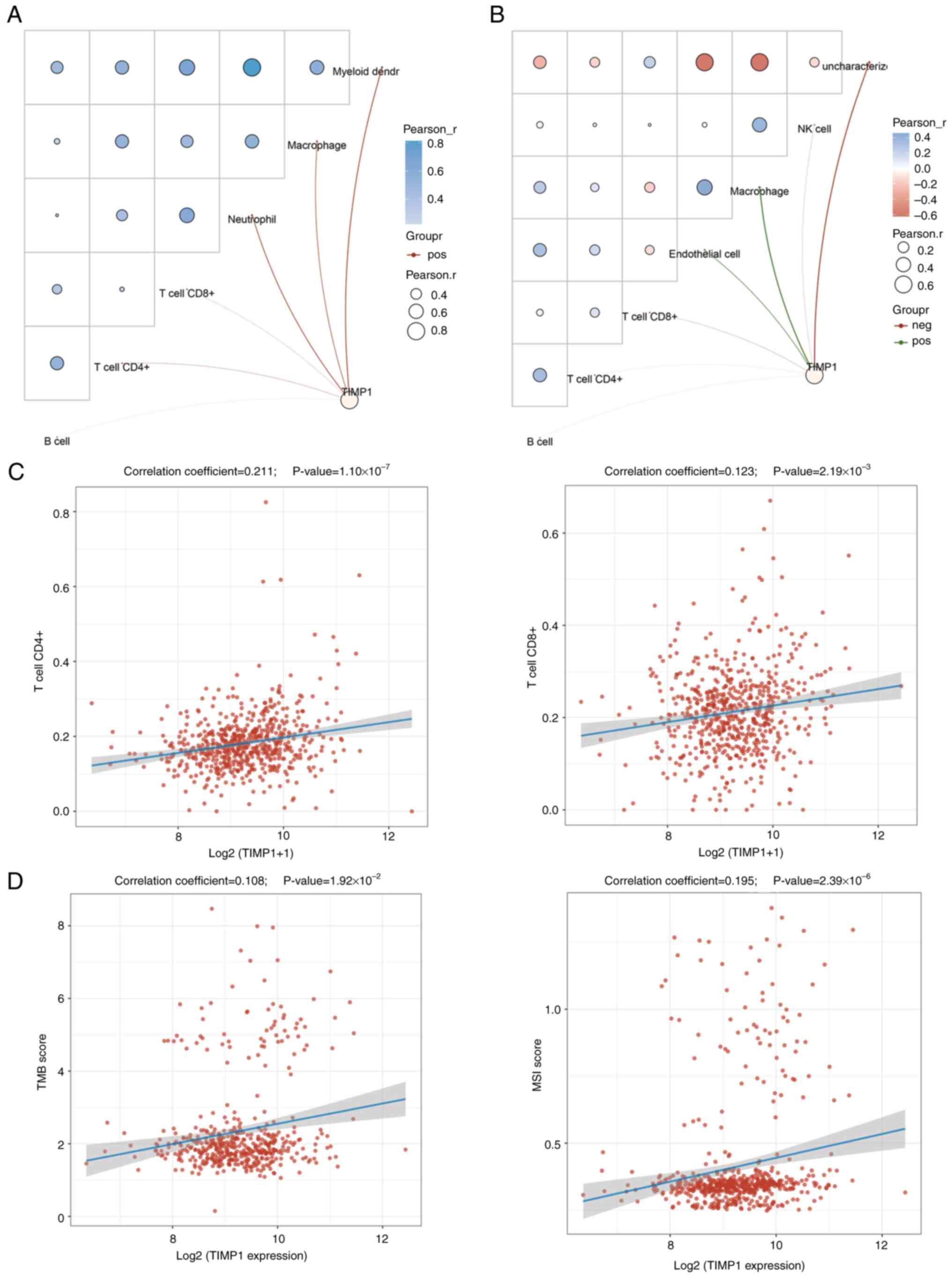
Subsequently, the role of TIMP1 in immunity was used
to evaluate its potential in CRC immunotherapy. Network connection
diagrams and heatmaps illustrated the link between TIMP1 expression
and immune scores, where red/blue intensity and ring size reflected
correlation strength (Fig. 4A and
B). Red lines indicate the positive correlations and green
lines the negative. Both TIMER and EPIC scores revealed the
positive correlation of TIMP1 with CD4+ and
CD8+ T cells. Spearman's correlation analysis using TCGA
CRC samples further confirmed this significant association
(Fig. 4C).
Previous studies have reported that TMB and MSI are
valuable biomarkers that provide insights into cancer prognosis and
treatment decisions (31–33). Both TMB and MSI serve roles in
cancer prognosis, treatment selection and immunotherapy development
(34). The findings of the present
study demonstrate a significant but weak positive correlation
between TIMP1 expression and TMB and MSI in CRC (Fig. 4D). This suggests that TIMP1 can
potentially serve as a biomarker to identify patients who may
benefit from immune checkpoint inhibitors, particularly in tumors
where high TMB and MSI predict a favorable response to
immunotherapy.
TIMP1 and pathways
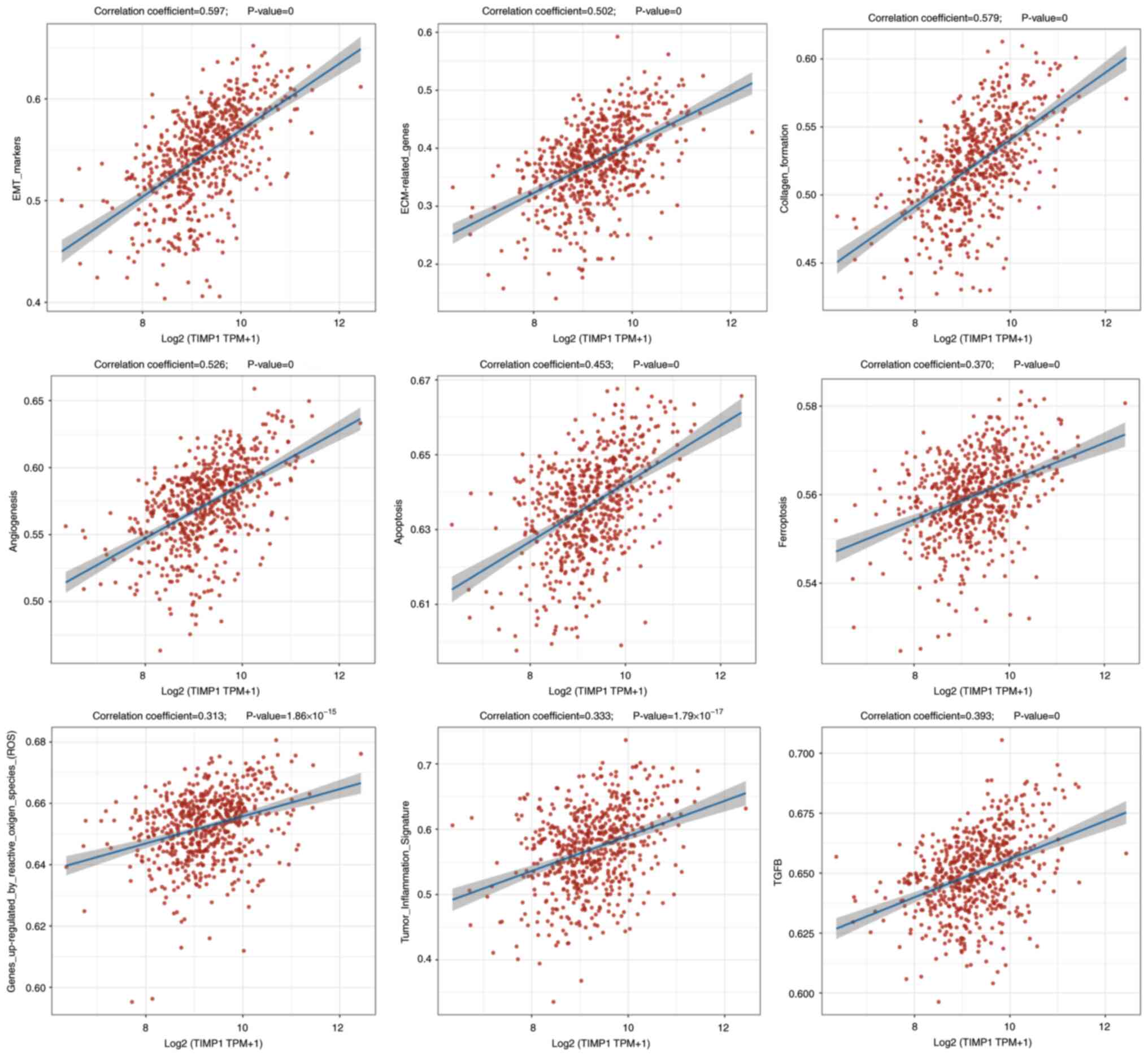
The ssGSEA algorithm was applied to compute the
enrichment fraction of each sample for specific pathways, thereby
exploring the sample-pathway relationship (27). The calculated fractions intuitively
reflected the interaction and association between samples and
pathways. The results indicated that in CRC, TIMP1 may be
positively correlated with epithelial-mesenchymal transition (EMT)
markers, extracellular matrix (ECM)-related genes, collagen
formation, angiogenesis, apoptosis, ferroptosis, the upregulation
of the expression levels of genes caused by reactive oxygen species
(ROS), tumor inflammation and the TGF-β signaling pathway (Fig. 5). Overall, these findings indicate
that TIMP1 may be associated with multiple pathways and may
regulate the progression of CRC.
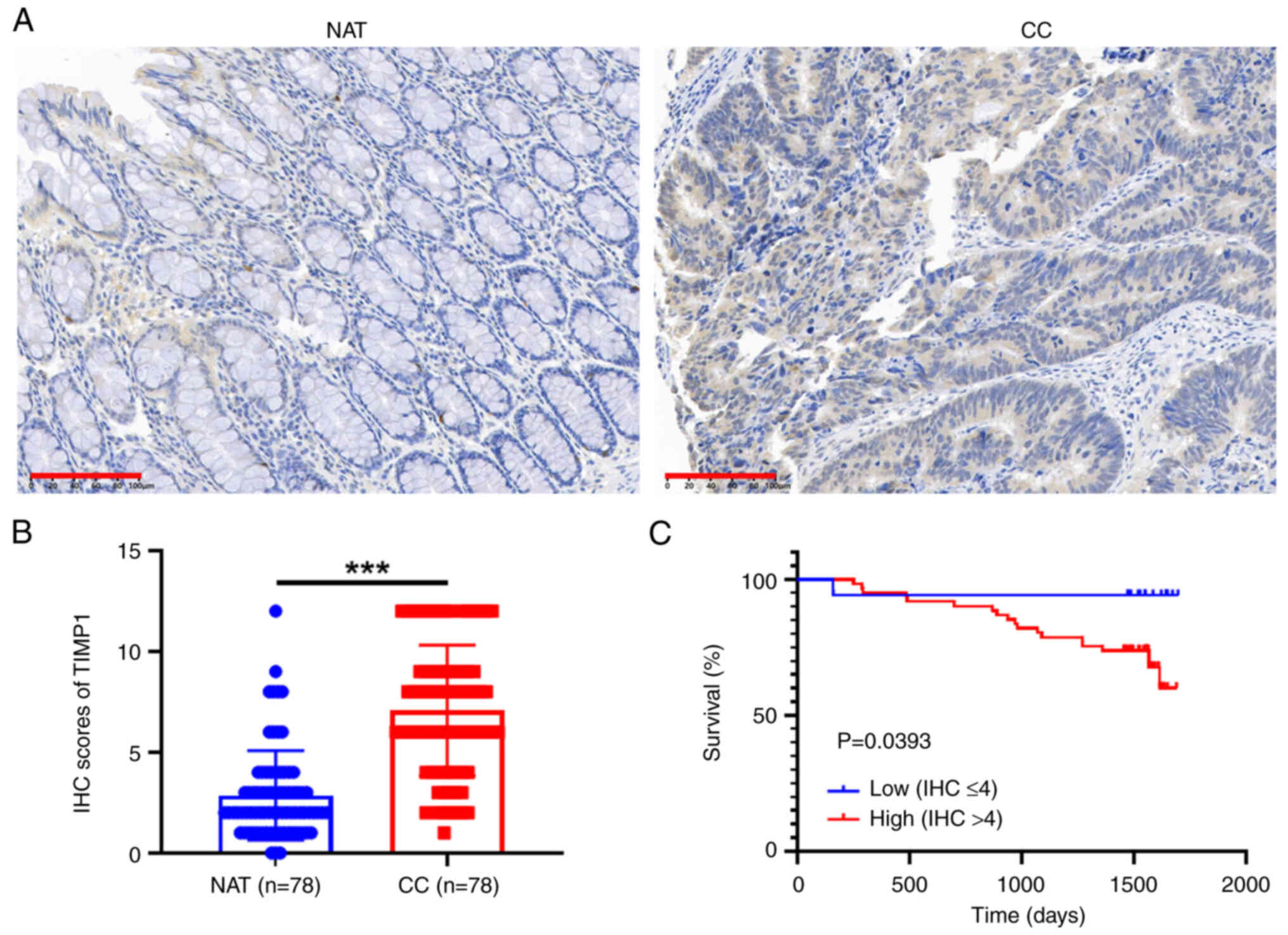
TIMP1 protein expression in CRC
tissues and prognosis
The IHC assay indicated that the expression levels
of TIMP1 were significantly higher in colon cancer tissues than
those in adjacent normal tissues (Fig.
6A and B). Notably, individuals with high TIMP1 expression (IHC
score >4) exhibited a significantly reduced survival rate
compared with those with low TIMP1 expression (IHC score ≤4) in
colon cancer (Fig. 6C).
Discussion
Most patients with CRC are diagnosed at an advanced
stage, which contributes to the poor prognosis commonly noted in
this patient population (35,36).
However, previous research indicates that an early diagnosis of CRC
can markedly improve patient outcomes (37). As such, the identification of
effective biomarkers for early CRC detection is crucial, as they
can serve a pivotal role in enhancing the prognosis of patients
with CRC.
TIMP1 belongs to the TIMP family, which includes
other members such as TIMP2, TIMP3 and TIMP4 (14). Research has indicated that TIMP1
expression is upregulated across multiple tumor types and this
upregulation is associated with poor prognosis and reduced survival
in patients with cancer (16). For
example, TIMP1 expression has been associated with gastric cancer
differentiation and poor prognosis in patients with gastric cancer
(38). Similarly, high TIMP1
expression in lung cancer tumors is associated with an unfavorable
prognosis (39). Macedo et
al (40) also reported that
elevated TIMP1 levels in patients with colon and gastric cancer are
associated with poor prognosis. These reports are consistent with
the findings of the present study, which revealed that both TIMP1
mRNA and protein levels were markedly elevated in CRC tissues, and
positively associated with worse overall and disease-free survival
in patients with CRC. Univariate and multivariate Cox regression
analyses further established high TIMP1 expression as an
independent prognostic indicator for overall survival in CRC. These
results indicate that TIMP1 expression is likely a predictor of CRC
prognosis.
Previous studies have established immunity as a
prognostic indicator for cancer progression (41–43).
Emerging mechanistic studies identify TIMP1 as a critical tumor
immune modulator (44,45). In thyroid cancer, TIMP1 promotes
cancer cell progression by inducing macrophage phenotypic
polarization via the PI3K/AKT signaling pathway (44). In glioblastoma, the expression of
TIMP1, induced by Sp1, is markedly elevated in tumor-infiltrating
lymphocytes and is associated with cancer progression (45). Moreover, TIMP1 expression in gliomas
is associated with tumor immune infiltration and immune
checkpoint-related gene expression (46). Wang et al (47) reported that intracellular
TIMP-1-CD63 signaling directs immune escape and metastasis
evolution in KRAS-mutated pancreatic cancer cells. In the present
study, network connection diagrams and heatmaps visualized the
association between TIMP1 expression and immune scores. Notably,
TIMP1 indicated a significant but weak correlation with
CD4+ and CD8+ T cells in CRC. Overall, these
findings suggest that TIMP1 may modulate the CRC tumor immune
environment by influencing CD4+ and CD8+ T
cell infiltration and function.
Immunotherapy, particularly the use of immune
checkpoint inhibitors (ICIs), has transformed cancer treatment by
leveraging the capacity of the immune system to combat tumors
(48). However, in contrast to
cancer types, such as non-small-cell lung cancer and melanoma,
which are known to be sensitive to immunotherapy, there is a lack
of reliable predictive biomarkers in multiple cancer types
(49). TMB and MSI serve as genomic
biomarkers to identify patients likely to benefit from ICIs, and
ICIs have shown promise in colon cancer treatment, notably in
patients with high TMB and MSI (34). The present study indicated a
significant association between TIMP1 expression and TMB and MSI in
CRC. This suggests that TIMP1 may be a potential predictive
biomarker for colon cancer immunotherapy.
Furthermore, previous studies have reported that
TIMP-1 can regulate tumor cell behavior by inducing signaling
pathways associated with cell growth, proliferation and survival
(14,16). For example, Song et al
(50) reported that inhibiting
TIMP1 expression reduced proliferation and metastasis, whilst
promoting apoptosis by targeting the FAK-PI3K/AKT and MAPK
pathways. The use of short hairpin RNA to knockdown TIMP1
expression was associated with a notable reduction in cell
proliferation and invasion in right-sided patient-derived organoids
from both left- and right-sided CRCs. This effect was achieved by
modulating the FAK/AKT signaling pathway (51). In the present study, the ssGSEA
algorithm revealed a positive correlation between TIMP1 expression
in CRC and multiple biological processes and pathways, including
EMT markers, ECM-related genes, collagen formation, angiogenesis,
apoptosis, ferroptosis, ROS-upregulated genes, tumor inflammation
signatures and the TGF-β pathway. Whilst further comprehensive
experimental validation is required to evaluate the pathways
associated with TIMP1, the existing data strongly suggest that
TIMP1 serves pivotal roles in driving CRC progression and
metastasis.
Moreover, whilst the present study highlights the
clinical significance of TIMP1, certain limitations are present.
Firstly, specific analyses rely on retrospective public-database
data; therefore, prospective studies are required to verify the
clinical relevance of the findings. Given the complexity of CRC and
the varied histological phenotypes, more detailed mechanistic and
clinical research is essential to explore the roles of TIMP1 across
CRC subtypes. In addition, the sample size of the present CRC
cohort is limited. In future work, the validation cohort will be
expanded by constructing a larger, multi-center TMA with balanced
representation of major CRC subtypes (such as CMS, MSI-H and
BRAF-mutant), on which subtype-stratified analyses will be
performed to determine if the role of TIMP1 is universal or
subtype-specific. Furthermore, comprehensive in vitro and
in vivo experiments are required to clarify the mechanistic
role of TIMP1 in tumor progression and its interaction with the
TME. Despite these limitations, the present study guides future
research, including clinical work and basic experiments, which will
be part of the subsequent research focus.
In conclusion, high TIMP1 expression in CRC is
associated with a poor prognosis for patients with CRC. TIMP1 may
also modulate the CRC immune microenvironment and facilitate CRC
progression and metastasis. The data suggest that TIMP1 represents
a promising diagnostic biomarker and therapeutic target for
CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Collaborative Custom
Development of RNA Probes (grant no. P112213323).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SX, DZ and TS contributed to the study conception
and design. JG contributed to performing experiments, acquisition
of data and interpretation of data. FC performed experiments. JG
and TS wrote the original manuscript. DZ and SX revised the
manuscript. JG and TS confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was granted approval by the
Institutional Review Board of the First Affiliated Hospital of
Soochow University (Suzhou, China; approval no. 2021-327). Written
informed consent was obtained from all participating patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Abedizadeh R, Majidi F, Khorasani HR,
Abedi H and Sabour D: Colorectal cancer: A comprehensive review of
carcinogenesis, diagnosis, and novel strategies for classified
treatments. Cancer Metastasis Rev. 43:729–753. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tao XY, Li QQ and Zeng Y: Clinical
application of liquid biopsy in colorectal cancer: Detection,
prediction, and treatment monitoring. Mol Cancer. 23:1452024.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mousavi SE, Ilaghi M, Hamidi Rad R and
Nejadghaderi SA: Epidemiology and socioeconomic correlates of
colorectal cancer in Asia in 2020 and its projection to 2040. Sci
Rep. 15:266392025. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y, Wang Y, Zhang B, Li P and Zhao Y:
Methods and biomarkers for early detection, prediction, and
diagnosis of colorectal cancer. Biomed Pharmacother.
163:1147862023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mo S, Tang P, Luo W, Zhang L, Li Y, Hu X,
Ma X, Chen Y, Bao Y, He X, et al: Patient-derived organoids from
colorectal cancer with paired liver metastasis reveal tumor
heterogeneity and predict response to chemotherapy. Adv Sci
(Weinh). 9:e22040972022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang W and Wang S: Relationships between
nutritional status and serum adipokine levels with chemotherapy
efficacy in late-stage colorectal cancer patients. Int J Colorectal
Dis. 40:252025. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prasetyanti PR and Medema JP: Intra-tumor
heterogeneity from a cancer stem cell perspective. Mol Cancer.
16:412017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu X, Liu R, Gao W, Wang X and Zhang Y:
Single-cell omics traces the heterogeneity of prostate cancer cells
and the tumor microenvironment. Cell Mol Biol Lett. 28:382023.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo L, Wang Y, Yang W, Wang C, Guo T, Yang
J, Shao Z, Cai G, Cai S, Zhang L, et al: Molecular profiling
provides clinical insights into targeted and immunotherapies as
well as colorectal cancer prognosis. Gastroenterology.
165:414–428.e7. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Singh H, Sahgal P, Kapner K, Corsello SM,
Gupta H, Gujrathi R, Li YY, Cherniack AD, El Alam R, Kerfoot J, et
al: RAS/RAF Comutation and ERBB2 copy number modulates HER2
heterogeneity and responsiveness to HER2-directed therapy in
colorectal cancer. Clin Cancer Res. 30:1669–1684. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jalali P, Aliyari S, Etesami M, Saeedi
Niasar M, Taher S, Kavousi K, Nazemalhosseini Mojarad E and Salehi
Z: GUCA2A dysregulation as a promising biomarker for accurate
diagnosis and prognosis of colorectal cancer. Clin Exp Med.
24:2512024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu S, Sun X, Tang H, Yu J, Wang B, Xiao R,
Qu J, Sun F, Deng Z, Li C, et al: Colorectal cancer with low
SLC35A3 is associated with immune infiltrates and poor prognosis.
Sci Rep. 14:3292024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Parisi E, Hidalgo I, Montal R, Pallise O,
Tarragona J, Sorolla A, Novell A, Campbell K, Sorolla MA, Casali A
and Salud A: PLA2G12A as a novel biomarker for colorectal cancer
with prognostic relevance. Int J Mol Sci. 24:108892023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jackson HW, Defamie V, Waterhouse P and
Khokha R: TIMPs: Versatile extracellular regulators in cancer. Nat
Rev Cancer. 17:38–53. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Caterina NC, Windsor LJ, Bodden MK,
Yermovsky AE, Taylor KB, Birkedal-Hansen H and Engler JA:
Glycosylation and NH2-terminal domain mutants of the tissue
inhibitor of metalloproteinases-1 (TIMP-1). Biochim Biophys Acta.
1388:21–34. 1388. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Justo BL and Jasiulionis MG:
Characteristics of TIMP1, CD63, and β1-Integrin and the functional
impact of their interaction in cancer. Int J Mol Sci. 22:93192021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Eckfeld C, Haussler D, Schoeps B, Hermann
CD and Kruger A: Functional disparities within the TIMP family in
cancer: Hints from molecular divergence. Cancer Metastasis Rev.
38:469–481. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tian Z, Ou G, Su M, Li R, Pan L, Lin X,
Zou J, Chen S, Li Y, Huang K and Chen Y: TIMP1 derived from
pancreatic cancer cells stimulates Schwann cells and promotes the
occurrence of perineural invasion. Cancer Let. 546:2158632022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guccini I, Revandkar A, D'Ambrosio M,
Colucci M, Pasquini E, Mosole S, Troiani M, Brina D,
Sheibani-Tezerji R, Elia AR, et al: Senescence reprogramming by
TIMP1 deficiency promotes prostate cancer metastasis. Cancer Cell.
39:68–82.e9. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B and Varambally
S: UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sveen A, Agesen TH, Nesbakken A, Rognum
TO, Lothe RA and Skotheim RI: Transcriptome instability in
colorectal cancer identified by exon microarray analyses:
Associations with splicing factor expression levels and patient
survival. Genome Med. 3:322011. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Z, Wang Y, Peng M and Yi L: UBASH3B
is a novel prognostic biomarker and correlated with immune
infiltrates in prostate cancer. Front Oncol. 9:15172019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thorsson V, Gibbs DL, Brown SD, Wolf D,
Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy
JA, et al: The immune landscape of cancer. Immunity.
48:812–830.e14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bonneville R, Krook MA, Kautto EA, Miya J,
Wing MR, Chen HZ, Reeser JW, Yu L and Roychowdhury S: Landscape of
microsatellite instability across 39 cancer types. JCO Precis
Oncol. 2017.PO.17.00073. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wei J, Huang K, Chen Z, Hu M, Bai Y, Lin S
and Du H: Characterization of glycolysis-associated molecules in
the tumor microenvironment revealed by pan-cancer tissues and lung
cancer single cell data. Cancers (Basel). 12:17882020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hanzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiao Z, Dai Z and Locasale JW: Metabolic
landscape of the tumor microenvironment at single cell resolution.
Nat Commun. 10:37632019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun L, Chen Y, Xia L, Wang J, Zhu J, Li J,
Wang K, Shen K, Zhang D, Zhang G, et al: TRIM69 suppressed the
anoikis resistance and metastasis of gastric cancer through
ubiquitin-proteasome-mediated degradation of PRKCD. Oncogene.
42:3619–3632. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kendre G, Murugesan K, Brummer T, Segatto
O, Saborowski A and Vogel A: Charting co-mutation patterns
associated with actionable drivers in intrahepatic
cholangiocarcinoma. J Hepatol. 78:614–626. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Di Mauro A, Santorsola M, Savarese G,
Sirica R, Ianniello M, Cossu AM, Ceccarelli A, Sabbatino F,
Bocchetti M, Carratu AC, et al: High tumor mutational burden
assessed through next-generation sequencing predicts favorable
survival in microsatellite stable metastatic colon cancer patients.
J Transl Med. 22:11072024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shi S, Wang Y, Wu J, Zha B, Li P, Liu Y,
Yang Y, Kong J, Gao S, Cui H, et al: Predictive value of PD-L1 and
TMB for short-term efficacy prognosis in non-small cell lung cancer
and construction of prediction models. Front Oncol. 14:13422622024.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hou W, Yi C and Zhu H: Predictive
biomarkers of colon cancer immunotherapy: Present and future. Front
Immunol. 13:10323142022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sasidharan Nair V, Saleh R, Taha RZ, Toor
SM, Murshed K, Ahmed AA, Kurer MA, Abu Nada M, Al Ejeh F and Elkord
E: Differential gene expression of tumor-infiltrating
CD4+ T cells in advanced versus early stage colorectal
cancer and identification of a gene signature of poor prognosis.
Oncoimmunology. 9:18251782020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Garborg K: Colorectal cancer screening.
Surg Clin North Am. 95:979–989. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tonini V and Zanni M: Why is early
detection of colon cancer still not possible in 2023? World J
Gastroenterol. 30:211–224. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zheng M, Wang P, Wang Y, Jia Z, Gao J, Tan
X, Chen H and Zu G: Clinicopathological and prognostic significance
of TIMP1 expression in gastric cancer: A systematic review and
meta-analysis. Expert Rev Anticancer Ther. 24:1169–1176. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dantas E, Murthy A, Ahmed T, Ahmed M,
Ramsamooj S, Hurd MA, Lam T, Malbari M, Agrusa C, Elemento O, et
al: TIMP1 is an early biomarker for detection and prognosis of lung
cancer. Clin Transl Med. 13:e13912023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Macedo FC, Cunha N, Pereira TC, Soares RF,
Monteiro AR, Bonito N, Valido F and Sousa G: A prospective cohort
study of TIMP1 as prognostic biomarker in gastric and colon cancer.
Chin Clin Oncol. 11:432022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zheng J, Peng L, Zhang S, Liao H, Hao J,
Wu S and Shen H: Preoperative systemic immune-inflammation index as
a prognostic indicator for patients with urothelial carcinoma.
Front Immunol. 14:12750332023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiong S, Dong L and Cheng L: Neutrophils
in cancer carcinogenesis and metastasis. J Hematol Oncol.
14:1732021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu R, Lu J, Liu J, Liao Y, Guo Y, Shi P,
Wang Z, Wang H and Lai J: Macrophages in prostate cancer: Dual
roles in tumor progression and immune evasion. J Transl Med.
23:6152025. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lin X, Zhao R, Bin Y, Huo R, Xue G and Wu
J: TIMP1 promotes thyroid cancer cell progression through
macrophage phenotypic polarization via the PI3K/AKT signaling
pathway. Genomics. 116:1109142024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu L, Yang S, Lin K, Yu X, Meng J, Ma C,
Wu Z, Hao Y, Chen N, Ge Q, et al: Sp1 induced gene TIMP1 is related
to immune cell infiltration in glioblastoma. Sci Rep. 12:111812022.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu J, Wei C, Wang C, Li F, Wang Z, Xiong
J, Zhou Y, Li S, Liu X, Yang G, et al: TIMP1/CHI3L1 facilitates
glioma progression and immunosuppression via NF-kappaB activation.
Biochim Biophys Acta Mol Basis Dis. 1870:1670412024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang CA, Hou YC, Hong YK, Tai YJ, Shen C,
Hou PC, Fu JL, Wu CL, Cheng SM, Hwang DY, et al: Intercellular
TIMP-1-CD63 signaling directs the evolution of immune escape and
metastasis in KRAS-mutated pancreatic cancer cells. Mol Cancer.
24:252025. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Butterfield LH and Najjar YG:
Immunotherapy combination approaches: Mechanisms, biomarkers and
clinical observations. Nat Rev Immunol. 24:399–416. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bai R, Lv Z, Xu D and Cui J: Predictive
biomarkers for cancer immunotherapy with immune checkpoint
inhibitors. Biomark Res. 8:342020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Song G, Xu S, Zhang H, Wang Y, Xiao C,
Jiang T, Wu L, Zhang T, Sun X, Zhong L, et al: TIMP1 is a
prognostic marker for the progression and metastasis of colon
cancer through FAK-PI3K/AKT and MAPK pathway. J Exp Clin Cancer
Res. 35:1482016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ma B, Ueda H, Okamoto K, Bando M, Fujimoto
S, Okada Y, Kawaguchi T, Wada H, Miyamoto H, Shimada M, et al:
TIMP1 promotes cell proliferation and invasion capability of
right-sided colon cancers via the FAK/Akt signaling pathway. Cancer
Sci. 113:4244–4257. 2022. View Article : Google Scholar : PubMed/NCBI
|